Abstract
Prevailing strategies in genome-wide association studies (GWAS) mostly rely on principles of medical genetics emphasizing one gene, one function, one phenotype concept. Here, we performed GWAS of blood lipids leveraging a new systemic concept emphasizing complexity of genetic predisposition to such phenotypes. We focused on total cholesterol, low- and high-density lipoprotein cholesterols, and triglycerides available for 29,902 individuals of European ancestry from seven independent studies, men and women combined. To implement the new concept, we leveraged the inherent heterogeneity in genetic predisposition to such complex phenotypes and emphasized a new counter intuitive phenomenon of antagonistic genetic heterogeneity, which is characterized by misalignment of the directions of genetic effects and the phenotype correlation. This analysis identified 37 loci associated with blood lipids but only one locus, FBXO33, was not reported in previous top GWAS. We, however, found strong effect of antagonistic heterogeneity that leaded to profound (quantitative and qualitative) changes in the associations with blood lipids in most, 25 of 37 or 68%, loci. These changes suggested new roles for some genes, which functions were considered as well established such as GCKR, SIK3 (APOA1 locus), LIPC, LIPG, among the others. The antagonistic heterogeneity highlighted a new class of genetic associations emphasizing beneficial and adverse trade-offs in predisposition to lipids. Our results argue that rigorous analyses dissecting heterogeneity in genetic predisposition to complex traits such as lipids beyond those implemented in current GWAS are required to facilitate translation of genetic discoveries into health care.
Keywords: Genome-wide association studies, Pleiotropy, Age-related phenotypes, Aging, Health span, Life span
The field of genetics provided unprecedented insights into the genetic mechanisms underlying predisposition to various health-related phenotypes, commonly referred to as traits. This progress was accelerated by invention of genome-wide association studies (GWAS) (1), which resulted in discovery of thousands of genetic variants associated with diseases and related traits (2). Despite progress in the GWAS era (3), linking genetic variants and traits is not straightforward, especially for complex (non-Mendelian) phenotypes characterizing human health span and life span (4). These connections are complicated by an inherent complexity of metabolic networks in human organisms adapted to different environments (5), which is supported by four principles of macromolecular organization including evolutionary conserved elementary components, organization in pathways and networks, pleiotropy, and redundancy (6), and by the lack of apparent and direct connections between factors maximizing fitness and health/life-span–related phenotypes (7,8). For example, one hypothesis of such connections is so-called antagonistic pleiotropy (9,10). Traditional GWAS, built on principles of medical genetics, follows the same strategy regardless of the nature of traits to be examined. Better understanding of genetic predisposition to complex traits requires shifting of this paradigm to the concept that “one gene, one function, one trait is the wrong way to view genetic variation in the human genome” (11). This change in the paradigm requires appropriate approaches, which are not yet a routine practice in currently prevailing GWAS.
Current article introduces a new systemic concept of GWAS of complex traits within the new paradigm relaxing the medical genetics hypothesis on “one gene, one function, one trait.” Originally, this paradigm was introduced to emphasize the role of pleiotropy (ie, one gene, multiple traits) in genetics of complex traits (12) following an intuitive assumption that the directions of genetic associations with correlated traits are aligned with the direction of correlation between these traits (11,13). We performed large-scale GWAS of four lipid traits within the new concept. These traits included total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG), which are a vivid example of complex traits. The new concept emphasizes the inherent heterogeneity in genetic predisposition to such traits and a new counter intuitive phenomenon of antagonistic genetic heterogeneity. Unlike the commonly known concept of antagonistic pleiotropy, implying that genes beneficial at the reproductive age can become disadvantageous in late life (9,10), antagonistic heterogeneity refers to a phenomenon characteristic for heterogeneous traits when the directions of genetic associations with different traits can be misaligned with correlation between them (14,15). Thus, besides the directions of genetic associations as in the case of antagonistic pleiotropy, the concept of antagonistic heterogeneity considers an additional characteristic of the trait correlation. For example, antagonistic heterogeneity can be manifested as opposite directions of the genetic effects (eg, the same genetic variant is associated with increased level of one trait and decreased level of the other trait) despite direct correlation between traits (eg, increased level of one trait tends to co-occur with increased level of the other trait). We show that dissecting complex relationships between single nucleotide polymorphisms (SNPs) and traits following this new concept results not merely in quantitative improvement of the association signals, but suggests qualitatively new roles of genes in complex traits, including genes, which functions are considered as well characterized by previous studies.
Methods
Study Cohorts
This manuscript was prepared using a limited access datasets obtained through dbGaP, accession numbers: phs000007.v28.p10 (FHS), phs000280.v3.p1 (ARIC), phs000285.v3.p2 (CARDIA), phs000209.v13.p3 (MESA), phs000287.v5.p1 (CHS), phs000428.v2.p2 (HRS), phs000200.v10.p3 (WHI). Phenotypic HRS data are available publicly and through restricted access from http://hrsonline.isr.umich.edu/index.php?p=data.
Data were obtained from seven studies (Supplementary Table 1) including the Atherosclerosis Risk in Communities (ARIC) study (16,17), Coronary Artery Risk Development in Young Adults (CARDIA) study (18), the Cardiovascular Health Study (CHS) (19), the Multi-Ethnic Study of Atherosclerosis (MESA) (20), the Framingham Heart Study (FHS) (21–23), the Health and Retirement Study (HRS) (24), and the Genomics and Randomized Trials Network (GARNET) substudy of the Women’s Health Initiative (WHI) (25,26) for individuals who identified themselves as of European ancestry. Taking into account complex structure of the FHS study, three cohorts comprising parental (FHS_C1), offspring (FHS_C2), and grandchildren (FHS_C3) generations were examined separately.
Phenotypes
The analyses focused on four lipid traits including HDL-C, LDL-C, TC, and TG (Supplementary Table 1). These health-related phenotypes were defined using data from the first examination at which all these lipids were measured. We used the same scale, mg/dl, harmonized across cohorts.
Genotypes
SNPs were available from Affymetrix 6.0 (1 M SNPs) chip in ARIC, CARDIA, and MESA, Illumina HumanCNV370v1 chip (370K SNPs) in CHS, Affymetrix 500K in FHS, Illumina HumanOmni 2.5 Quad chip (2.5 M SNPs) in HRS, and Illumina HumanOmni1-Quad_v1-0_B chip (1 M SNPs) in WHI. SNPs were included in the analyses after quality control in each study (call rate>95%, Hardy–Weinberg equilibrium p > 10–6). Given small overlap of SNPs between CHS, FHS, and other arrays, we imputed SNPs in CHS and FHS to have approximately 2.5 M SNPs overlapping with the Illumina HumanOmni 2.5 Quad chip. Nongenotyped SNPs were imputed (SHAPEIT2 (27) and Minimac3 (28)) according to the 1000 Genomes Phase 3 data reference panel in the NCBI build 37 (hg19) coordinate after removing low-quality SNPs. Only SNPs with high imputation quality (info>0.7) were retained for the analyses. SNPs with average minor allele frequency (MAF) >2% across all studies were selected for the analyses independently of their MAF in a specific study.
Mapping to Genes
SNPs were mapped to genes using NCBI SNP database (assembly GRCh37.p13). We reported either a plausible biological candidate gene in the locus or a gene within ~100kb of the reported SNP.
GWAS
GWAS was performed for each trait in each cohort separately. An additive genetic model with minor allele as an effect allele was adopted in all analyses throughout this article. We used the generalized estimating equation model with unstructured correlations (gee package in R) to correct for familial structure, when applicable, except the FHS. As the FHS included participants from large families, we used the linear mixed effects model (lme4 package in R) with random effects to correct for familial structure because the generalized estimating equation model was not efficient due to memory constraints.
The following basic adjustments were used for all models: (all studies) age and sex; (ARIC, CHS, and MESA) field center; (HRS) HRS cohorts, and (FHS) whether the DNA samples had been subject to whole-genome amplification (29). The analyses focused on individuals of European ancestry to offset population stratification. The results were reported for four models for each lipid trait. One model was with the basic adjustment alone (herein referred to as an unconditional model) whereas the other three models (herein referred to as conditional models) were additionally adjusted for one of the three remaining lipid traits, for example, the models for HDL-C were adjusted for (i) LDL-C, (ii) TC, or (iii) TG. No other adjustments were considered. Therefore, for each SNP we have the results from 16 models for four traits in the same sample.
Fixed-Effects Meta-analysis
We combined statistics across nine cohorts from each of the 16 models using the traditional GWAS fixed effects model with inverse-variance weighting (METAL software (30)). This meta-test accounts for the directions of the effects and it is more powerful than those combining p-values or Z-scores (31). The weighted average of the effect sizes was calculated as with variance , where is the inverse variance of effect size in the cohort for a given model. Wald test was then used to obtain p-value. Given these results, we selected SNPs that attained genome-wide significances, p≤pGW = 5 × 10–8, in at least one of 16 meta-tests.
Heterogeneity Coefficient
We used METAL software (30) to evaluate the heterogeneity coefficient I2. The I2 can be interpreted as the percentage of the total variability in a set of effect sizes due to between-sample variability.
Anterior and Posterior Antagonistic Heterogeneity
Antagonistic heterogeneity as a phenomenon is characterized by an inverse (ie, opposite) relationship between the directions of the trait associations and the trait correlation. For example, it can be manifested as opposite directions of the effects β (ie, positive and negative βs) in the associations with directly correlated traits (ie, when r > 0 implying that, for example, increased level of one trait tends to co-occur with increased level of the other trait). Having the results from the unconditional and conditional models, we can distinguish two cases of antagonistic heterogeneity, herein referred to as anterior and posterior antagonistic heterogeneity, respectively. The anterior antagonistic heterogeneity was assessed as misalignment of the directions of associations (regardless of their significance) of a SNP with lipid traits in either of six pairs (HDL-C&LDL-C, HDL-C&TC, HDL-C&TG, LDL-C&TC, LDL-C&TG, and TC&TG) in unconditional model and the direction of correlation between traits in that pair. The posterior antagonistic heterogeneity was assessed from the results from two conditional models. One model included a trait from a pair as an outcome and the other trait as covariate, whereas the other model swapped these traits, for example, a model for HDL-C adjusted by LDL-C and a model for LDL-C adjusted by HDL-C. The criterion for posterior antagonistic heterogeneity was the same as for the anterior one. However, because conditional analysis has power to amplify the association signals (see next), this criterion was strengthened by the requirement of the increase of significance in the conditional models compared to the unconditional models.
Antagonistic Heterogeneity Has Power to Amplify the Association Signals
Hallmark of antagonistic heterogeneity is that it has power to amplify the association signals leveraging misalignment of the directions of associations with traits and the direction of correlation between them. This property can be conveniently illustrated by pleiotropic statistic for associations of a SNP with two traits provided by an omnibus test (32–34). This statistic follows a chi-squared distribution with K degrees of freedom corresponding to the number of the considered traits (ie, K = 2 in this case),
| (1) |
from which a combined p-value for a pleiotropic association with traits is obtained. Here is a z-score vector of associations of a SNP with two traits, is an estimated effect size and is a standard error for the trait , and is the correlation matrix of traits (34). Prime symbol denotes transposition.
Because antagonistic heterogeneity is characterized by an inverse relationship between the effect directions and the trait correlation, the chi-square in Eq. (1) increases because in this case that corresponds to larger value of and, consequently, to smaller p-value.
Results
Study Overview
Analyses were performed for 29,902 individuals of European ancestry from 7 independent studies comprised of 9 cohorts, men and women combined, using an additive genetic model with minor allele as an effect allele. The systemic concept was implemented as synthesis of the traditional univariate (unconditional) GWAS of complex traits such as TC, HDL-C, LDL-C, and TG (Supplementary Table 1), and conditional GWAS using models adjusted by one of the three remaining lipid traits, that is, each trait was considered as an outcome and a covariate in different models resulting in four meta-statistics for each trait. For example, one statistic was provided by the unconditional GWAS of HDL-C and three statistics by GWAS of HDL-C conditional on TC, LDL-C, or TG, separately. Conditional analysis dissected antagonistic heterogeneity leveraging misalignment of the directions of associations with lipid traits and the directions of correlation between them. We used the simplest approach to characterize and dissect antagonistic heterogeneity by considering pair-wise combinations only, that is, HDL-C&LDL-C, HDL-C&TC, HDL-C&TG, LDL-C&TC, LDL-C&TG, and TC&TG with pair-wise correlations ranging from r = −.4 for HDL-C and TG to r = .9 for LDL-C and TC (see Supplementary Figure 1). Other details are given in “Methods.”
Unconditional and Conditional GWAS
Meta-analysis of the results from unconditional GWAS identified 29 loci with SNPs associated with lipid traits at genome-wide (GW) level of significance, p≤pGW = 5 × 10–8. Dissecting antagonistic heterogeneity, conditional GWAS identified 8 additional loci (ASAP3; PCSK9; ABCA1; LRP4; MVK; SBNO1; FBXO33; TOP1), totaling 37 loci. We used the strongest evidences for the associations from top lipid GWAS performed in the largest samples so far (35,36) to characterize known associated loci and statistical estimates for the respective SNPs. We found that only FBXO33 locus was not reported in these top GWAS. Our analyses, however, identified strong role of the new phenomenon of antagonistic heterogeneity that substantially changed the associations with lipid traits in 25 of 37 (67.6%) loci (Figure 1, bold italic font [red on-line] and non-italic font with † symbol [blue on-line]) and resulted in 19 associations in 15 of 24 known loci with lipid traits not reported in (35,36) (Table 1). To characterize these findings, we discuss below 98 associations with lipid traits for 50 lead SNPs representing these 37 loci.
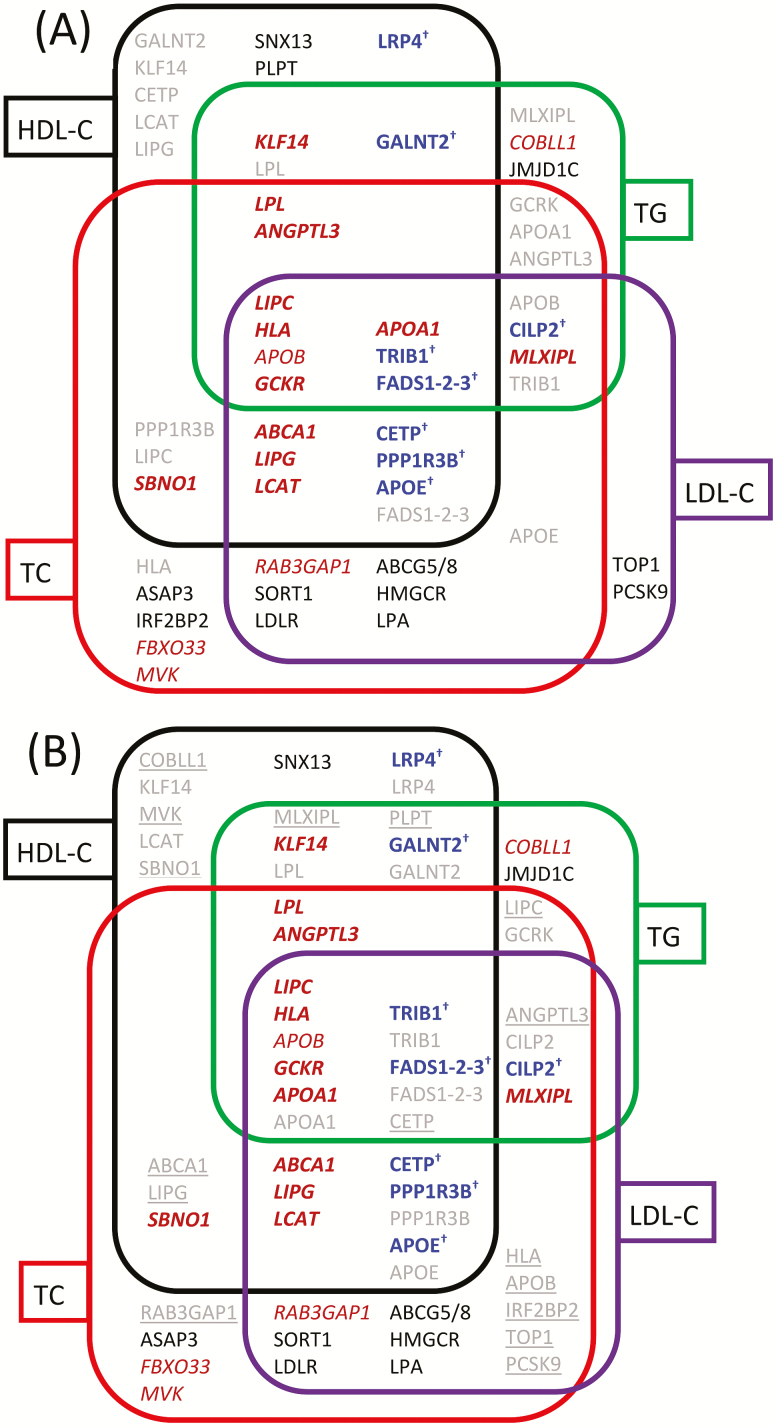
Figure 1.
Loci with predisposition to lipid traits. (A) Loci identified in our study. (B) Loci reported in (35,36) (black/gray) and in our study (bold italic font [red on-line] and non-italic font with † symbol [blue on-line]); the latter were given in figure (B) for convenience. Rounded rectangles indicate associations with HDL-C (black), TG (green), TC (red), and LDL-C (purple). Loci with variants associated with multiple lipid traits are located in intersects of the corresponding rounded rectangles. Bold font emphasizes loci strongly affected by antagonistic heterogeneity: non-italic font with † symbol (blue on-line) denotes loci from Supplementary Table 3 and bold italic font (red on-line) from Table 2. Nonbold italic font (red on-line) highlights loci showing new associations with minor effect of antagonistic genetic heterogeneity (with strength <20%) reported in Table 2. Black font indicates replicated loci with at most minor effect of the antagonistic heterogeneity reported in Supplementary Table 2. Gray font indicates loci affected by the antagonistic heterogeneity that resulted in quantitative and qualitative changes in the associations. If the same locus is reported in several tables, it was placed only once using information from the first table in the following order: Table 2, Supplementary Table 3, and Supplementary Table 2. Underlining in figure (B) indicates loci with SNPs, which did not attain genome-wide (p ≤ 5 × 10–8) significance in our unconditional analyses with the same lipid traits as in (35,36). HDL-C = High-density lipoprotein cholesterol; LDL-C = Low-density lipoprotein cholesterol; TC = Total cholesterol; TG = Triglycerides.
Table 1.
Loci Showing Novel Associations with Lipid Traits
| Current Study | Prior Studies | |||||
|---|---|---|---|---|---|---|
| Locus | Effect Sign | N new | New Associations | N tot | Effect Sign | N tot |
| ANGPTL3 | -?-- | 1 | HDL-C* | 3 | ?--- | 3 |
| APOB | -++? | 1 | HDL-C | 3 | ?++? | 2 |
| GCKR | -+++ | 2 | HDL-C*; LDL-C | 4 | ??++ | 2 |
| RAB3GAP1 | ?++? | 1 | LDL-C | 2 | ??+? | 1 |
| COBLL1 | ???- | 1 | TG | 1 | +??? | 1 |
| HLA | -+-- | 2 | HDL-C; TG | 4 | ?++? | 2 |
| MLXIPL | ?+-- | 2 | LDL-C; TC | 3 | +??- | 2 |
| KLF14 | +??- | 1 | TG | 2 | +??? | 1 |
| LPL | +?+- | 1 | TC* | 3 | +??- | 2 |
| ABCA1 | ---? | 1 | LDL-C* | 3 | -?-? | 2 |
| MVK | ??-? | 1 | TC | 1 | -??? | 1 |
| SBNO1 | +?+? | 1 | TC | 2 | +??? | 1 |
| FBXO33 | ??+? | 1 | TC | 1 | ???? | 0 |
| LIPC | +-++ | 1 | LDL-C | 4 | +?++ | 3 |
| LCAT | +-+? | 2 | LDL-C; TC | 3 | +??? | 1 |
| LIPG | -+-? | 1 | LDL-C | 3 | -?-? | 2 |
Note: Locus = locus name as used in previous studies. Effect Sign = directions of genetic associations with HDL-C, LDL-C, TC, and TG where “+” and “-” denote positive (increase) and negative (decrease) signs of statistical effects, respectively, and “?” indicates associations, which did not attain either GW (ie, p = pGW, in the current and prior studies) or suggestive effect (ie, p = 10–5; in the current study) significance. Nnew = count of new genetic associations with lipid traits in a given locus. Current Study, Ntot = count of total associations attained suggestive effect (ie, pGW<p≤10–5 for four associations denoted by an asterisk “*”) or genome-wide (p≤pGW) significance in a given locus in the current study. Prior Studies, Ntot = count of total genome-wide significant associations reported in a given locus in prior studies; the associations with traits denoted by the asterisk did not attain suggestive level of significance for the same SNPs in prior studies.
Sixteen loci in this table are a subset of 17 loci shown by italic font (red on-line) in Figure 1.
Replication of the Previously Reported Associations
Our analysis replicated 40 associations for 28 SNPs from 28 loci (Supplementary Table 2) with lipid traits (at p ≤ pGW) reported in (35,36). Of these SNPs, selected as one SNP per locus for a given trait, there were 30 associations for 20 SNPs reported in these GWAS or their proxies (with linkage disequilibrium [LD] r2 > 70%; 1000 Genomes Project), and 10 associations for 8 nonproxy SNPs showing associations with the same traits, regardless of the effect directions. These replicated associations were not strongly affected by the antagonistic heterogeneity. The strength of the effect of antagonistic heterogeneity was characterized by the ability of the conditional analysis to increase the significance of the estimates by decreasing p-values (pcond) compared to p-values from the unconditional analysis (puncond), that is, by the relative change of log-transformed p-values in percents: 100 × (log10(pcond)-log10(puncond))/log10(puncond). Ad hoc cutoff for the weak strength was set as less than 20%.
The Role of Antagonistic Heterogeneity and Novel Associations
Comparative analysis of the results from conditional and unconditional models showed that antagonistic heterogeneity affected most loci (25 of 37) including those reported as replicated in Supplementary Table 2. We found that the antagonistic heterogeneity strengthened 23 associations for 17 SNPs in 15 loci (with the strength ≥20%) with the same lipid traits as those reported in (35,36) (Supplementary Table 3). These associations were of suggestive effect (p ≤ 10–5) or genome-wide (p≤pGW = 5 × 10–8) significances in our unconditional analysis and all of them were of GW significance in our conditional analysis. The increased significance in the conditional analyses was observed for the same SNPs as reported in (35,36), their proxies (r2 > 70%), and nonproxy SNPs.
Table 2 shows the results for 35 associations for 21 SNPs in 18 loci. We found that 15 of these 35 associations (Table 2, asterisks) were with the same lipid traits as those reported in (35,36), although the reported SNPs were mostly in small LD with ours. All these 15 associations for 10 SNPs in 7 loci were strongly affected by the antagonistic heterogeneity with the strength >200% for 12 of 15 associations. Dissecting this strong effect in the conditional analyses, they attained either GW (11 associations) or suggestive-effect (4 associations) significances despite not having even suggestive-effect significances (ie, p > 10–5) in our univariate analysis. The remaining 20 associations for 17 SNPs in 16 loci were with lipid traits not reported in (35,36). They attained GW (15 associations) or pGW<p≤10–5 (5 associations) levels mostly because of strong effect of antagonistic heterogeneity (with strength >60% for 14 of 20 associations for 11 SNPs in 10 loci). We found that 10 of 35 associations attained GW (8 of 10 associations) or pGW<p≤10–5 levels in conditional analysis despite they were even not nominally significant (p > .05) in the unconditional analyses, that is, the significance which is often considered as noise. Dissecting antagonistic heterogeneity strengthened the associations via three modes by: (i) decreasing standard errors, (ii) increasing magnitude of the effect sizes, and (iii) both (Table 2 and Supplementary Tables 3 and 4).
Table 2.
New Genetic Associations with Lipid Traits and the Associations Strongly Affected by Antagonistic Genetic Heterogeneity
| Current Study | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unconditional | Conditional | Efficiency | Prior Studies | ||||||||||||||||
| ID | Locus | SNP | Chr | EA | Trait Pair | Main Trait | Beta | SE | p-value | Beta | SE | p-value | dlogP | Str | N | A | SNP | LD, % | Effect Sign |
| 1 | ANGPTL3 | rs4350231 | 1 | a | HDL-TG | HDL-C | −0.31 | 0.13 | 1.35E−02 | −0.57 | 0.12 | 8.07E−07 | 6.09 | 326 | 9 | AP | rs2131925 | 97.8 | ?--- |
| 2 | APOB | rs6544366 | 2 | t | HDL-TC | HDL-C | 0.78 | 0.15 | 1.20E−07 | 0.85 | 0.15 | 5.86E−09 | 1.31 | 19 | 6 | AP | rs1367117 | 10.7 | ?++? |
| 3 | GCKR | rs780094 | 2 | t | HDL-TG | HDL-C | 0.00 | 0.12 | 9.83E−01 | 0.58 | 0.11 | 2.05E−07 | 6.68 | 88144 | 7 | P | rs1260326 | 92.0 | ??++ |
| 4 | GCKR | rs780094 | 2 | t | LDL-TC | LDL-C | 0.92 | 0.28 | 1.12E−03 | −1.05 | 0.12 | 2.84E−19 | 15.60 | 529 | 9 | P | rs1260326 | 92.0 | ??++ |
| 5 | RAB3GAP1 | rs10928512 | 2 | g | HDL-LDL | LDL-C | 1.55 | 0.28 | 3.59E−08 | 1.65 | 0.28 | 3.12E−09 | 1.06 | 14 | 7 | AP | rs7570971 | 16.0 | ??+? |
| 6 | COBLL1 | rs7607980 | 2 | c | TC-TG | TG | −5.09 | 0.91 | 2.27E−08 | −4.96 | 0.87 | 1.19E−08 | 0.28 | 4 | 6 | P | rs12328675 | 96.2 | +??? |
| 7 | HLA | rs2394895 | 6 | c | HDL-TG | HDL-C | −0.51 | 0.14 | 1.71E−04 | −0.62 | 0.12 | 8.14E−07 | 2.32 | 62 | 9 | AP | rs3177928 | 5.7 | ?++? |
| 8 | HLA * | rs2394895 | 6 | c | LDL-TC | LDL-C | 0.32 | 0.32 | 3.21E−01 | 0.72 | 0.13 | 5.09E−08 | 6.80 | 1378 | 9 | AP | rs3177928 | 5.7 | ?++? |
| 9 | HLA * | rs2394895 | 6 | c | LDL-TC | TC | −0.41 | 0.35 | 2.38E−01 | −0.78 | 0.14 | 3.56E−08 | 6.82 | 1093 | 9 | AP | rs3177928 | 5.7 | ?++? |
| 10 | HLA | rs9267531 | 6 | g | HDL-TG | TG | −4.34 | 1.04 | 3.16E−05 | −5.28 | 0.95 | 2.81E−08 | 3.05 | 68 | 7 | AP | rs3177928 | 2.1 | ?++? |
| 11 | HLA * | rs9267531 | 6 | g | LDL-TC | LDL-C | −0.49 | 0.46 | 2.88E−01 | 0.86 | 0.19 | 3.53E−06 | 4.91 | 908 | 9 | P | rs3177928 | 2.1 | ?++? |
| 12 | MLXIPL | rs17145738 | 7 | t | LDL-TC | LDL-C | 0.22 | 0.43 | 6.04E−01 | 1.04 | 0.18 | 2.61E−09 | 8.36 | 3813 | 8 | AP | rs17145738 | 100 | +??- |
| 13 | MLXIPL | rs17145738 | 7 | t | LDL-TC | TC | −1.03 | 0.46 | 2.55E−02 | −1.20 | 0.19 | 2.55E−10 | 8.00 | 502 | 8 | AP | rs17145738 | 100 | +??- |
| 14 | KLF14 | rs13230111 | 7 | g | LDL-TG | TG | −2.83 | 0.67 | 2.29E−05 | −3.14 | 0.56 | 2.66E−08 | 2.94 | 63 | 5 | P | rs4731702 | 100 | +??? |
| 15 | LPL | rs263 | 8 | t | TC-TG | TC | 0.77 | 0.39 | 4.86E−02 | 1.88 | 0.38 | 5.22E−07 | 4.97 | 378 | 7 | AP | rs12678919 | 33.8 | +??- |
| 16 | ABCA1 | rs3890182 | 9 | a | LDL-TC | LDL-C | −0.83 | 0.42 | 4.97E−02 | 0.94 | 0.18 | 1.76E−07 | 5.45 | 418 | 6 | P | rs1883025 | 13.8 | -?-? |
| 17 | ABCA1 * | rs3890182 | 9 | a | LDL-TC | TC | −1.99 | 0.46 | 1.52E−05 | −1.28 | 0.19 | 3.27E−11 | 5.67 | 118 | 6 | P | rs1883025 | 13.8 | -?-? |
| 18 | APOA1 * | rs651821 | 11 | c | LDL-TC | LDL-C | 2.26 | 0.56 | 6.05E−05 | −1.17 | 0.23 | 5.43E−07 | 2.05 | 49 | 9 | P | rs964184 | 51.9 | +--- |
| 19 | APOA1 * | rs11216162 | 11 | a | HDL-TG | TG | 2.29 | 0.87 | 8.84E−03 | 3.96 | 0.80 | 6.53E−07 | 4.13 | 201 | 9 | AP | rs964184 | 9.9 | +--- |
| 20 | APOA1 * | rs11216162 | 11 | a | LDL-TC | LDL-C | −0.18 | 0.36 | 6.25E−01 | −1.11 | 0.15 | 4.53E−13 | 12.14 | 5943 | 9 | AP | rs964184 | 9.9 | +--- |
| 21 | APOA1 * | rs11216162 | 11 | a | LDL-TC | TC | 1.19 | 0.40 | 2.60E−03 | 1.30 | 0.17 | 8.27E−15 | 11.50 | 445 | 9 | AP | rs964184 | 9.9 | +--- |
| 22 | APOA1 * | rs7120963 | 11 | t | HDL-TG | HDL-C | 0.24 | 0.13 | 5.95E−02 | 0.68 | 0.12 | 4.91E−09 | 7.08 | 578 | 8 | AP | rs964184 | 25.2 | +--- |
| 23 | APOA1 * | rs7120963 | 11 | t | LDL-TC | LDL-C | 0.68 | 0.29 | 1.92E−02 | −0.84 | 0.12 | 8.73E−12 | 9.34 | 544 | 9 | P | rs964184 | 25.2 | +--- |
| 24 | MVK | rs7298565 | 12 | g | TC-TG | TC | −1.61 | 0.30 | 1.10E−07 | −1.62 | 0.29 | 2.55E−08 | 0.64 | 9 | 8 | P | rs7134594 | 96.0 | -??? |
| 25 | SBNO1 | rs1109559 | 12 | g | TC-TG | TC | 1.71 | 0.33 | 1.82E−07 | 1.80 | 0.31 | 9.04E−09 | 1.30 | 19 | 7 | AP | rs4759375 | 14.1 | +??? |
| 26 | SBNO1 * | rs1109559 | 12 | g | HDL-LDL | HDL-C | 0.54 | 0.13 | 3.78E−05 | 0.64 | 0.13 | 8.69E−07 | 1.64 | 37 | 8 | AP | rs4759375 | 14.1 | +??? |
| 27 | FBXO33 | rs2038280 | 14 | g | TC-TG | TC | 2.15 | 0.41 | 1.22E−07 | 2.20 | 0.39 | 1.32E−08 | 0.97 | 14 | 7 | AP | ???? | ||
| 28 | LIPC * | rs261332 | 15 | a | HDL-TG | TG | 3.27 | 0.82 | 6.17E−05 | 6.82 | 0.74 | 3.57E−20 | 15.24 | 362 | 8 | AP | rs1532085 | 0.3 | +?++ |
| 29 | LIPC | rs261332 | 15 | a | LDL-TC | LDL-C | 0.05 | 0.35 | 8.75E−01 | −1.90 | 0.15 | 3.49E−38 | 37.40 | 64325 | 9 | P | rs1532085 | 0.3 | +?++ |
| 30 | CETP * | rs289715 | 16 | a | LDL-TC | LDL-C | −0.93 | 0.44 | 3.54E−02 | −1.83 | 0.18 | 1.38E−23 | 21.41 | 1475 | 8 | AP | rs3764261 | 17.5 | +-+- |
| 31 | CETP * | rs289715 | 16 | a | LDL-TC | TC | 1.22 | 0.48 | 1.17E−02 | 1.99 | 0.20 | 3.25E−23 | 20.56 | 1065 | 8 | AP | rs3764261 | 17.5 | +-+- |
| 32 | LCAT | rs10468274 | 16 | a | LDL-TC | LDL-C | −0.05 | 0.37 | 8.90E−01 | −0.89 | 0.16 | 1.02E−08 | 7.94 | 15756 | 8 | AP | rs16942887 | 69.1 | +??? |
| 33 | LCAT | rs10468274 | 16 | a | LDL-TC | TC | 0.92 | 0.40 | 2.31E−02 | 1.04 | 0.17 | 8.71E−10 | 7.42 | 454 | 8 | AP | rs16942887 | 69.1 | +??? |
| 34 | LIPG | rs10438978 | 18 | t | LDL-TC | LDL-C | −0.06 | 0.38 | 8.65E−01 | 1.14 | 0.16 | 2.59E−13 | 12.52 | 19932 | 8 | P | rs7241918 | 96.4 | -?-? |
| 35 | LIPG * | rs10438978 | 18 | t | LDL-TC | TC | −1.69 | 0.41 | 3.76E−05 | −1.36 | 0.17 | 8.28E−16 | 10.66 | 241 | 8 | P | rs7241918 | 96.4 | -?-? |
Note: ID is a sequential single nucleotide polymorphism (SNP) number; Locus denotes locus name as used in previous studies; Chr = chromosome; EA denotes minor allele used as an effect allele in an additive genetic model; Trait pair = a pair of lipid traits examined; Main trait = leading trait in the pair; Beta denotes effect size and direction of the genetic association; SE = standard error; dlogP is the difference of the log-transformed p-values of unconditional and conditional analyses, ie, log10(puncond)-log10(pcond); Str denotes the strength of antagonistic heterogeneity defined as the relative change of log-transformed p-values (%), ie, 100 × (log10(pcond)-log10(puncond))/log10(puncond); Column N shows the number of cohorts in which antagonistic heterogeneity was replicated; Column A reports anterior (A) and posterior (P) antagonistic heterogeneity observed in our unconditional and conditional analyses, respectively; LD = linkage disequilibrium between SNPs in prior and current studies, r2; Effect Sign denotes directions of genetic associations with HDL-C, LDL-C, TC, and TG reported in (35,36) where “+” and “-” stay for positive (increase) and negative (decrease) signs of statistical effects, respectively, and “?” indicates associations, which did not attain GW significance (ie, p = pGW) in prior studies. More details are given in extended Supplementary Table 4.
*Associations reported in previous studies, which were strongly affected by the antagonistic genetic heterogeneity in our study.
The results of the unconditional and conditional analyses provided an opportunity to characterize anterior and posterior antagonistic heterogeneities, respectively (“Methods”). The anterior antagonistic heterogeneity was characteristic for 40 of 58 associations for 23 SNPs in 19 loci in Table 2 and Supplementary Tables 3 and 4 (see columns “A”). All these associations were replicated as posterior antagonistic heterogeneity. Conditional analysis identified 18 new associations characterized by posterior antagonistic heterogeneity for 12 SNPs in 11 loci. Some new cases were identified because of increased precision in determining the effect directions in the conditional models for non-significant associations in the unconditional ones, for example, β = −0.06, p = 8.65 × 10–1 (unconditional) versus β = 1.14, p = 2.59×10–13 (conditional) for rs10438978 (LIPG locus). For the others, the effect directions changed in opposite fashion despite the associations in unconditional models attained at least nominal (p < .05) significance, for example, β = 0.68, p = 1.92 × 10–2 (unconditional) versus β = −0.84, p = 8.73 × 10–12 (conditional) for rs7120963 (APOA locus). Stronger effects of antagonistic heterogeneity were observed for lipid traits with larger correlation (see Eq. (1) in “Methods”). The strength of the effect increased with the increase of magnitude of correlation in an exponential fashion (Figure 2).
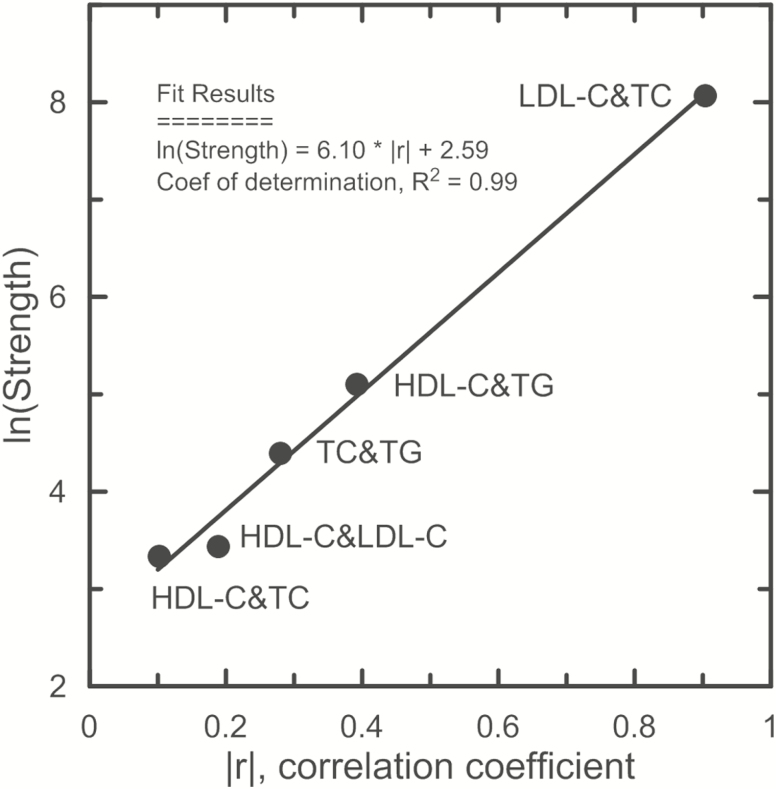
Figure 2.
Correlation between strength of the effect of antagonistic heterogeneity and phenotypic correlation. Symbols are mean values for the strength defined as the relative change of log-transformed p-values in unconditional (puncond) and conditional (pcond) analyses in percents, ie, 100 × (log10(pcond)-log10(puncond))/log10(puncond). The strength was evaluated for the associations reported in Supplementary Table 3 and Table 2, which demonstrated anterior and posterior antagonistic heterogeneity. Phenotypic correlation is given for each of five pairs of lipid traits, ie, HDL-C&LDL-C, HDL-C&TC, HDL-C&TG, LDL-C&TC, and TC&TG, as representatively assessed in the ARIC study (Supplementary Figure 1). LDL-C&TG pair is not presented in this figure because of lack of SNPs that demonstrated the anterior and posterior antagonistic heterogeneity. The y-axis shows natural logarithm (ln) of the strength. The x-axis shows magnitude (modulus) of the correlation coefficient r between phenotypes. Straight line in figure shows exponential increase of the strength with linear increase of the phenotypic correlation. Insert shows equation for the fit and the coefficient of determination (ie, goodness of fit). This figure indicates that different biological mechanisms may be implicated in regulation of even highly correlated traits. HDL-C = High-density lipoprotein cholesterol; LDL-C = Low-density lipoprotein cholesterol; TC = Total cholesterol; TG = Triglycerides.
Replication of Antagonistic Heterogeneity
We examined consistency of the directions of the effects in different studies that is widely regarded as replication (3). We show that the patterns of misalignment of the directions of associations of SNPs with traits and the directions of correlation between the traits, which is hallmark of the antagonistic heterogeneity, were replicated in 5 (one association), 6 (10 associations), and 7+ cohorts (47 associations) (see columns “N” in Table 2 and Supplementary Tables 3 and 4). Antagonistic heterogeneity was replicated in larger number of cohorts when the associations attained at least nominal significance. Replication of the antagonistic heterogeneity is further strengthened by consistent changes in the effects in conditional analysis compared to the unconditional one in most cohorts, including cases of nonsignificant associations (p > .05) in the unconditional analyses (Figure 3, Supplementary Tables 3 and 4). We also show that three modes of strengthening the associations by dissecting antagonistic heterogeneity (ie, decreasing standard errors, Figure 3B, increasing magnitude of the effect sizes, Figure 3A, and both, Figure 3C) were not due to dominant effect in one cohort but were replicated in most cohorts. Likewise, the change in the effect directions in the opposite fashion in the conditional models compared to the unconditional ones is replicated across cohorts (Figure 3D, Supplementary Tables 3 and 4).
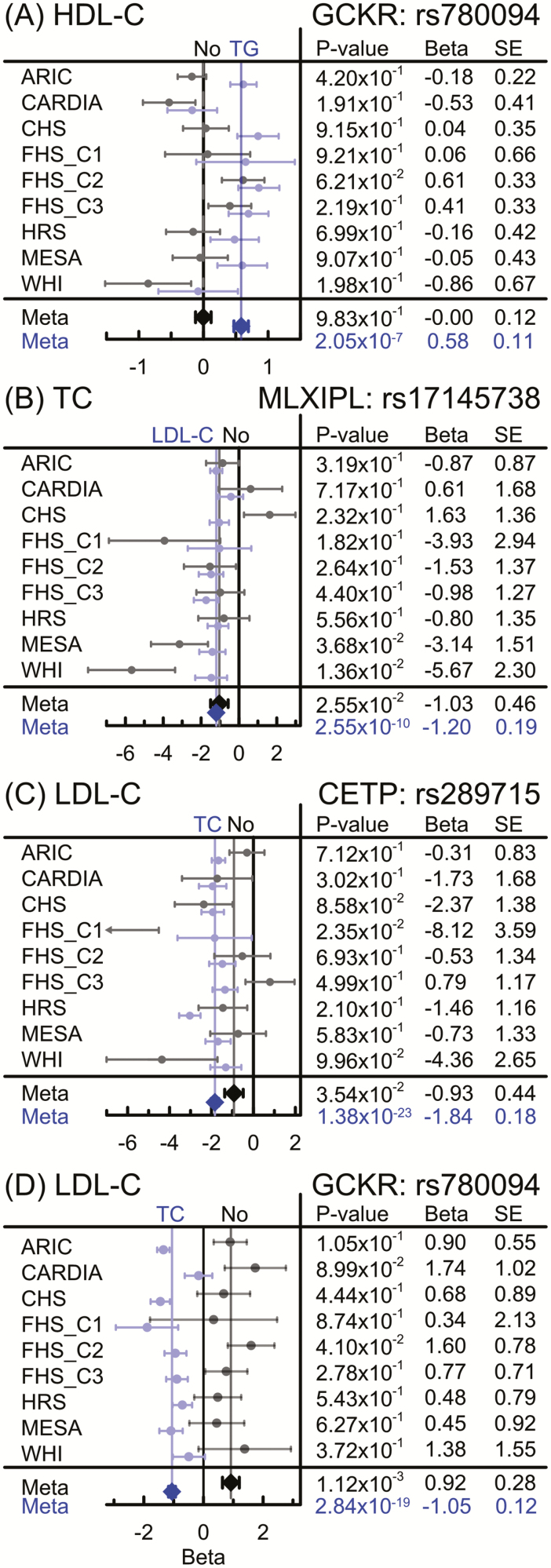
Figure 3.
Forest plots for selected associations with strong effect of the antagonistic genetic heterogeneity. Black and gray (blue on-line) show associations from the unconditional and conditional models, respectively, for minor alleles of: (A) rs780094 (GCKR locus) with HDL-C; conditioning is on TG, (B) rs17145738 (MLXIPL locus) with TC; conditioning is on LDL-C, (C) rs289715 (CETP locus) with LDL-C; conditioning is on TC, and (D) rs780094 (GCKR locus) with LDL-C; conditioning is on TC. Figures (A)–(C) illustrate three modes of strengthening the associations by dissecting the antagonistic genetic heterogeneity, ie, (A) increasing magnitude of the effect size, (B) decreasing the standard error, and (C) both. Figure (D) illustrates changes in the effect directions in opposite fashion in conditional and unconditional models. Bars show standard errors (SE). Cohorts: Atherosclerosis Risk in Communities Study (ARIC); Coronary Artery Risk Development in Young Adults (CARDIA); Cardiovascular Health Study (CHS); Framingham Heart Study (FHS) original cohort (FHS_C1); FHS offspring (FHS_C2); FHS 3rd generation cohort (FHS_C3), Health and Retirement Study (HRS); the Multi-Ethnic Study of Atherosclerosis (MESA), and Women’s Health Initiative (WHI). HDL-C = High-density lipoprotein cholesterol; LDL-C = Low-density lipoprotein cholesterol; TC = Total cholesterol; TG = Triglycerides.
Discussion
This article supports a promising avenue in studies of genetic predisposition to complex traits leveraging the concept relaxing the medical genetics hypothesis on “one gene, one function, one trait” (11). Relaxing this hypothesis is, particularly, inevitable in genetics of traits that make human bodies vulnerable to diseases in postreproductive life because of an inherent heterogeneity in genetic predisposition to such traits due to the undefined role of evolution in establishing their genetic mechanisms (7). This is relevant to lipid traits because: (i) they have not been selected against or in favor of their pathological dysregulation causing age-related diseases (37) and (ii) genes involved in regulation of lipid metabolism were selected in principally different conditions than those in modern societies (8,38,39). Accordingly, the lipid-associated genetic variants may show complex, even antagonistic, relationships to age-related traits (40,41). Here, we used the simplest approach to illustrate our concept by contrasting unconditional and conditional GWAS of four lipid traits. We show that most SNP associations identified in the current study (52 of 98) from loci that were reported in the largest GWAS of lipids (35,36) are not trivial and are strongly affected by the novel phenomenon of antagonistic heterogeneity, which is different from commonly regarded interpopulation ancestry-related heterogeneity. Dissecting the role of antagonistic heterogeneity leads to quantitative and qualitative changes in the associations with lipid traits in a population of the same individuals even for SNPs from genes/loci, which are considered as having well established functions (Table 1). Quantitative change refers to attaining GW significance, or substantial decrease of p-values, by dissecting the antagonistic heterogeneity for the associations with lipid traits reported in (35,36), which attained at least suggestive-effect significances (p ≤ 10–5) in our unconditional analysis. Qualitative change refers to novel associations with lipid traits at GW or suggestive-effect significances for SNPs, which either did not attain suggestive-effect significances in our univariate analysis or were not reported in (35,36). For 10 of these 52 associations such changes were so strong that GW (or suggestive-effect) significances were attained even when no nominally significant signals (p < .05) were identified in a traditional univariate analysis. Notably, this strong effect of antagonistic heterogeneity was observed for well-known lipid genes such as GCKR, SIK3 (APOA1 locus), LIPC, LIPG, etc. The findings of quantitative changes show that GWAS of such complex traits as lipids can be substantially improved just by leveraging more comprehensive analyses of inherently heterogeneous genetic predisposition to such traits. The observed qualitative changes suggest new roles for even those genes, which functions are considered as well established that strongly supports the view on relaxing the medical genetics hypothesis on “one gene, one function, one trait” in GWAS of complex health-related phenotypes (11,12).
The antagonistic genetic heterogeneity highlights a new class of associations emphasizing trade-offs in a potential role of a genetic variant in traits, which is manifested, in this study, as decrease of p-values in the conditional models compared to the unconditional ones. For example, attaining GW significance for the association of rs11216162 with LDL-C in the model conditional on TC (β = −1.11, SE = 0.15, p = 4.53 × 10–13) compared with the unconditional model (β = −0.18, SE = 0.36, p = 6.25 × 10–1) implies that the same carriers of the rs11216162 minor allele tend to have smaller concentrations of LDL-C and larger concentrations of TC (Supplementary Table 4). TC is a measure of the total amount of cholesterol in the blood. It includes “good” (HDL-C) and “bad” (LDL-C) cholesterol and a fraction of TG. Depending on whether the trade-off between TC and LDL-C for carriers of minor allele of rs11216162 is driven by the increased TC concentrations due to HDL-C or TG, it can be classified as the beneficial or adverse, respectively. Both types of these trade-offs are of unprecedented importance for translation to health care. The beneficial trade-off in this example would help identify the genetic predisposition to two beneficial factors of having low concentrations of LDL-C and, simultaneously, high concentrations of HDL-C for carriers of the same allele. The adverse trade-off opens an avenue in studies of the genetic mechanisms of potential side effects in medical treatment, which is especially important in the framework of personalized medicine (42) and geroscience (43,44). Side effect in this example would be manifested as predisposition to the beneficial effect of having low LDL-C concentrations and an adverse effect of having high TG concentrations for carriers of the same allele. The importance of these findings for translation strategies in health care is augmented by the ability of such analysis to identify: (i) more homogeneous populations (as evidenced by the decreased standard errors after dissecting antagonistic heterogeneity) and/or (ii) populations in which genetic effects can become stronger (as evidenced by the increased magnitudes of the effect sizes). Genetics of trade-offs strengthens the importance of identifying mechanistic pathways linking genetic variants with complex traits through intermediate factors including omics, biomarkers, physiological regulation, evolutionary adaptation, etc. Our findings show that implementation of genetic discoveries in health care requires substantially more comprehensive analyses of genetic predisposition to complex traits in each potentially promising locus beyond those implemented in current strategies in large-scale GWAS.
Funding
This research was supported by the National Institute on Aging (grant numbers P01 AG043352, R01 AG047310, and R01 AG061853). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. See also Supplementary Acknowledgment Text.
Author Contributions
A.M.K. conceived and designed the experiment and wrote the paper, Y.L. designed the experiment, performed statistical analyses, and contributed to drafting of the paper. A.N. and I.C. prepared data. I.C. examined genes in the identified loci.
Conflict of Interests
None reported.
Supplementary Material
References
- 1. Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi:10.1126/science.273.5281.1516 [DOI] [PubMed] [Google Scholar]
- 2. Welter D, MacArthur J, Morales J, et al. . The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. doi:10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marigorta UM, Rodríguez JA, Gibson G, Navarro A. Replicability and prediction: lessons and challenges from GWAS. Trends Genet. 2018;34:504–517. doi:10.1016/j.tig.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodríguez JA, Marigorta UM, Navarro A. Integrating genomics into evolutionary medicine. Curr Opin Genet Dev. 2014;29:97–102. doi:10.1016/j.gde.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 5. Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi:10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 6. Morange M. Gene function. C R Acad Sci III. 2000;323:1147–1153. [DOI] [PubMed] [Google Scholar]
- 7. Nesse RM, Williams GC.. Why we get sick: the new science of Darwinian medicine. 1st ed New York, NY: Times Books; 1994. [Google Scholar]
- 8. Vijg J, Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi:10.1146/annurev.med.56.082103.104617 [DOI] [PubMed] [Google Scholar]
- 9. Williams GC. Pleiotropy, natural-selection, and the evolution of senescence. Evolution. 1957;11:398–411. doi:10.1111/j.1558-5646.1957.tb02911.x [Google Scholar]
- 10. Williams PD, Day T. Antagonistic pleiotropy, mortality source interactions, and the evolutionary theory of senescence. Evolution. 2003;57:1478–1488. doi:10.1111/j.0014-3820.2003.tb00356.x [DOI] [PubMed] [Google Scholar]
- 11. Visscher PM, Wray NR, Zhang Q, et al. . 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22. doi:10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Visscher PM, Yang J. A plethora of pleiotropy across complex traits. Nat Genet. 2016;48:707–708. doi:10.1038/ng.3604 [DOI] [PubMed] [Google Scholar]
- 13. Lynch M, Walsh B.. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 14. Kulminski AM, Huang J, Loika Y, et al. . Strong impact of natural-selection-free heterogeneity in genetics of age-related phenotypes. Aging (Albany NY). 2018;10:492–514. doi:10.18632/aging.101407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulminski AM, Loika Y, Huang J, et al. . Pleiotropic meta-analysis of age-related phenotypes addressing evolutionary uncertainty in their molecular mechanisms. Front Genet. 2019;10:433. doi:10.3389/fgene.2019.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharrett AR. The Atherosclerosis Risk in Communities (ARIC) Study. introduction and objectives of the hemostasis component. Ann Epidemiol. 1992;2:467–469. [DOI] [PubMed] [Google Scholar]
- 17. Investigators TA. The Atherosclerosis Risk in Communities (Aric) Study - design and objectives. Am J Epidemiol. 1989;129:687–702. doi:10.1093/oxfordjournals.aje.a115184. [PubMed] [Google Scholar]
- 18. Hughes GH, Cutter G, Donahue R, et al. . Recruitment in the coronary artery disease risk development in young adults (Cardia) study. Control Clin Trials. 1987;8 (4 Suppl):68S–73S. [DOI] [PubMed] [Google Scholar]
- 19. Fried LP, Borhani NO, Enright P, et al. . The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. [DOI] [PubMed] [Google Scholar]
- 20. Bild DE, Bluemke DA, Burke GL, et al. . Multi-Ethnic Study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi:10.1093/aje/kwf113 [DOI] [PubMed] [Google Scholar]
- 21. Govindaraju DR, Adrienne Cupples L, Kannel WB, et al. . Genetics of the Framingham Heart Study population. Adv Genet. 2008;62:33–65. doi:10.1016/S0065-2660(08)00602-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Splansky GL, Corey D, Yang Q, et al. . The third generation cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. doi:10.1093/aje/kwm021 [DOI] [PubMed] [Google Scholar]
- 23. Cupples LA, Heard-Costa N, Lee M, Atwood LD; Framingham Heart Study Investigators Genetics analysis workshop 16 problem 2: the Framingham Heart Study data. BMC Proc. 2009;3 (Suppl 7):S3. doi:10.1186/1753-6561-3-s7-s3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juster FT, Suzman R. An overview of the health and retirement study. Journal of Human Resources. 1995;30:S7–S56. [Google Scholar]
- 25. Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study group. Controlled Clinical Trials. 1998;19:61–109. doi:10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 26. Anderson GL, Manson J, Wallace R, et al. . Implementation of the Women’s Health Initiative study design. Ann Epidemiol. 2003;13 (9 Suppl):S5–17. [DOI] [PubMed] [Google Scholar]
- 27. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi:10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- 28. Das S, Forer L, Schönherr S, et al. . Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi:10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikram MA, Seshadri S, Bis JC, et al. . Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi:10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi:10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res. 2012;40:3777–3784. doi:10.1093/nar/gkr1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu X, Tian L, Wei LJ. Combining dependent tests for linkage or association across multiple phenotypic traits. Biostatistics. 2003;4:223–229. doi:10.1093/biostatistics/4.2.223 [DOI] [PubMed] [Google Scholar]
- 33. Bolormaa S, Pryce JE, Reverter A, et al. . A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014;10:e1004198. doi:10.1371/journal.pgen.1004198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu X, Feng T, Tayo BO, et al. ; COGENT BP Consortium Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet. 2015;96:21–36. doi:10.1016/j.ajhg.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teslovich TM, Musunuru K, Smith AV, et al. . Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi:10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi:10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crespi BJ. The origins and evolution of genetic disease risk in modern humans. Ann N Y Acad Sci. 2010;1206:80–109. doi:10.1111/j.1749-6632.2010.05707.x [DOI] [PubMed] [Google Scholar]
- 38. Corella D, Ordovás JM. Aging and cardiovascular diseases: the role of gene-diet interactions. Ageing Res Rev. 2014;18:53–73. doi:10.1016/j.arr.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 39. Nesse RM, Ganten D, Gregory TR, Omenn GS. Evolutionary molecular medicine. J Mol Med (Berl). 2012;90:509–522. doi:10.1007/s00109-012-0889-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kulminski AM, Loika Y, Culminskaya I, et al. . Explicating heterogeneity of complex traits has strong potential for improving GWAS efficiency. Sci Rep. 2016;6:35390. doi:10.1038/srep35390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kulminski AM, Kernogitski Y, Culminskaya I, et al. . Uncoupling associations of risk alleles with endophenotypes and phenotypes: insights from the ApoB locus and heart-related traits. Aging Cell. 2017;16:61–72. doi:10.1111/acel.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520:609–611. doi:10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 43. Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: the ultimate preventative medicine. Science. 2015;350:1191–1193. doi:10.1126/science.aad3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franceschi C, Garagnani P. Suggestions from geroscience for the genetics of age-related diseases. PLoS Genet. 2016;12:e1006399. doi:10.1371/journal.pgen.1006399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.