Abstract
Gut microbes exhibit diurnal rhythmicity, and disruptions in this rhythmicity potentially impact host health. In this issue of Cell Host & Microbe, Reitmeier et al. (2020) employ timestamped gut microbiome sequencing data from human subjects coupled with machine learning to identify microbial rhythmicity patterns that predict Type 2 Diabetes incidence.
Circadian rhythms (CRs) and the trillions of microbes residing in the gut have been shown to independently regulate host metabolism (Frazier and Chang, 2020). CRs are naturally occurring, internally driven processes in the body that respond to environmental cues such as light, driving molecular and behavioral rhythms over a ~24 h period. Recent work has highlighted an interconnectedness between host circadian networks and gut microbes across species that is intricately intertwined with host metabolic function, suggesting an evolutionarily conserved trans-kingdom dynamic that aids in maintaining host health (Alvarez et al., 2020). Misalignment of host and microbial rhythms appears to play a causative role in driving metabolic dysregulation, resulting in diseases such as obesity and Type 2 Diabetes (T2D).
Gut microbes are sensitive to a myriad of environmental factors which can drive dysbiosis with negative host consequences, although few studies have identified specific microbes driving these outcomes. Recent studies in preclinical rodent models provided first evidence in mammals that ~20% of the gut microbial community exhibits diurnal rhythmicity under conditions of health, both in microbial membership/abundance and functional outputs, i.e., microbially derived metabolites (Leone et al., 2015; Thaiss et al., 2014; Zarrinpar et al., 2014). Environmentally-induced perturbations (i.e., diet or antibiotics), direct modulation of the host circadian system via light:dark cycle manipulations, or circadian gene network genetic disruption exacerbates gut dysbiosis, leading to loss of microbial rhythmicity and further contributing to metabolic dysregulation. Few interventions, aside from time-restricted feeding (Zarrinpar et al., 2014), can “restart” microbial rhythms with associated improvements in host metabolism. While several mechanisms have been put forth for how this rhythmic trans-kingdom dynamic occurs, whether it can be exploited to improve health remains elusive. Despite a myriad of preclinical data showing interdependence of host-microbe circadian rhythms, very few translational human studies have been performed, further contributing to a lack of understanding of how to harness microbial rhythms clinically to improve health outcomes.
The majority of preclinical studies rely on repeat stool collections obtained in series–i.e., every 4 h–or during animal harvest to obtain regional intestinal samples collected over a 24- to 48-h period. These methods are nearly impossible in humans, leading to reliance on easy-to-obtain repeat-sampling sites such as saliva (Collado et al., 2018; Takayasu et al., 2017) or on only a handful of subjects for timestamped stool collection, given the overwhelming difficulty and burden of sampling (Kaczmarek et al., 2017; Thaiss et al., 2014). These studies have provided proof-of-concept that gut microbes in humans do indeed exhibit rhythmicity, yet fall short in pushing the field forward to the broader population and into the clinic. A number of large-scale human studies have examined the gut microbiome in a myriad of healthy versus disease states (Shreiner et al., 2015) but despite extensive amounts of well-curated environmental metadata, including dietary intake, sleep habits, BMI, blood glucose, etc., few inroads have been made in applying observed differences in the microbiome to improve health outcomes. The question then becomes: can microbial rhythms be reconstructed from large-cohort datasets, and does this aid in identifying or defining health status in a given population?
In a study published in this month’s issue of Cell Host & Microbe, Reitmeier et al. (2020), reveal in two large, regionally similar, and deeply phenotyped German cohorts (KORA = 1,976 and FoCus = 1,363 subjects) that gut microbiota rhythmicity can be reconstructed by pooling single stool samples obtained over the course of the day using collection timestamp data. These cohorts contained a large proportion of both healthy subjects and patients with metabolic syndrome, including obesity and T2D. While obesity and T2D uniquely and differentially impacted the overall microbiota 16S rRNA gene profiles, both groups exhibited reduced microbial oscillations compared to healthy controls; those with pre-T2D exhibited an intermediate microbiota rhythmicity phenotype. Using the Detection of Differential Rhythmicity (DODR) R package, the authors identified 13 taxonomic features, including Bifidobacterium longum, Escherichia coli, Eubacterium rectale, Fecalibacterium prausnitzii, Intestinibacter bartletti, Clostridium celatum, and Romboutsia ilealis which lost rhythmicity in T2D relative to non-diabetic obese and healthy subjects (Figure 1). Surprisingly, treatment of T2D via Metformin did not restore microbial rhythms, particularly in the 13 predictive taxa, suggesting that pharmacological improvement of insulin sensitivity occurs independently of microbial oscillations and that these oscillations may not play a causal role in T2D or its resolution. In addition to 16S rRNA gene amplicon sequencing, shotgun metagenomics on a subset of samples revealed a significant association between 26 functional microbial pathways corresponding to the arrhythmic microbial signature in T2D incidence. Finally, application of a generalized linear model using the 13 arrhythmic taxa classified healthy versus T2D based solely on the arrhythmic microbial signature with high sensitivity and specificity (Figure 1). The prediction utility of the model was successfully validated in the regionally similar KORA 5-year followup, FoCus, and enable cohorts. While the predictive capacity of the model was sustained in regionally homogeneous samples, the 13 arrhythmic taxa were far less successful in identifying T2D incidence in cohorts from other European regions, suggesting that a refinement or expansion of the model is required to expand to a broader population.
Figure 1. Reconstruction of Rhythmic Microbial Oscillations Correlate With and Predict Healthy Versus T2D Incidence in the Large, Regionally Homogeneous Human KORA Cohort.
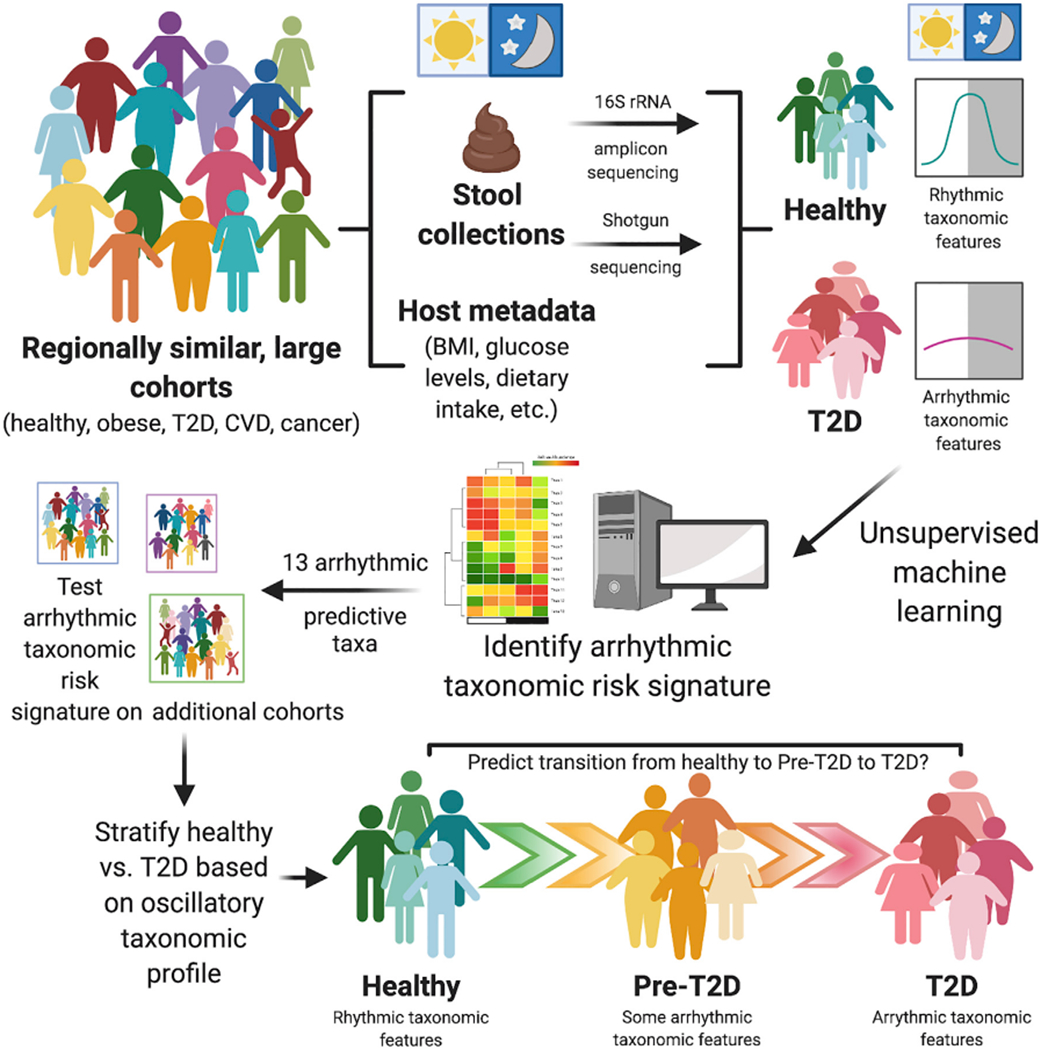
Reitmeier et al. (2020) demonstrate that gut microbiome rhythmicity can be retrospectively reconstructed using fecal collection timestamp data in single stool samples obtained from individual healthy subjects over the course of a day. A number of these microbial oscillations are absent or arrhythmic in Type 2 Diabetes (T2D) subjects. Thirteen taxa were identified as arrhythmic and predictive of T2D in the KORA cohort, which persisted in a 5-year follow-up study and was validated in other large cohorts from similar regions and may eventually allow for stratification of patients into categories of health, pre-T2D, and T2D in patient populations. While these 13 arrhythmic taxa were poor predictors of T2D in stool samples from other large cohorts outside this geographic region, this study provides a framework from which to build upon to incorporate into other timestamped datasets to aid in identifying complex diseases from gut microbiota samples in large heterogeneous cohorts.
This study serves as a first attempt to compile human stool microbiome samples using collection timestamp data to examine rhythmicity at such a large scale, providing a framework to build upon for existing and future studies examining circadian-microbiome dynamics in human subjects. First, reapplication of the KORA dataset to examine microbial rhythmicity was only possible because nearly all fecal samples were timestamped, emphasizing the necessity to employ time of defecation in all studies involving large human cohorts. The authors’ success in identifying this predictive T2D oscillatory signature was in part due to the extensive collection of behavioral and phenotypic metadata for each subject that could be correlated to microbiome outcomes. Coupled with sufficient effect sizes, this allowed for robust, unbiased statistical associations between oscillating microbial taxonomic features and metabolic disease incidence, which until now has been difficult to achieve. Finally, while associations in microbiome data can be misleading, the validation of the 13 arrhythmic microbial taxonomic features and their prediction of T2D in a prospective cohort directly demonstrates both the translational and clinical significance of this and other human microbiome studies. Building on the associative previous evidence in both humans and rodents that loss of microbial oscillations impacts metabolic diseases, this study demonstrates that changes in rhythmicity of microbial features can predict disease incidence, serving as biomarkers for diagnoses.
Reitmeier et al. (2020) have set the groundwork for future human studies to tackle important questions that remain regarding the functional and physiological impact of oscillating gut microbiota in metabolism. First, extrapolating these findings beyond a single geographic region is essential given the cohort-specific nature of the predictive model, which performed poorly in samples originating from other European regions. This will require studies with large sample sizes and detailed subject phenotyping with timestamp data. While lack of gut microbiota oscillations could predict T2D in this study, the causative role of microbial rhythmicity in human health versus disease remains elusive. Additionally, given that resolution of glucose dysregulation via Metformin did not restore lost oscillations, it remains unknown whether and how microbial rhythmicity can be jump-started pharmacologically. In order to move the needle to identify underlying mechanisms of microbial oscillations, more controlled studies, both human and animal, are required to parse out the functional dynamics and effects of oscillating microbes, which could then be exploited to not only predict but also alleviate disease. Whether similar inroads could be made in additional diseases, such as inflammatory bowel diseases or non-alcoholic fatty liver disease, remains to be determined. Most importantly, this study shows the promising prospect of analyzing longitudinally collected, sufficiently powered large-cohort microbiome datasets with associated timestamps in the context of health and disease, underscoring the importance of accurate record keeping that includes time of sample collection.
ACKNOWLEDGMENTS
K.F. is supported by NIH NIDDK F31 DK122714; V.L. is supported by NIH NIDDK K01 DK111785, NIH NIDDK R01 DK115221, and previous career development funds provided by the University of Chicago GI Research Foundation. Figure 1 was created with BioRender.com.
REFERENCES
- Alvarez Y, Glotfelty LG, Blank N, Dohnalova L, and Thaiss CA (2020). The Microbiome as a Circadian Coordinator of Metabolism. Endocrinology 161, bqaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Engen PA, Bandin C, Cabrera-Rubio R, Voigt RM, Green SJ, Naqib A, Keshavarzian A, Scheer FAJL, and Garaulet M (2018). Timing of food intake impacts daily rhythms of human salivary microbiota: a randomized, crossover study. FASEB J. 32, 2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier K, and Chang EB (2020). Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism. Trends Endocrinol. Metab 31, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek JL, Musaad SM, and Holscher HD (2017). Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am. J. Clin. Nutr 106, 1220–1231. [DOI] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, Neuhaus K, Grallert H, Linseisen J, Skurk T, et al. (2020). Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes. Cell Host Microbe 28, this issue, 258–272. [DOI] [PubMed] [Google Scholar]
- Shreiner AB, Kao JY, and Young VB (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol 31, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayasu L, Suda W, Takanashi K, lioka E, Kurokawa R, Shindo C, Hattori Y, Yamashita N, Nishijima S, Oshima K, et al. (2017). Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 24, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 150, 514–529. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Yooseph S, and Panda S (2014). Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 20, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]


