Abstract
Aims:
Metaplastic breast carcinoma (MBC) is a rare type of triple-negative breast cancer that displays vast histologic and genetic heterogeneity. Osseous differentiation can be found in different subtypes of MBC. Whether MBCs with osseous differentiation are underpinned by specific genetic alterations has yet to be defined. Here we investigate the repertoire of somatic mutations and copy number alterations (CNAs) in three MBCs with extensive osseous differentiation.
Methods and Results:
Tumor and normal DNA samples from three MBCs with extensive osseous differentiation were subjected to whole-exome sequencing. Somatic mutations, CNAs and mutational signatures were determined using a validated bioinformatics pipeline. Our analyses revealed clonal TP53 hotspot mutations associated with loss of heterozygosity of the wild-type allele coupled with mutations affecting genes related to the WNT and/or the PI3K/AKT/mTOR pathways in all cases analyzed. All cases displayed a dominant mutational signature 1 with two cases showing a secondary signature 3 in addition to other features of homologous recombination DNA repair defects (HRD). The Oncostatin M Receptor gene (OSMR), which plays a role in mesenchymal differentiation and bone formation, was found to be mutated in two MBCs with extensive osseous differentiation and in none of 35 previously published 35 MBCs.
Conclusion:
Our findings suggest that MBCs with osseous differentiation have somatic mutations similar to those of other forms of MBC.
Keywords: breast cancer, massively parallel sequencing, metaplastic breast carcinoma, PI3K pathway
INTRODUCTION
Metaplastic breast carcinoma (MBC) is a rare histologic special type of breast cancer, characterized by the differentiation of neoplastic epithelium into squamous or mesenchymal-like elements, which are frequently spindle, but can also include chondroid, osseous or rhabdoid elements.1 This phenotypic diversity is also observed at the transcriptomic level, given that MBCs can display varying intrinsic subtypes, integrative clustering and triple-negative breast cancer (TNBC) subtypes.2,3
Although MBCs are unlikely to be underpinned by a highly recurrent or pathognomonic somatic mutation or fusion gene,4–9 the histologic diversity of MBCs may be underpinned by distinct genetic alterations.4–9 Chondroid, spindle and squamous MBCs have been shown to display distinct genomic and transcriptomic profiles.4,6,7 In particular, spindle cell MBCs less frequently harbor gains of 7q11.22–237 and show a significantly higher prevalence of PIK3CA mutations, whereas chondroid MBCs lack PIK3CA mutations, but harbor CHERP mutations and more frequently a dominant mutational signature 3, a signature associated with homologous recombination deficiency (HRD)6. Although the genomic features of spindle cell, chondroid and squamous MBCs have been characterized by microarrays and massively parallel sequencing, 4,6–9 MBCs with osseous differentiation have been less well studied and whether MBCs with extensive osseous differentiation would be characterized by specific genetic alterations has yet to be investigated. Here we sought to determine whether MCBs with osseous differentiation resemble MBCs with other types of differentiation at the genetic level.
MATERIALS AND METHODS
Subjects and samples
Following approval by the Institutional Review Board of Memorial Sloan Kettering Cancer Center (MSKCC), representative formalin-fixed paraffin-embedded tissue blocks of metaplastic carcinomas of the breast with osseous differentiation were retrieved from the archives of the Department of Pathology of MSKCC or the consultation files of one of the authors (E.B.). Patient consent was obtained according to the research protocol approved by the Memorial Sloan Kettering Cancer Institutional Review Board. Samples were anonymized, reviewed by four pathologists (E.B., F.P., A.P.M.S. and J.S.R-F) and classified according to the latest World Health Organization (WHO) criteria.1 All cases subjected to whole-exome sequencing (WES) in this study (Supplementary Table 1) have not been previously reported. For the comparative analysis, we used the data from a previously published series of MBCs with mesenchymal differentiation.6
Whole Exome Sequencing analysis
DNA was extracted from microdissected representative tumor and normal breast tissue, as previously described4 and subjected to WES at the Integrated Genomics Operations (IGO) of Memorial Sloan Kettering Cancer Center (MSKCC), as also previously described.6 Details of the bioinformatics analysis employed for somatic mutation detection, gene copy number alteration (CAN) analysis, mutational signature decomposition and genomics features of homologous recombination DNA repair defect (HRD) are described in the Supplementary Methods.
Statistical analysis
Statistical analyses were conducted using R v3.1.2. Fisher’s exact tests were employed for comparisons between categorical variables, and Mann-Whitney U test were used for continuous variables. All tests were two-sided and P-values <0.05 were considered statistically significant.
RESULTS
The three cases of MBCs included in this study were found to display extensive osseous differentiation, given that the osseous component was the predominant metaplastic element, ranging from 40 to 60% (Supplementary Table 1). Two of the cases showed additional spindle cell components (cases MTC27 and MTC26) with markedly atypical spindle cells arranged in diverse architectural patterns including herringbone and storiform patterns. with areas of osseous differentiation intermingled (Figures 1A and 1B). MCT 27 and MTC28 also displayed minor chrondroid components in addition to the marked osseous differentiation (Figure 1C). Minor epithelial components were observed in all cases and were classified as of Nottingham histologic grade 3. Additional clinico-pathologic information is available in the supplementary materials (Supplementary Table 1). The carcinomas were either totally submitted or extensively sampled for histologic evaluation (Supplementary Table 1).
Figure 1 -. Histologic characteristics of metaplastic breast carcinomas (MBCs) with osseous differentiation.
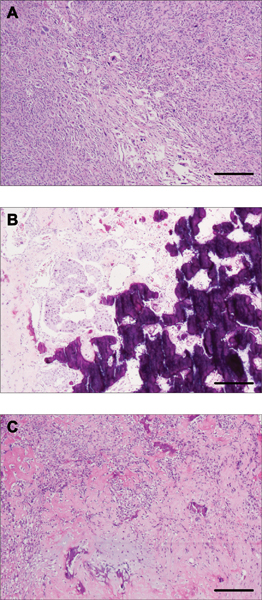
Representative photomicrographs of hematoxylin and eosin (H&E)-stained MBCs included in this study. (A) MTC26 displaying areas of spindle cell morphology with cells arranged in a storiform pattern. (B) MTC27 also displays the intervening epithelial differentiation around osseous areas. (C) MTC28 classified showing osseous and chondroid differentiation in a myxoid background. Scale bar, 500 μm (A, C), 200 μm (B).
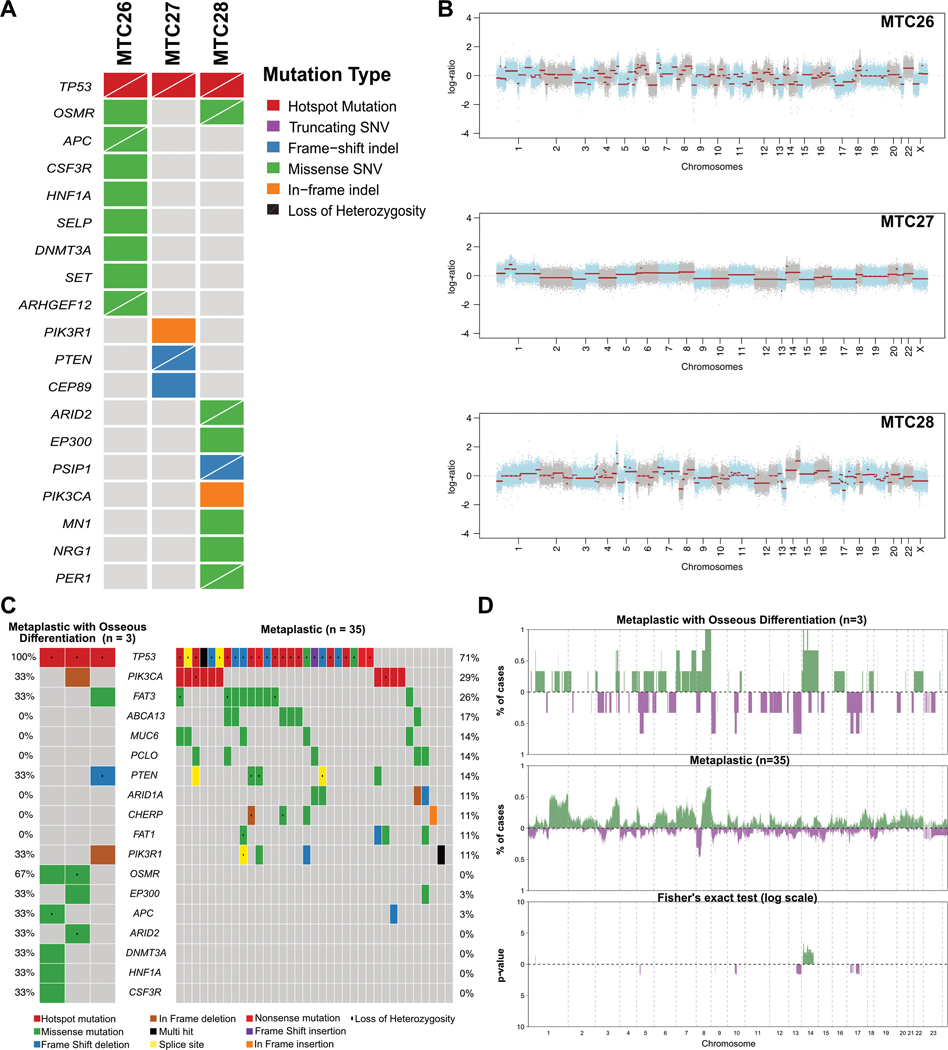
To determine whether MBC with osseous differentiation would be underpinned by a pathognomonic genetic alteration, these cases were subjected to WES (Supplementary Tables 2 and 3). This analysis revealed a median of 88 (range, 37–178) non-synonymous somatic mutations (Supplementary Table 2 and 3), 33 of which pathogenic or likely pathogenic, and an median of 9 (range, 3–11) mutations affecting cancer related genes10 per case (Figure 2A). Clonal TP53 hotspot mutations (R175H, P151S and R248Q) associated with loss of heterozygosity (LOH) of the wild-type allele were detected in all three cases analyzed. Other mutations in cancer related genes observed included a clonal APC (R1607T) missense mutation with LOH of the wild-type allele (Figure 2A and Supplementary Figure 1) in case (MTC26), as well as missense mutations in CSF3R (P549S), HNF1A (V448A), ARHGEF12 (G865A), DNMT3A (R167W) and SET (R77H). MTC27 harbored a PTEN clonal frameshift mutation (H61Tfs*36) associated with LOH of the wild-type allele and a clonal PIK3R1 in-frame deletion (D440_E443del). Case MTC28 displayed the highest mutational burden and a missense ARID2 mutation with LOH of the wild-type allele, as well as a missense mutation in NRG1 (S107T) (Figure 2A and Supplementary Figure 1). MTC26 and MTC28 harbored mutations affecting the Oncostatin M Receptor (OSMR) gene (Supplementary Table 3), genes whose protein product has been associated with bone homeostasis and development.11,12 In addition, in MTC26, a clonal and likely pathogenic affecting the HECT Domain E3 Ubiquitin Protein Ligase (HECTD1) gene (W1360L), another gene whose protein product plays a role in mesenchymal differentiation and bone homeostasis and development,13 was observed. An exploratory, hypothesis generating comparison between MBCs with osseous differentiation and 35 spindle cell, chondroid and squamous MBCs6 revealed OSMR was significantly more frequently mutated in MBCs with osseous differentiation (67% vs 0%, p=0.004), however this analysis ought to be interpreted with caution owing to the small sample size of this study.
Figure 2. Repertoire and comparison of somatic mutations and copy number alterations (gains/losses) of metaplastic breast carcinomas with osseous differentiation.
(A) Non-synonymous somatic mutations shared among cases and mutations affecting cancer-related genes in the MBC with osseous differentiation (n=3) subjected to whole-exome sequencing (WES). Cases are shown in columns and genes in rows. (B) Copy number plots depicting segmented Log2 ratios (y-axis) plotted according to genomic position (x-axis). Chromosomes are demarcated by alternating blue and gray colors (C) Comparison of non-synonymous somatic mutations prevalence in the MBC with osseous differentiation (n=3) and MBCs with no osseous differentiation (n=35). (D) Frequency plots and Fisher’s exact test comparison corrected for multiple testing of copy number gains and losses between the MBC with osseous differentiation (n=3) and MBCs with no osseous differentiation (n=35). Frequency (y-axis) of amplifications (green) and homozygous deletions (purple) is shown for each genomic region (x-axis). Inverse Log 10 values of the two-sided Fisher’s exact test p-values are plotted according to the genomic region (lower panel).
Copy number alteration analysis revealed a profile consistent with the previously reported in other MBCs and similar to TNBCs of no special type (IDC-NSTs), such as recurrent losses in 17p (3/3; 100%), and in 1p, 3p and 8p in 67% each (2/3), and recurrent gains of 1q and 8q in 67% of cases, each (2/3) (Figure 2B and 2D, Supplementary Figure 2). Amplification of AKT1 and TERT (MTC28) and CDKN2C (MTC27) were detected. Of note, BRCA1 showed heterozygous deletion in two cases (MTC26 and MTC28). An exploratory, hypothesis generating comparison of the CNA profiles of MBCs with osseous differentiation vs other previously reported MBCs6 did not reveal any differences in amplifications or homozygous deletions (Figure 2C).
We also sought to determine whether MBC with osseous differentiation would display genomic features suggestive of HRD. The three MBCs analyzed here displayed a dominant mutational signature 1 (MTC16: 30.3%, MTC27: 59.4% and MTC28: 27.9%). Cases MTC26 and MTC28 displayed a secondary signature 3 (26.7% and 20.3% of the mutational profiles of each case, respectively; Supplementary Figure S3). These two MBCs also displayed other features consistent with HRD, including a high LOH scores (17 and 8, respectively, vs 3 in MTC27), high large-scale state transitions (LST) scores14 (37 and 22, respectively, vs 4 in MTC27) and high telomeric allelic imbalance (NtAI) scores15 (26 and 24, respectively, vs 5 in MTC27; Supplementary Table 4). No bi-allelic mutations affecting HRD-related genes16 were detected in these two cases.
DISCUSSION
Here we demonstrate that in a way akin to other forms of MBC, MBCs with extensive osseous differentiation are also underpinned by clonal TP53 bi-allelic alterations and harbor mutations affecting genes known to be altered in MBCs,5–7 including those related to the Wnt and PI3K pathway family of genes. Despite our limited statistical power due to the small sample size, our study suggests that osseous differentiation in MBCs may not be underpinned by a pathognomonic mutation or copy number alteration.
An exploratory comparative analysis of MBCs with extensive osseous differentiation vs previously published MBCs of other histologic appearances revealed that mutations affecting OSMR were only numerically more frequently found in MBCs displaying osseous differentiation. Presently we have no data with statistical significance to support a difference in the prevalence of mutations affecting these genes in MBCs with extensive osseous differentiation, but we recognize the role of OSMR in bone development and homeostasis.11–13 In addition, we have also detected a clonal and likely pathogenic mutation affecting HECTD1, a gene whose silencing has been implicated in EMT, development of metastasis and reduced survival in breast cancer.17 Further studies to ascertain the role of OSMR and HECTD1 in the biology of MBCs are warranted.
This study has several limitations, including its small sample size, due to the rarity of MBCs with extensive osseous differentiation and the requirements for decalcification, which often result in nucleic acids of suboptimal quality being extracted from formalin-fixed paraffin embedded tissue samples. In addition, we were unable to extract RNA of sufficient quality to perform RNA sequencing analysis of the samples included in this study. Finally, we cannot rule out the presence of a pathognomonic mutation affecting non-coding regions of the genome or non-protein coding genes, or a pathognomonic fusion gene in MBCs with extensive osseous differentiation.
Despite the limitations, our study provides the characterization of the repertoire of somatic mutations, CNAs and mutational signatures in MBCs with extensive osseous differentiation, and demonstrates that these tumors share many of the genomics features of other forms of MBCs including frequent Wnt and PI3K pathways alterations. In addition, we provide evidence that osseous differentiation in MBCs is unlikely to be underpinned by a highly recurrently mutated protein coding gene or CNA.
Supplementary Material
Supplementary Figure 1. Cancer cell fractions of non-synonymous somatic mutations affecting cancer-related genes identified in metaplastic breast carcinomas with osseous differentiation by whole-exome sequencing.
Supplementary Figure 2. Mutational signatures of the metaplastic breast carcinomas with osseous differentiation
Supplementary Figure 3. Comparison of the frequencies of copy number alterations (amplifications/deletions) in metaplastic breast carcinomas with osseous differentiation and Metaplastic Breast Carcinomas with no osseous differentiation.
Supplementary Table 1. Clinicopathologic characteristics of in the metaplastic breast carcinomas with osseous differentiation included in this study.
Supplementary Table 2. Whole-exome Sequencing statistics.
Supplementary Table 3. Non-synonymous somatic mutations identified in the metaplastic breast carcinomas with osseous differentiation
Supplementary Table 4. Genomic features of homologous recombination deficiency and microsatellite instability in the metaplastic breast carcinomas with osseous differentiation included in this study.
ACKNOWLEDGEMENTS
This study was funded by the Breast Cancer Research Foundation. F.P. is funded in part by a K12 CA184746 grant and BW is funded by a Cycle for survival grant. Research reported in this paper was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No P30CA008748).
Footnotes
Conflict of interest: JSR-F reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Invicro and Genentech, outside the scope of the submitted work.
REFERENCES
- 1.Reis-Filho J, Gobbi H, McCart Reed A, et al. Metaplastic carcinoma In: WHO Classification of Tumours Editorial Board., ed. Breast Tumours. 5th ed. Lyon (France): International Agency for Research on Cancer; 2019:134–138. [Google Scholar]
- 2.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigelt B, Ng CKY, Shen R, et al. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity [corrected]. Mod Pathol. 2015;28(3):340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220(5):562–573. [DOI] [PubMed] [Google Scholar]
- 6.Ng CKY, Piscuoglio S, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas. Clin Cancer Res. 2017;23(14):3859–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piscuoglio S, Ng CKY, Geyer FC, et al. Genomic and transcriptomic heterogeneity in metaplastic carcinomas of the breast. NPJ breast cancer. 2017;3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krings G, Chen Y-Y. Genomic profiling of metaplastic breast carcinomas reveals genetic heterogeneity and relationship to ductal carcinoma. Mod Pathol. 2018;31(11):1661–1674. [DOI] [PubMed] [Google Scholar]
- 9.McCart Reed AE, Kalaw E, Nones K, et al. Phenotypic and molecular dissection of metaplastic breast cancer and the prognostic implications. J Pathol. 2019;247(2):214–227. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarty D, Gao J, Phillips S, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;1(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker EC, McGregor NE, Poulton IJ, et al. Oncostatin M promotes bone formation independently of resorption when signaling through leukemia inhibitory factor receptor in mice. J Clin Invest. 2010;120(2):582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malaval L, Liu F, Vernallis AB, Aubin JE. GP130/OSMR is the only LIF/IL-6 family receptor complex to promote osteoblast differentiation of calvaria progenitors. J Cell Physiol. 2005;204(2):585–593. [DOI] [PubMed] [Google Scholar]
- 13.Zohn IE, Anderson KV, Niswander L. The Hectd1 ubiquitin ligase is required for development of the head mesenchyme and neural tube closure. Dev Biol. 2007;306(1):208–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popova T, Manié E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–5462. [DOI] [PubMed] [Google Scholar]
- 15.Birkbak NJ, Wang ZC, Kim J-Y, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riaz N, Blecua P, Lim RS, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8(1):857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duhamel S, Goyette MA, Thibault MP, Filion D, Gaboury L, Côté JF. The E3 Ubiquitin Ligase HectD1 Suppresses EMT and Metastasis by Targeting the +TIP ACF7 for Degradation. Cell Rep. 2018;22(4):1016–1030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cancer cell fractions of non-synonymous somatic mutations affecting cancer-related genes identified in metaplastic breast carcinomas with osseous differentiation by whole-exome sequencing.
Supplementary Figure 2. Mutational signatures of the metaplastic breast carcinomas with osseous differentiation
Supplementary Figure 3. Comparison of the frequencies of copy number alterations (amplifications/deletions) in metaplastic breast carcinomas with osseous differentiation and Metaplastic Breast Carcinomas with no osseous differentiation.
Supplementary Table 1. Clinicopathologic characteristics of in the metaplastic breast carcinomas with osseous differentiation included in this study.
Supplementary Table 2. Whole-exome Sequencing statistics.
Supplementary Table 3. Non-synonymous somatic mutations identified in the metaplastic breast carcinomas with osseous differentiation
Supplementary Table 4. Genomic features of homologous recombination deficiency and microsatellite instability in the metaplastic breast carcinomas with osseous differentiation included in this study.