Abstract
RNA therapeutics are finally taking their place as a main drug category alongside small molecules and proteins. Here, we follow the twists and turns on their road to success and highlight areas of ongoing research.
Keywords: oligonucleotide, therapeutic, antisense, siRNA, miRNA, aptamer
The Twists and Turns on the Road
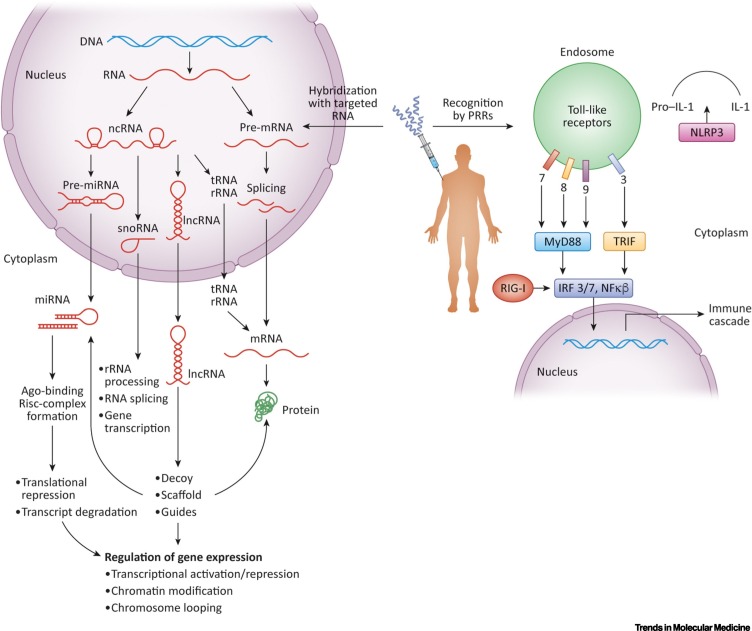
The central dogma of molecular biology proposed by Francis Crick in 1958 states that sequence information encoded in nucleic acids can be transferred to other nucleic acids and proteins but not from protein to protein or protein to nucleic acids [1]. tRNA, rRNA, mRNA, miRNA, small nucleolar RNAs (snoRNA), functional noncoding RNA (ncRNA), and long noncoding RNA (lncRNA) are essential in this transfer (Figure 1 ).
Figure 1.
RNAs Transfer Genetic Information.
Genetic information, stored in DNA, is transcribed into RNA and then translated into proteins. This information transfer involves various RNAs in addition to mRNA, such as miRNAs, rRNAs, small nucleolar RNAs (snoRNAs), tRNAs, noncoding RNA (ncRNA), and long noncoding RNAs (lncRNAs). snoRNAs add endogenous modifications to rRNAs and tRNAs, which are required for their function in the translation of mRNA into protein. mRNAs may undergo alternative splicing to make a wider array of proteins. ncRNAs regulate cellular nucleic acids by various mechanisms. Both antisense oligonucleotides (ASOs) and siRNAs can modulate the expression of the protein product of their target mRNA but may also be used to target any of the other RNAs in the cell. In these approaches, a synthetic oligonucleotide complementary to RNA or DNA is employed. While the intended target of these compounds is RNA or DNA, these compounds could also be recognized by pattern recognition receptors (PRRs) as pathogen associated molecular patterns (PAMPs). Nucleic acids from pathogens are recognized by PRRs, including Toll-like receptors (TLRs), RIG-like receptors (RLRs), and cytosolic DNA sensors as part of the innate immune system and induce immune signaling cascades to mount a host defense. Abbreviation: RISC, RNA-induced silencing complex.
Thus, modulating nucleic acids will affect proteins and can induce therapeutic effects. Starting with this premise, the road of RNA therapeutics forked into two separate directions: one where mRNA is introduced to cells to produce desired proteins (mRNA therapy) and one where synthetic nucleic acids are used to manipulate cellular RNAs (RNA therapeutics) [2]. Here, we concentrate on the latter approach. (See Figure 2 ).
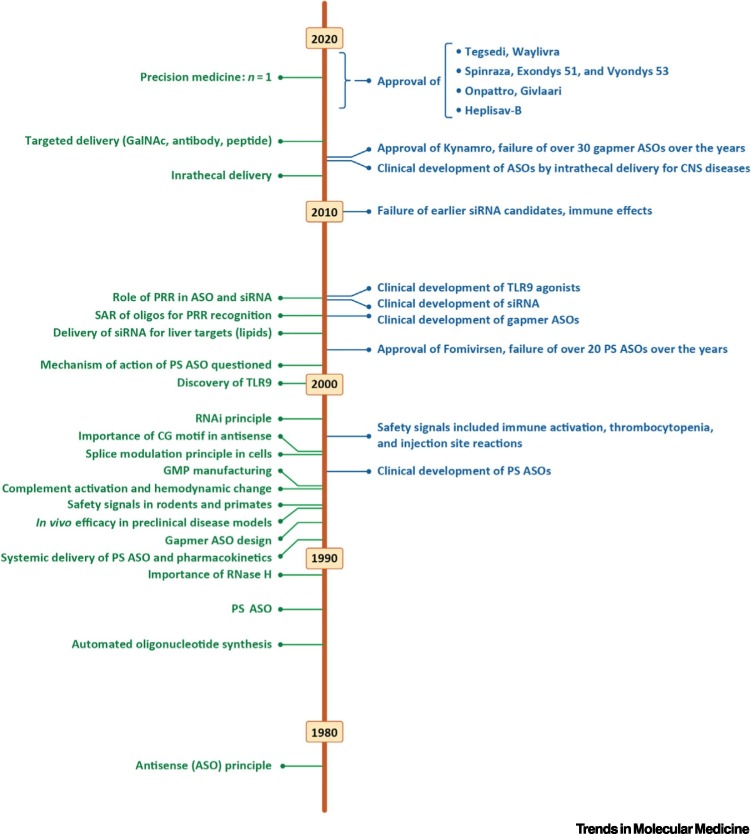
Figure 2.
Timelines of Key Developments in RNA Therapeutics.
Abbreviations: ASO, antisense oligonucleotide; CNS, central nervous system; GalNAc, N-acetylgalactosamine; GMP, good manufacturing practice; PRR, pattern recognition receptor; PS, phosphorothioate; TLR9, toll-like receptor 9.
RNA therapeutics first gained attention in 1978, when Paul Zamecnik reported that a synthetic 13-mer oligonucleotide complementary to the viral mRNA could inhibit replication of Rous sarcoma virus [3]. These findings established the viability of the antisense approach; however, further progress was slow. According to Paul Zamecnik, there was disbelief that an oligonucleotide could enter cells. Additionally, methodologies for sequencing DNA and synthesis of oligonucleotides were not yet practical or established. In the mid-1980s, improved synthesis methods developed by the Gait, Letsinger, and Caruthers laboratories revived efforts to use antisense oligonucleotides (ASOs) as therapeutics. While working in Gait’s laboratory in 1986, I received a call from Paul Zamecnik in which he described his recent data showing that ASOs could inhibit replication of HIV-1, a human retrovirus that had recently been discovered. He suggested that achieving drug-like properties for ASOs would require experimenting with the chemistry, a project he wanted me to take on. This phone call certainly changed the course of my career.
Our first objective was to improve nucleolytic stability so that the oligonucleotides could survive in biological fluids and reach target cells. To achieve this, we chemically modified the internucleotide linkages, but we soon realized that nucleolytic stability was only one parameter [4] and discovered that affinity to target RNA and RNase H activation were also important. Based on growing data, phosphorothioate (PS) modification became the choice for first generation ASOs and a number of companies quickly formed to advance therapeutic applications. Regrettably, during preclinical and clinical evaluations, side effects, including complement activation, the prolongation of activated partial thromboplastin time (aPTT) related to the polyanionic nature of PS ASOs, and immune activation, became apparent [5]. This led to the discontinuation of most of the PS antisense candidates and a broader scrutiny of the underlying mechanism of action. News started to appear with titles such as ‘antisense has growing pains’.
With more experience, we began to understand that each chemical modification confers particular properties to the modified oligonucleotide. However, to achieve the optimal therapeutic profile, the desirable characteristics of several chemical modifications needed to be combined. We developed hybrid oligonucleotides (also known as gapmers) with a central PS DNA region that allows for RNase H activity when bound to complementary RNA as well as two modified RNA wings that protect the central region [6]. This gapmer design is now the standard for ASOs and has been utilized in the approved drugs Tegsedi and Waylivra.
Many Roads Split Off
Chemical modifications that do not permit RNase H activity are used in splice switching ASOs that force the cellular machinery to splice pre-mRNA into mRNA in desired ways [7]. This property can be used for a variety of therapeutic purposes: restoration of cryptic splicing, reading frame restoration, exon inclusion or exclusion, forcing nonsense-mediated mRNA decay of resulting isoforms, changing ratios of naturally occurring splice isoforms, or reducing synthesis of nonproductive mRNA and thus increasing the synthesis of productive mRNA [8]. Several splice modulating therapeutics are now FDA approved, including Spinraza [9], Exondys 51, and Vyondys 53.
In parallel to these developments, novel functions of RNAs and the mechanisms of gene regulation were also discovered. In 1998, the Mello laboratory demonstrated that double-stranded RNA could interfere with gene expression in Caenorhabditis elegans. Further studies showed that long double-stranded RNAs as well as pre-miRNAs derived from the genome are cleaved by Dicer into 20–25 bp-long siRNAs and mature miRNAs, respectively. These products are then loaded onto the RNA-induced silencing complex (RISC) complex where the guide strand locates and hybridizes to complementary mRNA that is then cleaved by Ago2 [10]. Synthetic siRNAs, miRNA mimics, and miRNA antagomirs can all be designed to take advantage of this process. The first siRNA therapeutic (Onpattro) was approved in 2018, with a second, Givlaari, closely following. Many more are in clinical trials. miRNA mimics and ASOs targeting miRNA are in clinical studies, but until recently the outcomes have been disappointing, either due to lack of clinical activity or safety signal concerns. Antisense candidates targeting ncRNA and lncRNA have shown encouraging results in preclinical models [11].
Similar problems have beset aptamers, despite the fact that the aptamer Macugen was the first approved RNA therapeutic. Aptamers are protein-binding nucleic acids selected from a pool of random sequences in a reiterative process known as SELEX (systematic evolution of ligands by exponential enrichment) [12]. The nature of the SELEX process limits compatible chemical modifications and the postselection addition of chemical modifications is problematic due to resulting changes in the binding. However, recent progress in expanding the chemical repertoire will hopefully transfer into clinical success soon.
Tolls to Be Paid
All the RNA therapeutics discussed earlier are composed of modified nucleic acids. While the intended target of these compounds is RNA, they can also be recognized by pattern recognition receptors (PRRs). PRRs such as Toll-like receptors (TLRs), RIG-like receptors (RLRs), and cytosolic DNA sensors are part of the innate immune system (Figure 1). They are able to recognize nucleic acids from pathogens via nucleotide sequences, motifs, secondary structures, accessibility of 5′ and 3′ ends or palindromic regions. Of particular note to consider in RNA therapeutics are the unmethylated CpG dinucleotides normally present in bacterial DNA, which are recognized by TLR9. If these motifs are inadvertently contained in oligonucleotides, as was the case in many early therapeutic candidates, they can cause unintended immune activation, thereby impacting the intended mechanism of action and producing alarming safety signals [13].
However, these hard-earned insights have led to the development of synthetic DNA- and RNA-based compounds optimized to interact with PRRs [14]. Such compounds have shown promise as vaccine adjuvants, antiviral, anticancer, and anti-inflammatory agents. For example, the recently approved hepatitis B vaccine, HEPLISAV-B, contains a CpG oligonucleotide. This vaccine was shown to significantly increase immune response in older and diabetic populations and thus may make this adjuvant particularly well-suited for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines. Several TLR agonists (e.g., IMO-2125) are in clinical trials for intratumoral immunotherapy in combination with check point inhibitors.
For most RNA therapeutics, delivery remains the biggest issue. One notable exception is N-acetylgalactosamine conjugation, which enables successful delivery of ASOs and siRNA to hepatocytes. Another option is local delivery, for example, into the eye or brain for ASOs or intratumoral for CpG oligonucleotides. Progress is being made in using aptamers, antibody, and peptide-mediated delivery to muscles and other organs of choice, but clinical results are not yet available.
Concluding Remarks
Are we there yet? RNA drugs are composed of four nucleotides (A, C, G, and T or U) in a defined sequence. The nucleotides are connected with modified linkages and/or contain altered sugars and it is these synthetic nucleotides that provide the drug-like properties. Thus, in principle, the design of RNA therapeutics only requires the target sequence (except for aptamers). However, depending on the type of oligonucleotide drug and the target, a protein such as RNase H or Ago is required to modulate the expression of the associated protein. Additionally, the oligonucleotide therapeutic itself or the degradation products of the targeted nucleic acid may activate the innate immune system. Thus, the therapeutic outcome is not simply based on base pair recognition, but also protein recruitment and binding or the avoidance of unspecific protein binding.
However, once a lead candidate has been identified, the many steps remaining before clinical use, including manufacturing using good manufacturing practice (GMP) guidelines, pharmacokinetic (PK)/pharmacodynamic (PD) studies, and safety evaluations, are the same across targets. Thus, the development of RNA therapeutics can be simplified and accelerated to the point where individualized therapies can be designed and administered to patients within 1 year [15]. This is in contrast to small molecule and antibody drugs, where this pipeline must be optimized for each compound.
To date, 11 RNA therapeutics based on multiple mechanisms have been approved and over 100 therapeutic candidates are in clinical trials for diverse disease indications. I would say that after four decades we are finally there: RNA therapeutics have taken their place as a viable drug discovery platform!
Acknowledgments
I am grateful to Dr Petra Disterer for editorial assistance and Dr Daniel Blessing for comments on Figure 1.
References
- 1.Crick F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.Agrawal S., Gait M.J., editors. Advances in Nucleic Acid Therapeutics. Royal Society of Chemistry; 2019. [Google Scholar]
- 3.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. U. S. A. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal S. Oligodeoxynucleoside phosphoramidates and phosphorothioates as inhibitors of human immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7079–7083. doi: 10.1073/pnas.85.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galbraith W.M. Complement activation and hemodynamic changes following intravenous administration of phosphorothioate oligonucleotides in the monkey. Antisense Res. Dev. 1994;4:201–206. doi: 10.1089/ard.1994.4.201. [DOI] [PubMed] [Google Scholar]
- 6.Metelev, V. and Agrawal, S. Worester Foundation for Biomedical Research Inc., University of Massachusetts. Hybrid oligonucleotide phosphorothioates, US5652355, US6143881,US6346614
- 7.Sierakowska H. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim K.H. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-17093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel R.S. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 10.Setten R.L. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 11.Micheletti R. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimjee S.M. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal S., Kandimalla E.R. Role of Toll-like receptors in antisense and siRNA. Nat. Biotechnol. 2004;22:1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 14.Vanpouille-Box C. Pharmacological modulation of nucleic acid sensors - therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2019;18:845–867. doi: 10.1038/s41573-019-0043-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim J. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 2019;381:1644–1652. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]