Abstract
Adventitious root formation is essential for plant propagation, development, and response to various stresses. Reactive oxygen species (ROS) are essential for adventitious root formation. However, information on Respiratory Burst Oxidase Homolog (RBOH), a key enzyme that catalyzes the production ROS, remains limited in woody plants. Here, a total of 44 RBOH genes were identified from six Rosaceae species (Malus domestica, Prunus avium, Prunus dulcis 'Texas’, Rubus occidentalis, Fragaria vesca and Rosa chinensis), including ten from M. domestica. Their phylogenetic relationships, conserved motifs and gene structures were analyzed. Exogenous treatment with the RBOH protein inhibitor diphenyleneiodonium (DPI) completely inhibited adventitious root formation, whereas exogenous H2O2 treatment enhanced adventitious root formation. In addition, we found that ROS accumulated during adventitious root primordium inducing process. The expression levels of MdRBOH-H, MdRBOH-J, MdRBOH-A, MdRBOH-E1 and MdRBOH-K increased more than two-fold at days 3 or 9 after auxin treatment. In addition, cis-acting element analysis revealed that the MdRBOH-E1 promoter contained an auxin-responsive element and the MdRBOH-K promoter contained a meristem expression element. Based on the combined results from exogenous DPI and H2O2 treatment, spatiotemporal expression profiling, and cis-element analysis, MdRBOH-E1 and MdRBOH-K appear to be candidates for the control of adventitious rooting in apple.
1. Introduction
Adventitious rooting is essential for both woody perennial plants and herbaceous plants. Indeed, adventitious root formation is the cornerstone of woody perennial plant propagation, in which new plants are propagated vegetatively from elite genotypes [1]. Monocot plants such as cereals generate numerous crown roots, a type of adventitious root that dominates their root system [2]. Adventitious roots are also induced to permit survival under oxygen-deficient conditions in plants that are adapted to waterlogging and submersion [3, 4]. In recent years, our understanding of the mechanisms of adventitious root formation has improved greatly, especially in herbaceous plants for which adventitious rooting can be induced easily [5]. However, information on adventitious rooting is still lacking for many recalcitrant plants such as apple rootstocks [6–13]. More research on adventitious root formation will enrich our basic knowledge and aid in efforts to optimize propagation conditions for better rooting of recalcitrant species.
An increasing number of studies indicate that homeostasis and signaling of endogenous hormones are important for adventitious root formation [14, 15]. It has become clear that auxin plays a central role in adventitious root formation [16]. The other phytohormones, including cytokinin (CK), ethylene (ETH), gibberellic acid (GA), strigolactones (SLs), abscisic acid (ABA), salicylic acid (SA), brassinosteroids (BRs), and jasmonate (JA), have been shown to influence adventitious root development directly, by interacting with one another, or by interacting with auxin [5].
In addition, various small-molecule compounds such as nitric oxide (NO), hydrogen gas (H2), hydrogen sulfide (H2S), methane (CH4) and hydrogen peroxide (H2O2), are also involved in adventitious root formation and development [17]. According to the present evidence, H2O2 mainly acts to stimulate adventitious root formation. Treatment with 20–40 μM exogenous H2O2 significantly enhanced the adventitious rooting ability of cucumber [18], and 200 μM H2O2 increased adventitious root length and number in marigold and chrysanthemum [19, 20]. Moreover, it has been demonstrated that both calcium and calmodulin are two downstream signaling molecules in adventitious rooting induced by H2O2 in marigold [21]. In addition, H2O2 accumulated in cucumber plants during adventitious root formation [3]. Scavenging endogenous H2O2 through the application of diphenyleneiodonium (DPI), which inhibits the generation of reactive oxygen species (ROS) by RBOH decreased the number of adventitious root in cucumber and chrysanthemum [3, 20]. RBOH is a key enzyme that catalyzes the production of O2−, which is rapidly transformed in the plant into H2O2, a more stable form of ROS [22]. In general, information on the function of ROS produced by RBOHsin adventitious root formation is not as comprehensive as our understanding of their roles in plant stress response [23–26], root hair development [27, 28], main root growth [29–31] and lateral root development [32–34]. The expression levels of RBOH1 and RBOH3 increased more than 2.5-fold during adventitious root formation triggered by waterlogging in wheat [4]. Similarly, the expression levels of CsRBOHB and CsRBOHF3 were enhanced by ethylene and auxin, ultimately leading to adventitious root formation in cucumber [3]. These results indicate that RBOH, also known as NADPH oxidase, may play an important role in adventitious root formation. However, a role for RBOH family genes in adventitious rooting has not been reported in apple.
Here, we used bioinformatics methods to identify RBOH genes in the genomes of six Rosaceae species: Malus domestica, Prunus avium, Prunus dulcis ‘Texas’, Rubus occidentalis, Fragaria vesca, and Rosa chinensis. We analyzed their phylogenetic relationships, gene structures, conserved motifs and domains, and chromosome locations. We also analyzed the expression of apple MdRBOHs during adventitious rooting and used exogenous H2O2 and DPI treatments to assess the potential effects of RBOH on adventitious rooting. This study presents the molecular characteristics of RBOH genes from six Rosaceae species. It adds to our understanding of RBOH function in adventitious root formation and lays a foundation for optimizing parameters for better rooting of apple rootstocks.
2. Materials and methods
2.1. Identification and classification of RBOH genes
Sequences and genome-related information for M. domestica (apple), P. avium (sweet cherry), P. dulcis ‘Texas’ (almond), R. occidentalis (black raspberry), F. vesca (strawberry) and R. chinensis (rose) were downloaded from the Genome Database for Rosaceae (GDR, https://www.rosaceae.org/). Arabidopsis RBOH protein sequences were obtained from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/).
Using the Arabidopsis RBOH amino acid sequences as queries, candidate RBOHs were identified from the genome databases using two basic local alignment search tool (BLAST) methods implemented in TBtools (https://github.com/CJ-Chen/TBtools). After removing repetitive and redundant sequences, primary RBOH proteins were identified using BLAST methods in the UniProKB/Swiss-Prot database. Their conserved domains were analyzed using CD-search at the NCBI website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to determine whether each candidate sequence contained an NADPH_Ox (PF08414) domain.
The properties of each RBOH protein sequence, including its length (aa), molecular weight (MW), isoelectric point (pI) and predicted subcellular localization, were calculated using the ExPASy-Compute pI/MW tool (http://web.expasy.org/compute_pi/) and Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#).
2.2. Phylogenetic analysis
A species phylogenetic tree was obtained from the Common Taxonomy Tree (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) and visualized using EvolView (http://www.evolgenius.info/evolview/). Phylogenetic trees were constructed from RBOH protein sequences using MEGA7.0 software (Arizona State University, Tempe, AZ, USA) with the Maximum Likelihood method and 1000 bootstrap replicates [35].
2.3. Chromosomal distribution and synteny analysis
All RBOH genes were mapped to their respective chromosomes using Amazing Gene Location from GTF/GFF in TBtools. Synteny blocks within the apple genome and between the apple and Arabidopsis genomes were constructed using Quick MCScanX Wrapper and were visualized using the Dual Synteny Plotter in TBtools.
2.4. Sequence analysis
Motifs in RBOH proteins were identified using the MEME suite (http://meme-suite.org/). Conserved domains within the RBOH proteins were identified using Batch CD-Search (https://www.ncbi.nlm.nih.gov/cdd). The structures of the RBOH genes were constructed using the Amazing Optional Gene Viewer in TBtools. The cis-elements present in all promoter sequences (−2000 bp upstream) were identified using PlantCARE [36].
2.5. Plant materials and treatments
Stem cuttings of ‘Gala’ apples were sub-cultured in a normal medium of Murashige and Skoog (MS), 7.5 g L−1 agar and 30 g L−1 sugar (pH 5.8) with 0.5 mg L−1 IBA and 0.2 mg L−1 6-BA under a 16 h light/8 h dark photoperiod with day/night temperatures of 25±1°C and 20±1°C. When stem cuttings had grown to 2–3 cm in length, they were transferred into rooting medium containing 1/2 MS, 30 g L−1 sugar, 7.5 g L−1 agar, 0.5 mg L−1 IBA and 0.1 mg L−1 NAA (pH 5.8) [37]. For adventitious rooting assays, stem cuttings were transferred into rooting medium supplemented with various concentrations of H2O2 (1 mM, 5 mM and 25 mM) (Sigma–Aldrich, St. Louis, MO, USA) and DPI (5 μM, 10 μM and 20 μM) (Sigma–Aldrich, St. Louis, MO, USA). There were three biological replicates for each treatment, and each biological replicate consisted of six tissue culture cuttings.
Apple tissues and organs, including young roots, stems, leaves, and shoot apices, were harvested from two-year-old own-rooted ‘Gala’ seedlings grown in the field.
2.6. Histological analysis
We collected 0.5 cm sections of ‘Gala’ stem cuttings at 0, 6, 9, and 12 d after subculture on rooting medium. The samples were fixed in a 50:5:5 (v/v/v) ethanol/formaldehyde/acetic acid (FAA) solution for 2 days at room temperature. Then the sections were dehydrated in an ethanol series (50%, 70%, 85%, 95%, and 100%), infiltrated with xylene, and embedded in paraffin. Longitudinal sections with a thickness of 10 μm were cut with a rotary microtome (KD-2258, KEDEE, China) and stained with toluidine blue [1]. For nitro-blue-tetrazolium (NBT) staining of apple stem cuttings, the cross sections were cut after the staining according to the method described previously [3].
2.7. Gene expression analysis
For relative expression analysis of RBOH genes during adventitious rooting, 0.5 cm sections of ‘Gala’ stem cuttings were frozen in liquid nitrogen at 0, 3, 6, and 9 d after subculture on rooting medium. Total RNA was extracted from approximately 0.5 g of frozen sample using the TIANGEN Plant RNA Kit (TIANGEN biotech CO., LTD, Beijing, China, DP305). Then 1 μg of total RNA was used for first-strand cDNA synthesis using the PrimeScript™ RTase (TaKaRa Biotechnology, Dalian, China). Quantitative RT-PCR was performed using SYBR green reagents (RR820A, Takara, Dalian, China) on an Applied Biosystems 7500 real-time PCR system. The apple EF1α gene was used as the control. Relative expression levels were calculated according to the 2−ΔΔCT method [38]. Three independent biological replicates and technical replicates were conducted. Primer sequences used in this study are listed in S1 Table.
3. Results
3.1. Identification and characterizaition of RBOH family genes
BLASTp were conducted in the genome databases of six Rosaceae species using the protein sequence of AtRBOHs as the query sequences. After removing repetitive and redundant sequences, a total of 52 RBOH genes were originally identified using BLAST methods in the UniProKB/Swiss-Prot database (S1 Table). Based on the presence of apparently complete NADPH_Ox (PF08414) domain and EF-hand (CD00051), 44 genes were subsequently selected and annotated as being RBOH genes. Genes without complete predicted NADPH_Ox and EF-hand domains were removed (MD12G1213800, Pav_co4068891.1, Pav_sc0000671.1, Pav_sc0001450.1, Prudul26A013915P2, Prudul26A019458P1, RcHm_v2.0_Chr2g0103591, RcHm_v2.0_Chr2g0103601) (S2 Table). Finally, a total of 44 candidate RBOHs were identified from six Rosaceae genomes: ten from M. domestica (MdRBOH), four from P. avium (PavRBOH), seven from P. dulcis ‘Texas’ (PdRBOH), seven from R. occidentalis (RoRBOH), seven from F. vesca (FvRBOH) and nine from R. chinensis (RcRBOH). The RBOHs were named based on their phylogenetic relationships with their Arabidopsis counterparts (Table 1). The deduced RBOH protein sequences ranged from 758 (RoRBOH-K) to 972 (PdRBOH-D) amino acids in length. Their molecular weights and pIs ranged from 86.88 (RcRBOH-A2) to 109.59 (PavRBOH-F) kDa and from 7.59 (MdRBOH-H) to 9.35 (PdRBOH-F), respectively (Table 1). Subcellular localization predications indicated that the ROBHs were all located in the cell membrane (Table 1). In addition, transiently expressed MdRBOH-E1_GFP or MdRBOH-F1/F2_GFP, or MdRBOH-J_GFP fusion protein in tobacco leaves supporting the notion that RBOHs are localized to the cell membrane (S1 Fig).
Table 1. RBOH genes identified in this study.
Subcellular localization predications indicated that the ROBHs were all located in the cell membrane.
| Species | Gene name | Locus | chromosome | start position | end position | Length (aa) | MW (kD) |
|---|---|---|---|---|---|---|---|
| Malus domestica | MdRBOH-D2 | MD16G1145100 | Chr16 | 11156971 | 11161436 | 965 | 108.64 |
| MdRBOH-D1 | MD13G1134500 | Chr13 | 10304950 | 10310061 | 962 | 108.44 | |
| MdRBOH-A | MD06G1128500 | Chr06 | 27087364 | 27094696 | 941 | 105.38 | |
| MdRBOH-F1 | MD06G1093000 | Chr06 | 22375873 | 22385466 | 961 | 109.27 | |
| MdRBOH-F2 | MD14G1113700 | Chr14 | 18347131 | 18359289 | 959 | 109.37 | |
| MdRBOH-E1 | MD02G1099900 | Chr02 | 7918841 | 7925357 | 912 | 103.16 | |
| MdRBOH-E2 | MD15G1222800 | Chr15 | 18082033 | 18086949 | 946 | 106.97 | |
| MdRBOH-H | MD11G1118400 | Chr11 | 10784434 | 10788882 | 859 | 98.11 | |
| MdRBOH-J | MD03G1106300 | Chr03 | 9106580 | 9110548 | 823 | 93.42 | |
| MdRBOH-K | MD14G1211700 | Chr14 | 29690183 | 29695068 | 785 | 89.50 | |
| Prunus avium | PavRBOH-A | Pav_sc0000589 | Chr05 | 10841240 | 10843909 | 943 | 105.4 |
| PavRBOH-F | Pav_sc0000886 | Chr05 | 8458927 | 8463180 | 964 | 109.59 | |
| PavRBOH-E2 | Pav_sc0000129 | Chr07 | 15367462 | 15367766 | 799 | 90.27 | |
| PavRBOH-H | Pav_sc0000323 | Chr06 | 5597348 | 5598268 | 860 | 98.43 | |
| Prunus dulcis Texas | PdRBOH-A | Prudul26A009078P1 | Chr05 | 12324110 | 12329280 | 942 | 105.27 |
| PdRBOH-D | Prudul26A009232P1 | Chr01 | 19719419 | 19724815 | 972 | 108.95 | |
| PdRBOH-B | Prudul26A005532P1 | Chr06 | 27292497 | 27296750 | 894 | 101.9 | |
| PdRBOH-E | Prudul26A013915P1 | Chr07 | 17253193 | 17258382 | 965 | 103.46 | |
| PdRBOH-F | Prudul26A017061P1 | Chr05 | 10368633 | 10377258 | 913 | 109.55 | |
| PdRBOH-H | Prudul26A019458P2 | Chr06 | 6163889 | 6168519 | 857 | 98.23 | |
| PdRBOH-K | Prudul26A030363P1 | Chr05 | 16131323 | 16146293 | 811 | 92.5 | |
| Rubus occidentalis | RoRBOH-J | Ro03_G02103 | Chr03 | 7378990 | 7384860 | 841 | 95.98 |
| RoRBOH-E | Ro01_G03041 | Chr01 | 4216430 | 4224173 | 925 | 105.10 | |
| RoRBOH-F | Ro05_G16791 | Chr05 | 7260878 | 7272779 | 957 | 109.06 | |
| RoRBOH-K | Ro05_G03331 | Chr05 | 2246266 | 2252031 | 758 | 86.94 | |
| RoRBOH-A | Ro05_G01806 | Chr05 | 4074465 | 4082558 | 919 | 103.69 | |
| RoRBOH-D | Ro04_G08892 | Chr04 | 7473145 | 7479394 | 933 | 105.15 | |
| RoRBOH-B | Ro06_G18529 | Chr06 | 27530616 | 27538049 | 862 | 98.80 | |
| Fragaria vesca | FvRBOH-A | FvH4_5g10010.1 | Chr05 | 5700687 | 5708036 | 917 | 103.18 |
| FvRBOH-D | FvH4_4g22100.1 | Chr04 | 25038971 | 25043196 | 935 | 105.46 | |
| FvRBOH-B | FvH4_6g06490.1 | Chr06 | 3796200 | 3801416 | 879 | 100.64 | |
| FvRBOH-E | FvH4_1g09270.1 | Chr01 | 4945413 | 4949975 | 867 | 97.94 | |
| FvRBOH-F | FvH4_5g03310.1 | Chr05 | 1974927 | 1983225 | 945 | 107.43 | |
| FvRBOH-H | FvH4_3g34420.1 | Chr03 | 29860660 | 29865289 | 865 | 98.37 | |
| FvRBOH-K | FvH4_5g13850.1 | Chr05 | 7816138 | 7820214 | 818 | 93.12 | |
| Rosa chinensis | RcRBOH-A1 | RcHm_v2.0_Chr7g0188251 | Chr07 | 7870102 | 7877173 | 925 | 103.87 |
| RcRBOH-A2 | RcHm_v2.0_Chr7g0202531 | Chr07 | 20116816 | 20123208 | 766 | 86.88 | |
| RcRBOH-D | RcHm_v2.0_Chr4g0428361 | Chr04 | 53408287 | 53412982 | 936 | 105.85 | |
| RcRBOH-B1 | RcHm_v2.0_Chr3g0455621 | Chr03 | 5279721 | 5284973 | 878 | 101.05 | |
| RcRBOH-B2 | RcHm_v2.0_Chr3g0455641 | Chr03 | 5311755 | 5316513 | 881 | 101.46 | |
| RcRBOH-E | RcHm_v2.0_Chr2g0095831 | Chr02 | 8765834 | 8770818 | 923 | 104.91 | |
| RcRBOH-F | RcHm_v2.0_Chr7g0196471 | Chr07 | 14440777 | 14448994 | 953 | 108.37 | |
| RcRBOH-H | RcHm_v2.0_Chr5g0062021 | Chr05 | 67592381 | 67596621 | 847 | 97.08 | |
| RcRBOH-K | RcHm_v2.0_Chr7g0183081 | Chr07 | 4122936 | 4127024 | 790 | 90.48 |
3.2. Phylogenetic and comparative analysis of RBOH genes in various plant species
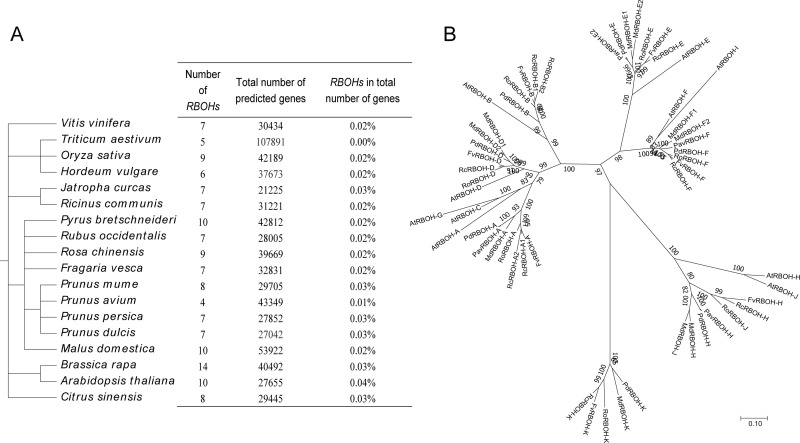
To investigate the evolutionary history of the RBOH family, we compared the number of RBOHs among 18 plant species [39–44]. As shown in Fig 1A, no individual species had more than 14 RBOH members. The percentage of RBOHs with respect to the total number of genes in each genome was low and differed little among species. This indicates that the number of the RBOH gene family in these 18 species was not associated with an increase in the total number of predicted genes.
Fig 1. Phylogenetic and comparative analysis of RBOH genes from various plants.
(A) A summary of the species phylogeny and the number of RBOH genes in each species. (B) Phylogenetic analysis of RBOH proteins from six Rosaceae species and Arabidopsis.
To further evaluate the evolutionary relationships among RBOH family members from the six Rosaceae species and Arabidopsis, 54 RBOH predicted protein sequences were used to construct a phylogenetic tree (Fig 1B). The RBOHs clustered into seven subfamilies, which are indicated by different colors in Fig 1B. With the exception of the RBOH-K subfamily, the other six subfamilies contained at least one AtRBOH member. In addition, multiple sequence alignments of the 54 RBOH proteins were performed (S2 Fig). It can be found that the amino acid sequences of RBOH genes identified in this study and AtRBOH family members are highly conserved.
3.3. Chromosomal distribution and duplication of RBOH genes
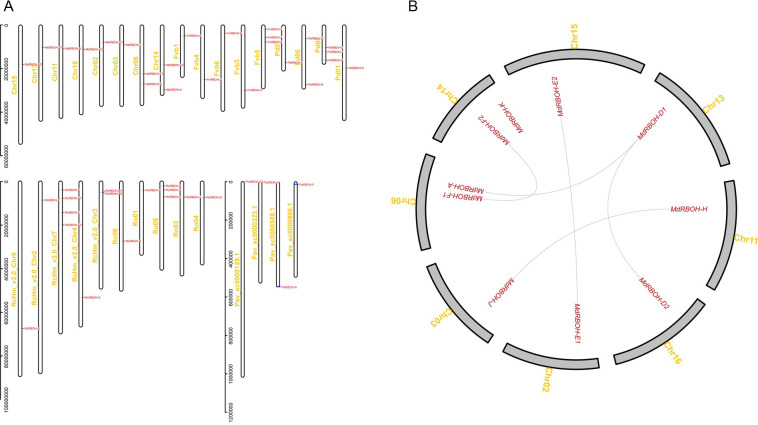
The genomic distribution of the 44 RBOH genes is shown in Fig 2A. Only one tandem duplication event was identified according to the methods of Holub [45]. It occurred in the Rosa chinensis RBOH family, and the tandem duplicated gene pair was RcRBOH-B1 and RcRBOH-B2. The other 42 RBOH genes were irregularly distributed on their species’ chromosomes, and no gene clusters were found. In apple, the 10 MdRBOHs were mapped to 8 of the 17 chromosomes. Seven FvRBOHs, seven PdRBOHs, nine RcRBOHs, seven RoRBOHs and four PaRBOHs were located on five strawberry chromosomes, four almond chromosomes, five rose chromosomes, five black raspberry chromosomes, and four sweet cherry chromosomes, respectively.
Fig 2. Chromosomal distribution and duplication of RBOH genes.
(A) Chromosomal distribution of the RBOH genes among six Rosaceae species. (B) Synteny analysis of the apple RBOH genes. Syntenic apple RBOH genes are linked by lines.
The synteny analysis (Fig 2B) and phylogenetic tree (Fig 1B) indicated that five gene duplication events had occurred in the apple RBOH family. The duplicated gene pairs were MdRBOH-D1 and MdRBOH-D2, MdRBOH-D1 and MdRBOH-A, MdRBOH-E1 and MdRBOH-E2, MdRBOH-F1 and MdRBOH-F2, and MdRBOH-H and MdRBOH-J.
3.4. Motif composition, conserved domains and gene structures of RBOH genes
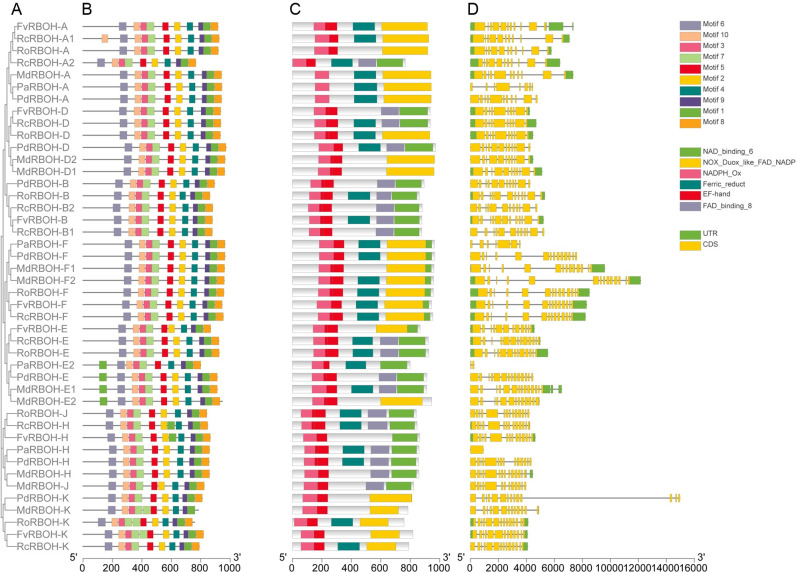
MEME analysis revealed ten conserved motifs within the 44 RBOH proteins (Fig 3B). In general, these ten motifs were present in all 44 RBOH proteins with a similar order and distribution. However, the N-terminal domain of the PaRBOH-E2, PdRBOH-E, MdRBOH-E1, and MdRBOH-E2, which belong to the same subfamily, all contains an additional motif 1. These results indicated that differences in the composition of the N-terminal domain may confer different biological functions to this subfamily.
Fig 3. Phylogenetic relationships, motif composition, conserved domains, and gene structures of RBOH proteins and their corresponding genes from six Rosaceae species.
(A) The phylogenetic tree was drawn based on the full-length protein sequences of RBOHs using MEGA 7.0. (B) Motifs in RBOH proteins. Ten motifs are indicated by different colored boxes. (C) Conserved domains of the RBOH proteins. Six conserved domains are indicated with different colored boxes. (D) Gene structures of the RBOHs. Introns, exons and untranslated regions (UTRs) are indicated by gray lines, yellow rectangles and green rectangles, respectively.
We further analyzed the conserved domains of the 44 RBOHs (Fig 3C). They included NAD_binding_6 (pfam08030), NOX_Duox_like_FAD_NADP (CD06186), NADPH_Ox (pfam08414), Ferric_reduct (pfam01794), EF-hand (CD00051) and FAD_binding_8 (pfam08022) domains. Every RBOH contained the NADPH_Ox and EF-hand domains, which are characteristic of plant RBOH proteins (Fig 3C).
Gene structure analysis showed that the RBOHs had 1 to 14 exons and that members of the same subfamily shared the same or similar intron–exon patterns (Fig 3D). RBOH exons also tended to be quite short. In general, RBOHs in the same subfamily exhibited similar conserved domains, intron–exon organizations, and motif characteristics, which supports their classification in the same subfamily and their close evolutionary relationship.
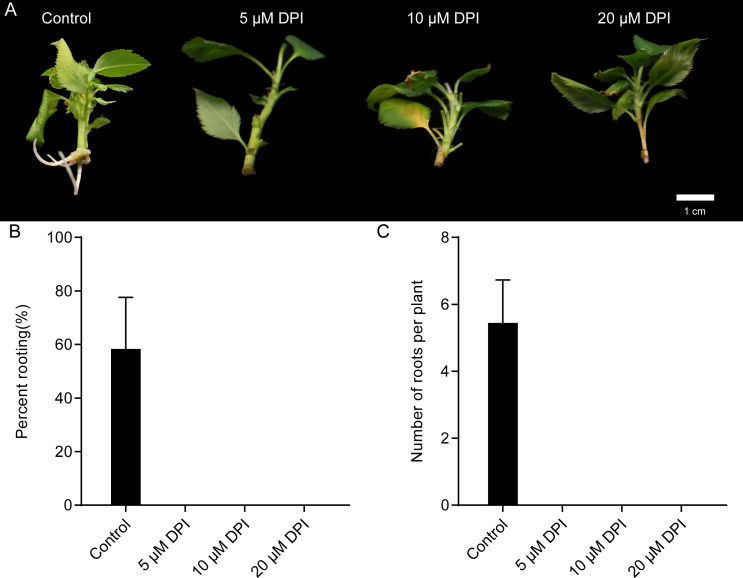
3.5. Effects of DPI on auxin-induced adventitious root formation
To investigate the potential role of RBOHs in apple adventitious root formation, DPI, which inhibits the generation of reactive oxygen species (ROS) by RBOH, was added to the rooting medium. After 18 days of treatment, adventitious roots were counted. Treatment of apple with DPI suppressed adventitious root formation (Fig 4). Treatment with 5 μM DPI completely suppressed adventitious root formation but did not damage the stem cuttings. On the other hand, treatment with 10 or 20 μM DPI completely suppressed adventitious root formation and also damaged the stem cuttings. These results indicate that the production of ROS by RBOH plays an important role in auxin-induced adventitious root formation in apple.
Fig 4. Effects of DPI on adventitious root formation of apple stem cuttings.
(A) Adventitious root formation in apple stem cuttings after 18 days in rooting medium (control) or rooting medium supplemented with DPI (5 μM, 10 μM, and 20 μM). Percent rooting (B) and adventitious root number per cutting (C) were counted at 18 days. Bars indicate mean and Standard Deviation (SD) of three biological replicates; n = 6 individuals per replicate.
3.6. Effects of exogenous H2O2 on apple adventitious root formation
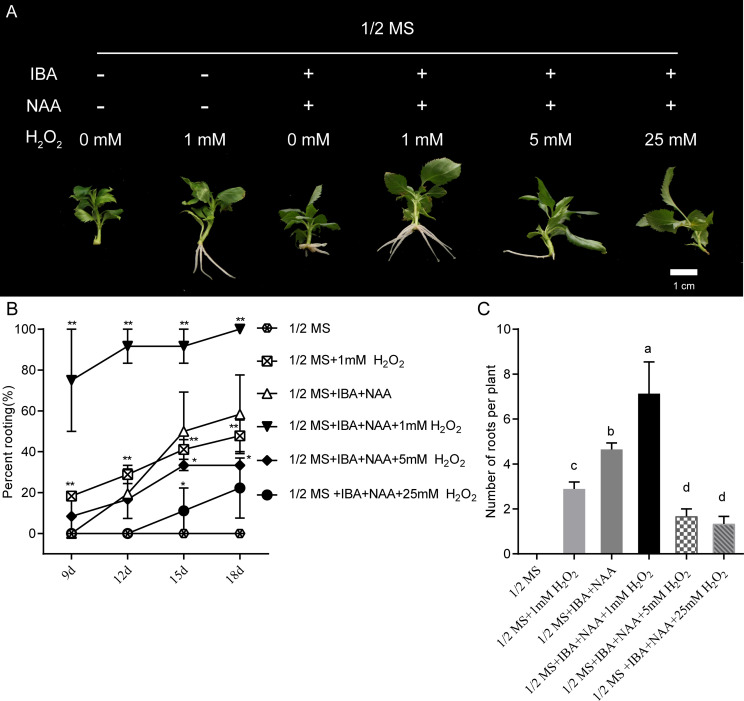
As shown in Fig 4, inhibiting the generation of ROS by the application of DPI completely suppressed adventitious root formation. Therefore, we next applied exogenous H2O2 to assess its effects on adventitious root formation. Various concentrations of H2O2 were applied to apple stem cuttings grown on auxin-free 1/2 MS medium or on 1/2 MS medium supplemented IBA and NAA. Exogenous H2O2 affected apple adventitious root formation in a dose-dependent manner (Fig 5). Treatment of apple stem cuttings with 1 mM H2O2 significantly enhanced adventitious rooting, but treatment with 5 mM or 25 mM H2O2 significantly inhibited adventitious root formation (Fig 5A). Treatment with 1 mM H2O2 not only caused significantly faster rooting, but also significantly increased adventitious root number compared with the other treatments (Fig 5B and 5C). Interestingly, 1 mM H2O2 treatment rescued the adventitious rooting defect of stem cuttings that were grown on auxin-free 1/2 MS medium (Fig 5).
Fig 5. Effects of exogenous H2O2 on adventitious root formation of apple stem cuttings.
(A) Adventitious root formation in apple stem cuttings after 18 days in rooting medium or rooting medium supplemented with H2O2 (1 mM, 5 mM, and 25 mM). “+” represents presence, “−” represents absence. The concentrations of IBA and NAA were 0.5 mg L−1 and 0.1 mg L−1, respectively. (B) Percent rooting. Bars indicate mean and SE from three biological replicates; n = 6 individuals per replicate. Asterisks indicate significant differences between stem cuttings subcultured in 1/2 MS and stem cuttings subcultured in 1/2 MS + IBA +NAA (Student’s t-test, **P < 0.01, *P < 0.05). (C) Adventitious root number per cutting. Adventitious root number per cutting was counted at 18 days. Bars indicate mean and SE from three biological replicates; n = 6 individuals per replicate. The statistical analysis was conducted using Duncan’s multiple range test (P < 0.05). The different lowercase letters indicate significant differences.
3.7. Superoxide accumulation during adventitous root formation
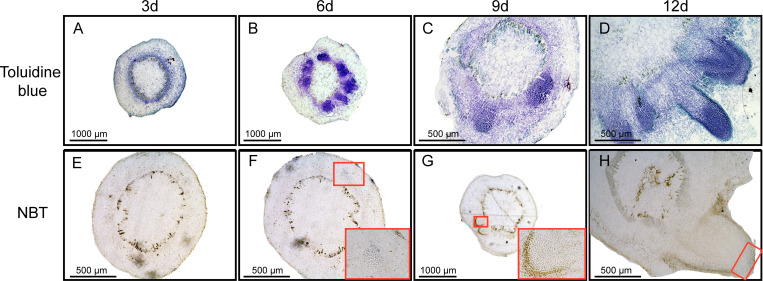
Cross sections of toluidine blue-stained stem cuttings showed that dome-shaped adventitious primordia were clearly visible in stem cross-sections at 6–9 days (Fig 6B and 6C). After 12 days, adventitious roots began to appear (Fig 6D). After 6 days, the amounts of O2- (as shown by NBT) accumulated mainly in the differentiating adventitious root primordia (Fig 6F). After 9 days, the O2- production was almost undetected when adventitious root primordia were completely differentiated (Fig 6G). Interestingly, the amounts of O2- accumulated in the root tip during the adventitious root growth after 12 days (Fig 6H).
Fig 6. Reactive oxygen species are participated in adventitious root formation in apple stem cuttings.
Stem cross-sections of ‘Gala’ apples taken at 3, 6, 9, and 12 days after subculture on rooting medium and stained with toluidine blue (A-D) and Nitro blue tetrazolium (NBT; O2-) (E-H), respectively. Scale bars (1000 μm, 500 μm, and 200 μm) are indicated in the figures. Pictures in red boxes show a 20⊆ view of the stem cuttings in (F, G). The rectangle indicates NBT-stained root tip in (H).
3.8. Expression patterns of MdRBOH genes during adventitous root formation
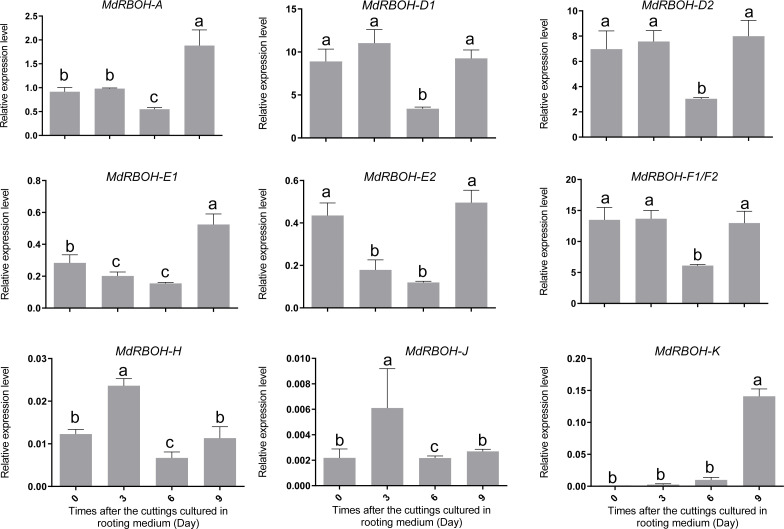
To investigate whether MdRBOH genes are involved in apple adventitious root formation, we examined the expression profiles of ten MdRBOH genes during the induction of adventitious rooting. According to the histological features in Fig 6A–6D, we used stem cuttings collected at 0, 3, 6, and 9 days after sub-culturing in rooting medium for gene expression analysis. As shown in Fig 7, the expression of MdRBOH-H and MdRBOH-J increased more than two-fold on day 3 relative to day 0. In addition, the expression of MdRBOH-A, MdRBOH-E1 and MdRBOH-K increased more than two-fold on day 9 relative to day 0. Of these five RBOH genes, MdRBOH-A, MdRBOH-E1, and MdRBOH-K showed an expression pattern that most closely tracked the appearance of adventitious root primordia. These results suggest that MdRBOH-A, MdRBOH-E1, and MdRBOH-K may be involved in the requirement for ROS generation during adventitious root formation in apple.
Fig 7. Relative expression levels of MdRBOH genes during the adventitious rooting process.
Relative transcript levels of 10 MdRBOH genes were measured in stem tissue after the cuttings had been transferred to rooting medium for 0, 3, 6, or 9 days. Transcript levels were measured by qRT-PCR using apple EF1α as the reference gene. Data are means ± SD of three biological replicates. Refer to S1 Table for primers. The statistical analysis was conducted using Duncan’s multiple range test (P < 0.05). The different lowercase letters indicate significant differences.
4. Discussion
RBOH enzymes catalyze ROS production and play multiple critical roles in plant development, biotic and abiotic stress responses, signal transduction and hormone responses [25, 46–52]. Hence, RBOH genes have been widely identified in diverse plant species [39–44]. In this study, 44 putative RBOH genes were identified from six horticultural Rosaceae species; their numbers ranged from a minimum of 4 to a maximum of 10 in the Rosaceae genomes (Table 1). Among the RBOHs surveyed in a wider group of plants, the maximum number was found in the soybean genome (17), while the minimum number was found in sweet cherry (4). A comparison of RBOH numbers in 18 species (Fig 1A) indicated that the expansion of the RBOH gene family was not associated with an increase in the total number of predicted genes in a genome. We concluded that no direct relationship exists between genome size and the number of RBOH genes.
It has been reported that whole-genome duplication (WGD) may have had an important role in the expansion of the RBOH gene family in Brassica rapa because two gene pairs derived from a whole-genome triplication and three gene pairs derived from segmental duplications [39]. Whole-genome and segmental duplications occurred widely in the apple genome during the process of the apple domestication [53, 54]. As shown in Fig 2A, 8 of 10 MdRBOH genes were located on four homologous pair of chromosomes (chromosomes 6 and 14, 2 and 15, 3 and 11, 13 and 16) [54]. For example, chromosomes 2 and 15 are homologous pairs, and both contain an MdRBOH gene (named as MdRBOH-E1 and MdRBOH-E2) (Fig 2A). Similar findings were also observed for chromosomes homologous pairs 13–16, 3–11, 6–14, and these genes were named as MdRBOH-D1 and MdRBOH-D2, MdRBOH-H and MdRBOH-J, MdRBOH-F1 and MdRBOH-F2. These findings clearly indicated that the duplication of these MdRBOH genes is related to apple whole-genome duplications. In addition, MdRBOH-D1 and MdRBOH-A gene pairs was generated by segmental duplications. None of the RBOHs in the other five Rosaceae genomes appeared to have arisen from segmental duplication, probably because a WGD occurred during the process of apple domestication but did not occur in the other five species we investigated [53]. Notably, RcRBOH-B1 and RcRBOH-B2 appeared to be a tandem duplicated gene pair, based on a comprehensive analysis of their sequence properties and their genome locations.
Several studies have shown that ROS play a crucial role in the adventitious rooting process of herbaceous plants, such as cucumber, rice and Chrysanthemum [3, 18–20, 55]. However, the role of ROS in the adventitious rooting of woody plants has received less attention. In this study, exogenous H2O2 application significantly enhanced adventitious rooting of apple stem cuttings cultured in 1/2 MS medium supplemented with auxin (Fig 5). To our surprise, exogenous H2O2 treatment also rescued the adventitious rooting defect normally exhibited by apple stem cuttings grown on auxin-free 1/2 MS medium (Fig 5). It has been well established that auxin is necessary for adventitious root formation in apple [1, 6, 37]. To explain these observations, we speculate that auxin-induced adventitious root formation in apple is dependent upon on ROS production. Consistent with our inference, previous work has shown that exogenous NAA enhances ROS accumulation during adventitious rooting triggered by waterlogged conditions [3]. In addition, auxin-induced ROS are known to participate directly in root gravitropism, cell elongation, and quiescent center formation [56–58]. In our study, auxin-induced adventitious root formation was completely inhibited by preventing O2− production using DPI (Fig 4). These results are supported by observations in cucumber seedling explants, in which ethylene-, ABA- or auxin-induced adventitious root formation was eliminated by DPI [3, 18, 59]. In addition, our findings that the amounts of O2- accumulated mainly in the differentiating adventitious root primordia (Fig 6F), but almost undetected when adventitious root primordia were completely differentiated (Fig 6G) suggest that ROS are essential for adventitious root primordia induction. In agreement with our findings, O2- production increased sharply after waterlogging inducing adventitious rooting in cucumber, and O2- production also was elevated for only a short time [3]. Collectively, these results indicate that auxin-induced adventitious root formation is probably mediated by ROS. Hence, it will be interesting to learn the molecular basis of the crosstalk between auxin and ROS during the induction of adventitious root formation in woody plants.
Although ROS are clearly necessary for adventitious root formation, the functions of RBOH genes in adventitious rooting remain largely unexplored. It has been reported that the expression of RBOH1 and RBOH3 increased more than 2.5-fold during adventitious root formation triggered by waterlogging in wheat [4]. Similarly, the expression levels of CsRBOHB and CsRBOHF3 were enhanced by ethylene and auxin, ultimately leading to adventitious root formation in cucumber [3]. In addition, RBOHs act downstream of auxin treatment in root hair elongation in rice and Arabidopsis [27, 28], and lateral root development in Arabidopsis [33]. In this study, the expression levels of MdRBOH-A, MdRBOH-E1 and MdRBOH-K increased more than two-fold at day 9 after auxin treatment, coincident with the appearance of adventitious root primordia (Fig 7), indicating that these RBOHs may play a role in adventitious root formation. Based on the above results, we conclude that RBOHs act downstream of auxin to trigger ROS production in root hair, lateral root, and adventitious root development. However, the molecular link between auxin and RBOHs remain largely unknown. Until recently, the molecular link between auxin and ROS-mediated polar root hair growth has been established in Arabidopsis. They found that auxin-auxin response factors (ARF5) module to activation of the bHLH transcription factor RSL4, which directly regulates two RBOHs genes impacting on apoplastic ROS homeostasis, thereby stimulating root hair cell elongation [27]. In our study, MdRBOH-E1 and MdRBOH-K were mainly expressed in the stem, whereas MdRBOH-A was highly expressed in both roots and stems (S3 Fig). Furthermore, analysis of cis-acting elements revealed that the MdRBOH-E1 promoter contained an auxin-responsive element and the MdRBOH-K promoter contained a meristem expression element (S3 Table). Taken together, this evidence suggests that MdRBOH-E1 and MdRBOH-K are candidates for the control of the adventitious rooting process. Further research will be needed to confirm their functions in adventitious root formation, and further experiments are required to identify the molecular link between auxin and MdRBOH-E1 and MdRBOH-K genes expression in apple during adventitious rooting process.
5. Conclusions
The present study adds to our understanding of the evolutionary history of RBOHs by systematically analyzing their protein phylogeny, gene structures, conserved motifs, syntenic relationships, and expression patterns. We identified ten, four, seven, seven, seven and nine RBOH genes from apple, sweet cherry, almond, black raspberry and rose, respectively. These RBOHs were clustered into seven subfamilies that could be distinguished on the basis of their similar gene structures and conserved motifs. Interestingly, whole-genome duplications were found to be responsible for the expansion of the RBOH gene family in apple. The results of exogenous H2O2 and DPI treatment demonstrated that RBOH-derived ROS play an important role in adventitious root formation in apple. Expression analysis suggested potential functions of MdRBOH-E1 and MdRBOH-K in adventitious rooting.
Supporting information
GFP was fused to the N terminus of MdRBOHs (MdRBOH-E1-GFP, MdRBOH-F1/F2-GFP and MdRBOH-J-GFP). The fluorescence signal of N.benthamiana leaves was detected by confocal microscope 72 hours after infiltration.
(TIF)
(TIF)
Transcript levels were measured by qRT-PCR using apple EF1α as the reference gene. Data are means ± SD of three biological replicates. The statistical analysis was conducted using Duncan’s multiple range test (P < 0.05). The different lowercase letters indicate significant differences.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the National Natural Science Foundation of China (grant number 31801824), Specialist Project of Fruit Tree Innovation Team, Technical System of Modern Agricultural Industry in Shandong Province (SDAIT-06-05), Breeding Plan of Shandong Provincial Qingchuang Research Team (2019), and Scientific Research Funds for High-level Personnel of Qingdao Agricultural University (663/1118036 and 663/1119046). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Xu X.; Li X.; Hu X.; Wu T.; Wang Y.; Xu X.; et al. High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 Independent of PINs and ARFs in Malus xiaojinensis. Front Plant Sci 2017, 8, 1059 10.3389/fpls.2017.01059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inukai Y.; Sakamoto T.; Ueguchi-Tanaka M.; Shibata Y.; Gomi K.; Umemura I.; et al. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. 10.1105/tpc.105.030981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi X.; Li Q.; Ma X.; Qian C.; Wang H.; Ren N.; et al. Waterlogging-induced adventitious root formation in cucumber is regulated by ethylene and auxin through reactive oxygen species signalling. Plant Cell Environ 2019, 42, 1458–1470. 10.1111/pce.13504 [DOI] [PubMed] [Google Scholar]
- 4.Nguyen T.-N.; Tuan P.A.; Mukherjee S.; Son S.; Ayele B.T. Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat. J Exp Bot 2018, 69, 4065–4082. 10.1093/jxb/ery190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellini C.; Pacurar D.I.; Perrone I. Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 2014, 65, 639–666. 10.1146/annurev-arplant-050213-035645 [DOI] [PubMed] [Google Scholar]
- 6.Lei C.; Fan S.; Li K.; Meng Y.; Mao J.; Han M.;et al. iTRAQ-based proteomic analysis reveals potential regulation networks of IBA-induced adventitious root formation in apple. Int J Mol Sci 2018, 19, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K.; Liang Y.; Xing L.; Mao J.; Liu Z.; Dong F.; et al. Transcriptome analysis reveals multiple hormones, wounding and sugar signaling pathways mediate adventitious root formation in apple rootstock. Int J Mol Sci 2018, 19, 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K.; Liu Z.; Xing L.; Wei Y.; Mao J.; Meng Y.; et al. miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol Biochem 2019, 139, 66–81. 10.1016/j.plaphy.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 9.Li F.; Sun C.; Li X.; Yu X.; Luo C.; Shen Y.; et al. The effect of graphene oxide on adventitious root formation and growth in apple. Plant Physiol Biochem 2018, 129, 122–129. 10.1016/j.plaphy.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 10.Mao J.; Zhang D.; Meng Y.; Li K.; Wang H.; Han M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol Plant 2019, 166, 663–676. 10.1111/ppl.12817 [DOI] [PubMed] [Google Scholar]
- 11.Zhang W.; Fan J.; Tan Q.; Zhao M.; Zhou T.; Cao F. The effects of exogenous hormones on rooting process and the activities of key enzymes of Malus hupehensis stem cuttings. PLoS One 2017, 12, e0172320–e0172320. 10.1371/journal.pone.0172320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naija S.; Elloumi N.; Jbir N.; Ammar S.; Kevers C. Anatomical and biochemical changes during adventitious rooting of apple rootstocks MM 106 cultured in vitro. C R Biol 2008, 331, 518–525. 10.1016/j.crvi.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 13.You C.-X.; Zhao Q.; Wang X.-F.; Xie X.-B.; Feng X.-M.; Zhao L.-L.; et al. A dsRNA-binding protein MdDRB1 associated with miRNA biogenesis modifies adventitious rooting and tree architecture in apple. Plant Biotechnol J 2014, 12, 183–192. 10.1111/pbi.12125 [DOI] [PubMed] [Google Scholar]
- 14.Osmont K.S.; Sibout R.; Hardtke C.S. Hidden branches: developments in root system architecture. Annu Rev Plant Biol 2007, 58, 93–113. 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- 15.Malamy J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 2005, 28, 67–77. 10.1111/j.1365-3040.2005.01306.x [DOI] [PubMed] [Google Scholar]
- 16.Steffens B.; Rasmussen A. The physiology of adventitious roots. Plant Physiol 2016, 170, 603–617. 10.1104/pp.15.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y.; Wang C.; Wang N.; Wei L.; Li W.; Yao Y.; et al. Roles of small-molecule compounds in plant adventitious root development. Biomolecules 2019, 9, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S.; Xue L.; Xu S.; Feng H.; An L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regulation 2007, 52, 173–180. [Google Scholar]
- 19.Liao W.; Huang G.; Yu J.; Zhang M.; Shi X. Nitric oxide and hydrogen peroxide are involved in indole-3-butyric acid-induced adventitious root development in marigold. J Hortic Sci Biotechnol 2011, 86, 159–165. [Google Scholar]
- 20.Liao W.-B.; Xiao H.-L.; Zhang M.-L. Effect of nitric oxide and hydrogen peroxide on adventitious root development from cuttings of ground-cover Chrysanthemum and associated biochemical changes. J Plant Growth Regul 2010, 29, 338–348. [Google Scholar]
- 21.Liao W.; Zhang M.; Huang G.; Yu J. Ca2+ and CaM are involved in NO- and H2O2-induced adventitious root development in marigold. J Plant Growth Regul 2012, 31, 253–264. [Google Scholar]
- 22.Anderson A.; Bothwell J.H.; Laohavisit A.; Smith A.G.; Davies J.M. NOX or not? Evidence for algal NADPH oxidases. Trends Plant Sci 2011, 16, 579–581. 10.1016/j.tplants.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N.; Miller G.; Morales J.; Shulaev V.; Torres M.A.; Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 2011, 14, 691–699. 10.1016/j.pbi.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 24.Torres M.A.; Dangl J.L.; Jones J.D.G. Arabidopsis gp91phox homologues AtRBOHD and AtRBOHF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A 2002, 99, 517–522. 10.1073/pnas.012452499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak J.M.; Mori I.C.; Pei Z.-M.; Leonhardt N.; Torres M.A.; Dangl J.L.; et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 2003, 22, 2623–2633. 10.1093/emboj/cdg277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller K.; Carstens A.C.; Linkies A.; Torres M.A.; Leubner-Metzger G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 2009, 184, 885–897. 10.1111/j.1469-8137.2009.03005.x [DOI] [PubMed] [Google Scholar]
- 27.Mangano S.; Denita-Juarez S. P.; Choi H. S.; Marzol E.; Hwang Y.; Ranocha P.; et al. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci U S A 2017, 114, 20, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E.; Kim Y.; Hong W.; Lee C.; Jeon J.; Jung K. Genome-wide analysis of root hair preferred RBOH genes suggests that three RBOH genes are associated with auxin-mediated root hair development in rice. J. Plant Biol 2019, 62, 229–238. [Google Scholar]
- 29.Tsukagoshi H.; Busch W.; Benfey P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 2010, 143, 606–616. 10.1016/j.cell.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 30.Dunand C.; Crèvecoeur M.; Penel C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 2007, 174, 2, 332–341. 10.1111/j.1469-8137.2007.01995.x [DOI] [PubMed] [Google Scholar]
- 31.Yamauchi T.; Yoshioka M.; Fukazawa A.; Mori H.; Nishizawa N. K.; Tsutsumi N.; et al. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 4, 775–790. 10.1105/tpc.16.00976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller K.; Linkies A.; Leubner-Metzger G.; Kermode A. R. Role of a respiratory burst oxidase of Lepidium sativum (cress) seedlings in root development and auxin signalling. J Exp Bot 2012, 63, 18, 6325–6334. 10.1093/jxb/ers284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orman-Ligeza B.; Parizot B.; de Rycke R.; Fernandez A.; Himschoot E.; Van Breusegem F.; et al. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 18, 3328–3339. 10.1242/dev.136465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montiel J.; Arthikala M. K.; Quinto C. Phaseolus vulgaris RbohB functions in lateral root development. Plant Signal Behav 2013, 8, 1, e22694 10.4161/psb.22694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S.; Stecher G.; Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lescot M.; Dehais P.; Thijs G.; Marchal K.; Moreau Y.; Van de Peer Y.; et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002, 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao Z.; Ji N.; Zhang X.; Zhang Y.; Wang Y.; Wu T.; et al. The lose of juvenility elicits adventitious rooting recalcitrance in apple rootstocks. Plant Cell, Tissue Organ Cult. 2014, 119, 51–63. [Google Scholar]
- 38.Livak K.J.; Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 39.Li D.; Wu D.; Li S.; Dai Y.; Cao Y. Evolutionary and functional analysis of the plant-specific NADPH oxidase gene family in Brassica rapa L. R Soc Open Sci 2019, 6, 181727–181727. 10.1098/rsos.181727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navathe S.; Singh S.; Singh V.K.; Chand R.; Mishra V.K.; Joshi A.K. Genome-wide mining of respiratory burst homologs and its expression in response to biotic and abiotic stresses in Triticum aestivum. Genes Genomics 2019, 41, 1027–1043. 10.1007/s13258-019-00821-x [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y.; Zou Z. Genomics analysis of genes encoding respiratory burst oxidase homologs (RBOHs) in jatropha and the comparison with castor bean. PeerJ 2019, 7, e7263–e7263. 10.7717/peerj.7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y.; Li Y.; He Y.; Hu W.; Zhang Y.; Wang X.; et al. Identification of NADPH oxidase family members associated with cold stress in strawberry. FEBS Open Bio 2018, 8, 593–605. 10.1002/2211-5463.12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu-Puga Á.; González-Gordo S.; Rodríguez-Ruiz M.; Palma J.M.; Corpas F.J. NADPH Oxidase (Rboh) activity is up regulated during sweet pepper (Capsicum annuum L.) fruit ripening. Antioxidants (Basel) 2019, 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X.; Li G.; Manzoor M.A.; Wang H.; Abdullah M.; Su X.; et al. In silico genome-wide analysis of respiratory burst oxidase homolog (RBOH) family genes in five fruit-producing trees, and potential functional analysis on lignification of stone cells in chinese white pear. Cells 2019, 8, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holub E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2001, 2, 516–527. 10.1038/35080508 [DOI] [PubMed] [Google Scholar]
- 46.Lambeth J.D. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 2004, 4, 181–189. 10.1038/nri1312 [DOI] [PubMed] [Google Scholar]
- 47.Singh R.; Singh S.; Parihar P.; Mishra R.K.; Tripathi D.K.; Singh V.P.; et al. Reactive oxygen species (ROS): beneficial companions of plants' developmental processes. Front Plant Sci 2016, 7, 1299–1299. 10.3389/fpls.2016.01299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres M.A.; Dangl J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 2005, 8, 397–403. 10.1016/j.pbi.2005.05.014 [DOI] [PubMed] [Google Scholar]
- 49.Foreman J.; Demidchik V.; Bothwell J.H.F.; Mylona P.; Miedema H.; Torres M.A.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. 10.1038/nature01485 [DOI] [PubMed] [Google Scholar]
- 50.Choi W.-G.; Miller G.; Wallace I.; Harper J.; Mittler R.; Gilroy S. Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J 2017, 90, 698–707. 10.1111/tpj.13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marino D.; Dunand C.; Puppo A.; Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci 2012, 17, 9–15. 10.1016/j.tplants.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 52.Sagi M.; Fluhr R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 2006, 141, 336–340. 10.1104/pp.106.078089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasco R.; Zharkikh A.; Affourtit J.; Dhingra A.; Cestaro A.; Kalyanaraman A.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 2010, 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- 54.Han Y.; Zheng D.; Vimolmangkang S.; Khan M.A.; Beever J.E.; Korban S.S. Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. J Exp Bot 2011, 62, 5117–5130 10.1093/jxb/err215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffens B.; Sauter M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 2009, 21, 184–196. 10.1105/tpc.108.061887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schopfer P. Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J 2001, 28, 679–688. 10.1046/j.1365-313x.2001.01187.x [DOI] [PubMed] [Google Scholar]
- 57.Joo J.H.; Bae Y.S.; Lee J.S. Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol 2001, 126, 1055–1060. 10.1104/pp.126.3.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heyman J.; Cools T.; Vandenbussche F.; Heyndrickx K.S.; Van Leene J.; Vercauteren I.; et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 2013, 342, 860–863. 10.1126/science.1240667 [DOI] [PubMed] [Google Scholar]
- 59.Li C.; Bian B.; Gong T.; Liao W. Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. J Plant Physiol 2018, 229, 185–194. 10.1016/j.jplph.2018.07.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP was fused to the N terminus of MdRBOHs (MdRBOH-E1-GFP, MdRBOH-F1/F2-GFP and MdRBOH-J-GFP). The fluorescence signal of N.benthamiana leaves was detected by confocal microscope 72 hours after infiltration.
(TIF)
(TIF)
Transcript levels were measured by qRT-PCR using apple EF1α as the reference gene. Data are means ± SD of three biological replicates. The statistical analysis was conducted using Duncan’s multiple range test (P < 0.05). The different lowercase letters indicate significant differences.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.