Abstract
The number of proposed prognostic models for coronavirus disease 2019 (COVID-19) is growing rapidly, but it is unknown whether any are suitable for widespread clinical implementation.
We independently externally validated the performance of candidate prognostic models, identified through a living systematic review, among consecutive adults admitted to hospital with a final diagnosis of COVID-19. We reconstructed candidate models as per original descriptions and evaluated performance for their original intended outcomes using predictors measured at the time of admission. We assessed discrimination, calibration and net benefit, compared to the default strategies of treating all and no patients, and against the most discriminating predictors in univariable analyses.
We tested 22 candidate prognostic models among 411 participants with COVID-19, of whom 180 (43.8%) and 115 (28.0%) met the endpoints of clinical deterioration and mortality, respectively. Highest areas under receiver operating characteristic (AUROC) curves were achieved by the NEWS2 score for prediction of deterioration over 24 h (0.78, 95% CI 0.73–0.83), and a novel model for prediction of deterioration <14 days from admission (0.78, 95% CI 0.74–0.82). The most discriminating univariable predictors were admission oxygen saturation on room air for in-hospital deterioration (AUROC 0.76, 95% CI 0.71–0.81), and age for in-hospital mortality (AUROC 0.76, 95% CI 0.71–0.81). No prognostic model demonstrated consistently higher net benefit than these univariable predictors, across a range of threshold probabilities.
Admission oxygen saturation on room air and patient age are strong predictors of deterioration and mortality among hospitalised adults with COVID-19, respectively. None of the prognostic models evaluated here offered incremental value for patient stratification to these univariable predictors.
Short abstract
Oxygen saturation on room air and patient age are strong predictors of deterioration and mortality, respectively, among hospitalised adults with COVID-19. None of the 22 prognostic models evaluated in this study adds incremental value to these univariable predictors. https://bit.ly/2Hg24TO
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), causes a spectrum of disease ranging from asymptomatic infection to critical illness. Among people admitted to hospital, COVID-19 has a reported mortality of 21%–33%, with 14%–17% requiring admission to high dependency or intensive care units (ICUs) [1–4]. Exponential surges in transmission of SARS-CoV-2 coupled with the severity of disease among a subset of those affected pose major challenges to health services by threatening to overwhelm resource capacity [5]. Rapid and effective triage at the point of presentation to hospital is therefore required to facilitate adequate allocation of resources and to ensure that patients at higher risk of deterioration are managed and monitored appropriately. Prognostic models may have additional value in patient stratification for emerging drug therapies [6, 7].
As a result, there has been global interest in developing prediction models for COVID-19 [8]. These include models aiming to predict a diagnosis of COVID-19, and prognostic models aiming to predict disease outcomes. At the time of writing, a living systematic review has already catalogued 145 diagnostic or prognostic models for COVID-19 [8]. Critical appraisal of these models using quality assessment tools developed specifically for prediction modelling studies suggests that the candidate models are poorly reported and at high risk of bias and overestimation of their reported performance [8, 9]. However, independent evaluation of candidate prognostic models in unselected datasets has been lacking. It remains unclear how well these proposed models perform in practice, or whether any are suitable for widespread clinical implementation. We aimed to address this knowledge gap by systematically evaluating the performance of proposed prognostic models among consecutive patients hospitalised with a final diagnosis of COVID-19 at a single centre when using predictors measured at the point of hospital admission.
Methods
Identification of candidate prognostic models
We used a published living systematic review to identify all candidate prognostic models for COVID-19 indexed in PubMed, Embase, Arxiv, medRxiv or bioRxiv until May 5, 2020, regardless of underlying study quality [8]. We included models that aim to predict clinical deterioration or mortality among patients with COVID-19. We also included prognostic scores commonly used in clinical practice [10–12], but not specifically developed for COVID-19 patients, given that these models may also be considered for use by clinicians to aid risk-stratification for patients with COVID-19. For each candidate model identified, we extracted predictor variables, outcome definitions (including time horizons), modelling approaches and final model parameters from original publications. We contacted authors for additional information where required. We excluded scores for which the underlying model parameters were not publicly available, because we were unable to reconstruct them, along with models for which included predictors were not available in our dataset. The latter included models that require computed tomography imaging or arterial blood gas sampling, because these investigations were not routinely performed among unselected patients with COVID-19 at our centre.
Study population
Our study is reported in accordance with transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidance for external validation studies [13]. We included consecutive adults admitted to University College Hospital London with a final diagnosis of PCR-confirmed (including all sample types) or clinically diagnosed COVID-19, between February 1 and April 30, 2020. Because we sought to use data from the point of hospital admission to predict outcomes, we excluded patients transferred in from other hospitals and those with hospital-acquired COVID-19 (defined as the first PCR swab sent >5 days from date of hospital admission, as a proxy for the onset of clinical suspicion of SARS-CoV-2 infection). Clinical COVID-19 diagnoses were made on the basis of manual record review by an infectious disease specialist, using clinical features, laboratory results and radiological appearances, in the absence of an alternative diagnosis. During the study period, PCR testing was performed on the basis of clinical suspicion, and no SARS-CoV-2 serology investigations were routinely performed.
Data sources and variables of interest
Data were collected by direct extraction from electronic health records, complemented by manual curation. Variables of interest in the dataset included demographics (age, sex, ethnicity), comorbidities (identified through manual record review), clinical observations, laboratory measurements, radiology reports and clinical outcomes. Each chest radiograph was reported by a single radiologist, who was provided with a short summary of the indication for the investigation at the time of request, reflecting routine clinical conditions. Chest radiographs were classified using British Society of Thoracic Imaging criteria, and using a modified version of the Radiographic Assessment of Lung Edema (RALE) score [14, 15]. For each predictor, measurements were recorded as part of routine clinical care. Where serial measurements were available, we included the measurement taken closest to the time of presentation to hospital, with a maximum interval between presentation and measurement of 24 h.
Outcomes
For models that used ICU admission or death, or progression to “severe” COVID-19 or death, as composite endpoints, we used a composite “clinical deterioration” endpoint as the primary outcome. We defined clinical deterioration as the initiation of ventilatory support (continuous positive airway pressure, non-invasive ventilation, high-flow nasal cannula oxygen, invasive mechanical ventilation or extracorporeal membrane oxygenation) or death, equivalent to World Health Organization (WHO) Clinical Progression Scale ≥6 [16]. This definition does not include standard oxygen therapy. We did not apply any temporal limits on 1) the minimum duration of respiratory support or 2) the interval between presentation to hospital and the outcome. The rationale for this composite outcome was to make the endpoint more generalisable between centres, given that hospital respiratory management algorithms may vary substantially. Defining the outcome based on level of support, as opposed to ward setting, also ensured that it is appropriate in the context of a pandemic, when treatments that would usually only be considered in an ICU setting may be administered in other environments owing to resource constraints. Where models specified their intended time horizon in their original description, we used this timepoint in the primary analysis to ensure unbiased assessment of model calibration. Where the intended time horizon was not specified, we assessed the model to predict in-hospital deterioration or mortality, as appropriate. All deterioration and mortality events were included, regardless of their clinical aetiology.
Participants were followed up clinically to the point of discharge from hospital. We extended follow-up beyond discharge by cross-checking National Health Service (NHS) Spine records to identify reported deaths post-discharge, thus ensuring >30 days’ follow-up for all participants.
Statistical analyses
For each prognostic model included in the analyses, we reconstructed the model according to the authors’ original descriptions, and sought to evaluate the model discrimination and calibration performance against our approximation of their original intended endpoint. For models that provide online risk calculator tools, we validated our reconstructed models against original authors’ models by cross-checking our predictions against those generated by the web-based tools for a random subset of participants.
For all models, we assessed discrimination by quantifying the area under the receiver operating characteristic curve (AUROC) [17]. For models that provided outcome probability scores, we assessed calibration by visualising calibration of predicted versus observed risk using LOESS-smoothed plots, and by quantifying calibration slopes and calibration-in-the-large (CITL). A perfect calibration slope should be 1; slopes <1 indicate that risk estimates are too extreme, while slopes >1 reflect risk estimates not being extreme enough. Ideal CITL is 0; CITL>0 indicates that predictions are systematically too low, while CITL<0 indicates that predictions are too high. For models with points-based scores, we assessed calibration visually by plotting model scores versus actual outcome proportions. For models that provide probability estimates, but where the model intercept was not available, we calibrated the model to our dataset by calculating the intercept when using the model linear predictor as an offset term, leading to perfect CITL. This approach, by definition, overestimated calibration with respect to CITL, but allowed us to examine the calibration slope in our dataset.
We also assessed the discrimination of each candidate model for standardised outcomes of 1) our composite endpoint of clinical deterioration and 2) mortality across a range of pre-specified time horizons from admission (7 days, 14 days, 30 days and any time during hospital admission) by calculating time-dependent AUROCs (with cumulative sensitivity and dynamic specificity) [18]. The rationale for this analysis was to harmonise endpoints to facilitate more direct comparisons of discrimination between the candidate models.
To further benchmark the performance of candidate prognostic models, we then computed AUROCs for a limited number of univariable predictors considered to be of highest importance a priori, based on clinical knowledge and existing data, for predicting our composite endpoints of clinical deterioration and mortality (7 days, 14 days, 30 days and any time during hospital admission). The a priori predictors of interest examined in this analysis were age, clinical frailty scale, oxygen saturation at presentation on room air, C-reactive protein level and absolute lymphocyte count [8, 19].
Decision curve analysis allows the clinical utility of candidate models to be assessed, and is dependent on both model discrimination and calibration [20]. We performed decision curve analyses to quantify the net benefit achieved by each model for predicting the original intended endpoint across a range of risk:benefit ratios [20]. In this approach, the risk:benefit ratio is analogous to the cut point for a statistical model above which an intervention or treatment would be considered beneficial (deemed the “threshold probability”). Net benefit was calculated as sensitivity×prevalence–(1–specificity)×(1–prevalence)×w where w is the odds at the threshold probability and the prevalence is the proportion of patients who experienced the outcome [20]. We calculated net benefit across a range of clinically relevant threshold probabilities, ranging from 0 to 0.5, because the risk:benefit ratio may vary for any given intervention. We compared the utility of each candidate model against strategies of treating all and no patients, and against the best-performing univariable predictor for in-hospital clinical deterioration, or mortality, as appropriate. To ensure that fair, head-to-head net benefit comparisons were made between multivariable probability-based models, points score models and univariable predictors, we calibrated each of these to the validation dataset for the purpose of decision curve analysis. Probability-based models were recalibrated to the validation data by refitting logistic regression models with the candidate model linear predictor as the sole predictor. We calculated “delta” net benefit as net benefit when using the index model minus net benefit when 1) treating all patients and 2) using the most discriminating univariable predictor. Decision curve analyses were done using the rmda package in R [21].
We handled missing data using multiple imputation by chained equations [22], using the mice package in R [23]. All variables and outcomes in the final prognostic models were included in the imputation model to ensure compatibility [22]. A total of 10 imputed datasets were generated; discrimination, calibration and net benefit metrics were pooled using Rubin's rules [24].
All analyses were conducted in R (version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria).
Sensitivity analyses
We recalculated discrimination and calibration parameters for each candidate model using 1) a complete case analysis (in view of the large amount of missingness for some models), 2) excluding patients without PCR-confirmed SARS-CoV-2 infection and 3) excluding patients who met the clinical deterioration outcome within 4 h of arrival at hospital. We also examined for non-linearity in the a priori univariable predictors using restricted cubic splines, with three knots. Finally, we estimated optimism for discrimination and calibration parameters for the a priori univariable predictors using bootstrapping (1000 iterations), using the rms package in R [25].
Ethical approval
The pre-specified study protocol was approved by East Midlands - Nottingham 2 Research Ethics Committee (REF: 20/EM/0114; IRAS: 282900).
Results
Summary of candidate prognostic models
We identified 37 studies describing prognostic models, of which 19 studies (including 22 unique models) were eligible for inclusion (supplementary figure S1 and table 1). Of these, five models were not specific to COVID-19, but were developed as prognostic scores for emergency department attendees [27], hospitalised patients [12, 44], people with suspected infection [10] or community-acquired pneumonia [11]. Of the 17 models developed specifically for COVID-19, most (10 out of 17) were developed using datasets originating in China. Overall, discovery populations included hospitalised patients and were similar to the current validation population with the exception of one study that discovered a model using community data [28], and another that used simulated data [29]. A total of 13 out of 22 models use points-based scoring systems to derive final model scores, with the remainder using logistic regression modelling approaches to derive probability estimates. A total of 12 out of 22 prognostic models had the primary aim of predicting clinical deterioration, while the remaining 10 sought to predict mortality alone. When specified, time horizons for prognosis ranged from 1 to 30 days. Candidate prognostic models not included in the current validation study are summarised in supplementary table S1.
TABLE 1.
Characteristics of studies describing prognostic models included in systematic evaluation
| Authors | Score name | Country of derivation | Development population | Pre-existing or COVID-specific? | Model outcome | Predictors | Original modelling approach | How are predictors combined? |
| Subbeet al. [26] | MEWS# | UK | Hospital inpatients | Pre-existing (hospital patients) | Mortality, ICU admission or cardiac arrest (no specified timepoint) | Systolic blood pressure, pulse rate, respiratory rate, temperature, AVPU score | Clinical consensus | Points-based score |
| Olssonet al. [27] | REMS# | Sweden | Patients presenting to emergency department | Pre-existing (emergency department patients) | Mortality (in-hospital) | Blood pressure, respiratory rate, pulse rate, Glasgow coma scale, oxygen saturation, age | Logistic regression | Points-based score |
| Seymouret al. [10] | qSOFA | USA | Electronic health record encounters | Pre-existing (suspected infection) | Mortality (in-hospital) | Systolic hypotension (≤100 mmHg, tachypnoea (≥22 beats·min−1), altered mentation | Logistic regression | Points-based score |
| Limet al. [11] | CURB65 | UK, New Zealand, Netherlands | Patients with community-acquired pneumonia | Pre-existing (community-acquired pneumonia) | Mortality (30 days) | Confusion, urea >7 mmol·L−1, respiratory rate >30 breaths·min−1, low systolic (<90 mmHg) or diastolic (<60 mmHg) blood pressure, age >65 years | Logistic regression | Points-based score |
| Royal College of Physicians [12] | NEWS2+ | UK | Hospital admissions | Pre-existing (hospital patients) | Mortality, ICU admission or cardiac arrest (24 h) | Respiratory rate, oxygen saturation, systolic blood pressure, pulse rate, level of consciousness or new confusion, temperature | Clinical consensus | Points-based score |
| Bello-Chavollaet al. [28] | BelloChavolla | Mexico | Confirmed COVID-19 patients presenting in primary care | COVID-specific | Mortality (30 day) | Age ≥65 years, diabetes, early-onset diabetes, obesity, age <40 years, chronic kidney disease, hypertension, immunosuppression (rheumatoid arthritis, lupus, HIV or immunosuppressive drugs) | Cox regression | Points-based score |
| Carameloet al. [29] | Caramelo¶ | Simulated data | Simulated data | COVID-specific | Mortality (period unspecified) | Age, hypertension, diabetes, CVD, chronic respiratory disease, cancer | Logistic regression | Logistic regression |
| Carret al. [30] | Carr_final, Carr_threshold | UK | Inpatients with confirmed COVID-19 | COVID-specific | ICU admission or death (14 days from symptom onset) | NEWS2, CRP, neutrophils, estimated glomerular filtration rate, albumin, age | Regularised logistic regression with LASSO estimator | Regularised logistic regression |
| Colombiet al. [31] | Colombi_clinical¶ (clinical model only) | Italy | Inpatients with confirmed COVID-19 | COVID-specific | ICU admission or in-hospital mortality (period unspecified) | Age >68 years, CVD, CRP >76 mg·L−1, LDH >347 U·L−1, platelets >180×109 L−1 | Logistic regression | Logistic regression |
| Gallowayet al. [32] | Galloway | UK | Inpatients with confirmed COVID-19 | COVID-specific | ICU admission or death during admission | Modified RALE score >3, oxygen saturation <93%, creatinine >100 μmol·L−1, neutrophils >8×109 L−1, age >40 years, chronic lung disease, CRP >40 mg·L−1, albumin <34 g·L−1, male, non-white ethnicity, hypertension, diabetes | Logistic regression (LASSO) | Points-based score |
| Guoet al. [33] | Guo | China | Inpatients with confirmed COVID-19 | COVID-specific | Deterioration within 14 days of admission | Age >50 years, underlying chronic disease (not defined), neutrophil/lymphocyte ratio >5, CRP >25 mg·L−1, D-dimer >800 ng·mL−1 | Cox regression | Points-based score |
| Hallet al. [34] | TACTIC | UK | Inpatients with confirmed COVID-19 | COVID-specific | Admission to ICU or death during admission | Modified RALE score >3, age >40 years, male, non-white ethnicity, diabetes, hypertension, neutrophils >8×109 L−1, CRP >40 mg·L−1 | Logistic regression (LASSO) | Points-based score |
| Huet al. [35] | Hu | China | Inpatients with confirmed COVID-19 | COVID-specific | Mortality (in-hospital) | Age, CRP, lymphocytes, D-dimer (μg/mL) | Logistic regression | Logistic regression |
| Huanget al. [36] | Huang | China | Inpatients with confirmed COVID-19 | COVID-specific | Progression to severe COVID (defined as respiratory rate ≥30 breaths·min−1, oxygen saturation ≤93% in the resting state or PaO2/oxygen concentration FiO2 ≤300 mmHg), 3–7 days from admission | CRP >10 mg·L−1, LDH >250 U·L−1, respiratory rate >24 breaths·min−1, comorbidity (hypertension, coronary artery disease, diabetes, obesity, COPD, chronic kidney disease, obstructive sleep apnoea) | Logistic regression | Logistic regression |
| Jiet al. [37] | Ji | China | Inpatients with confirmed COVID-19 | COVID-specific | Progression to severe COVID-19 at 10 days (defined as respiratory rate ≥30 breaths·min−1, resting oxygen saturation ≤93%, PaO2/FiO2 ≤300 mmHg, requirement of mechanical ventilation or worsening of lung CT findings) | Age >60 years, lymphocytes ≤1×109 L−1) LDH <250, 250–500, >500 U·L−1, comorbidity (hypertension, diabetes, CVD, chronic lung disease or HIV) | Cox regression | Points-based score |
| Luet al. [38] | Lu | China | Inpatients with suspected or confirmed COVID-19 | COVID-specific | Mortality (12 days) | Age ≥60 years, CRP ≥34 mg·L−1 | Cox regression | Points-based score |
| Shiet al. [39] | Shi | China | Inpatients with confirmed COVID-19 | COVID-specific | Death or “severe” COVID-19 (not defined) over unspecified period | Age >50 years, male, hypertension | Not specified | Points-based score |
| Xieet al. [40] | Xie | China | Inpatients with confirmed COVID-19 | COVID-specific | Mortality (in-hospital) | Age, lymphocytes, LDH, oxygen saturation | Logistic regression | Logistic regression |
| Yanet al. [41] | Yan | China | Inpatients with suspected COVID-19 | COVID-specific | Mortality (period unspecified) | LDH >365 U·L−1, CRP >41.2 mg·L−1, lymphocyte percentage >14.7% | Decision-tree model with XG boost | Points-based score |
| Zhanget al. [42] | Zhang_poor, Zhang_death | China | Inpatients with confirmed COVID-19 | COVID-specific | Mortality and poor outcome (ARDS, intubation or ECMO, ICU admission) as separate models; no timepoint specified | Age, sex, neutrophils, lymphocytes, platelets, CRP, creatinine | Logistic regression (LASSO) | Logistic regression |
MEWS: modified early warning score; qSOFA: quick sequential (sepsis-related) organ failure assessment; REMS: rapid emergency medicine score; NEWS: national early warning score; TACTIC: therapeutic study in pre-ICU patients admitted with COVID-19; ICU: intensive care unit; AVPU: alert/responds to voice/responsive to pain/unresponsive; COVID: coronavirus disease; CVD: cardiovascular disease; CRP: C-reactive protein; LDH: lactate dehydrogenase; RALE: Radiographic Assessment of Lung Edema; COPD: chronic obstructive pulmonary disease; CT: computed tomography; ARDS: acute respiratory distress syndrome; ECMO: extracorporeal membrane oxygenation. #: MEWS and REMS were evaluated among people with COVID-19 by Hu et al. [43], and thus were included in the present study; ¶: no model intercept was available so the intercepts for these models were calibrated to the validation dataset, using the model linear predictors as offset terms; +: using oxygen scale 1 for all participants, except for those with target oxygen saturation ranges of 88%–92%, e.g. in hypercapnic respiratory failure, when scale 2 is used, as recommended [12].
Overview of study cohort
During the study period, 521 adults were admitted with a final diagnosis of COVID-19, of whom 411 met the eligibility criteria for inclusion (supplementary figure S2). The median age of the cohort was 66 years (interquartile range (IQR) 53–79 years), and the majority were male (252 of 411; 61.3%). Table 2 shows the baseline demographics, comorbidities, laboratory results and clinical measurements of the study cohort, of which most (370 of 411; 90.0%) had PCR-confirmed SARS-CoV-2 infection (315 of 370 (85.1%) were positive on their first PCR test). A total of 180 participants (43.8%) met the endpoints of clinical deterioration and 115 participants (28.0%) met the endpoints of mortality, above the minimum requirement of 100 events recommended for external validation studies [45]. The risks of clinical deterioration and death declined with time since admission (median days to deterioration 1.4 (IQR 0.3–4.2); median days to death 6.6 (IQR 3.6–13.1); supplementary figure S3). Most of the variables required for calculating the 22 prognostic model scores were available among the vast majority of participants. However, admission lactate dehydrogenase was only available for 183 out of 411 (44.5%) and D-dimer was only measured for 153 out of 411 (37.2%), resulting in significant missingness for models requiring these variables (supplementary figure S4).
TABLE 2.
Baseline characteristics of hospitalised adults with COVID-19 included in systematic evaluation cohort
| Variable | Subjects with available data | Overall |
| Demographics | ||
| Age years | 411 (100) | 66.0 (53.0–79.0) |
| Sex | 411 (100) | |
| Female | 159 (38.7) | |
| Male | 252 (61.3) | |
| Ethnicity | 390 (94.9) | |
| Asian | 52 (13.3) | |
| Black | 56 (14.4) | |
| White | 234 (60.0) | |
| Mixed | 7 (1.8) | |
| Other | 41 (10.5) | |
| Clinical frailty scale | 411 (100) | 2.0 (1.0–6.0) |
| Comorbidities | ||
| Hypertension | 411 (100) | 172 (41.8) |
| Chronic cardiovascular disease | 410 (99.8) | 108 (26.3) |
| Chronic respiratory disease | 411 (100) | 99 (24.1) |
| Diabetes | 411 (100) | 105 (25.5) |
| Obesity# | 411 (100) | 83 (20.2) |
| Chronic kidney disease | 410 (99.8) | 40 (9.8) |
| Laboratory measurements | ||
| C-reactive protein mg·L−1 | 403 (98.1) | 96.7 (45.2–178.7) |
| Lymphocytes ×109 | 410 (99.8) | 0.9 (0.6–1.4) |
| Lactate dehydrogenase U·L−1 | 183 (44.5) | 395.0 (309.0–511.0) |
| D-dimer ng·mL−1 | 153 (37.2) | 1070.0 (640.0–2120.0) |
| SARS-CoV-2 PCR | 411 (100) | 370 (90.0) |
| Physiological measurements | ||
| Respiratory rate breaths·min−1 | 410 (99.8) | 24.0 (20.0–28.0) |
| Heart rate beats·min−1 | 410 (99.8) | 94.0 (81.2–107.0) |
| Systolic blood pressure mmHg | 411 (100) | 131.0 (115.0–143.0) |
| Oxygen saturation % on air | 403 (98.1) | 91.0 (86.0–95.0) |
| Outcome | ||
| Deteriorated | 411 (100) | 180 (43.8) |
| Died | 411 (100) | 115 (28.0) |
Laboratory and physiological measurements reflect parameters at the time of hospital admission. Data are presented as n (%) or median (interquartile range). COVID-19: coronavirus disease 2019; SARS-CoV-2: severe acute respiratory syndrome coronavirus-2. #: clinician-defined obesity.
Evaluation of prognostic models for original primary outcomes
Table 3 shows discrimination and calibration metrics, where appropriate, for the 22 evaluated prognostic models in the primary multiple imputation analysis. The highest AUROCs were achieved by the NEWS2 score for prediction of deterioration over 24 h (0.78, 95% CI 0.73–0.83) and the Carr “final” model for prediction of deterioration over 14 days (0.78, 95% CI 0.74–0.82). Of the other prognostic scores currently used in routine clinical practice, CURB65 had an AUROC of 0.75 for 30-day mortality (95% CI 0.70–0.80), while qSOFA discriminated in-hospital mortality with an AUROC of 0.6 (95% CI 0.55–0.65).
TABLE 3.
Validation metrics of prognostic scores for COVID-19, using primary multiple imputation analysis (n=411)
| Score | Primary outcome | AUROC (95% CI) | Calibration slope (95% CI) | Calibration-in-the-large (95% CI) |
| NEWS2 | Deterioration (1 day) | 0.78 (0.73–0.83) | ||
| Ji | Deterioration (10 days) | 0.56 (0.5–0.62) | ||
| Carr_final | Deterioration (14 days) | 0.78 (0.74–0.82) | 1.04 (0.8–1.28) | 0.33 (0.11–0.55) |
| Carr_threshold | Deterioration (14 days) | 0.76 (0.71–0.81) | 0.85 (0.65–1.05) | −0.34 (−0.57– −0.12) |
| Guo | Deterioration (14 days) | 0.67 (0.61–0.73) | ||
| Zhang_poor | Deterioration (in-hospital) | 0.74 (0.69–0.79) | 0.33 (0.22–0.43) | 0.56 (0.3–0.81) |
| Galloway | Deterioration (in-hospital) | 0.72 (0.68–0.77) | ||
| TACTIC | Deterioration (in-hospital) | 0.7 (0.65–0.75) | ||
| Colombi_clinical | Deterioration (in-hospital) | 0.69 (0.63–0.74) | 0.53 (0.35–0.71) | 0 (−0.23–0.23) |
| Huang | Deterioration (in-hospital) | 0.67 (0.61–0.73) | 0.18 (0.1–0.26) | −4.26 (−4.61– −3.91) |
| Shi | Deterioration (in-hospital) | 0.61 (0.56–0.66) | ||
| MEWS | Deterioration (in-hospital) | 0.6 (0.54–0.65) | ||
| Lu | Mortality (12 days) | 0.72 (0.67–0.76) | ||
| CURB65 | Mortality (30 days) | 0.75 (0.7–0.8) | ||
| BelloChavolla | Mortality (30 days) | 0.66 (0.6–0.72) | ||
| REMS | Mortality (in-hospital) | 0.76 (0.71–0.81) | ||
| Xie | Mortality (in-hospital) | 0.76 (0.69–0.82) | 0.83 (0.51–1.15) | 0.41 (0.16–0.66) |
| Hu | Mortality (in-hospital) | 0.74 (0.68–0.79) | 0.33 (0.2–0.45) | −1.07 (−1.37– −0.77) |
| Caramelo | Mortality (in-hospital) | 0.71 (0.66–0.76) | 0.53 (0.36–0.69) | 0 (−0.25–0.25) |
| Zhang_death | Mortality (in-hospital) | 0.7 (0.65–0.76) | 0.29 (0.19–0.4) | 0.89 (0.6–1.19) |
| qSOFA | Mortality (in-hospital) | 0.6 (0.55–0.65) | ||
| Yan | Mortality (in-hospital) | 0.58 (0.49–0.67) |
For each model, performance is evaluated for its original intended outcome, shown in “Primary outcome” column. AUROC: area under the receiver operating characteristic curve.
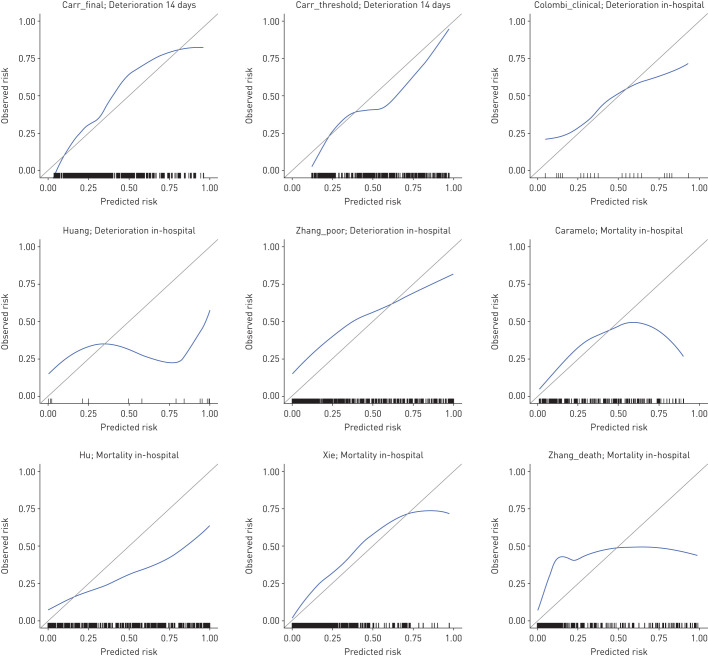
For all models that provide probability scores for either deterioration or mortality, calibration appeared visually poor with evidence of overfitting and either systematic overestimation or underestimation of risk (figure 1). Supplementary figure S5 shows associations between prognostic models with points-based scores and actual risk. In addition to demonstrating reasonable discrimination, the NEWS2 and CURB65 models demonstrated approximately linear associations between scores and actual probability of deterioration at 24 h and mortality at 30 days, respectively.
FIGURE 1.
Calibration plots for prognostic models estimating outcome probabilities. For each plot, the blue line represents a LOESS-smoothed calibration curve from the stacked multiple imputed datasets and rug plots indicate the distribution of data points. No model intercept was available for the Caramelo or Colombi “clinical” models; the intercepts for these models were calibrated to the validation dataset using the model linear predictors as offset terms. The primary outcome of interest for each model is shown in the plot sub-heading.
Time-dependent discrimination of candidate models and a priori univariable predictors for standardised outcomes
Next, we sought to compare the discrimination of these models for both clinical deterioration and mortality across the range of time horizons, benchmarked against preselected univariable predictors associated with adverse outcomes in COVID-19 [8, 19]. We recalculated time-dependent AUROCs for each of these outcomes, stratified by time horizon to the outcome (supplementary figures S6 and S7). These analyses showed that AUROCs generally declined with increasing time horizons. Admission peripheral oxygen saturation on room air was the strongest predictor of in-hospital deterioration (AUROC 0.76, 95% CI 0.71–0.81), while age was the strongest predictor of in-hospital mortality (AUROC 0.76, 95% CI 0.71–0.81).
Decision curve analyses to assess clinical utility
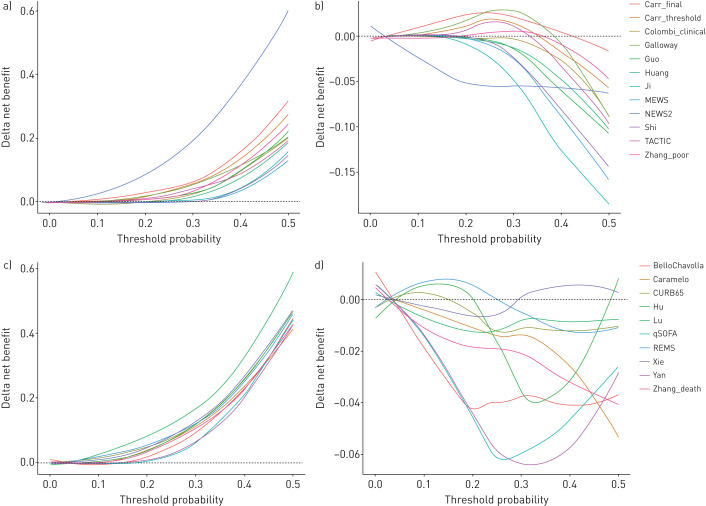
We compared net benefit for each prognostic model (for its original intended endpoint) to the strategies of treating all patients, treating no patients and using the most discriminating univariable predictor for either deterioration (i.e. oxygen saturation on air) or mortality (i.e. patient age) to stratify treatment (supplementary figure S8). Although all prognostic models showed greater net benefit than treating all patients at the higher range of threshold probabilities, none of these models demonstrated consistently greater net benefit than the most discriminating univariable predictor, across the range of threshold probabilities (figure 2).
FIGURE 2.
Decision curve analysis showing delta net benefit of each candidate model, compared to treating all patients and best univariable predictors. a) Deterioration models versus treat all; b) deterioration models versus peripheral oxygen saturation (SpO2) on air alone; c) mortality models versus treat all; d) mortality models versus age alone. For each analysis, the endpoint is the original intended outcome and time horizon for the index model. Each candidate model and univariable predictor was calibrated to the validation data during analysis to enable fair, head-to-head comparisons. Delta net benefit is calculated as net benefit when using the index model minus net benefit when 1) treating all patients and 2) using the most discriminating univariable predictor. The most discriminating univariable predictor is admission SpO2 on room air for deterioration models and patient age for mortality models. Delta net benefit is shown with LOESS-smoothing. Black dashed line indicates threshold above which index model has greater net benefit than the comparator. Individual decision curves for each candidate model are shown in supplementary figure S8.
Sensitivity analyses
Recalculation of model discrimination and calibration metrics for predicting the original intended endpoint using 1) a complete case analysis, 2) excluding patients without PCR-confirmed SARS-CoV-2 infection and 3) excluding patients who met the clinical deterioration outcome within 4 h of arrival to hospital revealed similar results to the primary multiple imputation approach, though discrimination was noted to be lower overall when excluding early events (supplementary table S2). Visual examination of associations between the most discriminating univariable predictors and log odds of deterioration or death using restricted cubic splines showed no evidence of non-linear associations (supplementary figure S9). Finally, internal validation using bootstrapping showed near zero optimism for discrimination and calibration parameters for the univariable models (supplementary table S3).
Discussion
In this observational cohort study of consecutive adults hospitalised with COVID-19, we systematically evaluated the performance of 22 prognostic models for COVID-19. These included models developed specifically for COVID-19, along with existing scores in routine clinical use prior to the pandemic. For prediction of clinical deterioration or mortality, AUROCs ranged from 0.56–0.78. NEWS2 performed reasonably well for prediction of deterioration over a 24-h interval, achieving an AUROC of 0.78, while the Carr “final” model [30] also had an AUROC of 0.78, but tended to systematically underestimate risk. All COVID-specific models that derived an outcome probability of either deterioration or mortality showed poor calibration. We found that peripheral oxygen saturation on room air (AUROC 0.76) and patient age (AUROC 0.76) were the most discriminating single variables for prediction of in-hospital deterioration and mortality respectively. These predictors have the added advantage that they are immediately available at the point of presentation to hospital. In decision curve analysis, which is dependent upon both model discrimination and calibration, no prognostic model demonstrated clinical utility consistently greater than using these univariable predictors to inform decision-making.
While previous studies have largely focused on novel model discovery, or evaluating a limited number of existing models, this is the first study to our knowledge to evaluate systematically identified candidate prognostic models for COVID-19. We used a comprehensive living systematic review [8] to identify eligible models and sought to reconstruct each model as per the original authors’ description. We then evaluated performance against its intended outcome and time horizon, wherever possible, using recommended methods of external validation incorporating assessments of discrimination, calibration and net benefit [17]. We used a robust approach of electronic health record data capture, supported by manual curation, to ensure a high-quality dataset, and inclusion of unselected and consecutive COVID-19 cases that met our eligibility criteria. We used robust outcome measures of mortality and clinical deterioration, aligning with the WHO Clinical Progression Scale [16].
A weakness of the current study is that it is based on retrospective data from a single centre, and therefore cannot assess between-setting heterogeneity in model performance. Second, owing to the limitations of routinely collected data, predictor variables were available for varying numbers of participants for each model, with a large proportion of missingness for models requiring lactate dehydrogenase and D-dimer measurements. We therefore performed multiple imputation, in keeping with recommendations for development and validation of multivariable prediction models, in our primary analyses [46]. Findings were similar in the complete case sensitivity analysis, thus supporting the robustness of our results. Future studies would benefit from standardising data capture and laboratory measurements prospectively to minimise predictor missingness. Third, a number of models could not be reconstructed in our data. For some models, this was due the absence of predictors in our dataset, such as those requiring computed tomography imaging, because this is not currently routinely recommended for patients with suspected or confirmed COVID-19 [15]. We were also not able to include models for which the parameters were not publicly available. This underscores the need for strict adherence to reporting standards in multivariable prediction models [13]. Finally, we used admission data only as predictors in this study, because most prognostic scores are intended to predict outcomes at the point of hospital admission. We note, however, that some scores are designed for dynamic inpatient monitoring, with NEWS2 showing reasonable discrimination for deterioration over a 24-h interval, as originally intended [44]. Future studies may integrate serial data to examine model performance when using such dynamic measurements.
Despite the vast global interest in the pursuit of prognostic models for COVID-19, our findings show that none of the COVID-19-specific models evaluated in this study can currently be recommended for routine clinical use. In addition, while some of the evaluated models that are not specific to COVID-19 are routinely used and may be of value among inpatients [12, 44], people with suspected infection [10] or community-acquired pneumonia [11], none showed greater clinical utility than the strongest univariable predictors among patients with COVID-19. Our data show that admission oxygen saturation on room air is a strong predictor of clinical deterioration and suggest that it should be evaluated in future studies to stratify inpatient management and for remote community monitoring. We note that all novel prognostic models for COVID-19 assessed in the current study were derived from single-centre data. Future studies may seek to pool data from multiple centres to robustly evaluate the performance of existing and newly emerging models across heterogeneous populations, and develop and validate novel prognostic models, through individual participant data meta-analysis [47]. Such an approach would allow assessments of between-study heterogeneity and the likely generalisability of candidate models. It is also imperative that discovery populations are representative of target populations for model implementation, with inclusion of unselected cohorts. Moreover, we strongly advocate for transparent reporting in keeping with TRIPOD standards (including modelling approaches, all model coefficients and standard errors) along with standardisation of outcomes and time horizons, to facilitate ongoing systematic evaluations of model performance and clinical utility [13].
We conclude that baseline oxygen saturation on room air and patient age are strong predictors of deterioration and mortality, respectively. None of the prognostic models evaluated in this study offer incremental value for patient stratification to these univariable predictors when using admission data. Therefore, none of the evaluated prognostic models for COVID-19 can be recommended for routine clinical implementation. Future studies seeking to develop prognostic models for COVID-19 should consider integrating multi-centre data to increase generalisability of findings, and should ensure benchmarking against existing models and simpler univariable predictors.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-03498-2020.Supplement (2.2MB, pdf)
Shareable PDF
Footnotes
The UCLH COVID-19 Reporting Group comprised the following individuals, who were involved in data curation as non-author contributors: Asia Ahmed, Ronan Astin, Malcolm Avari, Elkie Benhur, Anisha Bhagwanani, Timothy Bonnici, Sean Carlson, Jessica Carter, Sonya Crowe, Mark Duncan, Ferran Espuny-Pujol, James Fullerton, Marc George, Georgina Harridge, Ali Hosin, Rachel Hubbard, Adnan Hubraq, Prem Jareonsettasin, Zella King, Avi Korman, Sophie Kristina, Lawrence Langley, Jacques-Henri Meurgey, Henrietta Mills, Alfio Missaglia, Ankita Mondal, Samuel Moulding, Christina Pagel, Liyang Pan, Shivani Patel, Valeria Pintar, Jordan Poulos, Ruth Prendecki, Alexander Procter, Magali Taylor, David Thompson, Lucy Tiffen, Hannah Wright, Luke Wynne, Jason Yeung, Claudia Zeicu, Leilei Zhu.
Author contributions: R.K. Gupta and M. Noursadeghi conceived the study. R.K. Gupta conducted the analysis and wrote the first draft of the manuscript. All other authors contributed towards data collection, study design and/or interpretation. All authors have critically appraised and approved the final manuscript prior to submission. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. Members of The UCLH COVID-19 Reporting Group contributed towards data curation and are non-author contributors/collaborators for this study.
This article has an editorial commentary: https://doi.org/10.1183/13993003.03728-2020
This article has supplementary material available from erj.ersjournals.com
The conditions of regulatory approvals for the present study preclude open access data sharing to minimise risk of patient identification through granular individual health record data. The authors will consider specific requests for data sharing as part of academic collaborations subject to ethical approval and data transfer agreements in accordance with GDPR regulations.
Support statement: The study was funded by National Institute for Health Research (DRF-2018-11-ST2-004 to R.K. Gupta; NF-SI-0616-10037 to I. Abubakar), the Wellcome Trust (207511/Z/17/Z to M. Noursadeghi) and has been supported by the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC), in particular by the NIHR UCLH/University College London (UCL) BRC Clinical and Research Informatics Unit. This paper presents independent research supported by the NIHR. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. The funder had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: M. Marks has nothing to disclose.
Conflict of interest: T.H.A. Samuels has nothing to disclose.
Conflict of interest: A. Luintel has nothing to disclose.
Conflict of interest: T. Rampling has nothing to disclose.
Conflict of interest: H. Chowdhury has nothing to disclose.
Conflict of interest: M. Quartagno has nothing to disclose.
Conflict of interest: A. Nair reports non-financial support from AIDENCE BV and grants from NIHR UCL Biomedical Research Centre, outside the submitted work.
Conflict of interest: M. Lipman has nothing to disclose.
Conflict of interest: I. Abubakar has nothing to disclose.
Conflict of interest: M. van Smeden has nothing to disclose.
Conflict of interest: W.K. Wong has nothing to disclose.
Conflict of interest: B. Williams has nothing to disclose.
Conflict of interest: M. Noursadeghi reports grants from Wellcome Trust and National Institute for Health Research Biomedical Research Centre at University College London NHS Foundation Trust, during the conduct of the study.
Conflict of interest: R.K. Gupta has nothing to disclose.
References
- 1.Richardson S, Hirsch JS, Narasimhan M, et al. . Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 2020; 323: 2052–2059. doi: 10.1001/jama.2020.6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Docherty AB, Harrison EM, Green CA, et al. . Features of 20 133 UK patients in hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ 2020; 369: m1985. doi: 10.1136/bmj.m1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G, Pesenti A, Cecconi M. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy. JAMA 2020; 323: 1545. doi: 10.1001/jama.2020.4031 [DOI] [PubMed] [Google Scholar]
- 4.Imperial College COVID-19 Response Team Report 17 - Clinical characteristics and predictors of outcomes of hospitalised patients with COVID-19 in a London NHS Trust: a retrospective cohort study. www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-17-clinical/. Date last accessed: 14 May 2020. Date last updated: 29 April 2020.
- 5.Li R, Rivers C, Tan Q, et al. . The demand for inpatient and ICU beds for COVID-19 in the US: lessons from Chinese cities. medRxiv 2020; preprint [ 10.1101/2020.03.09.20033241]. [DOI] [Google Scholar]
- 6.Beigel JH, Tomashek KM, Dodd LE. Remdesivir for the treatment of COVID-19 – preliminary report. N Engl J Med 2020; 383: 994. doi: 10.1056/NEJMc2022236 [DOI] [PubMed] [Google Scholar]
- 7.Horby P, Lim WS, Emberson J, et al. . Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. N Engl J Med 2020; in press [ 10.1056/NEJMoa2021436]. [DOI] [Google Scholar]
- 8.Wynants L, Van Calster B, Collins GS, et al. . Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. BMJ 2020; 369: m1328. doi: 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolff RF, Moons KGM, Riley RD, et al. . PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med 2019; 170: 51–58. doi: 10.7326/M18-1376 [DOI] [PubMed] [Google Scholar]
- 10.Seymour CW, Liu VX, Iwashyna TJ, et al. . Assessment of clinical criteria for sepsis. JAMA 2016; 315: 762. doi: 10.1001/jama.2016.0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim WS, van den Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 2003; 58: 377–382. doi: 10.1136/thorax.58.5.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royal College of Physicians National Early Warning Score (NEWS). www.rcplondon.ac.uk/projects/outputs/national-early-warning-score-news-2. Date last accessed: 1 July 2020. Date last updated: 19 December 2017.
- 13.Collins GS, Reitsma JB, Altman DG, et al. . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015; 350: g7594. doi: 10.1136/bmj.g7594 [DOI] [PubMed] [Google Scholar]
- 14.Wong HYF, Lam HYS, Fong AH-T, et al. . Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology 2019; 296: E72–E78. doi: 10.1148/radiol.2020201160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.British Society of Thoracic Imaging COVID-19 Resources. www.bsti.org.uk/covid-19-resources/. Date last accessed: 1 July 2020
- 16.WHO Working Group on the Clinical Characterisation and Management of COVID-19 Infection Marshall JC, Murthy S, et al. . A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis 2020; 20: e192–e197. doi: 10.1016/S1473-3099(20)30483-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riley RD, van der Windt D, Croft P, et al. . Prognosis research in healthcare: concepts, methods, and impact. Oxford, Oxford University Press, 2019. [Google Scholar]
- 18.Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol 2017; 17: 53. doi: 10.1186/s12874-017-0332-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewitt J, Carter B, Vilches-Moraga A, et al. . The effect of frailty on survival in patients with COVID-19 (COPE): a multicentre, European, observational cohort study. Lancet 2020; 5: E444–E451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers AJ, van Calster B, Steyerberg EW. A simple, step-by-step guide to interpreting decision curve analysis. Diagnostic Progn Res 2019; 3: 18. doi: 10.1186/s41512-019-0064-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown M. rmda: Risk Model Decision Analysis. https://mdbrown.github.io/rmda/. Date last updated: 20 March 2018.
- 22.White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med 2011; 30: 377–399. doi: 10.1002/sim.4067 [DOI] [PubMed] [Google Scholar]
- 23.van Buuren S, Groothuis-Oudshoorn K. mice: multivariate imputation by chained equations in R. J Stat Softw 2011; 45: 1–67. [Google Scholar]
- 24.Rubin DB. Multiple imputation for nonresponse in surveys. New York, Wiley-Interscience, 2004. [Google Scholar]
- 25.Harrell FE., Jr rms: Regression Modeling Strategies. https://hbiostat.org/R/rms/. Date last updated: 21 August 2020.
- 26.Subbe CP, Kruger M, Rutherford P, et al. . Validation of a modified early warning score in medical admissions. QJM 2001; 94: 521–526. doi: 10.1093/qjmed/94.10.521 [DOI] [PubMed] [Google Scholar]
- 27.Olsson T, Terent A, Lind L. Rapid Emergency Medicine Score: a new prognostic tool for in-hospital mortality in nonsurgical emergency department patients. J Intern Med 2004; 255: 579–587. doi: 10.1111/j.1365-2796.2004.01321.x [DOI] [PubMed] [Google Scholar]
- 28.Bello-Chavolla OY, Bahena-López JP, Antonio-Villa NE, et al. . Predicting mortality due to SARS-CoV-2: a mechanistic score relating obesity and diabetes to COVID-19 outcomes in Mexico. J Clin Endocrinol Metab 2020; 105: 2752–2761. doi: 10.1210/clinem/dgaa346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caramelo F, Ferreira N, Oliveiros B. Estimation of risk factors for COVID-19 mortality - preliminary results. medRxiv 2020; preprint [ 10.1101/2020.02.24.20027268]. [DOI] [Google Scholar]
- 30.Carr E, Bendayan R, Bean D, et al. . Evaluation and improvement of the National Early Warning Score (NEWS2) for COVID-19: a multi-hospital study. medRxiv 2020; preprint [ 10.1101/2020.04.24.20078006]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colombi D, Bodini FC, Petrini M, et al. . Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology 2020: 296: E86–E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galloway JB, Norton S, Barker RD, et al. . A clinical risk score to identify patients with COVID-19 at high risk of critical care admission or death: an observational cohort study. J Infect 2020; 81: 282–288. doi: 10.1016/j.jinf.2020.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Liu Y, Lu J, et al. . Development and validation of an early warning score (EWAS) for predicting clinical deterioration in patients with coronavirus disease 2019. medRxiv 2020; preprint [ 10.1101/2020.04.17.20064691]. [DOI] [Google Scholar]
- 34.Hall F, Jayne D; Cambridge Clinical Trials Unit TACTIC Trial. www.cctu.org.uk/portfolio/COVID-19/TACTIC. Date last accessed: 1 July 2020.
- 35.Chen X, Liu Z. Early prediction of mortality risk among severe COVID-19 patients using machine learning. medRxiv 2020; preprint [ 10.1101/2020.04.13.20064329]. [DOI] [Google Scholar]
- 36.Huang H, Cai S, Li Y, et al. . Prognostic factors for COVID-19 pneumonia progression to severe symptom based on the earlier clinical features: a retrospective analysis. medRxiv 2020; preprint [ 10.1101/2020.03.28.20045989]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji D, Zhang D, Xu J, et al. . Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis 2020; 71: 1393–1399. doi: 10.1093/cid/ciaa414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Hu S, Fan R, et al. . ACP risk grade: a simple mortality index for patients with confirmed or suspected severe acute respiratory syndrome coronavirus 2 disease (COVID-19) during the early stage of outbreak in Wuhan, China. medRxiv 2020; preprint [ 10.1101/2020.02.20.20025510]. [DOI] [Google Scholar]
- 39.Shi Y, Yu X, Zhao H, et al. . Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care 2020; 24: 108. doi: 10.1186/s13054-020-2833-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J, Hungerford D, Chen H, et al. . Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID-19. medRxiv 2020; preprint [ 10.1101/2020.04.28.20082222]. [DOI] [Google Scholar]
- 41.Yan L, Zhang H-T, Goncalves J, et al. . An interpretable mortality prediction model for COVID-19 patients. Nat Mach Intell 2020; 2: 283–288. doi: 10.1038/s42256-020-0180-7 [DOI] [Google Scholar]
- 42.Zhang H, Shi T, Wu X, et al. . Risk prediction for poor outcome and death in hospital in-patients with COVID-19: derivation in Wuhan, China and external validation in London, UK. medRxiv 2020; preprint. doi: 10.1101/2020.04.28.20082222 [DOI] [Google Scholar]
- 43.Hu H, Yao N, Qiu Y. Comparing rapid scoring systems in mortality prediction of critically ill patients with novel coronavirus disease. Acad Emerg Med 2020; 27: 461–468. 10.1111/acem.13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith GB, Prytherch DR, Meredith P, et al. . The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation 2013; 84: 465–470. doi: 10.1016/j.resuscitation.2012.12.016 [DOI] [PubMed] [Google Scholar]
- 45.Collins GS, Ogundimu EO, Altman DG. Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med 2016; 35: 214–226. doi: 10.1002/sim.6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moons KGM, Altman DG, Reitsma JB, et al. . Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015; 162: W1–W73. doi: 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 47.Debray TPA, Riley RD, Rovers MM, et al. . Individual participant data (IPD) meta-analyses of diagnostic and prognostic modeling studies: guidance on their use. PLoS Med 2015; 12: e1001886. doi: 10.1371/journal.pmed.1001886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-03498-2020.Supplement (2.2MB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-03498-2020.Shareable (527.9KB, pdf)