Abstract
A complex network of cellular receptors, RNA targeting pathways, and small-molecule signaling provides robust plant immunity and tolerance to viruses. To maximize their fitness, viruses must evolve control mechanisms to balance host immune evasion and plant-damaging effects. The genus Potyvirus comprises plant viruses characterized by RNA genomes that encode large polyproteins led by the P1 protease. A P1 autoinhibitory domain controls polyprotein processing, the release of a downstream functional RNA-silencing suppressor, and viral replication. Here, we show that P1Pro, a plum pox virus clone that lacks the P1 autoinhibitory domain, triggers complex reprogramming of the host transcriptome and high levels of abscisic acid (ABA) accumulation. A meta-analysis highlighted ABA connections with host pathways known to control RNA stability, turnover, maturation, and translation. Transcriptomic changes triggered by P1Pro infection or ABA showed similarities in host RNA abundance and diversity. Genetic and hormone treatment assays showed that ABA promotes plant resistance to potyviral infection. Finally, quantitative mathematical modeling of viral replication in the presence of defense pathways supported self-control of polyprotein processing kinetics as a viral mechanism that attenuates the magnitude of the host antiviral response. Overall, our findings indicate that ABA is an active player in plant antiviral immunity, which is nonetheless evaded by a self-controlled RNA virus.
Keywords: abscisic acid, antiviral immune evasion, mathematical modeling, viral polyprotein processing, Potyvirus, RNA metabolism
Molecular networks provide robust plant immunity against pathogens, including viruses. Here, meta-analyses indicate that abscisic acid (ABA) connects hormone signaling with RNA metabolic pathways and contributes to plant antiviral immunity, which is evaded by an RNA virus with self-controlled polyprotein-processing kinetics. This study reveals a regulatory mechanism that controls viral infection dynamics and the magnitude of host antiviral responses.
Introduction
Recent surveys underscore the great abundance and diversity of plant viruses that pose major challenges to host defense pathways and, thus, to plant health and food security (Pasin et al., 2019). Multiple layers of defense and complex networks of receptors, small molecules, and transcriptional regulators provide robust plant immunity and tolerance to pathogens (Nakahara and Masuta, 2014; Tsuda and Somssich, 2015; Jeon et al., 2017; Nobori et al., 2018; Paudel and Sanfaçon, 2018; Murphy et al., 2020). Salicylic acid (SA) is a defense phytohormone that mediates resistance to biotrophic pathogens and interacts antagonistically or synergistically with other host small molecules (Berens et al., 2017). SA biosynthesis and signaling are activated during the hypersensitive response (HR) to viruses and are integrated in emerging models of plant immunity to viruses (Nakahara and Masuta, 2014; Alazem and Lin, 2015; Murphy et al., 2020). Viruses in turn have evolved strategies to repress or evade antiviral HR and SA-dependent resistance (Nakahara and Masuta, 2014). In accordance with these models, members of the families Caulimoviridae, Bromoviridae, Potyviridae, Tombusviridae, and Virgaviridae are known to interfere with SA pathways by mechanisms already reviewed (Nakahara and Masuta, 2014; Alazem and Lin, 2015; Yang and Li, 2018; Murphy et al., 2020; Shen et al., 2020).
Evidence indicates that RNA silencing and small-molecule signaling cooperate to promote plant resistance to viruses (Alazem and Lin, 2015, 2020; Yang and Li, 2018; Murphy et al., 2020; Yang et al., 2020). Whereas early studies focused on the interplay of RNA silencing and SA, an increasing body of data supports the contribution of other phytohormones, such as abscisic acid (ABA), to antiviral responses (Pacheco et al., 2012; Chen et al., 2013, 2017; Westwood et al., 2013; Seo et al., 2014; Alazem et al., 2017; Zhang et al., 2019; Alazem and Lin, 2020). Defects in RNA-silencing components that contribute to antiviral immunity were shown to enhance sensitivity to ABA, as reported for the Arabidopsis thaliana mutant lines of HYPONASTIC LEAVES 1 (HYL1), HUA ENHANCER 1 (HEN1), ARGONAUTE1 (AGO1), and DICER-LIKE (DCL) proteins (Zhang et al., 2008), reviewed in Alazem and Lin (2020). In addition to RNA silencing, several pathways control RNA maturation and turnover in plant cells, and their impact on virus infection dynamics has just begun to be appreciated (Mäkinen et al., 2017; Chantarachot and Bailey-Serres, 2018; Boudreault et al., 2019; Xu et al., 2020). For instance, RNA decay was recently reported to act in concert with RNA silencing to promote antiviral immunity in plants (Li and Wang, 2018, 2019). Studies have highlighted connections between osmotic stress responses, ABA signaling, and RNA decay (Kawa and Testerink, 2017; Soma et al., 2017; Wawer et al., 2018); however, the biological significance of the interplay between ABA and RNA metabolic pathways (beyond RNA silencing) in plant antiviral immunity remains largely unexplored.
Viruses are obligate parasites that adopt distinct translational strategies to maximize the coding capacity of their small genomes. Polyprotein synthesis from long open reading frames is one of the most common strategies used by RNA viruses. Large polypeptide precursors are synthesized, and the release of functional viral gene products is mediated by polyprotein processing. Many viral agents of human and veterinary infectious diseases, and almost half of known plant viruses, rely on proteolytic cleavage events as key co- and post-translational modifications throughout their infection cycle (Yost and Marcotrigiano, 2013; Rodamilans et al., 2018; Mann and Sanfaçon, 2019). Virus-encoded proteases are multifunctional hubs that coordinate virus replication and host–virus interactions to promote infectivity and virulence (Martínez et al., 2016; Lei and Hilgenfeld, 2017; Rodamilans et al., 2018). The spatial-temporal regulation of protease activity and the processing of viral polyproteins can further determine viral persistence and disease lethality (Lackner et al., 2004; Yost and Marcotrigiano, 2013; Isken et al., 2019).
Polyproteins are synthesized by members of the picornavirus supergroup that includes Potyviridae, the largest family of plant RNA viruses (Revers and García, 2015). Most potyvirids have narrow host ranges, and their leader P1 proteases show considerable sequence variability linked to host adaptation (Cui and Wang, 2019). P1 self-cleaves from viral polyproteins and the mature P1 lacks trans-cleavage activity (Cui and Wang, 2019). P1 self-cleavage defects impair the RNA-silencing suppressor activity of the downstream protein element HCPro and preclude virus infectivity (Verchot and Carrington, 1995; Pasin et al., 2014; Shan et al., 2015). Deletion mutagenesis and in vitro assays enabled mapping of the core C-terminal protease domain of P1 and showed that its N terminus negatively regulates the self-cleavage activity (Pasin et al., 2014; Shan et al., 2018). The N terminus is therefore an autoinhibitory domain that controls P1 self-cleavage and HCPro release. In vitro assays indicated that P1 autoinhibition is alleviated by an unknown host factor. Consistent with these results, removal of the P1 N terminus boosted in vivo P1 self-cleavage and local viral amplification in a non-permissive host (Shan et al., 2018). In viral infections of permissive hosts, removal of the P1 autoinhibitory domain accelerated early replication and enhanced symptom severity, which were accompanied by a later drop in viral load (Pasin et al., 2014).
Here, we used a plum pox virus (PPV) (genus Potyvirus) isolate adapted to infect herbaceous hosts to investigate the contribution of viral leader proteases and polyprotein processing to virus–host interactions and pathogen fitness. P1Pro is a PPV clone that lacks the autoinhibitory domain of the P1 leader protease and has an uncontrolled release of the viral-silencing suppressor HCPro. HCPro release and proteolytic activity of the truncated P1 version encoded by P1Pro was confirmed in vitro and in planta, including in non-host plant species (Shan et al., 2018). P1Pro infection was found to elicit HR and to induce the accumulation of pathogenesis-related (PR) proteins, including PR-2 and PR-3. This induction was considerably reduced but not eliminated in a transgenic N. benthamiana nahG line that expresses an SA hydroxylase gene (Pasin et al., 2014). We reasoned that SA and uncharacterized defense components could participate in P1Pro-activated responses. In this study, host molecular changes triggered by P1Pro were further characterized using a transcriptomic approach. Our results support the role of SA as a major defense phytohormone involved in antiviral responses. Analysis of SA-defective plants uncovered ABA as an additional antiviral component that contributes to potyviral resistance. To provide an understanding of the ABA effects on antiviral immunity, public datasets of A. thaliana transcriptomes and phosphoproteomes were analyzed. ABA treatments affected the phosphorylation status of RNA metabolic components, including those involved in RNA cap binding, maturation, decay, and translation initiation. Genetic and hormone treatment assays showed that ABA promotes plant resistance to PPV infection. Together, our results suggest that an ABA-dependent, genome-wide perturbation of host RNA metabolism can promote an antiviral response that is evaded by a self-controlled RNA virus. Finally, we developed a mathematical model supporting the mechanistic relationships among the controlled processing of viral polyproteins, the strength of the silencing suppression, the activation of host hormonal responses, and overall virus fitness.
Results
P1Pro Infection Elicits SA Accumulation
The P1Pro clone is a PPV deletion mutant lacking the N-terminal autoinhibitory domain that controls the self-cleavage of the P1 leader protease (Figure 1A). Wild-type and SA-defective (nahG) lines of N. benthamiana were used as hosts to study defense responses elicited by P1Pro infection; mock- or PPV-treated plants were used as controls. Analysis of upper uninoculated leaves showed significantly increased free SA in P1Pro-infected wild-type host samples compared with mock- or PPV-treated plants (wild-type or nahG) or nahG plants inoculated with P1Pro (p < 0.01; Figure 1B). The result confirmed SA as a major contributor to P1Pro-activated responses.
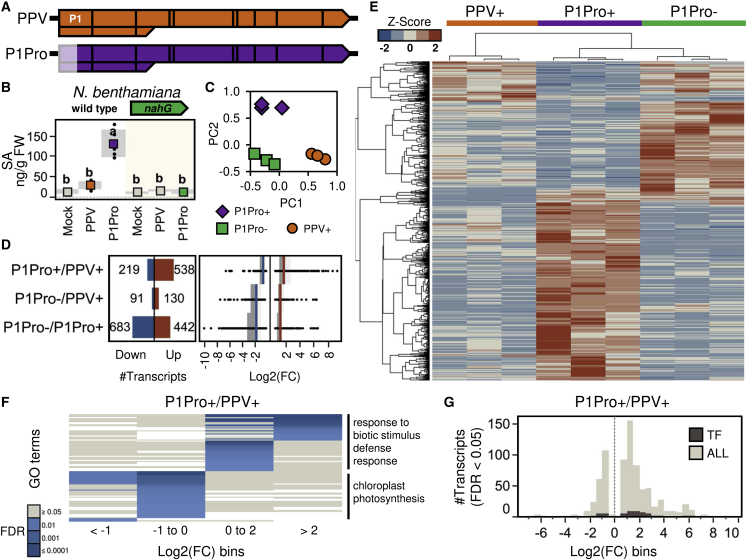
Figure 1.
Transcriptomic Changes in Host Plants Infected with PPV and Its Deletion Mutant P1Pro.
(A) Genome representation of wild-type PPV or P1Pro, a PPV clone that lacks the N-terminal autoinhibitory domain of the P1 protease. The green fluorescent protein (GFP) gene, located between the NIb and CP cistrons of both viral clones, is omitted for clarity.
(B) Quantification of SA in plants inoculated with PPV or P1Pro clones; wild-type Nicotiana benthamiana and a transgenic line expressing a salicylate hydroxylase gene (nahG) were used as host plants. The plot shows mean ± SD (boxes and shadows, respectively; n = 5); letters indicate p < 0.01, one-way ANOVA and Tukey's honestly significant difference (HSD) test. Conditions in color (orange, purple, and green) were analyzed by RNA-seq with three biological replicates per condition.
(C) Principal component analysis of the samples analyzed by transcriptomics.
(D) Total number (left) and fold change (right) of differentially expressed transcripts (FDR < 0.05); fold change medians are indicated by colored lines.
(E) Normalized read counts of differentially expressed transcripts and their clustering analysis; for each condition, the values of different biological replicates are shown. PPV+ and P1Pro+ denote wild-type N. benthamiana plants agroinoculated with PPV and P1Pro clones, respectively; P1Pro, nahG-expressing N. benthamiana plants agroinoculated with P1Pro.
(F) Enrichment of functional categories in genes differentially regulated by P1Pro with respect to PPV in the wild-type host (P1Pro+/PPV+ comparison); representative gene ontology (GO) terms across fold change windows are indicated (right).
(G) Fold change distribution (bins of 0.5) of differentially expressed transcripts (ALL) or only those that encoded transcription factors (TFs) identified in the P1Pro+/PPV+ comparison.
P1Pro Infection Reprograms the Host Transcriptome
The transcriptomes of wild-type N. benthamiana plants inoculated with PPV or P1Pro clones (PPV+ and P1Pro+ samples, respectively) were analyzed by Illumina RNA sequencing (RNA-seq). N. benthamiana nahG plants inoculated with P1Pro (P1Pro−) were further analyzed to identify genes whose expression is altered by P1Pro infection but does not depend on SA signaling (Figure 1B). For each condition, three biological replicates were analyzed. Sequencing reads were mapped to the PPV genome and to the nahG transgene as preliminary quality control (Supplemental Figure 1A; see Supplemental Information). Coverage of the full-length P1 sequence was obtained for PPV samples, whereas P1Pro samples showed a coverage gap in the 5′ end corresponding to the P1 truncation. Samples from the transgenic N. benthamiana line showed high nahG coverage and confirmed the host genotypes used. Reads were further mapped to an N. benthamiana transcriptome version (Nakasugi et al., 2014). To assess sample consistency and host-response similarities, count values were subjected to an unsupervised multivariate analysis. Representation of principal components showed a clear separation between the three conditions analyzed (i.e., PPV+, P1Pro+, and P1Pro−) and correct grouping of their biological replicates (Figure 1C). Hierarchical clustering and bootstrap analysis corroborated the results with approximately unbiased confidence of ≥ 99% (Supplemental Figure 1B). To identify statistically significant differentially expressed genes, transcript count values were used in two-way comparisons to compute false discovery rate (FDR) values. The PPV+, P1Pro+, and P1Pro− datasets were thus analyzed to yield three different two-way comparisons (see Supplemental Data 1A). Transcripts were filtered based on FDR < 0.05 and, to further explore sample similarities, count values of transcripts that were differentially expressed in at least one comparison were used to calculate pairwise correlation coefficients between samples. Consistent with the principal component analysis, samples from the same condition showed a high correlation (r > 0.96), whereas correlations were lower between conditions, ranging from 0.76 to 0.94 (Supplemental Figure 1C).
The magnitude of expression profile changes showed that, in wild-type hosts and using the PPV clone as a reference, P1Pro infections were dominated by gene upregulation (538 of 757 genes in the P1Pro+/PPV+ comparison; Figure 1D and Supplemental Data 1A). The median fold change of the P1Pro+/PPV+ comparison was greater than those of the P1Pro−/PPV+ and P1Pro−/P1Pro+ comparisons. Hierarchical clustering using read counts of the differentially expressed transcripts showed major sample clusters that corresponded to the three conditions analyzed (Figure 1E). Heatmap visualization and the clustering of individual transcripts further highlighted specific groups of genes that were up- and, in a few cases, downregulated by P1Pro versus PPV infections in the wild-type host (P1Pro+ versus PPV+; Figure 1E). The expression profiles of these gene groups were largely divergent in P1Pro infections of the nahG plants (P1Pro+ versus P1Pro−). These findings indicate that, in P1Pro+ samples, the abundance of a large set of transcripts significantly deviates from that of wild-type virus infections or P1Pro-infected hosts with defective SA signaling.
P1Pro Infection Alters the Expression of SA-Related Defense Genes and Transcription Factors Associated with Diverse Signaling Pathways
To confirm the role of SA and identify additional defense components that may be associated with P1Pro infection, we analyzed transcripts that showed differential expression in the P1Pro+/PPV+ comparison. Functional annotations were made using the proteome of the model plant Arabidopsis thaliana (Supplemental Data 1A) and its associated gene ontology (GO) terms, or a custom sequence list that covered reported PR families (Supplemental Data 1B). Specific GO categories were overrepresented and showed partial overlaps in windows of genes that were significantly up- or downregulated by P1Pro (Figure 1F). Upregulated genes were enriched in stress response terms, including "response to biotic stimulus” and "defense response"; conversely, downregulated genes were associated with the terms "chloroplast" and "photosynthesis" (Figure 1F). P1Pro upregulated several genes associated with SA signaling, including NPR3 homologs and a wide array of PR genes from different families (Supplemental Figure 2). Pipecolic acid is a metabolite that can act synergistically with SA to promote plant immunity (Hartmann and Zeier, 2019; Wang et al., 2019); homologs of its core signaling components ALD1, WRKY33, and SARD1 were also upregulated in P1Pro samples. RNA-silencing components, including AGO1 and AGO2, were downregulated in P1Pro samples (Supplemental Figure 2).
Transcription factors (TFs) were specifically examined because they transmit information signals to reprogram host transcriptomes and modulate immune responses (Tsuda and Somssich, 2015). In addition to WRKY33 and SARD1, members of several TF families with known roles in plant immunity, including the AP2/ERF, bHLH, MYB, NAC, and WRKY families, were differentially expressed in P1Pro versus PPV infections (Supplemental Figure 2). Like the 757 differentially expressed transcripts (ALL) identified in the P1Pro+/PPV+ comparison, TFs were distributed across different expression windows, and the majority were upregulated (Figure 1G and Supplemental Figure 2). We hypothesized that selective enrichment of GO terms associated with TFs would provide insight into signals activated by P1Pro. Combined functional analyses of the complete list of ALL transcripts and the TF subset supported the role of SA in P1Pro responses (GO:0009751, GO:0009627) and highlighted putative contributions of the phytohormones jasmonic acid, ethylene, auxin, and gibberellin, as well as components involved in abiotic stress signaling (GO:0009628, GO:0009414, GO:0047484; Supplemental Figure 3).
Salicylate-Dependent and -Independent Immune Responses Are Elicited by P1Pro Infection
Functional analysis indicates that SA and unknown phytohormone(s) might contribute to P1Pro responses. SA depletion in nahG-expressing plants is predicted to alter transcriptional regulation of genes linked to SA signaling, whereas the expression of SA-independent genes should be unaffected. The transcriptomes of PPV+ and P1Pro+ samples were compared with those of P1Pro-inoculated N. benthamiana nahG plants (P1Pro−). Expression profiles of wild-type or nahG plants infected with P1Pro (P1Pro+ and P1Pro−, respectively) were significantly divergent (Figure 1C–1E and Supplemental Figure 1B). Relationships of lists of differentially expressed genes were analyzed (Figure 2A and Supplemental Data 1A). The intersection of the P1Pro+/PPV+ and P1Pro−/P1Pro+ sets included 524 genes (bars labeled with 470 plus 54 in Figure 2A); of these, 48 were identified as PR genes. Compared with the union set, this functional enrichment was significantly higher than expected by chance (Q = 3.54 × 10−7), indicating that the regulation of PR gene expression depends both on the virus clones and functional SA signaling in the host.
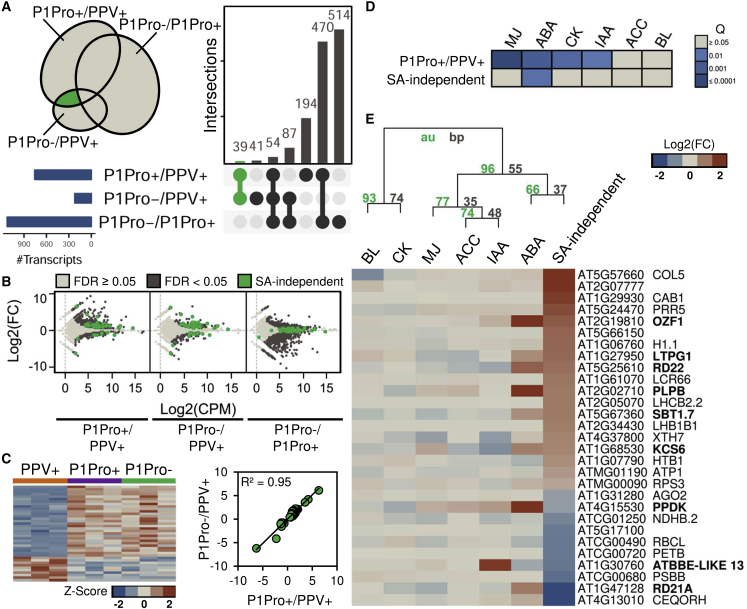
Figure 2.
SA-Dependent and -Independent Transcriptomic Changes in P1Pro-Infected Plants.
(A) Comparisons of lists of differentially expressed genes (FDR < 0.05). The Venn diagram shows spatial relationships between gene lists; the numbers of overlapping genes are indicated (right). The green intersection highlights a subset of genes that are altered in P1Pro-infected wild-type plants but not in nahG-transgenic plants (where SA signaling is downregulated; i.e., SA-independent genes).
(B) Fold change and coverage plots of transcriptomic data; SA-independent genes are colored green.
(C) Left, normalized read counts (heatmap; n = 3) and, right, correlation plot of fold changes for the SA-independent genes in wild-type or nahG-transgenic plants infected with P1Pro (P1Pro+ and P1Pro−, respectively), using the PPV+ condition as a reference.
(D) The SA-independent set significantly overlaps ABA marker genes. A. thaliana homologs of the differentially expressed genes in N. benthamiana were identified by BLASTX searches. Lists obtained were confronted against hormone marker genes from public A. thaliana microarrays (MJ, methyl jasmonate; ABA; CK, zeatin; IAA, indole-3-acetic acid; ACC, aminocyclopropane-1-carboxylic acid; BL, brassinolide). The color scale shows overlap significance based on the hypergeometric test.
(E) Hierarchical clustering and fold changes of the SA-independent gene set identified by our RNA-seq and microarrays from hormone-treated A. thaliana plants. Cluster probabilities are indicated (au, approximately unbiased; bp, bootstrap probability). Symbols of genes whose expression is significantly altered (p < 0.01) in ABA-treated A. thaliana seedlings are in boldface.
The intersection of P1Pro+/PPV+ and P1Pro−/PPV+ sets revealed 39 elements outside the P1Pro−/P1Pro+ list, implying that their regulation depends on the virus clones used but does not require SA signaling (i.e., SA-independent genes; green in Figure 2A; Supplemental Table 1 and Supplemental Data 1A). Visual inspection of the fold change plots of P1Pro+/PPV+ and P1Pro−/PPV+ comparisons confirmed that elements of the SA-independent set were distributed in the space associated with significantly up- or downregulated transcripts (FDR < 0.05); conversely, they lay close to the x axis (i.e., fold change ∼ 0 and FDR ≥ 0.05) in the P1Pro−/P1Pro+ plot (Figure 2B). SA-independent transcripts showed similar read counts in the P1Pro+ and P1Pro− conditions and diverged from PPV+ samples, and their fold changes using PPV+ values as reference were strongly correlated (R2 = 0.95; Figure 2C).
Compared with N. benthamiana, rich public datasets are available for the model plant A. thaliana. The SA-independent transcripts we identified were mapped to 29 A. thaliana homologs, and these were compared with hormone marker genes from a transcriptomic study of A. thaliana (Nemhauser et al., 2006). This meta-analysis showed that the SA-independent genes showed significantly greater overlap with ABA markers than expected by chance (nine genes, Q = 0.010; Figure 2D). Fold change values from the A. thaliana transcriptomic study and our RNA-seq analysis, as well as hierarchical clustering, supported grouping of the ABA and SA-independent sets (Figure 2E). Among the A. thaliana ABA-responsive genes (boldface, Figure 2E) and compared with our data, the expression values of the two markers were discordant, whereas the expression change of the remainder coincided (OZF1, LTPG1, RD22, PLPB, SBT1.7, KCS6, and ATBBE-LIKE 13).
Removal of the P1 Autoinhibitory Domain Triggers ABA Accumulation in Viral Infection but Is Itself Insufficient to Activate Local HR
In N. benthamiana compared with mock-treated or PPV-infected plants, P1Pro infection triggered HR that was accompanied by enhanced accumulation of the OZF1 homolog NbOZF1 and ABA (p < 0.01; Figure 3A). In A. thaliana, expression of the OZF1 zinc finger protein gene is activated by ABA in vegetative tissues (Lee et al., 2012), and its protein was recently shown to be a preferential target of multiple secreted effectors from Pseudomonas syringae (Cao et al., 2019). We asked whether the truncated P1 version encoded by the P1Pro clone and lacking the N-terminal autoinhibitory domain was sufficient to activate effector-triggered immunity (ETI), thereby causing HR. Transient expression of the P1 N-terminal deletion construct, either alone or as a part of a polyprotein including HCPro, was not sufficient to activate HR, as was apparent in N. benthamiana leaves infiltrated with a bacterial effector construct (avr; Figure 3B).
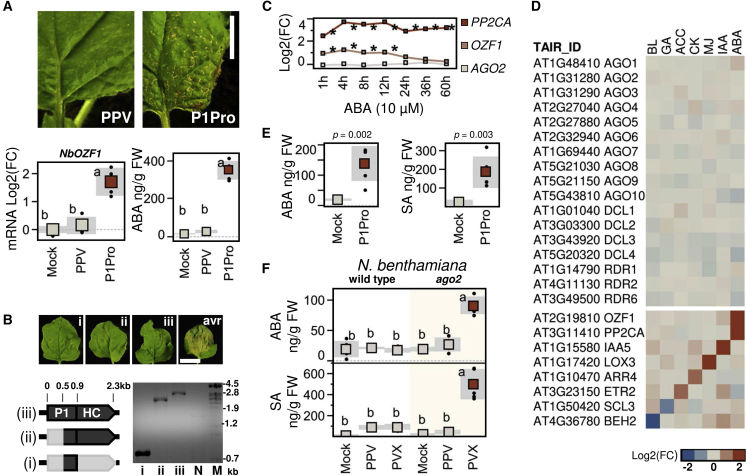
Figure 3.
The Self-Controlled, Wild-Type PPV Evades the Activation of ABA Signaling during Plant Infection.
(A) Symptoms of upper uninoculated leaves of wild-type plants inoculated with PPV or P1Pro. Scale bar corresponds to 1 cm. Plots show the qRT–PCR quantification of NbOZF1 (mean ± SD, n = 4) and ABA amounts (mean ± SD, n = 6); letters indicate p < 0.01, one-way ANOVA and Tukey's HSD test.
(B) Transient expression of (i) N-terminally truncated P1, (ii) N-terminally truncated P1 and HCPro, and (iii) full-length P1 and HCPro constructs; avr indicates the HopAB2 construct that was used as a positive control of HR. Images of leaves were taken at 6 days post-agroinoculation (dpa). Scale bar corresponds to 2 cm. The gel shows the RT–PCR analysis of infiltrated leaves using primers that anneal to the 5′ and 3′ UTR of the constructs. N, non-infiltrated sample; M, DNA size marker.
(C) Time course analysis of OZF1 and AGO2 fold changes in the public RNA-seq datasets of ABA-treated A. thaliana seedlings; PP2CA serves as an ABA-responsive control; ∗FDR < 0.01.
(D) Transcriptional regulation of the main RNA-silencing genes by phytohormones. Transcript fold changes from the public microarray data of A. thaliana seedlings treated for 3 h with BL, gibberellic acid (GA), ACC, CK, MJ, IAA, or ABA are shown. OZF1, PP2CA, IAA5, LOX3, ARR4, ETR2, SCL3, and BEH2 are included as hormone-responsive control genes.
(E)N. benthamiana ago2 plants were inoculated with P1Pro or mock treated. ABA and SA amounts at 7 dpi are plotted (mean ± SD, n = 5); p values are shown (Student's t-test).
(F) PPV and potato virus X (PVX) were delivered to wild-type or ago2-mutant plants; ABA and SA amounts are plotted (mean ± SD, n = 5); letters indicate p < 0.01, one-way ANOVA and Tukey's HSD test. Viruses were delivered to N. benthamiana plants by mechanical inoculation and transient expression constructs by Agrobacterium infiltration.
AGO2 Depletion Is Sufficient to Enhance ABA Levels during Potato Virus X Infection but Not upon Infection with the Self-Controlled, Wild-Type PPV
ABA has been proposed to promote antiviral defenses through the transcriptional regulation of RNA-silencing genes, including AGO2 (Alazem and Lin, 2020). AGO2 was identified in the SA-independent set of differentially expressed transcripts in our RNA-seq data. Compared with PPV samples, P1Pro infection showed high ABA levels (Figure 3A) and reduced AGO2 transcript and protein levels (Figure 2E and Supplemental Figure 2; Supplemental Tables 1 and 2). Microarray analysis nonetheless indicated that ABA was insufficient to enhance AGO2 expression in A. thaliana seedlings at 3 h post-treatment (Figure 2E). Time series RNA-seq data from A. thaliana seedlings treated with ABA showed that OZF1 and PP2CA, a known component of ABA signaling in vegetative tissues, were upregulated by 10 μM ABA within 1 h post-treatment (FDR < 0.01). Conversely, AGO2 expression showed no significant changes during the 60 h assay (Figure 3C). Microarray analysis of hormone-treated A. thaliana plants further showed that transcript abundance of the main RNA-silencing components, i.e., AGO, RDR, and DCL, was largely unaltered by ABA or other phytohormones (Figure 3D; note that the effects of SA were not analyzed).
To further test whether AGO2 regulation and ABA signaling were related, we measured ABA amount in an ago2 knockout line of N. benthamiana. The ago2 plants inoculated with P1Pro showed high ABA and SA levels (Figure 3E), indicating that induction of ABA and SA accumulation does not require AGO2. Based on our working model, host defenses are activated by uncontrolled silencing suppression activity of the P1Pro clone. We asked whether, in a complementary approach, ago2 depletion might be sufficient to trigger ABA accumulation during wild-type virus infections. Wild-type and ago2 plants were therefore inoculated with PPV. Potato virus X (PVX) was also included because a recombinant PVX clone expressing HCPro of PPV was previously shown to trigger ABA accumulation in N. benthamiana plants; this effect was dependent on the silencing suppressor activity of HCPro (Pacheco et al., 2012). Mock-treated wild-type and ago2 plants showed similar phytohormone amounts, indicating that AGO2 absence is itself insufficient to activate ABA or SA. Irrespective of the host genotype used, no significant changes in ABA levels were detected in PPV infections. The ABA and SA amounts in the PVX-infected ago2 host were significantly higher than in the other conditions (Figure 3F). These results showed that (1) AGO2 and major RNA-silencing genes are not primary transcriptional targets of ABA signaling; (2) the perturbation of RNA silencing triggers ABA accumulation during P1Pro or PVX infections; and (3) by contrast, viral control of silencing suppressor activity avoids the activation of ABA signaling during PPV infection in both wild-type and RNA-silencing-defective hosts.
ABA Regulates the Protein Phosphorylation Dynamics of RNA Metabolic Components
ABA coordinates plant responses to osmotic stress, and its perception is mediated by PYR/PYL receptors that selectively bind and inhibit PP2C protein phosphatases in the presence of ABA. This in turn allows the activation of SnRK2 kinases and the phosphorylation of downstream TFs (Cutler et al., 2010). We asked whether ABA signaling could affect host antiviral RNA-silencing components at a post-translational level. Public data from A. thaliana phosphoproteomes were analyzed (Umezawa et al., 2013; Wang et al., 2013, 2020; Mergner et al., 2020). We considered a subset of proteins redundantly identified in at least two independent phosphoproteomic datasets from plants treated with ABA or subjected to osmotic stress (Supplemental Data 1C). This responsive subset, which accounted for 9.21% of known phosphoproteins (Figure 4A), was also characterized by a high phosphorylation level of predicted phosphosites in untreated conditions (Figure 4B), and included none of the major RNA-silencing components (Supplemental Figure 4). We concluded that the transcriptional and post-translational regulation of antiviral RNA silencing is largely independent of ABA signaling (Figure 3D and Supplemental Figure 4).
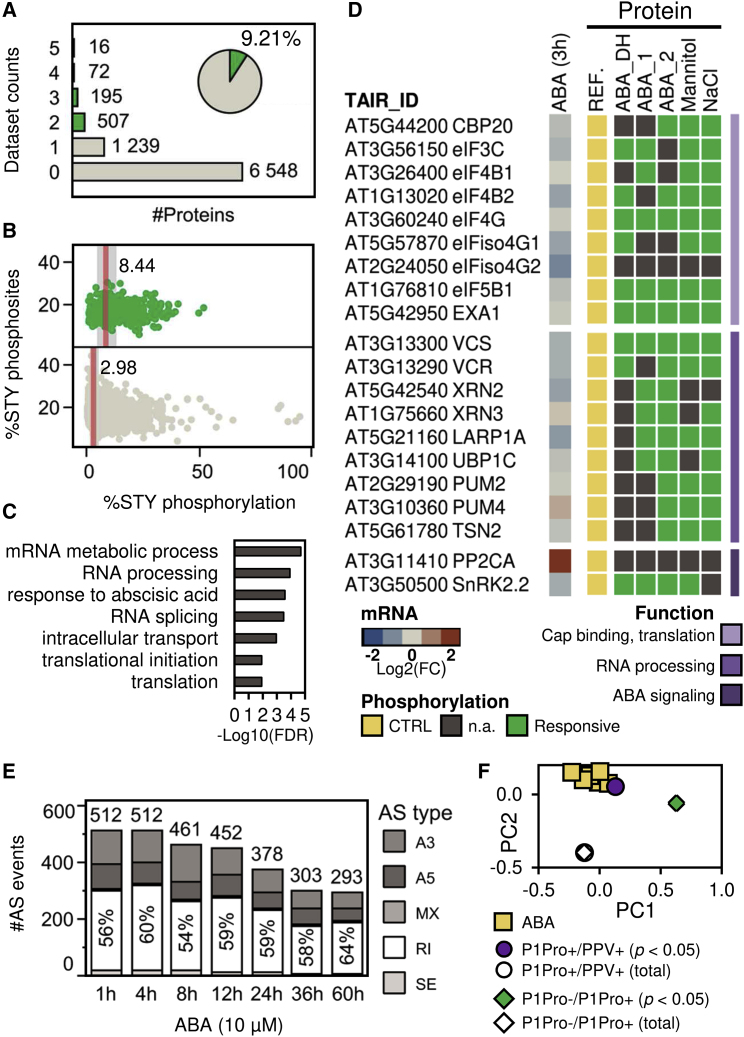
Figure 4.
ABA Modulates the Phosphorylation Dynamics of RNA Cap-Binding and Processing Complex Components and Host Transcriptomic Diversity.
Public phosphoproteomic datasets, including A. thaliana proteins whose phosphorylation status is significantly altered by ABA and dehydration (ABA_DH), ABA (ABA_1, ABA_2), mannitol, or NaCl were analyzed; a comprehensive phosphoproteome was used as a control of known phosphoproteins (REF.; see Supplemental Data 1C).
(A) Overlap of phosphoproteomic datasets. Proteins identified in at least two treatment datasets were considered the responsive subset (green).
(B) Predicted serine, threonine, and tyrosine (STY) phosphosites and their phosphorylation status in proteins identified in the responsive subset (green, top) or a reference phosphoproteome (bottom). Median values are indicated.
(C) Representative GO terms enriched in the responsive subset proteins.
(D) Transcriptional and post-translational regulation of the components of cap-binding and RNA processing complexes. For each A. thaliana gene accession, the heatmap shows microarray expression values from ABA-treated seedlings at 3 h (left) and their protein phosphorylation status and functional annotations (right). Proteins identified in the treatment (responsive) and control phosphoproteome datasets (CTRL) are shown; n.a., no change or phosphopeptide identified. PP2CA and SnRK2.2 are included as ABA-responsive transcriptional or post-translational markers, respectively.
(E) Time course analysis of alternative splicing (AS) events significantly altered (adjusted p < 0.05) in ABA-treated A. thaliana seedlings. Event numbers and the ratios of AS types are shown; SE, skipping exon; RI, retained intron; MX, mutually exclusive exon; A5, alternative 5′ splice site; A3, alternative 3′ splice site.
(F) Principal component analysis of SE, MX, A5, A3, and the RI ratios of events shown in (E) (ABA; each point of the time-course analysis is plotted) or altered in N. benthamiana P1Pro+/PPV+ and P1Pro−/P1Pro+ comparisons (adjusted p < 0.05). Type ratios of all AS events quantified in the N. benthamiana transcriptome analysis, i.e., without applying a p value threshold, are also plotted as a database control (total).
The functional analysis of the identified subset revealed enrichment in GO terms related to RNA metabolism, including RNA processing, splicing, and translation (Figure 4C). Multiple proteins involved in plant RNA metabolism are known (Kawa and Testerink, 2017; Chantarachot and Bailey-Serres, 2018). The phosphorylation status of several components of RNA cap-binding complexes that coordinate host mRNA stability and translation initiation was altered by ABA and osmotic stress (Figure 4D). These treatments also altered the phosphorylation of RNA decay and mRNA turnover regulators.
ABA and P1Pro Infection Alter Plant RNA Diversity
Close inspection of the identified phosphoproteome subset showed proteins involved in pre-mRNA splicing, such as serine/arginine-rich splicing factors, components of the cap-binding complex and those necessary for spliceosomal complex assembly (Supplemental Figure 5A). Consistent with these findings and a regulatory role of ABA in plant RNA maturation and diversity (Laloum et al., 2018), the use of a robust bioinformatic workflow (see Supplemental Information) and time series RNA-seq datasets from A. thaliana enabled the quantification of alternative splice (AS) events significantly altered by ABA (Supplemental Data 1D). Classification of the AS types identified skipping exon (SE), retained intron (RI), mutually exclusive exon (MX), and alternative 5′ or 3′ splice site (A5, A3) events, and showed that RI was the predominant type (Figure 4E). Differentially expressed genes and those presenting AS events showed low overlap and different dynamics; a large increase in differentially expressed genes was observed at 4 h, whereas the number of AS genes peaked as early as 1 h and then declined gradually (Supplemental Figure 5B; Supplemental Data 1E). In ABA-dependent responses, the modulation of RNA maturation and diversity thus provides a rapid effect complementary to mechanisms that regulate RNA abundance.
We used a similar bioinformatics workflow to test whether P1Pro infection affects AS and host transcriptomic diversity. An augmented assembly of the N. benthamiana transcriptome was generated by merging reported transcriptome datasets (Nakasugi et al., 2014; Kourelis et al., 2019) and transcripts assembled from our RNA-seq data. This augmented transcriptome was used to examine transcript abundance and the diversity of P1Pro or PPV samples from inoculated N. benthamiana plants (Supplemental Figure 6). Gene-level differential expression and functional analyses were consistent with and reinforced the findings described above. In the wild-type host, using PPV as a reference (PPV+ samples), P1Pro infection significantly altered host expression profiles, which were again dominated by the upregulation of genes functionally related to defense response, SA and ABA signaling, and protein phosphorylation (Supplemental Figure 6A–6C; Supplemental Data 1F). Transcriptomic comparison of PPV+ and P1Pro+ samples and P1Pro-inoculated N. benthamiana nahG plants (P1Pro−) revealed a comprehensive list of 84 genes whose regulation was altered by P1Pro independent of SA signaling (i.e., SA-independent genes; Supplemental Figure 6A and 6D; Supplemental Data 1G). This SA-independent gene list comprised homologs of known A. thaliana ABA-responsive genes and validated the previous identification of OZF1 and AGO2 (Supplemental Figure 6D). Information from quantified transcripts was then used to identify splice events that were differentially regulated by P1Pro infection (Supplemental Data 1H). By comparing P1Pro and PPV samples from wild-type plants (P1Pro+/PPV+), 156 AS events classified as SE, RI, MX, A5, and A3 were identified with an adjusted p value of < 0.05 (Supplemental Figure 6E). Among these AS types, RI was predominant, and its frequency in the differential AS events was higher (51%; p < 0.05 set) than that of a reference list composed of all events quantified in the P1Pro+/PPV+ comparison (45%; Total set).
P1Pro infection triggers ABA accumulation (Figure 3A), and an unsupervised multivariate analysis was used to assess similarities between AS linked to P1Pro infection and AS linked to ABA (Figure 4F). The representation of principal components showed that the distributions of AS types identified in the P1Pro+/PPV+ or time series ABA/mock comparisons were grouped and were unrelated to those associated with host genotype (P1Pro−/P1Pro+ comparison) or to a bias of the reference transcriptomic dataset. Hierarchical clustering and bootstrap analysis corroborated the association of AS differentially regulated by ABA or P1Pro (Supplemental Figure 6F). These results strongly suggest that AS triggered by P1Pro is biologically relevant; thus, P1Pro infection has a broad impact on the plant transcriptome by simultaneously affecting host RNA abundance and diversity.
Host Genetic ABA Hypersensitivity and RNA Metabolic Perturbation Promote Potyvirus Resistance
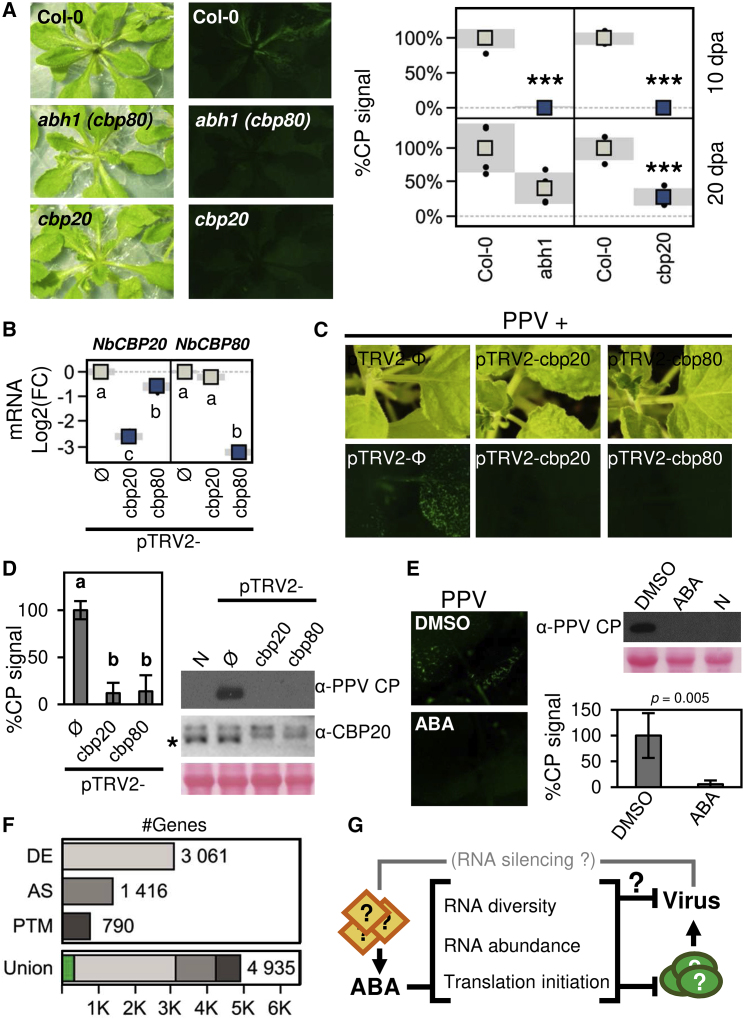
PPV was used in a screen of A. thaliana mutant lines of core ABA signaling components to test whether genetic perturbation of ABA signaling can alter plant susceptibility to virus infection (Supplemental Figure 7A). Mutant lines of the nuclear cap-binding complex components ABA HYPERSENSITIVE1/CAP BINDING PROTEIN 80 (ABH1/CBP80) and CAP BINDING PROTEIN 20 (CBP20) were also analyzed (Hugouvieux et al., 2001; Papp et al., 2004). ABA had no effect on CBP20 or CBP80 expression (Supplemental Figure 5C), however, CBP20 phosphorylation was altered by the ABA treatment or osmotic stress (Figure 4D). Deficiencies in the nuclear cap-binding complex cause ABA hypersensitivity and broad RNA metabolic defects but have no known effect on antiviral RNA silencing (Daszkowska-Golec, 2018; Kørner et al., 2018). Following PPV agro-inoculation, ABA-hyposensitive lines showed no obvious differences in virus accumulation, possibly due to genetic redundancy (Supplemental Figure 7A). By contrast, ABA-hypersensitive lines showed reduced virus accumulation as assessed by immunoblot analysis of the PPV coat protein (CP). Time course infection analysis showed that PPV levels were significantly lower in abh1(cbp80) and cbp20 lines than in wild-type plants at 10 days post-agroinoculation (dpa), and PPV amounts increased at later times (Figure 5A and Supplemental Figure 7B). The results suggest that cap-binding complex mutants are susceptible hosts in which the onset of PPV infection is significantly delayed.
Figure 5.
ABA and the Perturbation of Host RNA Metabolism Promote Plant Resistance to PPV Infection.
(A) Genetic depletion of cap-binding complex promotes PPV resistance. Images show green fluorescence in A. thaliana abh1(cbp80) and cbp20 lines inoculated with the GFP-tagged PPV. At the indicated dpa, virus accumulation was assessed by anti-PPV coat protein (CP) immunoblots. CP signal values are plotted (mean ± SD, n = 4); ∗∗∗p < 0.001 by Student's t-test.
(B–D) Silencing of cap-binding complex genes reduces PPV accumulation in N. benthamiana. The pTRV2-cbp20 or pTRV2-cbp80 VIGS vectors were delivered to target NbCBP20 and NbCBP80 transcripts, respectively; pTRV2-Φ, empty vector control. In (B), the plots of endogenous transcript quantification show mean ± SD (boxes and shadows, respectively; n = 4); letters indicate p < 0.01, one-way ANOVA and Tukey's HSD test. TRV-treated plants were inoculated with GFP-tagged PPV, and samples were collected from upper uninoculated leaves after 6 days; green fluorescence images are shown in (C). (D) Immunoblots show the accumulation of virus (anti-PPV CP) and CBP20. The asterisk marks a major band detected using the anti-CBP20 serum. This band is absent from pTRV2-cbp20 and pTRV2-cbp80 samples. N, untreated sample. The bar plot shows CP signal values (mean ± SD, n = 4); letters indicate p < 0.01, one-way ANOVA and Tukey's HSD test.
(E) ABA treatments delay PPV infection. Plants were mechanically inoculated with the GFP-tagged PPV and treated with 25 μM ABA or a mock solution (DMSO). Green fluorescence images were taken and samples for immunoblot analysis were collected from upper uninoculated leaves at 6 dpi. Anti-PPV CP immunoblot and signal quantification values are shown (mean ± SD, n = 4); p value is shown (Student's t-test). Ponceau red-stained blots are shown as a loading control.
(F) Genome-wide impact of ABA. The plot shows A. thaliana genes differentially expressed (DE), alternatively spliced (AS), or post-translationally regulated (PTM) by ABA, as identified in Supplemental Figure 5C and Figure 4A. The cumulative number of regulated genes and their overlap (green) are shown at the bottom (see Supplemental Data 1E).
(G) A regulatory model of ABA and RNA metabolic pathways in antiviral immunity. Modulation of RNA diversity, abundance, and turnover and translation initiation by ABA signaling could impair host factors with pro-viral functions or directly act on host or viral components to promote plant resistance. Perturbation of RNA silencing may be involved in ABA activation during virus infection.
We evaluated the role of the cap-binding complex in N. benthamiana by virus-induced gene silencing (VIGS) of the NbCBP80 and NbCBP20 homologs, using the pTRV2-cbp80 or the pTRV2-cbp20 constructs, respectively; gene knockdown was confirmed by qRT–PCR assays (Figure 5B). PPV was inoculated mechanically to avoid possible confounding effects of Agrobacterium. At 6 dpi, fluorescence imaging of the viral reporter GFP showed a clear signal in upper uninoculated leaves of control plants, whereas GFP fluorescence was barely detectable in pTRV2-cbp20 or pTRV2-cbp80 conditions (Figure 5C). Immunoblotting confirmed that PPV accumulation was significantly reduced in pTRV2-cbp20 and pTRV2-cbp80 samples (p < 0.01, Figure 5D and Supplemental Figure 7C). In agreement with the mRNA quantification, immunoblotting with anti-CBP20 antibodies showed that the signal of a major band in control samples dropped in pTRV2-cbp20 samples (p = 0.001, Supplemental Figure 7C). In A. thaliana, the CBP20 protein is stabilized by CBP80, and its accumulation is impaired in the abh1(cbp80) mutant line (Daszkowska-Golec, 2018). Consistent with this finding, anti-CBP20 immunoblotting showed that the major CBP20 band could not be detected in either pTRV2-cbp20 or pTRV2-cbp80 samples (Figure 5D). The collective results of A. thaliana and N. benthamiana assays indicate that defects in the nuclear cap-binding complex promote plant resistance to PPV.
ABA Treatment Enhances Potyvirus Resistance
To provide additional evidence of ABA antiviral effects, we treated PPV-inoculated N. benthamiana plants with control (DMSO) or ABA solutions (50 or 25 μM). At 6 dpi, upper uninoculated leaves of control plants showed a clear signal for the viral reporter GFP, whereas GFP fluorescence was barely detectable in ABA-treated plants (Figure 5E). CP immunoblotting confirmed significantly reduced PPV accumulation in plant samples treated with ABA solutions compared with controls (Figure 5E and Supplemental Figure 7D). The results in Figures 3 and 5 thus indicate that the self-controlled PPV prevents or limits the activation of ABA signaling, which in turn promotes antiviral immunity once activated by a genetic or exogenous means.
In Silico Simulations Support Controlled Polyprotein Processing as an Enhancement Mechanism for Virus Accumulation
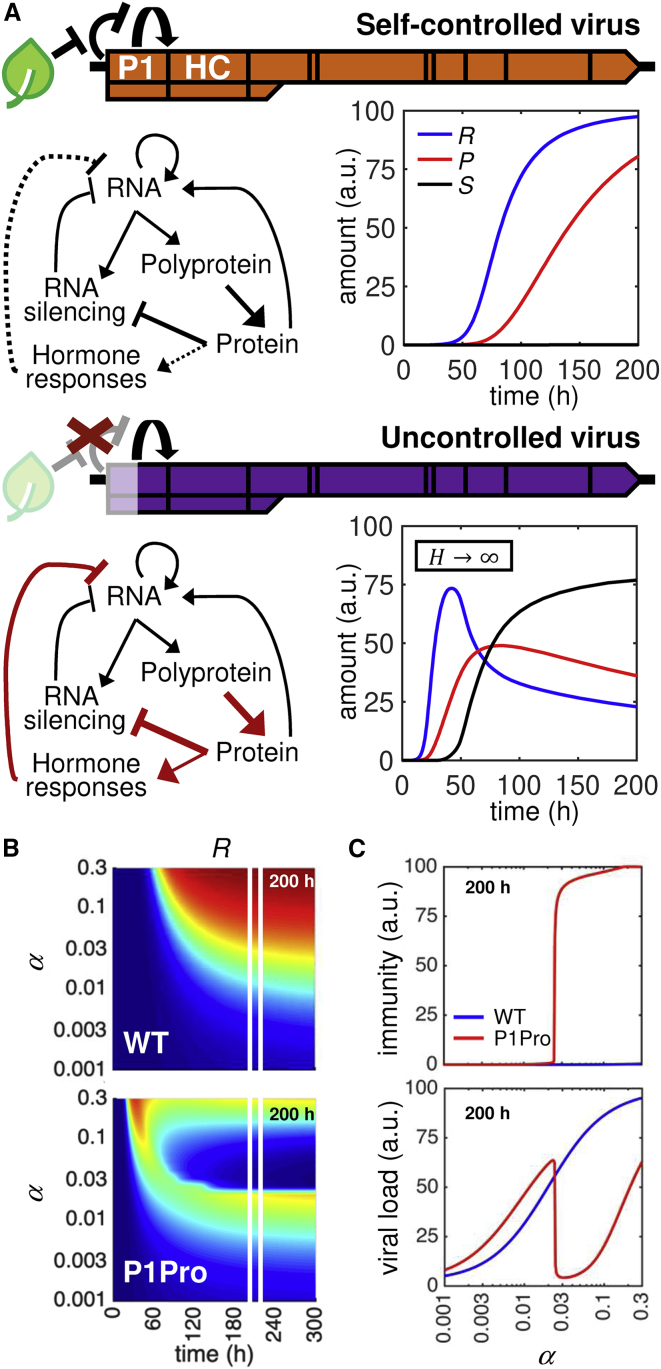
Host RNA-silencing and hormone-mediated defenses, as well as self-controlled processing of viral polyproteins, were integrated into a mathematical model to assess their contributions to potyvirus fitness (Figure 6A). We developed a model based on four ordinary differential equations, which included the viral RNA, the viral polyprotein and processed proteins, and host antiviral immunity mediated by RNA silencing and hormone signaling as variables (Supplemental Table 3). In the wild-type virus scenario, polyprotein processing was assumed to depend on a host factor (H) that, in a finite, limiting amount, relieves the autoinhibition of P1 cleavage and HCPro release (i.e., self-controlled virus; Figure 6A, top; WT, Supplemental Figure 8). The conditional, controlled release of viral proteins reduced the effectiveness of silencing suppression and the viral amplification rate but allowed the evasion of hormone-mediated defenses and high viral loads at later times. In the second scenario, uncontrolled self-cleavage was simulated in the limit of infinite H amount (Figure 6A, bottom; P1Pro, Supplemental Figure 8). Unrestricted, continuous polyprotein processing promoted high viral loads but also the activation of defense response at early times; these were followed by a drop in viral load at later times.
Figure 6.
Mathematical Model of Within-Host Viral Replication—Interplay between Viral Polyprotein Processing and Host RNA Silencing and Hormone Responses.
(A) A mathematical model was developed and the numerical simulations of viral RNA (R, blue line), mature protein (P, red), and immune response (S, black) levels are shown for different scenarios. Top, the autoinhibition of P1 self-cleavage and HCPro release is relieved by a host cofactor (H) at a finite, limiting amount. The controlled release of HCPro, an RNA-silencing suppressor, allows the attenuation of pathogen virulence and the evasion of hormone-mediated defense responses. This regulatory mechanism assures high long-term viral loads. Bottom, removal of the P1 autoinhibitory domain (simulated with H→∞) leads to an uncontrolled release of HCPro and the suppression of RNA silencing but activates hormone responses that restrict viral amplification.
(B) Time course simulations for varying strength of the RNA-silencing suppression (α). Viral RNA (R) accumulation relative to the maximum (dark blue for 0% and dark red for 100%) is shown for the wild-type PPV (WT, top) and the uncontrolled self-cleavage (P1Pro, bottom) scenarios. The 200-h time point is marked and plotted in (C).
(C) Viral RNA load (bottom) and immune response (top) levels at 200 h of the dynamics shown in (B). The wild-type PPV (WT, blue) and the uncontrolled self-cleavage (P1Pro, red) scenarios are shown; α, strength of the RNA-silencing suppression.
In the model, uncontrolled self-cleavage triggers defense activation through the enhancement of silencing suppression and viral amplification rates. The effect of viral replication itself on the activation of defense response were explored by varying the RNA synthesis rate (ksyn) or RNA binding constant (θ) parameters of the viral replicase (Supplemental Figure 9). Simulations showed that the θ value has negligible effects on viral replication dynamics (Supplemental Figure 9A). By contrast, an increase in RNA synthesis rate triggered a rapid viral amplification and defense response activation, followed by a drop in viral load at later times (Supplemental Figure 9B and 9C). These results mimicked those of the uncontrolled self-cleavage simulation, albeit to a lesser extent due to the preservation of controlled polyprotein processing. Although oversimplified, this pathosystem model supports the controlled processing of viral polyproteins as a regulatory mechanism for virus accumulation that defines the timing and magnitude of host defense response activation and, consequently, the pathogen fitness.
A Working Model to Explain P1 Variability in the Potyviridae
P1 self-cleavage requirements vary greatly in the Potyviridae family; in addition, some potyvirids lack P1, and thus its control function over the viral silencing suppressor (Cui and Wang, 2019). We tested whether the adjustment of silencing suppressor activity would itself be sufficient to compensate for the lack of controlled P1 self-cleavage. The effects of varying the strength of RNA-silencing suppression (α) on infection dynamics were explored with (WT) or without (P1Pro) controlled polyprotein processing. Simulations of viral RNA (R) accumulation showed that a low α value enhances P1Pro loads (in our model, α ∼ 0.01; Figure 6B). A reduction in RNA-silencing suppression strength permitted P1Pro to avoid the activation of immune responses and to outperform the wild-type, self-controlled virus in terms of accumulation (α < 0.02; Figure 6C and Supplemental Figure 10). In other words, the acquisition of mutations that reduce suppression strength is predicted to be sufficient to escape hormone-mediated defenses and overcome the lack of P1 or self-cleavage control. These results suggest that uncontrolled self-cleavage (or P1 absence) would be advantageous in hosts with stringent RNA silencing or in which the virus replicates deficiently; conversely, the regulation of P1 cleavage and silencing suppressor activity would maximize viral fitness in adapted hosts.
Discussion
Viruses are obligate parasites that, to maximize their fitness, must evolve control mechanisms to escape host immunity, while simultaneously avoiding acute responses that might lead to early host death. We previously identified an autoinhibitory domain in PPV that controls P1 self-cleavage, which in turn modulates the release of a functional RNA-silencing suppressor and viral replication (Pasin et al., 2014).
Here, a genome-wide analysis showed that P1Pro, a PPV clone that lacks the P1 autoinhibitory domain, triggered complex transcriptomic changes that included the SA and possibly the pipecolate pathways, central components of plant systemic acquired resistance and antiviral immunity (Hartmann and Zeier, 2019; Wang et al., 2019; Murphy et al., 2020). Infection of SA signaling-defective hosts led to the identification of additional players in P1Pro-induced responses, and meta-analysis using our transcriptomic datasets and those of hormone-treated A. thaliana plants highlighted the contribution of ABA. Our findings here indicate that multiple host signals, such as SA, ABA, and (possibly) pipecolate, overlap to provide robust defense responses to P1Pro, which are evaded by the wild-type PPV.
The P1Pro infections showed HR accompanied by an increase in ABA levels and the expression of OZF1, which encodes an ABA-responsive transcription factor thought to participate in PTI/ETI (Lee et al., 2012; Cao et al., 2019). How can P1Pro infection trigger ABA-dependent responses? Various viral components, such as viral nucleic acids, proteins, and their activities, can act as pathogenesis determinants and elicit ETI (García and Pallás, 2015). In our assays, the truncated P1 version that lacks the autoinhibitory domain and is encoded by the P1Pro clone was itself insufficient to elicit local HR. This indicates that additional factors of P1Pro infection contribute to systemic ETI, which is inactive during infection with wild-type PPV, including the full-length, self-controlled P1. A single mutation of the polymerase domain of Plantago asiatica mosaic virus was shown to promote viral replication, and the resultant necrotic symptoms were caused by overaccumulation of an elicitor domain located in the viral helicase domain (Komatsu et al., 2011). Uncontrolled infection may also lead to autoimmune responses due to the activation of otherwise silent host components. Excess silencing suppression could cause disease by relieving microRNA repression of resistance genes (Li et al., 2012; Shivaprasad et al., 2012) or by altering the biogenesis of virus-activated small interfering RNAs (Cao et al., 2014).
Several lines of evidence indicate that RNA silencing and ABA signaling are interlinked, and ABA was found to affect the transcript level of RNA-silencing genes, as recently reviewed by Alazem and Lin (2020). Despite this body of data, our meta-analysis of available microarray and RNA-seq datasets revealed that the main RNA-silencing genes are unlikely to be primary transcriptional targets of ABA signaling. Treatment of 10 μM ABA was sufficient to upregulate the expression of known ABA marker genes in A. thaliana seedlings (Nemhauser et al., 2006; Song et al., 2016); by contrast, the transcript levels of RNA-silencing genes were largely unaffected. These discrepancies might be explained by differences in the amount and timing of phytohormone treatments or by plant growth stages, which were recently shown to influence ABA perception (Berens et al., 2019). Differences in experimental and technical focus and setups should also be considered. Transcriptome-wide microarray or RNA-seq analyses might provide a more accurate, unbiased estimate of transcript abundance than qRT–PCR. In this regard, the expression of host genes commonly used as references in qRT–PCR assays might fluctuate after ABA treatment (e.g., ACT2 or ACT8; Supplemental Figure 5C) and interfere with the quantification of targeted transcripts.
AGO2 is a main RNA-silencing component for plant resistance to RNA viruses (Carbonell and Carrington, 2015). AGO2 and ABA were found to accumulate differentially in P1Pro versus PPV infections. In our assays, AGO2 genetic depletion was itself insufficient to induce ABA in control conditions or to prevent ABA and SA accumulation during P1Pro infection. A recombinant PVX clone with high silencing suppressor activity was shown to induce ABA (Pacheco et al., 2012). Here, in a complementary approach, PVX infection of the ago2 mutant line was characterized by significantly augmented ABA levels. In contrast to P1Pro or PVX infection results, the PPV clone with a controlled P1 self-cleavage and silencing suppressor release could avoid ABA induction in the wild-type or RNA-silencing-defective hosts.
Viral silencing suppressors are known to target endogenous factors that regulate antiviral RNA silencing and cell RNA metabolism and to affect viral fitness and the transcriptomic profiles of infected hosts (Cervera et al., 2018; Valli et al., 2018; Li and Wang, 2019). One intriguing possibility is that an excess of silencing suppression and viral replication or translation products prime ABA signaling directly or as a result of cellular metabolic/ionic homeostasis perturbation or HR. This would safeguard plants against the depletion of antiviral RNA-silencing components and provide an additional layer of immunity. In tobacco, the simultaneous perception of a viral silencing suppressor and unbalanced cellular ionic homeostasis activates HR and SA signaling (Jeon et al., 2017). It would be interesting to determine whether this perception also triggers ABA, which coordinates plant response to osmotic stress.
If ABA signaling does not regulate the transcript abundance of RNA-silencing genes, what is the functional role of ABA in antiviral immunity? A network of protein phosphatases and kinases mediates ABA signaling and plant responses to osmotic stress (Cutler et al., 2010; Umezawa et al., 2013; Wang et al., 2013, 2020), and this network might post-translationally regulate host proteins that are directly or indirectly necessary for plant immunity or susceptibility. Translation inhibition, RNA maturation and turnover, and their regulation by phosphorylation events have critical roles in the antiviral immune responses of human cells (Narayanan and Makino, 2013; McCormick and Khaperskyy, 2017; Boudreault et al., 2019). In plants, phosphorylation events promote RNA decay in response to bacteria and abiotic stress (Xu and Chua, 2012; Soma et al., 2017; Yu et al., 2019), inhibit or disrupt the translation of host transcripts (Bruns et al., 2019; Cho et al., 2019), and have been suggested to alter RNA maturation by affecting the activity or subcellular distribution of splicing factors (Laloum et al., 2018). Consistent with these studies, results of our phosphoproteomic meta-analysis revealed ABA-dependent responsiveness of the phosphorylation status of host components that regulate RNA stability, turnover, maturation, and translation initiation. These included (1) factors known to be needed for potyvirus (eIFiso4G1, but not eIFiso4G2) or potexvirus infection (EXA1) (Nicaise et al., 2007; Yusa et al., 2019); (2) regulators of the emerging antiviral pathway of RNA decay, such as those involved in mRNA decapping (i.e., VCS and VCR) and 5′-3′ mRNA decay (XRN2, XRN3, LARP1A); (3) determinants of mRNA turnover, such as UBP1C, PUM2, PUM4, and TUDOR-SN PROTEIN 2 (TSN2); and (4) components necessary for mRNA splicing, including coilin, a protein recently linked to SA signaling activation in response to virus infection (Mäkinen et al., 2017; Chantarachot and Bailey-Serres, 2018; Li and Wang, 2019; Shaw et al., 2019; Xu et al., 2020).
Although the functional consequences of these phosphorylation events remain to be determined in the context of virus infection, we analyzed alternative splicing of the plant transcriptome, aiming to establish a link between P1Pro-activated responses and ABA-dependent perturbation of RNA metabolism. Alternative splicing is a phenomenon reported in virus (and potyvirus) infection (Martin et al., 2016; Boudreault et al., 2019) and can be used as a proxy for measuring changes in host RNA maturation. Type frequencies of AS events differentially regulated by P1Pro with respect to PPV are similar to those in plant transcriptomes after ABA treatment. This similarity was significantly higher than expected by chance and was unrelated to the AS type frequency of the reference transcriptome used in the analysis, supporting its biological relevance. We anticipate that future use of improved transcriptomic assemblies and high sequencing depths will help better characterize the extent of AS events in N. benthamiana infected with P1Pro and other viruses.
Having established links between P1Pro-activated responses, high ABA levels, and ABA-dependent changes in host RNA abundance and diversity, we evaluated the effects of the genetic depletion or downregulation of ABA signaling components on PPV infection. These included components of the nuclear cap-binding complex that were identified in our phosphoproteomic meta-analysis, whose dysregulation is known to cause ABA hypersensitivity and broad RNA metabolic defects (Hugouvieux et al., 2001; Papp et al., 2004; Laubinger et al., 2008; Daszkowska-Golec, 2018). We showed that plant resistance to PPV was enhanced by exogenous ABA application and, in a complementary approach, by the knockout of CBP20 or CBP80 in A. thaliana, or their downregulation in N. benthamiana.
In A. thaliana, our analyses showed that ABA signaling affects the transcript abundance, splice dynamics, or protein phosphorylation status of ca. 5000 genes (Figure 5F). Supported by our transcriptomic data, the meta-analysis of A. thaliana datasets, and the infection results of hosts defective in the nuclear cap-binding complex, we propose that ABA could promote antiviral immunity through an extensive, multi-level effect on host RNA metabolism (Figure 5G). This would alter the availability of host pro-viral factor or act directly on viral components to promote plant resistance. For effective resistance, protein phosphatases and kinases of the core ABA or downstream pathways might further target and impair the function of host proteins (e.g., translation initiation factors; see above) and viral proteins necessary for virus infection. The phosphorylation of viral proteins regulates viral cell-to-cell movement, suppressor activity, and virion assembly, among other processes (Hung et al., 2014; Hoover et al., 2016; Shen et al., 2018; Zhang et al., 2018; Hervás et al., 2020).
Based on these observations, we built a mathematical model to explain the effect of controlled polyprotein processing on viral replication and the induction of plant defense. The quantitative modeling of a simplified pathosystem and in silico simulations corroborated our working hypothesis that controlled viral polyprotein cleavage can act as a key mechanism that maximizes pathogen fitness by balancing viral protein release and replication with the magnitude of RNA silencing and hormone responses. Simulations further showed that a decrease in the strength of silencing suppression could partly compensate for a lack of self-control. Lack of P1 or its uncontrolled self-cleavage described in some potyvirids would therefore be explained by a low replicative capacity that maintains viral accumulation below a threshold needed for the activation of immune responses.
Can P1 or its protease domain have additional, as-yet-undefined roles in potyviral infection? P1 from the tobacco etch virus was shown to bind ribosomal subunits and proposed to stimulate the translation of viral transcripts (Martínez and Daròs, 2014). P1 translation stimulatory activity was not considered in our mathematical modeling because additional experiments are required to determine whether the full-length protein or the protease domain itself (i.e., that encoded by P1Pro) is necessary for P1 interaction with ribosomal subunits and also with polysomal complexes, which include transcripts undergoing active translation (Chantarachot and Bailey-Serres, 2018). According to our working model, P1 self-cleavage regulates viral replication through controlled HCPro release. HCPro was found to localize with RNA turnover components, including the homologs of VCS and UBP1C (Hafrén et al., 2015) and ABA-responsive phosphoproteins identified in our meta-analysis. HCPro interacts physically with and inhibits XRN4 to counteract RNA decay antiviral defense (Li and Wang, 2018). HCPro from the sugarcane mosaic virus is reported to bind host violaxanthin de-epoxidase, an enzyme that regulates ABA precursor abundance (Chen et al., 2017); the importance of this interaction in ABA signaling is currently unknown. Additional HCPro interactions with functionally diverse plant proteins were reviewed by Valli et al. (2018). Controlled P1 self-cleavage could thus fine-tune multiple HCPro functions, including its silencing suppression and RNA decay-antagonizing activities.
Self-control mechanisms have been reported in plant viruses. To attenuate virus accumulation, the tobamovirus movement protein and the sobemovirus P1 promote silencing spread (Vogler et al., 2008; Lacombe et al., 2010), whereas cucumber mosaic virus 2b suppressor activity is antagonized by the viral CP (Zhang et al., 2017). A host-dependent self-control mechanism would allow viruses to sense the status of host physiology and its fluctuations due to environmental factors. The geminivirus-silencing suppressor βC1 was recently shown to activate and be degraded by host autophagy to reduce viral virulence (Ismayil et al., 2020). Cellular peptidylprolyl isomerases or protein chaperones regulate polyprotein cleavage kinetics and the persistence of human and animal viruses, such as the hepatitis C virus, yellow fever virus, and bovine viral diarrhea virus (Kaul et al., 2009; Bozzacco et al., 2016; Isken et al., 2019). More recently, a functional link between polyprotein processing and host RNA metabolism was reported in alphaviruses. Infection by alphaviruses with high polyprotein processing efficiency was found to depend on G3BP, a host regulatory protein involved in RNA translation and turnover (Götte et al., 2020). Such host-dependent regulation of polyprotein cleavage would be especially beneficial for establishing persistent infections that can allow virus adaptation to perennial and long-lived hosts, such as woody trees (Peng et al., 2001; Salvador et al., 2008; Shan et al., 2015, 2018; Su et al., 2016; Kang et al., 2018).
Finally, in human cells, the activation of interferon antiviral immunity results in translation inhibition and the indiscriminate degradation of host and viral RNA by host ribonucleases (Narayanan and Makino, 2013; McCormick and Khaperskyy, 2017). In prokaryotic cells, the activation of a CRISPR/Cas system with nonspecific ribonuclease activity was recently reported to trigger the indiscriminate degradation of host transcripts and provide robust phage immunity through the induction of cellular dormancy (Meeske et al., 2019). In plants, ABA promotes growth arrest and accumulates at high levels in dormant tissues in vegetative organs (Yao and Finlayson, 2015; Martín-Fontecha et al., 2018). The translation initiation disruption and translation repression of a large portion of plant genes were reported in response to dehydration (Kawaguchi et al., 2004), and it is plausible that a similar phenomenon occurs during ABA signaling. ABA-dependent control of host RNA metabolism and translation could contribute to cellular metabolic arrest and dormancy in an antiviral response that shows a striking resemblance to interferon- and CRISPR/Cas-mediated antiviral immunity of human and prokaryotic cells, respectively. ABA may therefore be an overlooked, active player that controls viral infection dynamics in plants.
Materials and Methods
Plant Materials
Wild-type Nicotiana benthamiana, the N. benthamiana nahG-transgenic line expressing an SA hydroxylase gene (Ying et al., 2010), and the N. benthamiana ago2 mutant line (Ludman et al., 2017) were grown in a greenhouse with a 16-h light/8-h dark photoperiod in a 19°C–23°C temperature range. The Arabidopsis thaliana abh1(cbp80) and cbp20 single-mutant lines (Hugouvieux et al., 2001; Papp et al., 2004) were grown in vitro on half-strength Murashige and Skoog medium (pH 5.7) with 2-(N-morpholino)ethanesulfonic acid and vitamins, 1% (w/v) sucrose, and 0.8% (w/v) Bacto Agar and were maintained in a growth chamber (16-h light/8-h dark photoperiod, 21°C ± 1°C, 60% relative humidity).
Plasmids, Virus Inoculation, and VIGS Assays
A full-length cDNA copy of a PPV isolate adapted to Nicotiana spp., tagged with a GFP gene, and inserted into the pSN-PPV binary plasmid has been reported (Supplemental Table 4), as has pSN-PPV P1Pro[V164] (herein P1Pro), a pSN-PPV-based vector with a truncated P1 sequence (Pasin et al., 2014). Binary vectors used in transient expression assays were reported or generated as detailed in Supplemental Table 4. A tobacco rattle virus (TRV) system was used to silence endogenous transcripts, and the SGN VIGS tool was used to select suitable regions (Fernandez-Pozo et al., 2015). The assembly of pTRV2-cbp80 and pTRV2-cbp20 and the sequences of the primers and the fragments cloned are detailed in Supplemental Tables 4–7. The Agrobacterium C58C1-313 strain (Pasin et al., 2017) was used for plant agro-inoculation and VIGS vector delivery. PPV clones were delivered by Agrobacterium-mediated infection to soil- and in vitro-grown N. benthamiana and A. thaliana plants, respectively (Pasin et al., 2014). VIGS assays were conducted as previously described (Aguilar et al., 2019). Agrobacterium strains that harbored pTRV1 and pTRV2 or its derivatives were mixed and infiltrated into 2-week-old N. benthamiana plants. After 10 days, young plant leaves were inoculated mechanically with PPV. For mechanical inoculations, crude extracts were prepared in 100 mM phosphate buffer (pH 7.2) from agro-infected N. benthamiana plants and used as PPV inoculum sources. PPV inoculum was applied with carborundum and gently rubbed on young N. benthamiana leaves. PVX was inoculated as previously described (Ludman et al., 2017).
Transcriptomic Analysis
N. benthamiana plants were inoculated by agro-infection (see above), and upper uninoculated leaves were collected at 14 dpa. For each experimental condition, three biological replicates (each sample was a pool of ≥2 infected plants) were used for transcriptomic analysis (RNA-seq). Plant tissues were ground in liquid nitrogen in a mortar, and total RNA was extracted with the FavorPrep plant total RNA mini kit (Favorgen), including on-column DNAse I treatment. Following a second DNAse I digestion, samples were purified by organic extraction and sodium acetate precipitation. Ribosomal RNA molecules were depleted using the RiboMinus plant kit (Invitrogen). Recovered RNA was quantified using a fluorescent RNA binding dye (QuantiFluor RNA system, Promega) and analyzed in a Bioanalyzer with the RNA 6000 Pico kit (Agilent). Libraries were prepared using the TruSeq Stranded mRNA kit (Illumina), omitting the mRNA enrichment step, and were sequenced (2 × 100-nt paired-end reads) on an Illumina HiSeq 2000 platform at the Centro Nacional de Análisis Genómico (CNAG-CRG, Spain) to yield ≥1.8 Gb/sample. See Supplemental Information for details of transcriptomic dataset analysis and gene functional annotations.
A study of hormone-treated A. thaliana plants has been reported (Nemhauser et al., 2006), and its microarray data (AT-00110) were obtained from Genevestigator (Hruz et al., 2008). RNA-seq results and transcript fold changes from ABA time series experiments (Song et al., 2016) were retrieved from the NCBI Gene Expression Omnibus (GEO) (GSE80565), and RNA-seq reads were used to analyze AS events (see Supplemental Information).
Quantitative Proteomic and Phosphoproteomic Datasets
Sample preparation, isobaric tag labeling, liquid chromatography-tandem mass spectrometry (LC-MS/MS), and data analysis of N. benthamiana nahG plants inoculated with PPV or P1Pro have been described previously (Pasin et al., 2014), and the data are available at ProteomeXchange with identifier PXD017769.
The datasets of A. thaliana proteins with significantly altered phosphorylation status in response to ABA or dehydration stress (ABA_DH), ABA (ABA_1, ABA_2), mannitol, or NaCl have been reported (Umezawa et al., 2013; Wang et al., 2013, 2020). Wild-type and snrk2.2/2.3/2.6 triple-mutant results were considered, and protein lists are included in Supplemental Data 1D. The phosphoproteome from Mergner et al. (2020) was used as a reference list and as the background in GO term enrichment analysis.
qRT–PCR Assays
Total RNA was purified from the upper uninoculated leaf samples using the Plant Total RNA mini kit (Geneaid). To remove DNA contaminants, 1 μg purified RNA was treated with ezDNAse enzyme (Thermo Fisher) before cDNA synthesis using M-MuLV reverse transcriptase (New England BioLabs) and random hexamers. The cDNA samples were used in qPCR reactions that included gene-specific primers (Supplemental Tables 5 and 6) and 5× HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne) and were run on a 7300 real-time PCR System (Applied Biosystems). Expression was normalized using NbPSMD1 as a reference gene (Supplemental Table 6), and fold changes relative to the control condition were calculated by the ΔΔCT method (Rodamilans et al., 2014) and expressed as log2 values. The RT–PCR assays shown in Figure 4B were performed using the 2174_F/1631_R primers (Supplemental Table 5).
Protein Detection
GFP fluorescence was visualized under an epifluorescence microscope (MZ FLIII, Leica; excitation and emission wavelengths 470/40 and 525/50 nm, respectively) and photographed with an Olympus DP70 digital camera. Total protein extracts from plant samples were prepared and resolved by SDS–PAGE as described previously (Pasin et al., 2014). Immunodetection was conducted using rabbit anti-PPV CP (Pasin et al., 2014) and anti-CBP20 sera (Raczynska et al., 2014) as primary antibodies.
Hormone Quantification and Treatments
Frozen leaf material (∼100 mg fresh weight) was extracted in 20:80 (v/v) methanol:isopropanol with 1% glacial acetic acid and isotope-labeled internal standards. Supernatants were analyzed by ultra-performance LC-MS/MS, and free SA and ABA were quantified as described previously (Müller and Munné-Bosch, 2011). For treatment, (±)-ABA (A1049, Sigma) was dissolved in DMSO, and ABA solutions containing 0.005% Silwet-77 were sprayed onto plants once on the day before virus inoculation and twice daily thereafter. A DMSO-containing solution was used as a control. Upper uninoculated leaves were collected at 6 dpi.
Statistics
A two-tailed Student's t-test was used for comparisons of two groups. A one-way ANOVA with Tukey's honestly significant difference test was used to assess significant differences between more than two groups. Significance levels of p values are indicated in the figures.
Mathematical Modeling
Sigmoidal equations were used to describe different biochemical processes and were then assembled to construct a system of ordinary differential equations to describe the dynamics (see Supplemental Information). The host was assumed to be a single, uniform compartment in which biochemical processes occurred. Parameter values were chosen according to available experimental data (Supplemental Table 3). Numerical simulations to obtain the dynamic behavior of viral infection were carried out with MATLAB (MathWorks). Different scenarios were modeled by changing key parameter values.
Funding
This work was supported by funds to J.A.G. from the Ministerio de Ciencia e Innovación (Spain), grants BIO2016-80572-R and PID2019-109380RB-I00 / AEI / 10.13039/501100011033 (AEI-FEDER). K.F. is funded by grant K124705 from the National Research Development and Innovation Office (Hungary), and S.M.-B. by grant 2017 SGR 980 from the Generalitat de Catalunya (Spain). Galaxy is a platform supported by NIH grant HG006620. F.P. was the recipient of a post-doctoral fellowship from Academia Sinica (Taiwan).
Author Contributions
F.P. and J.A.G. conceived the study. F.P. designed and coordinated the research. F.P., H.S., and B.G. performed the experiments. F.P., D.S.L., and J.A.G. analyzed the data. K.F. and M.L. performed the N. benthamiana ago2 experiments. S.M.-B., M.M., and D.H.F. carried out the hormone quantification. G.R. developed the mathematical model and performed the numerical simulations. J.A.G., S.M.-B., and K.F. secured funding. F.P. wrote the paper with the collaboration of J.A.G. and G.R. All authors approved the final version of the manuscript.
Acknowledgments
We are grateful to Bernardo Rodamilans, István Papp, Brian D. Gregory, Eduardo González-Grandío, Pilar Cubas, Francisco Tenllado, Hui-Shan Guo, Gregory B. Martin, Katarzyna Dorota Raczyńska, and Artur Jarmołowski for the supply of materials. We thank Galaxy (Afgan et al., 2018) for computational resources and Catherine Mark for editorial assistance. No conflict of interest declared.
Published: July 7, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Accession Numbers
The RNA-seq raw reads from this study can be found at NCBI under GEO accession number GSE146746 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146746). Proteomic data from PPV- or P1Pro-inoculated N. benthamiana plants are available at the Proteomics Identification Database with identifier PXD017769. The A. thaliana microarray datasets from hormone-treated plants are available at Genevestigator (AT-00110). The RNA-seq fold change values and raw reads from ABA time series experiments are available at GEO (GSE80565) and the Sequence Read Archive (SRP073711). Major A. thaliana genes mentioned herein are as follows: AGO2, AT1G31280; OZF1, AT2G19810; CBP20, AT5G44200; CBP80, AT2G13540; PP2CA, AT3G11410; SnRK2.2, AT3G50500.
Supplemental Information
References
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar E., Del Toro F.J., Brosseau C., Moffett P., Canto T., Tenllado F. Cell death triggered by the P25 protein in Potato virus X-associated synergisms results from endoplasmic reticulum stress in Nicotiana benthamiana. Mol. Plant Pathol. 2019;20:194–210. doi: 10.1111/mpp.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., Lin N.-S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015;16:529–540. doi: 10.1111/mpp.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., Lin N.-S. Interplay between ABA signaling and RNA silencing in plant viral resistance. Curr. Opin. Virol. 2020;42:1–7. doi: 10.1016/j.coviro.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Alazem M., He M.-H., Moffett P., Lin N.-S. Abscisic acid induces resistance against Bamboo mosaic virus through Argonaute2 and 3. Plant Physiol. 2017;174:339–355. doi: 10.1104/pp.16.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens M.L., Berry H.M., Mine A., Argueso C.T., Tsuda K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017;55:401–425. doi: 10.1146/annurev-phyto-080516-035544. [DOI] [PubMed] [Google Scholar]
- Berens M.L., Wolinska K.W., Spaepen S., Ziegler J., Nobori T., Nair A., Krüler V., Winkelmüller T.M., Wang Y., Mine A. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. U S A. 2019;116:2364–2373. doi: 10.1073/pnas.1817233116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault S., Roy P., Lemay G., Bisaillon M. Viral modulation of cellular RNA alternative splicing: a new key player in virus-host interactions? Wiley Interdiscip. Rev. RNA. 2019;10:e1543. doi: 10.1002/wrna.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzacco L., Yi Z., Andreo U., Conklin C.R., Li M.M.H., Rice C.M., MacDonald M.R. Chaperone-assisted protein folding is critical for yellow fever virus NS3/4A cleavage and replication. J. Virol. 2016;90:3212–3228. doi: 10.1128/JVI.03077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns A.N., Li S., Mohannath G., Bisaro D.M. Phosphorylation of Arabidopsis eIF4E and eIFiso4E by SnRK1 inhibits translation. FEBS J. 2019;286:3778–3796. doi: 10.1111/febs.14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Du P., Wang X., Yu Y.-Q., Qiu Y.-H., Li W., Gal-On A., Zhou C., Li Y., Ding S.-W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2014;111:14613–14618. doi: 10.1073/pnas.1407131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F.Y., Khan M., Taniguchi M., Mirmiran A., Moeder W., Lumba S., Yoshioka K., Desveaux D. A host-pathogen interactome uncovers phytopathogenic strategies to manipulate plant ABA responses. Plant J. 2019;100:187–198. doi: 10.1111/tpj.14425. [DOI] [PubMed] [Google Scholar]
- Carbonell A., Carrington J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015;27:111–117. doi: 10.1016/j.pbi.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera H., Ambrós S., Bernet G.P., Rodrigo G., Elena S.F. Viral fitness correlates with the magnitude and direction of the perturbation induced in the host’s transcriptome: the tobacco etch potyvirus-tobacco case study. Mol. Biol. Evol. 2018;35:1599–1615. doi: 10.1093/molbev/msy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantarachot T., Bailey-Serres J. Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 2018;176:254–269. doi: 10.1104/pp.17.01468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang L., Li D., Wang F., Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2013;110:1963–1971. doi: 10.1073/pnas.1221347110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yan Z., Xia Z., Cheng Y., Jiao Z., Sun B., Zhou T., Fan Z. A violaxanthin deepoxidase interacts with a viral suppressor of RNA silencing to inhibit virus amplification. Plant Physiol. 2017;175:1774–1794. doi: 10.1104/pp.17.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-Y., Lu M.-Y.J., Shih M.-C. The SnRK1-eIFiso4G1 signaling relay regulates the translation of specific mRNAs in Arabidopsis under submergence. New Phytol. 2019;222:366–381. doi: 10.1111/nph.15589. [DOI] [PubMed] [Google Scholar]
- Cui H., Wang A. The biological impact of the hypervariable N-terminal region of potyviral genomes. Annu. Rev. Virol. 2019;6:255–274. doi: 10.1146/annurev-virology-092818-015843. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Daszkowska-Golec A. Emerging roles of the nuclear cap-binding complex in abiotic stress responses. Plant Physiol. 2018;176:242–253. doi: 10.1104/pp.17.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N., Rosli H.G., Martin G.B., Mueller L.A. The SGN VIGS tool: user-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant. 2015;8:486–488. doi: 10.1016/j.molp.2014.11.024. [DOI] [PubMed] [Google Scholar]
- García J.A., Pallás V. Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 2015;11:21–30. doi: 10.1016/j.coviro.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Götte B., Utt A., Fragkoudis R., Merits A., McInerney G.M. Sensitivity of alphaviruses to G3BP deletion correlates with efficiency of replicase polyprotein processing. J. Virol. 2020;94 doi: 10.1128/JVI.01681-19. e01681–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A., Lõhmus A., Mäkinen K. Formation of Potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 2015;11:e1005314. doi: 10.1371/journal.ppat.1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Zeier J. N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 2019;50:44–57. doi: 10.1016/j.pbi.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Hervás M., Navajas R., Chagoyen M., García J.A., Martínez-Turiño S. Phosphorylation-related cross-talk between distant regions of the core region of the coat protein contributes to virion assembly of Plum pox virus. Mol. Plant Microbe Interact. 2020;33:653–667. doi: 10.1094/MPMI-10-19-0305-R. [DOI] [PubMed] [Google Scholar]
- Hoover H.S., Wang J.C.-Y., Middleton S., Ni P., Zlotnick A., Vaughan R.C., Kao C.C. Phosphorylation of the brome mosaic virus capsid regulates the timing of viral infection. J. Virol. 2016;90:7748–7760. doi: 10.1128/JVI.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J.M., Schroeder J.I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Hung C.-J., Huang Y.-W., Liou M.-R., Lee Y.-C., Lin N.-S., Meng M., Tsai C.-H., Hu C.-C., Hsu Y.-H. Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Mol. Plant Microbe Interact. 2014;27:1211–1225. doi: 10.1094/MPMI-04-14-0112-R. [DOI] [PubMed] [Google Scholar]
- Isken O., Postel A., Bruhn B., Lattwein E., Becher P., Tautz N. CRISPR/Cas9-mediated knockout of DNAJC14 verifies this chaperone as a pivotal host factor for RNA replication of pestiviruses. J. Virol. 2019;93:e01714–e01718. doi: 10.1128/JVI.01714-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismayil A., Yang M., Haxim Y., Wang Y., Li J., Han L., Wang Y., Zheng X., Wei X., Nagalakshmi U. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell. 2020;32:1124–1135. doi: 10.1105/tpc.19.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon E.J., Tadamura K., Murakami T., Inaba J.-I., Kim B.M., Sato M., Atsumi G., Kuchitsu K., Masuta C., Nakahara K.S. rgs-CaM detects and counteracts viral RNA silencing suppressors in plant immune priming. J. Virol. 2017;91 doi: 10.1128/JVI.00761-17. e00761–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-H., Atallah O.O., Sun Y.-D., Folimonova S.Y. Functional diversification upon leader protease domain duplication in the Citrus tristeza virus genome: role of RNA sequences and the encoded proteins. Virology. 2018;514:192–202. doi: 10.1016/j.virol.2017.11.014. [DOI] [PubMed] [Google Scholar]
- Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lopez M.Z., Lohmann V., Luban J. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawa D., Testerink C. Regulation of mRNA decay in plant responses to salt and osmotic stress. Cell. Mol. Life Sci. 2017;74:1165–1176. doi: 10.1007/s00018-016-2376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 2004;38:823–839. doi: 10.1111/j.1365-313X.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Hashimoto M., Maejima K., Shiraishi T., Neriya Y., Miura C., Minato N., Okano Y., Sugawara K., Yamaji Y. A necrosis-inducing elicitor domain encoded by both symptomatic and asymptomatic Plantago asiatica mosaic virus isolates, whose expression is modulated by virus replication. Mol. Plant Microbe Interact. 2011;24:408–420. doi: 10.1094/MPMI-12-10-0279. [DOI] [PubMed] [Google Scholar]
- Kørner C.J., Pitzalis N., Peña E.J., Erhardt M., Vazquez F., Heinlein M. Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nat. Plants. 2018;4:157–164. doi: 10.1038/s41477-018-0117-x. [DOI] [PubMed] [Google Scholar]
- Kourelis J., Kaschani F., Grosse-Holz F.M., Homma F., Kaiser M., van der Hoorn R.A.L. A homology-guided, genome-based proteome for improved proteomics in the alloploid Nicotiana benthamiana. BMC Genomics. 2019;20:722. doi: 10.1186/s12864-019-6058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner T., Müller A., Pankraz A., Becher P., Thiel H.-J., Gorbalenya A.E., Tautz N. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J. Virol. 2004;78:10765–10775. doi: 10.1128/JVI.78.19.10765-10775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., Bangratz M., Vignols F., Brugidou C. The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 2010;61:371–382. doi: 10.1111/j.1365-313X.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- Laloum T., Martín G., Duque P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018;23:140–150. doi: 10.1016/j.tplants.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Ratsch G., Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Jung H.J., Kang H., Kim S.Y. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012;53:673–686. doi: 10.1093/pcp/pcs023. [DOI] [PubMed] [Google Scholar]
- Lei J., Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang A. RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 2018;14:e1007228. doi: 10.1371/journal.ppat.1007228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang A. RNA-targeted antiviral immunity: more than just RNA silencing. Trends Microbiol. 2019;27:792–805. doi: 10.1016/j.tim.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U S A. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludman M., Burgyán J., Fátyol K. CRISPR/Cas9 mediated inactivation of Argonaute 2 reveals its differential involvement in antiviral responses. Sci. Rep. 2017;7:1010. doi: 10.1038/s41598-017-01050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen K., Lõhmus A., Pollari M. Plant RNA regulatory network and RNA granules in virus infection. Front. Plant Sci. 2017;8:2093. doi: 10.3389/fpls.2017.02093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K., Sanfaçon H. Expanding repertoire of plant positive-strand RNA virus proteases. Viruses. 2019;11:66. doi: 10.3390/v11010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Singh J., Hill J.H., Whitham S.A., Cannon S.B. Dynamic transcriptome profiling of bean common mosaic virus (BCMV) infection in common bean (Phaseolus vulgaris L.) BMC Genomics. 2016;17:613. doi: 10.1186/s12864-016-2976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez F., Daròs J.-A. Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 2014;88:10725–10737. doi: 10.1128/JVI.00928-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez F., Rodrigo G., Aragonés V., Ruiz M., Lodewijk I., Fernández U., Elena S.F., Daròs J.-A. Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genomics. 2016;17:87. doi: 10.1186/s12864-016-2394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Fontecha E.S., Tarancón C., Cubas P. To grow or not to grow, a power-saving program induced in dormant buds. Curr. Opin. Plant Biol. 2018;41:102–109. doi: 10.1016/j.pbi.2017.10.001. [DOI] [PubMed] [Google Scholar]
- McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- Meeske A.J., Nakandakari-Higa S., Marraffini L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019;570:241–245. doi: 10.1038/s41586-019-1257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergner J., Frejno M., List M., Papacek M., Chen X., Chaudhary A., Samaras P., Richter S., Shikata H., Messerer M. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature. 2020;579:409–414. doi: 10.1038/s41586-020-2094-2. [DOI] [PubMed] [Google Scholar]
- Müller M., Munné-Bosch S. Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods. 2011;7:37. doi: 10.1186/1746-4811-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.M., Zhou T., Carr J.P. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr. Opin. Virol. 2020;42:8–17. doi: 10.1016/j.coviro.2020.02.008. [DOI] [PubMed] [Google Scholar]
- Nakahara K.S., Masuta C. Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr. Opin. Plant Biol. 2014;20:88–95. doi: 10.1016/j.pbi.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Nakasugi K., Crowhurst R., Bally J., Waterhouse P. Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS One. 2014;9:e91776. doi: 10.1371/journal.pone.0091776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Interplay between viruses and host mRNA degradation. Biochim. Biophys. Acta. 2013;1829:732–741. doi: 10.1016/j.bbagrm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nicaise V., Gallois J.-L., Chafiai F., Allen L.M., Schurdi-Levraud V., Browning K.S., Candresse T., Caranta C., Le Gall O., German-Retana S. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 2007;581:1041–1046. doi: 10.1016/j.febslet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Nobori T., Mine A., Tsuda K. Molecular networks in plant-pathogen holobiont. FEBS Lett. 2018;592:1937–1953. doi: 10.1002/1873-3468.13071. [DOI] [PubMed] [Google Scholar]
- Pacheco R., García-Marcos A., Manzano A., de Lacoba M.G., Camañes G., García-Agustín P., Díaz-Ruíz J.R., Tenllado F. Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant-virus interactions that lead to cell death. Mol. Plant Microbe Interact. 2012;25:709–723. doi: 10.1094/MPMI-11-11-0305. [DOI] [PubMed] [Google Scholar]
- Papp I., Mur L.A., Dalmadi A., Dulai S., Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- Pasin F., Simón-Mateo C., García J.A. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 2014;10:e1003985. doi: 10.1371/journal.ppat.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin F., Bedoya L.C., Bernabé-Orts J.M., Gallo A., Simón-Mateo C., Orzaez D., García J.A. Multiple T-DNA delivery to plants using novel mini binary vectors with compatible replication origins. ACS Synth. Biol. 2017;6:1962–1968. doi: 10.1021/acssynbio.6b00354. [DOI] [PubMed] [Google Scholar]
- Pasin F., Menzel W., Daròs J.-A. Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnol. J. 2019;17:1010–1026. doi: 10.1111/pbi.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel D.B., Sanfaçon H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018;9:1575. doi: 10.3389/fpls.2018.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C.-W., Peremyslov V.V., Mushegian A.R., Dawson W.O., Dolja V.V. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 2001;75:12153–12160. doi: 10.1128/JVI.75.24.12153-12160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W.S., Jarmolowski A. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:1224–1244. doi: 10.1093/nar/gkt894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers F., García J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015;92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Rodamilans B., San León D., Mühlberger L., Candresse T., Neumüller M., Oliveros J.C., García J.A. Transcriptomic analysis of Prunus domestica undergoing hypersensitive response to Plum pox virus infection. PLoS One. 2014;9:e100477. doi: 10.1371/journal.pone.0100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodamilans B., Shan H., Pasin F., García J.A. Plant viral proteases: beyond the role of peptide cutters. Front. Plant Sci. 2018;9:666. doi: 10.3389/fpls.2018.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador B., Saénz P., Yangüez E., Quiot J.B., Quiot L., Delgadillo M.O., García J.A., Simón-Mateo C. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 2008;9:147–155. doi: 10.1111/j.1364-3703.2007.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.-K., Kwon S.-J., Cho W.K., Choi H.-S., Kim K.-H. Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 2014;4:5905. doi: 10.1038/srep05905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H., Pasin F., Valli A., Castillo C., Rajulu C., Carbonell A., Simón-Mateo C., García J.A., Rodamilans B. The Potyviridae P1a leader protease contributes to host range specificity. Virology. 2015;476:264–270. doi: 10.1016/j.virol.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Shan H., Pasin F., Tzanetakis I.E., Simón-Mateo C., García J.A., Rodamilans B. Truncation of a P1 leader proteinase facilitates potyvirus replication in a non-permissive host. Mol. Plant Pathol. 2018;19:1504–1510. doi: 10.1111/mpp.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J., Yu C., Makhotenko A.V., Makarova S.S., Love A.J., Kalinina N.O., MacFarlane S., Chen J., Taliansky M.E. Interaction of a plant virus protein with the signature Cajal body protein coilin facilitates salicylic acid-mediated plant defence responses. New Phytol. 2019;224:439–453. doi: 10.1111/nph.15994. [DOI] [PubMed] [Google Scholar]
- Shen W., Bobay B.G., Greeley L.A., Reyes M.I., Rajabu C.A., Blackburn R.K., Dallas M.B., Goshe M.B., Ascencio-Ibáñez J.T., Hanley-Bowdoin L. Sucrose nonfermenting 1-related protein kinase 1 phosphorylates a geminivirus Rep protein to impair viral replication and infection. Plant Physiol. 2018;178:372–389. doi: 10.1104/pp.18.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Shi Y., Dai Z., Wang A. The RNA-dependent RNA polymerase NIb of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses. 2020;12:77. doi: 10.3390/v12010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.-M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma F., Mogami J., Yoshida T., Abekura M., Takahashi F., Kidokoro S., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants. 2017;3:16204. doi: 10.1038/nplants.2016.204. [DOI] [PubMed] [Google Scholar]
- Song L., Huang S.-S.C., Wise A., Castanon R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R. A transcription factor hierarchy defines an environmental stress response network. Science. 2016;354:aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Fu S., Qian Y., Zhang L., Xu Y., Zhou X. Discovery and small RNA profile of Pecan mosaic-associated virus, a novel potyvirus of pecan trees. Sci. Rep. 2016;6:26741. doi: 10.1038/srep26741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Somssich I.E. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–947. doi: 10.1111/nph.13286. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- Valli A.A., Gallo A., Rodamilans B., López-Moya J.J., García J.A. The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018;19:744–763. doi: 10.1111/mpp.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J., Carrington J.C. Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J. Virol. 1995;69:1582–1590. doi: 10.1128/jvi.69.3.1582-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler H., Kwon M.-O., Dang V., Sambade A., Fasler M., Ashby J., Heinlein M. Tobacco mosaic virus movement protein enhances the spread of RNA silencing. PLoS Pathog. 2008;4:e1000038. doi: 10.1371/journal.ppat.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.-J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U S A. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Han K., Peng J., Zhao J., Jiang L., Lu Y., Zheng H., Lin L., Chen J., Yan F. NbALD1 mediates resistance to turnip mosaic virus by regulating the accumulation of salicylic acid and the ethylene pathway in Nicotiana benthamiana. Mol. Plant Pathol. 2019;20:990–1004. doi: 10.1111/mpp.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Hsu C.-C., Du Y., Zhu P., Zhao C., Fu X., Zhang C., Paez J.S., Macho A.P., Tao W.A. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. U S A. 2020;117:3270–3280. doi: 10.1073/pnas.1919901117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer I., Golisz A., Sulkowska A., Kawa D., Kulik A., Kufel J. mRNA decapping and 5’-3’ decay contribute to the regulation of ABA signaling in Arabidopsis thaliana. Front. Plant Sci. 2018;9:312. doi: 10.3389/fpls.2018.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood J.H., McCann L., Naish M., Dixon H., Murphy A.M., Stancombe M.A., Bennett M.H., Powell G., Webb A.A.R., Carr J.P. A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol. Plant Pathol. 2013;14:158–170. doi: 10.1111/j.1364-3703.2012.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chua N.-H. Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012;31:1975–1984. doi: 10.1038/emboj.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Mazur M.J., Tao X., Kormelink R. Cellular RNA hubs: friends and foes of plant viruses. Mol. Plant Microbe Interact. 2020;33:40–54. doi: 10.1094/MPMI-06-19-0161-FI. [DOI] [PubMed] [Google Scholar]
- Yang Z., Li Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018;32:88–99. doi: 10.1016/j.coviro.2018.08.003. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang Y., Yang J., Yao S., Zhao K., Wang D., Qin Q., Bian Z., Li Y., Lan Y. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe. 2020;28:89–103.e8. doi: 10.1016/j.chom.2020.05.001. [DOI] [PubMed] [Google Scholar]
- Yao C., Finlayson S.A. Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 2015;169:611–626. doi: 10.1104/pp.15.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying X.-B., Dong L., Zhu H., Duan C.-G., Du Q.-S., Lv D.-Q., Fang Y.-Y., Garcia J.A., Fang R.-X., Guo H.-S. RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell. 2010;22:1358–1372. doi: 10.1105/tpc.109.072058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S.A., Marcotrigiano J. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr. Opin. Virol. 2013;3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li B., Jang G.-J., Jiang S., Jiang D., Jang J.-C., Wu S.-H., Shan L., He P. Orchestration of processing body dynamics and mRNA decay in Arabidopsis immunity. Cell Rep. 2019;28:2194–2205.e6. doi: 10.1016/j.celrep.2019.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa A., Neriya Y., Hashimoto M., Yoshida T., Fujimoto Y., Hosoe N., Keima T., Tokumaru K., Maejima K., Netsu O. Functional conservation of EXA1 among diverse plant species for the infection by a family of plant viruses. Sci. Rep. 2019;9:5958. doi: 10.1038/s41598-019-42400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-F., Yuan L.-J., Shao Y., Du W., Yan D.-W., Lu Y.-T. The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 2008;31:562–574. doi: 10.1111/j.1365-3040.2008.01786.x. [DOI] [PubMed] [Google Scholar]
- Zhang X.-P., Liu D.-S., Yan T., Fang X.-D., Dong K., Xu J., Wang Y., Yu J.-L., Wang X.-B. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 2017;13:e1006522. doi: 10.1371/journal.ppat.1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong K., Xu K., Zhang K., Jin X., Yang M., Zhang Y., Wang X., Han C., Yu J. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 2018;218:1570–1585. doi: 10.1111/nph.15065. [DOI] [PubMed] [Google Scholar]
- Zhang T., Liu P., Zhong K., Zhang F., Xu M., He L., Jin P., Chen J., Yang J. Wheat yellow mosaic virus NIb interacting with host light induced protein (LIP) facilitates its infection through perturbing the abscisic acid pathway in wheat. Biology. 2019;8:E80. doi: 10.3390/biology8040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.