ABSTRACT
Selective intrauterine growth restriction (sIUGR) is a severe complication in monochorionic (MC) twin pregnancies, and it carries increased risks of poor prognosis. Current data suggest that vascular anastomoses and unequal placental sharing may be the key contributor to discordant foetal growth. While MC twins derive from a single zygote and have almost identical genetic information, the precise mechanisms remain unknown. DNA hydroxymethylation is a newly discovered epigenetic feature associated with gene regulation and modification. Here, we investigate discordant hydroxymethylation patterns between two placental shares of sIUGR and analyse the potential role of aberrant hydroxymethylation of angiopoietin-like 4 (ANGPTL4) in placental dysplasia. Hydroxymethylation DNA immunoprecipitation (hMeDIP)-chip and mRNA sequencing were performed to identify hydroxymethylation-associated genes. Real-time qPCR, western blotting, and immunohistochemistry were used to confirm ANGPTL4 expression. The mechanisms regulating ANGPTL4 were investigated by cell migration assay, invasion assay, viability assay, and apoptotic ratio assays, western blotting and hMeDIP-qPCR. Decreased ANGPTL4 was detected in the smaller placental shares of sIUGR. ANGPTL4 knockdown suppressed trophoblast invasiveness and migration, which possibly occurred through hypoxia inducible factor 1α (HIF-1α) and HIF-1 signalling pathway. Hypoxia leads to aberrant expression of ANGPTL4 and HIF-1α, positively correlated with their aberrant hydroxymethylation levels in promoter regions. Aberrant hydroxymethylation of ANGPTL4 may contribute to placental impairment by the HIF-1 signalling pathway in smaller placental shares of sIUGR.
KEYWORDS: Selective intrauterine growth restriction, DNA hydroxymethylation, hypoxia, angiopoietin-like 4, placenta
Introduction
Selective intrauterine growth restriction (sIUGR) affects approximately 10–15% of monochorionic (MC) twin pregnancies, and it is associated with an increased risk of perinatal mortality and morbidity [1]. Although previous studies have reported that the pathogenesis of sIUGR relies on discordant placental sharing and vascular anastomoses, the underlying molecular mechanisms remain obscure [2]. MC twins, which derive from a single zygote, harbour almost identical genetic information and share a similar intrauterine environment, so epigenetic events may be involved in these mechanisms [3].
DNA hydroxymethylation is a stable epigenetic modification that plays unique regulatory roles in various physiopathological processes [4]. 5-Hydroxymethylcytosine (5 hmC, the marker of hydroxymethylation) is distributed in various different tumours or different development stages, suggesting that it may have key roles in different cancers and disorders [5–7]. The distribution of 5 hmC can be found at the enhancer, promoter, transcription start site (TSS), gene body, 3ʹUTR or intragenic region, so the function and mechanism of 5 hmC need to be further investigated [8].
The placenta is a temporary organ that plays a critical role in sustaining pregnancy and foetal growth. Placental dysplasia is considered to be a central feature of pregnancy-associated disorders such as preeclampsia (PE) or foetal growth restriction (FGR) [9,10]. Accumulating evidence has shown that epigenetic factors such as microRNAs, imprinted genes or DNA methylation may take part in placental impairment [2,11,12]. In contrast to singleton pregnancy, sIUGR twins are considered an optimal model for exploring epigenetic molecular mechanisms due to the exclusion of effects from different genetic backgrounds among individuals.
Our preliminary data suggested that the level of genome‐wide DNA hydroxymethylation was significantly decreased in smaller placental shares compared to the larger ones in sIUGR, as shown by ultra‐performance liquid chromatography/tandem mass spectrometry (UPLC‐MS/MS) technology [2]. Thus, we hypothesized that discordant DNA hydroxymethylation statuses might be important for placental dysfunction and growth discordance between two foetuses of sIUGR.
Angiopoietin-like 4 (ANGPTL4) is a hypoxia responsive gene regulating vascular permeability, angiogenesis, and inflammation [13]. ANGPTL4 is also a downstream target of hypoxia inducible factor 1α (HIF-1α), and it is regulated by HIF-1α in various cell types [14]. However, few studies have reported the regulatory relationship of DNA hydroxymethylation between HIF-1α and ANGPTL4 in placental insufficiency and foetal growth. In this study, we first identified differential hydroxymethylation patterns between two placental shares of sIUGR and then tried to reveal and validate an epigenetic-mediated mechanism mainly focused on hypoxia-induced aberrant hydroxymethylation of ANGPTL4 and HIF-1α. The results may highlight a potential mechanism between hydroxymethylation modification and discordant foetal growth or even placenta-associated disorders.
Results
Clinical data
Clinical characteristics are summarized in Table 1. Both the smaller and larger foetal weights were significantly lower in the sIUGR group than in the normal group (p = 0.000 < 0.01). The intertwin difference in birth weight was significantly larger in the sIUGR group than in the normal group (p = 0.000 < 0.01). The gestational age of delivery was much earlier in the sIUGR group than in the normal group (p = 0.000 < 0.01). No significant difference was found between the maternal age in the sIUGR and normal groups (p = 0.995 > 0.05).
Table 1.
Characteristics of sIUGR and normal MCDA twin pregnancies.
| sIUGR(n = 13) | Normal(n = 18) | p value | |
|---|---|---|---|
| Maternal age (years) | 31.85 ± 5.37 | 31.83 ± 5.39 | 0.995 |
| Gestational age (weeks) | 32.58 ± 2.19 | 36.02 ± 1.17 | 0.000** |
| Birth weight of larger twin (kg) | 1.83 ± 0.34 | 2.47 ± 0.27 | 0.000** |
| Birth weight of smaller twin (kg) | 1.22 ± 0.30 | 2.33 ± 0.30 | 0.000** |
| Birth weight discordance (%) | 33.74%±8.50% | 5.71%±3.68% | 0.000** |
Data are shown as the mean ± SD. **p < 0.01
sIUGR: selective intrauterine growth restriction. Normal: normal MCDA twins.
Distributions of DhMRs and identifying hydroxymethylation-associated genes
Hydroxymethylation DNA immunoprecipitation (hMeDIP)-chip and mRNA sequencing (mRNA-seq) were performed to examine the differential distributions of 5hmC and hydroxymethylation-associated genes between two placental shares in four sIUGR twin pairs and four normal MCDA twin pairs (Figure 1(a,b)). Compared with the normal group, 181 upregulated genes and 614 downregulated genes were identified in smaller placental shares of the sIUGR group (Figure 1(c), supplement figure S1). Furthermore, we subdivided promoter regions into three classes based on the CpG ratio, GC content, and length of the CpG-rich region as previously reported: high CpG-density promoters (HCPs), low CpG-density promoters (LCPs) and intermediate CpG-density promoters (ICPs) [15]. Compared with the normal group, there were 729 differential hydroxymethylation regions (DhMRs) in HCPs, 239 DhMRs in ICPs and 198 DhMRs in LCPs identified in the sIUGR group (supplement Table S1). Combination analysis of Gene Ontology (GO) term from hMeDIP-chip data and mRNA-seq data were used to determine possible hydroxymethylation-associated genes (Figure S3, supplement Table S2).
Figure 1.
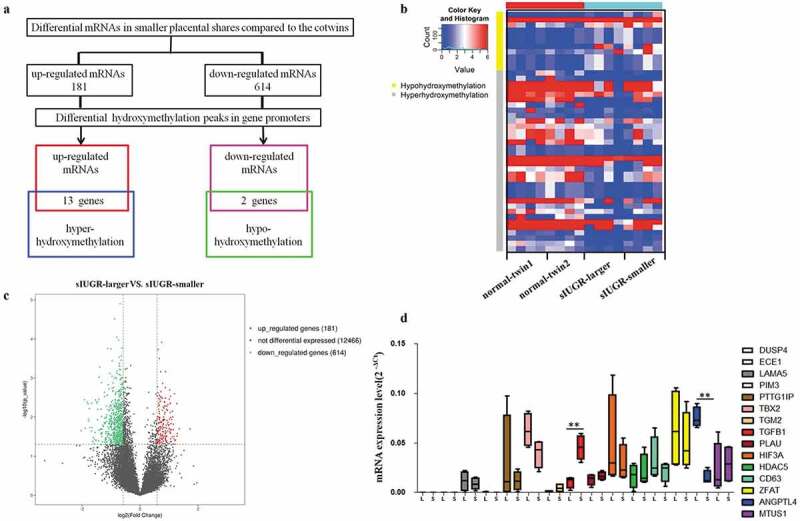
Hydromxymethylation-associated genes detected in sIUGR twin pairs through hMeDIP-chip and mRNA sequencing. (a) Schematic representation for identifying hydroxymethylation-associated genes. The putative targets obtained by overlapping the differentially expressed mRNAs and differentially hydroxymethylation peaks in promoter region. (b) The differentially expressed mRNAs and differentially hydroxymethylation peaks between two placental shares in sIUGR group and normal MCDA group shown on the heatmap. (c) The differentially expressed mRNAs between larger and smaller placental shares of sIUGR plotted in the volcano plot. One hundred eighty-one upregulated genes and 614 downregulated genes identified in smaller placental shares of the sIUGR group. (d) Expression of hydroxymethylation-regulated associated genes detected in the smaller placental shares compared to the larger ones in sIUGR twin pairs by quantitative RT-PCR.
Fifteen differentially hydroxymethylation-associated genes were found, including 13 upregulated mRNAs associated with high 5 hmC enrichment and 2 downregulated mRNAs associated with low 5 hmC enrichment (Figure 1(a)). Furthermore, Reverse transcription-qPCR(RT-qPCR) was performed to validate the expression of these genes in each of the placental shares from the sIUGR group and the normal group. Among these genes, ANGPTL4 expression was significantly reduced in smaller placental shares of the sIUGR group (Figure 1(d)). No significant difference in ANGPTL4 expression was identified between the two placental shares of the normal group (supplement figure S2).
Hydroxymethylation-associated ANGPTL4 is decreased in smaller placental shares of sIUGR
RT-qPCR and western blotting were used to further verify the expression of ANGPTL4 in the placentas of sIUGR and the normal group. The results showed that the mRNA and protein expression levels of ANGPTL4 were both decreased in the smaller placental shares compared to the larger ones in the sIUGR group, but no differences were observed between the two placental shares of the normal group (Figure 2(a,b)). Immunohistochemistry (IHC) analysis of placental villous tissues was performed to investigate the localization of ANGPTL4 in placental villous tissue. A weaker positive signal for ANGPTL4 was detected in syncytiotrophoblast (SCT) from the smaller placental shares compared to what was observed in the larger ones (Figure 2(c)) in the sIUGR group. In contrast, similar ANGPTL4 expression was observed in SCT in the two placental shares of the normal group (Figure 2(d)).
Figure 2.
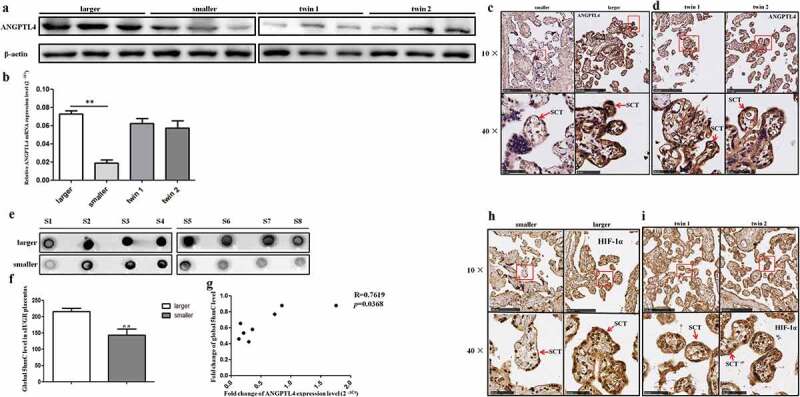
Hydromxymethylation-associated ANGPTL4 identified in sIUGR twin pairs. (a, b) Decreased ANGPTL4 detected in smaller placental shares of the sIUGR group compared to the normal MCDA group by western blotting and quantitative RT-PCR. (**p < 0.01). (c, d) Single staining of maternal villi from sIUGR (smaller & larger) and normal MCDA group (twin1 & twin2) using ANGPTL4 antibody was visualized. Above scale bar = 250 μm. Below scale bar = 50 μm. (e, f) Global 5hmC levels detected in two placental shares of sIUGR group by dot blot. S1-8 was eight pairs of sIUGR placentas (smaller & larger). (g) Correlation analysis of ANGPTL4 expression with global 5hmC level in smaller placental shares compared to larger ones in the sIUGR group. (h, i) Single staining of maternal villi from sIUGR (smaller & larger) and normal MCDA group (twin1 & twin2) using HIF-1α antibody was visualized. Above scale bar = 250 μm. Below scale bar = 50 μm. Compared to the normal MCDA group, weaker positive signal for ANGPTL4 and HIF-1α both detected in smaller placental shares than larger ones in the sIUGR group.
In addition, the global 5 hmC level was measured in eight pairs of placental tissues in the sIUGR group by dot blot and was correlated with the level of ANGPTL4 mRNA expression (p = 0.037 < 0.05) (Figure 2(e–g)). These data suggested that decreased ANGPTL4 expression may be associated with hypo-hydroxymethylation levels in the smaller placental shares of sIUGR.
We also examined the expression and localization of HIF-1α by IHC. As shown in Figure 2(h,i), the signal for HIF-1α was much weaker in SCT from the smaller placental shares than it was in the larger placental shares of the sIUGR group, while no difference was found between two placental shares of the normal group (Figure 2(h,i)).
Reduced ANGPTL4 suppresses trophoblast invasion and migration
To address the role of ANGPTL4 in human trophoblasts, the HTR8/SVneo cell line (HTR8) was transfected with ANGPTL4 siRNA. RT-qPCR results confirmed that ANGPTL4 expression was decreased after siRNA transfection, and siRNA3 was the most effective siRNA (Figure 3(a)). Subsequently, ANGPTL4 siRNA3 was used directly to reduce ANGPTL4 expression in HTR8 cells.
Figure 3.
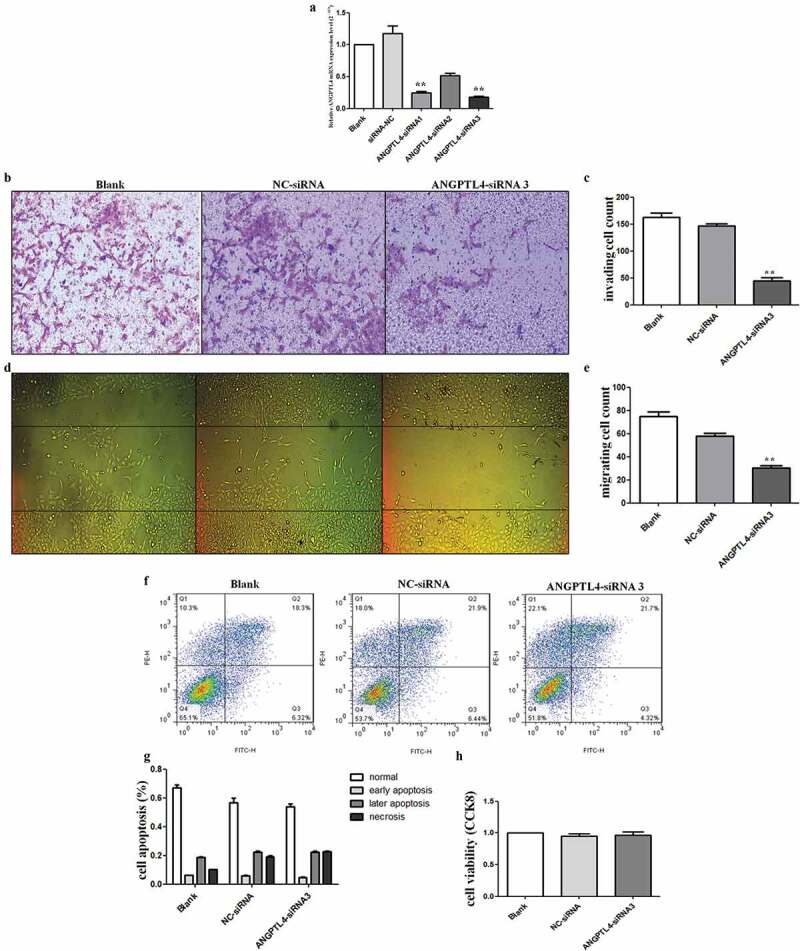
Decreased ANGPTL4 directly inhibited trophoblast invasion and migration in vitro. (a) The expression of ANGPTL4 when using ANGPTL4 siRNA. Decreased ANGPTL4 expression detected when using three siRNAs, of which siRNA3 was the most effective. (b, c) Transwell assays showed that invaded cells reduced when using siRNA3. (**p < 0.01). (d, e) Wound healing assays shown that migrated cells decreased when using siRNA3. (**p < 0.01). (f, g) No significant difference in the apoptopsis and necrosis detected when using siRNA3 by Flow cytometry (FCM). (h) No significant difference in cell viability identified when using siRNA3 by CCK8 assay.
Liu et al have reported that silenced ANGPTL4 weakened trophoblast cells migration and invasion [16]. Consistent with Liu et al results, our data showed that reduced ANGPTL4 drastically inhibited the invasive and migration abilities of HTR8 cells when compared with blank and negative control-siRNA (NC) cells (Figure 3(b–e)). However, HTR8 cells transfected with the siRNA exhibited no significant changes in cell viability and apoptosis (Figure 3(f–h)). These results indicated that decreased ANGPTL4 expression suppressed trophoblast invasiveness and migration.
ANGPTL4 is the downstream target of HIF-1α and is regulated by the HIF-1 signalling pathway
Previous data have suggested that a hypoxic environment might be related to placental dysfunction. HIF-1α, as a hypoxia-induced factor, is also an upstream regulator of ANGPTL4. Moreover, IHC data revealed that the localization and expression of HIF-1α was positively correlated with ANGPTL4 (Figure 2(c,d,h,i)). We next investigated the regulatory relationship between HIF-1α and ANGPTL4, and we determined the underlying mechanisms causing decreased ANGPTL4 to effect trophoblast functions by using a HIF-1 agonist (DMOG), a HIF-1 inhibitor (digoxin), and a HIF-1α siRNA.
Western blot results showed that HIF-1α and ANGPTL4 were both highly expressed when using DMOG, and they exhibited low expression levels when using digoxin regardless of normoxic or hypoxic conditions (Figure 4(a–d)). In addition, HIF-1α and ANGPTL4 were also decreased when using HIF-1α siRNA compared with blank or NC-siRNA (Figure 4(e,f)).
Figure 4.
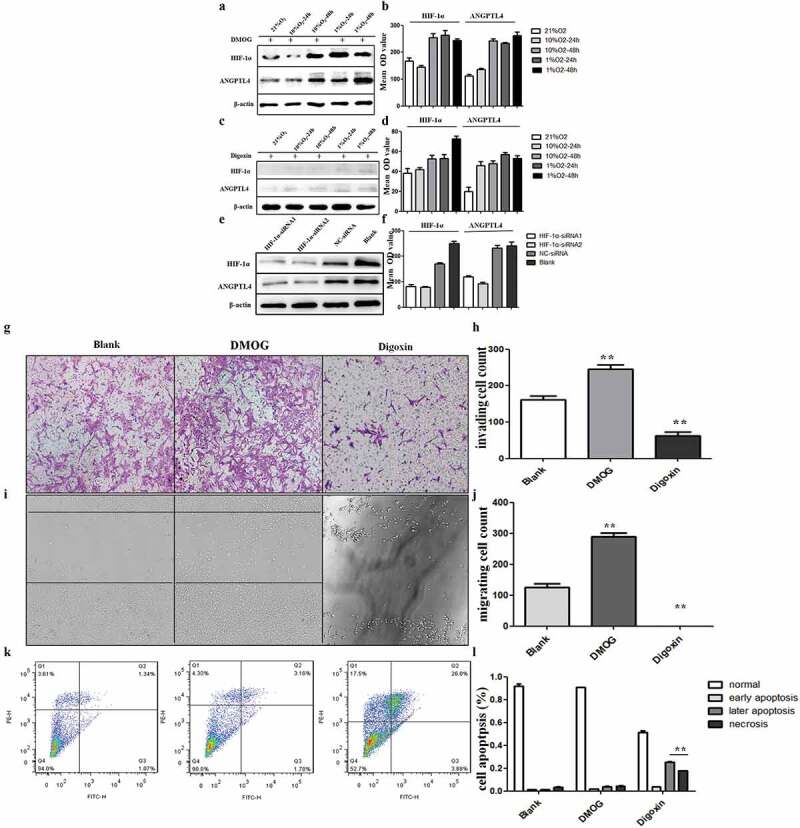
ANGPTL4 is a downstream target of HIF-1α and regulated by HIF-1 signalling pathway in vitro. (a–f) Western blot analysis of HIF-1α and ANGPTL4 expression in HTR8 cells with HIF-1 agonist (DMOG), HIF-1 inhibitor (digoxin) and HIF-1α siRNA for 48 hours. Expressions of HIF-1α and ANGPTL4 both increased when using DMOG, while decreased by using digoxin or HIF-1α siRNA. (g, h) Transwell assays showed that invaded cells increased in DMOG group, and reduced in digoxin group. (**p < 0.01). (i, j) Wound healing assays shown that migrated cells increased in DMOG group, and reduced in digoxin group. (**p < 0.01). (k, l) FCM results showed that late apoptopsis and necrosis of HTR8 cells increased significantly when using digoxin. (**p < 0.01).
We next explored the effects of the aberrant HIF-1 signalling pathway on trophoblast functions. The results revealed that migration and invasive abilities were both enhanced by DMOG, whereas the migration and invasive capacities were weakened (Figure 4(g–j)) by digoxin. Moreover, late apoptosis and necrosis ratios in trophoblasts were significantly increased when using digoxin compared to the ratios of blank trophoblasts (Figure 4(k,l)). Taken together, ANGPTL4 expression may be regulated by the HIF-1 signalling pathway in normoxic or hypoxic conditions. Aberrant HIF-1 signalling pathway-affected trophoblast functions may target ANGPTL4.
Hypoxia leads to aberrant expression of ANGPTL4 and HIF-1α, positively correlated with their aberrant hydroxymethylation levels in promoter regions
The above results have demonstrated that aberrant HIF-1α-regulated ANGPTL4 may affect trophoblast functions. We next constructed a hypoxic trophoblastic model in vitro and further explored the relationship between the expression of ANGPTL4 and HIF-1α and their 5hmC levels in promoter regions by hMeDIP-qPCR and western blotting.
Compared to the normoxic conditions (21% O2), the expression levels of ANGPTL4 and HIF-1α were both upregulated in 10% O2-48 hr and downregulated in 1% O2-48 hr (p < 0.05) (Figure 5(a,b)). Meanwhile, the 5 hmC levels of ANGPTL4 and HIF-1α were also increased in 10% O2-48 hr and decreased in 1% O2-48 hr (p < 0.05) (Figure 5(c,d)). These data suggested that the 5 hmC levels of ANGPTL4 and HIF-1α were positively correlated with their expression in 10% O2-48 hr and 1% O2-48 hr; aberrant ANGPTL4 and HIF-1α expression may be related to their aberrant 5 hmC levels of promoter regions in hypoxic conditions.
Figure 5.
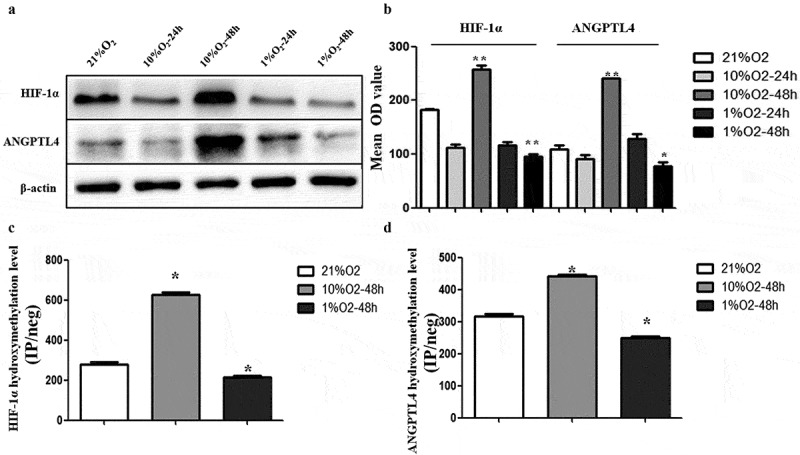
Hypoxia leads to aberrant expression of ANGPTL4 and HIF-1α, positively correlated with their aberrant hydroxymethylation levels in promoter regions. (a, b) Western blot results showed the protein levels of HIF-1α and ANGPTL4 under normoxic conditions with 21%O2 and under hypoxic conditions with 10% and 1% O2 for 24 hours and 48 hours. Expression of HIF-1α and ANGPTL4 both highly expressed in 10%O2-48 hours and reduced expressed in 1%O2-48 hours. (c, d) The 5hmC levels in the promoters of HIF-1α and ANGPTL4 under normoxic and hypoxic conditions were detected by hMeDIP-qPCR. The percentage of the input is shown as indicated. The results showed that the 5hmC enrichment of HIF-1α and ANGPTL4 increased in 10%O2-48 hours and reduced in 1%O2-48 hours. (*p < 0.05, **p < 0.01).
Discussion
In MC twins with sIUGR, vascular anastomoses and unequal placental territories are thought to be the important reasons for discordant foetal growth and different clinical outcomes [17]. The presence of placental anastomoses and discordant placental shares between two foetuses may interfere with the natural progression of the smaller foetus, leading to the failure to achieve its growth potential. However, the underlying molecular mechanism of sIUGR remains poorly understood. Because almost all MC twins were monozygotic twins, both foetuses of a twin pair harboured identical genomic DNA and grew in a similar intrauterine environment [18]. It is a good model for evaluating the epigenetic-modified mechanism in foetal development and placental dysfunction.
DNA hydroxymethylation is reported to be an important epigenetic modification in different mammalian cells that contributes to the regulation of genomic structure and function [19,20]. In singleton pregnancy, altered 5 hmC levels have been observed in women who are obese, have PE or have gestational diabetes mellitus (GDM) [21–23]. Cardenas et al [24] reported that prenatal mercury exposure was related to decreased 5 hmC levels in cord blood. These results demonstrated that maternal complications or adverse intrauterine environments may influence 5 hmC levels during foetal development. In our study, decreased global 5 hmC levels were identified in smaller placental shares compared to larger ones in cases of sIUGR. Furthermore, a possible hydroxymethylation-associated gene, ANGPTL4, was identified and verified in sIUGR placentas and hypoxic trophoblasts in vitro. This finding suggests that reduced 5 hmC levels and 5 hmC-associated ANGPTL4 may be related to discordant foetal development and placental dysplasia.
The placenta is a mediator of communication between foetal development and maternal environment. The placenta regulates maternal-foetal nutrient and waste exchange, facilitates interactions with the maternal immune system, and acts as a neuroendocrine organ for developing foetuses [25]. Trophoblasts are the main component of the placenta, dysfunction of which will lead to placental dysplasia. In singleton pregnancy, ANGPTL4 repression regulated by PVT1 may play an essential role in PE by affecting trophoblast functions [26]. ANGPTL4 may also be involved in fatty acid-induced angiogenesis in vitro [27]. However, a discrepancy among different studies was found, in that ANGPTL4 has been reported to have a role in angiogenesis, anti-angiogenesis or even pathological angiogenesis [28–30]. In our data, reduced ANGPTL4 significantly inhibited the invasiveness and migration of trophoblasts, whereas no effect was found in cell viability and apoptosis.
Different environmental factors, which have the potential to alter placental epigenomic profiles, might contribute to placental dysplasia [31]. Environmental oxygen is a potent driver of vascular development in the maternal-foetal interface [32]. Insufficient adaptive responses to too much or too little oxygen may lead to cellular injury [33]. HIF-1α is an important hypoxia-induced factor, and it was confirmed to regulate proliferation, apoptosis and tolerance to hypoxia [34]. Previous studies have reported that HIF-1α was associated with the pathology of PE and FGR [35,36]. Hypoxia-induced HIF-1α promoted the invasion of trophoblast cells in vitro [37]. ANGPTL4, as a downstream target of HIF-1α, has been verified to be regulated by HIF-1α in osteosarcoma cells [14]. Our data showed that the expression and localization of ANGPTL4 were positively related to HIF-1α in the smaller placental shares of sIUGR. In addition, HIF-1α knockdown can directly downregulate the expression of HIF-1α and ANGPTL4 in vitro. The expression of HIF-1α and ANGPTL4 and trophoblast functions were both affected when using a HIF-1 agonist and inhibitor. Therefore, multiple lines of evidence support the role of HIF-1α in affecting trophoblast functions via regulating ANGPTL4 expression. Interestingly, treatment with an HIF-1 inhibitor induced trophoblast apoptosis and necrosis, which was not observed following ANGPTL4 knockdown. It is possible that HIF-1α affects different cell functions via different downstream targets. In fact, HIF-1α induces more than 100 target genes to adapt to hypoxic conditions by different regulatory mechanisms [38].
Furthermore, we verified the possible correlation between 5hmC enrichment and the expression changes of ANGPTL4 and HIF-1α under different hypoxic conditions. Previous studies have confirmed positive relationships between the levels of 5 hmC and gene expression in the brain, chondrocytes and embryonic stem cells [39–41]. Consistent with previous data, our results show that 5 hmC levels and expression of HIF-1α/ANGPTL4 were both increased in 10% O2-48 hr and decreased in 1% O2-48 hr, which means that the 5 hmC levels in promoter regions were positively related to gene expression in different oxygen concentrations. These results suggest that hypoxia-induced aberrant expression of ANGPTL4 and HIF-1α may be related to their aberrant 5 hmC levels in promoter regions, which contribute to trophoblast dysfunction and placental dysplasia and further result in disrupted foetal development.
There is a limitation in this study in that we have only one single time point in every placental sample, which is the time for delivery in gestational weeks. In fact, the placental expression profiles and the dynamic regulation of cytosine are known to vary at different trimesters [25]. This suggests that the relationship between 5 hmC and gene expression may be complicated and unique in different gestational weeks.
In summary, this study first demonstrated that the global 5hmC level was decreased in smaller placental shares compared to larger ones in cases of sIUGR. Reduced hydroxymethylation-associated ANGPTL4 was validated in placentas and trophoblasts in vitro. Hypoxia-induced aberrant expression of ANGPTL4 and HIF-1α may be related to their aberrant 5hmC levels in promoter regions, which contribute to trophoblast dysfunction. Therefore, our findings revealed that aberrant hydroxymethylation-associated ANGPTL4 may be involved in the pathogenesis of sIUGR and may further provide a new epigenetic mechanism in which discordant DNA hydroxymethylation may be associated with phenotypic discrepancies between monozygotic twin foetuses.
Materials and methods
Patients and placental sample collection
From December 2015 to December 2017, a total of 31 MCDA twin pregnancies were enrolled, including 13 pairs of sIUGR twins (sIUGR group) and 18 pairs of normal MCDA twins (normal group). The study was approved by the ethics committee in the hospital. The collection of placental tissues was performed with the patients’ informed consent.
The chorionicity was determined by ultrasound at 11–14 weeks as previously described [42], and it was confirmed by placental examination after delivery [43]. sIUGR was defined when the birth weight of one foetus was below the 10th percentile, and the intertwin discordance was greater than 25% [44]. The normal group included normal MCDA twin pregnancies without sIUGR or other complications. Pregnancies with severe maternal complications, twin to twin transfusion syndrome (TTTS), twin-anaemia polycythaemia sequence (TAPS), structural defects and foetal death were excluded.
In sIUGR twin pregnancies, smaller placental shares were the placental sections excised at the region of the umbilical cord insertion point from the restricted foetus (<10th percentile), while larger placental shares were the placental sections of the co-twins.
Placental tissue pieces of approximately 2 × 2 × 1 cm3 were excised immediately after delivery by caesarean section from placentas at the region around the individual umbilical cord insertion point, rinsed with ice-cold phosphate-buffered saline (PBS) and stored appropriately for subsequent experiments.
Cell culture, reagents and transfection
HTR8/SVneo (HTR8) was obtained from American Type Culture Collection (ATCC). Hypoxia was induced in a hypoxic incubator with 1% or 10% O2 for 24 and 48 hr. DMOG (Catalogue No. A4506), a HIF-1 agonist, and digoxin (Catalogue No. B7684), a HIF-1 inhibitor, were purchased from APExBIO. DMOG and digoxin were both diluted at a concentration of 10 μmol/L and cultured with HTR8 cells for 24 and 48 hr. ANGPTL4 and HIF-1α knockdown was achieved using small interfering RNA (siRNAs; Gene Pharma) introduced via Lipofectamine 3000 (Invitrogen, Catalogue No. L3000015) according to the manufacturer’s instructions. The sequences of the oligonucleotides are listed in supplement Table S3.
hMeDIP-Chip and data processing
Four pairs of sIUGR placentas and four pairs of normal MCDA placentas were used in hMeDIP-Chip. Genomic DNA was extracted from placentas by a DNeasy Blood and Tissue Kit (Qiagen, Catalogue No. 69506) and sonicated to fragments of 200–1000 bp with a Bioruptor sonicator (Diagenode).
Differential enrichment peaks (DEP) were calculated by M’ value as follows: M’ = Average (log2 hMeDIPsmaller foetus/Inputsmaller foetus) – Average (log2 hMeDIPlarger foetus/Inputlarger foetus). Gene ontology (GO) terms and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways were also analysed via their online databases (http://www.geneontology.org/, http://www.kegg.jp/, respectively) [45].
mRNA sequencing
Four pairs of sIUGR placentas and four pairs of normal MCDA placentas were used in mRNA sequencing. Total mRNA from placentas was isolated by using a KAPA Stranded RNA-Seq Library Preparation Kit (Illumina platforms) (KAPA Biosystems, Catalogue No. KK8401). The completed libraries were qualified on an Agilent 2100 Bioanalyzer, quantified and sequenced on an Illumina HiSeq 4000 (Illumina) according to the manufacturer’s instructions.
After sequencing, base calling was carried out using Off-Line Base caller software (version V1.8.0; Illumina). The trimmed reads that passed sequence quality examination and trimmed 5ʹ,3ʹ-adaptor bases using cutadapt [46], were aligned to the reference genome by HISAT 2 software (v2.0.4) [47]. StringTie (v1.2.3) [48] and R package Ballgown (v2.6.0) [49] were used to calculate the transcript abundance and the FPKM value [50] for gene and transcript levels. The differentially expressed genes and transcripts were filtered by using the R package Ballgown.
RNA extraction and reverse transcription-qPCR validation
Thirteen pairs of placental samples from sIUGR twins and eighteen pairs of placental samples from normal MCDA twins were used to perform RT-qPCR to detect the mRNA expression of candidate genes. Total RNA was extracted using RNAiso Plus (Takara, Catalogue No. 9109) according to the manufacturer’s protocol. cDNA was synthesized from total RNA using PrimeScript RT Master Mix (TaKaRa, Catalogue No. RR036A), and then, real-time PCR was performed with SYBR Premix Ex Taq II (Takara, Catalogue No. RR820A) on an ABI 7500 Real-Time PCR System (Thermo Fisher Scientific) using the 2−ΔΔCt method. A list of primers is available in supplement Table S4.
Western blot, dot blot and immunohistochemistry
Thirteen pairs of sIUGR placentas and eighteen pairs of normal MCDA placentas were used to perform western blotting to detect the protein expression of ANGPTL4 and HIF-1α according to the standard protocol [51]. Eight pairs of sIUGR placentas and eight pairs of normal MCDA placentas were used in immunohistochemistry (IHC). Meanwhile, eight pairs of sIUGR placentas were also used in dot blot to detect the global 5hmC level in sIUGR group. Genomic DNA was extracted from placental tissues by a MiniBEST FFPE DNA Extraction Kit (TaKaRa, Catalogue No. 9782), and the concentrations were measured using a NanoDrop™ 2000 Spectrophotometer (Thermo Scientific). Primary antibodies against ANGPTL4 (1:250) (Abcam, Catalogue No. ab196746), HIF-1α (1:200) (Abcam, Catalogue No. ab82832) and 5hmC (1:500) (Abcam, Catalogue No. ab214728) were used and were then visualized by ImmobilonTM Western chemiluminescent HRP substrate (Millipore, Catalogue No. WBKLS0500). Quantification of images was performed using ImageJ software. Staining for IHC was visualized by DAB (Beyotime, Catalogue No. P0203). High-resolution digital images were acquired with a NanoZoomer S360 Slide Scanner (Hamamatsu) and were then analysed by NDP.view2 Viewing software (Hamamatsu).
hMeDIP-qPCR
A hMeDIP assay combined with qPCR was used to quantitatively evaluate the hydroxymethylation status of ANGPTL4 and HIF-1α in normoxic and hypoxic trophoblasts. hMeDIP was performed according to a previously published method [52]. The primer sequences are listed in supplement Table S5. This experiment was performed in triplicate. The relative changes in the extent of promoter hydroxymethylation were determined by calculating the amount of promoter in immunoprecipitated DNA after normalization to the input DNA: %(hMeDNA-IP/Input) = 2(Ctinput-CthMeDNA-IP) ×Input dilution factor×100%.
Cell migration analysis
Migration analysis was performed with the wound healing assay to investigate the trophoblast migration ability in vitro. Cells were seeded and treated the next day with ANGPTL4 siRNA, DMOG and digoxin. Subsequently, a wound was made in each well with the use of a sterile micropipette tip, and cells were cultured for an additional 48 hours. Photographs were taken and used for analysis by ImageJ software.
Cell invasion assay
A transwell invasion assay was used to investigate trophoblast invasion ability in vitro. A 24-transwell plate (Co-Star) was coated with 100 μl of BD Matrigel (BD Biosciences, Catalogue No. 356234). Cells were suspended in FBS-free medium, and 100 μl were transferred into the upper chamber of the transwell. The lower chamber was filled with 600 μl of medium containing 10% FBS. After incubation for 48 hours, the Matrigel was removed with a cotton swab, and invaded cells were stained with crystal violet.
Cell viability analysis
CCK8 assays were used to detect trophoblast viability (Beyotime, Catalogue No. C0038). Cells were placed in 96-well plates and treated with ANGPTL4 siRNA, DMOG or digoxin. Then, the cells were cultured with 10 μl of CCK8 reagent in the incubator for 2 hours at 37°C. Cell viability was measured by a microplate reader at a wavelength of 450 nm.
Cell apoptotic ratio assay
Cell apoptosis assays were performed by an Annexin V-FITC Cell Apoptosis kit (Beyotime, Catalogue No. C1062 L) to detect trophoblast apoptosis and necrosis ratio according to the manual instructions. In brief, cells were treated with ANGPTL4 siRNA, DMOG or digoxin, washed with PBS twice, washed with 1× binding buffer once and then suspended in 1× binding buffer. Cells were double-stained with Annexin V-FITC and PI for 15 minutes in the dark at room temperature and then analysed by flow cytometry (BD).
Statistical analysis
All statistical values were calculated using SPSS 22.0 (Chicago, IL, USA) and GraphPad Prism 5. Experimental groups were analysed by independent sample t-tests when comparing between 2 groups and one-way ANOVA when comparing among multiple groups. Correlations were analysed using Spearman’s rank correlation test. Data are presented as the mean ± SD. All p values are two-sided. A p value of < 0.05 was considered to indicate statistical significance.
Supplementary Material
Funding Statement
This work was supported by the China Postdoctoral Science Foundation under Grant [2018M643128]; President Foundation of Nanfang Hospital, Southern Medical University under Grant [2019Z005]; Science and Technology Program of Guangdong Province under Grant [2015B050501006]; and National Natural Science Foundation of China under Grant [81671464].
Disclosure Statement
The authors declare they have no competing financial interests or conflict of interest.
Supplementary Materials
Supplemental data for this article can be accessed here.
References
- [1].Rustico MA, Consonni D, Lanna M, et al. Selective intrauterine growth restriction in monochorionic twins: changing patterns in umbilical artery Doppler flow and outcomes. Ultrasound Obstet Gynecol. 2017;49(3):387–393. [DOI] [PubMed] [Google Scholar]
- [2].He Z, Lu H, Luo H, et al. The promoter methylomes of monochorionic twin placentas reveal intrauterine growth restriction-specific variations in the methylation patterns. Sci Rep. 2016;6:20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wen H, Chen L, He J, et al. MicroRNA expression profiles and networks in placentas complicated with selective intrauterine growth restriction. Mol Med Rep. 2017;16(5):6650–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shukla A, Sehgal M, Singh TR.. Hydroxymethylation and its potential implication in DNA repair system: A review and future perspectives. Gene. 2015;564(2):109–118. [DOI] [PubMed] [Google Scholar]
- [5].Zhao J, Ma XL, Ma JX, et al. TET3 mediates alterations in the epigenetic marker 5hmC and Akt pathway in steroid-associated osteonecrosis. J Bone Miner Res. 2017;32(2):319–332. [DOI] [PubMed] [Google Scholar]
- [6].Fischer AP, Miles SL. Silencing HIF-1α induces TET2 expression and augments ascorbic acid induced 5-hydroxymethylation of DNA in human metastatic melanoma cells. Biochem Biophys Res Commun. 2017;490(2):176–181. [DOI] [PubMed] [Google Scholar]
- [7].Hsieh TH, Liu YR, Chang TY, et al. Global DNA methylation analysis reveals miR-214-3p contributes to cisplatin resistance in pediatric intracranial nongerminomatous malignant germ cell tumors. Neuro Oncol. 2018;20(4):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shi DQ, Ali I, Tang J, et al. New insights into 5hmC DNA modification: generation, distribution and function. Front Genet. 2017;8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen X, Chen K, Feng Y, et al. The potential role of pregnancy-associated plasma protein-A2 in angiogenesis and development of preeclampsia. Hypertens Res. 2019;42(7):970–980. [DOI] [PubMed] [Google Scholar]
- [10].Gunatillake T, Chui A, Fitzpatrick E, et al. Decreased placental glypican expression is associated with human fetal growth restriction. Placenta. 2019;76:6–9. [DOI] [PubMed] [Google Scholar]
- [11].Li L, Huang X, He Z, et al. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J Cell Mol Med. 2019;23(6):4422–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Luo YM, Fang Q, Shi HJ, et al. Imprinting and promoter usage of insulin-like growth factor II in twin discordant placenta. Obstet Gynecol Int. 2010;2010:498574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang X, Cheng Y, Su G. A review of the multifunctionality of angiopoietin-like 4 in eye disease. Biosci Rep. 2018;38(5):BSR20180557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang T, Kastrenopoulou A, Larrouture Q, et al. Angiopoietin-like 4 promotes osteosarcoma cell proliferation and migration and stimulates osteoclastogenesis. BMC Cancer. 2018;18(1):536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang CZ, Xu JH, Zhong W, et al. Sox9 transcriptionally regulates Wnt signaling in intestinal epithelial stem cells in hypomethylated crypts in the diabetic state. Stem Cell Res Ther. 2017;8(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu L, Zhuang X, Jiang M, et al. ANGPTL4 mediates the protective role of PPARγ activators in the pathogenesis of preeclampsia. Cell Death Dis. 2017;8(9):e3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bennasar M, Eixarch E, Martinez JM, et al. Selective intrauterine growth restriction in monochorionic diamniotic twin pregnancies. Semin Fetal Neonatal Med. 2017;22(6):376–382. [DOI] [PubMed] [Google Scholar]
- [18].Xiang Z, Yang Y, Chang C, et al. The epigenetic mechanism for discordance of autoimmunity in monozygotic twins. J Autoimmun. 2017;83:43–50. [DOI] [PubMed] [Google Scholar]
- [19].Etchegaray JP, Chavez L, Huang Y, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17(5):545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Spiers H, Hannon E, Schalkwyk LC, et al. 5-hydroxymethylcytosine is highly dynamic across human fetal brain development. BMC Genomics. 2017;18(1):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mitsuya K, Parker AN, Liu L, et al. Alterations in the placental methylome with maternal obesity and evidence for metabolic regulation. PLoS One. 2017;12(10):e186115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ma M, Zhou QJ, Xiong Y, et al. Preeclampsia is associated with hypermethylation of IGF-1 promoter mediated by DNMT1. Am J Transl Res. 2018;10(1):16–39. [PMC free article] [PubMed] [Google Scholar]
- [23].Sun M, Song MM, Wei B, et al. 5-Hydroxy-methylcytosine-mediated alteration of transposon activity associated with the exposure to adverse in utero environments in human. Hum Mol Genet. 2016;25(11):2208–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cardenas A, Rifas-Shiman SL, Godderis L, et al. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ Health Perspect. 2017;125(8):87022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Green BB, Houseman EA, Johnson KC, et al. Hydroxymethylation is uniquely distributed within term placenta, and is associated with gene expression. Faseb J. 2016;30(8):2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu Y, Lian Y, Zhang Y, et al. The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. J Cell Mol Med. 2018;22(2):1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Basak S, Duttaroy AK. Effects of fatty acids on angiogenic activity in the placental extravillious trophoblast cells. Prostaglandins Leukot Essent Fatty Acids. 2013;88(2):155–162. [DOI] [PubMed] [Google Scholar]
- [28].Chomel C, Cazes A, Faye C, et al. Interaction of the coiled-coil domain with glycosaminoglycans protects angiopoietin-like 4 from proteolysis and regulates its antiangiogenic activity. Faseb J. 2009;23(3):940–949. [DOI] [PubMed] [Google Scholar]
- [29].Hu K, Babapoor-Farrokhran S, Rodrigues M, et al. Hypoxia-inducible factor 1 upregulation of both VEGF and ANGPTL4 is required to promote the angiogenic phenotype in uveal melanoma. Oncotarget. 2016;7(7):7816–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Babapoor-Farrokhran S, Jee K, Puchner B, et al. Angiopoietin-like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proc Natl Acad Sci U S A. 2015;112(23):E3030–E3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee SA, Ding C. The dysfunctional placenta epigenome: causes and consequences. Epigenomics. 2012;4(5):561–569. [DOI] [PubMed] [Google Scholar]
- [32].Falkowski PG, Katz ME, Milligan AJ, et al. The rise of oxygen over the past 205 million years and the evolution of large placental mammals. Science. 2005;309(5744):2202–2204. [DOI] [PubMed] [Google Scholar]
- [33].Samanta D, Prabhakar NR, Semenza GL. Systems biology of oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2017;9(4):e1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ge L, Wang Y, Cao Y, et al. MiR-429 improved the hypoxia tolerance of human amniotic cells by targeting HIF-1alpha. Biotechnol Lett. 2018;40(11–12):1477–1486. [DOI] [PubMed] [Google Scholar]
- [35].Verma S, Pillay P, Naicker T, et al. Placental hypoxia inducible factor −1alpha & CHOP immuno-histochemical expression relative to maternal circulatory syncytiotrophoblast micro-vesicles in preeclamptic and normotensive pregnancies. Eur J Obstet Gynecol Reprod Biol. 2018;220:18–24. [DOI] [PubMed] [Google Scholar]
- [36].Robb KP, Cotechini T, Allaire C, et al. Inflammation-induced fetal growth restriction in rats is associated with increased placental HIF-1alpha accumulation. PLoS One. 2017;12(4):e175805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Highet AR, Khoda SM, Buckberry S, et al. Hypoxia induced HIF-1/HIF-2 activity alters trophoblast transcriptional regulation and promotes invasion. Eur J Cell Biol. 2015;94(12):589–602. [DOI] [PubMed] [Google Scholar]
- [38].Oh ET, Kim CW, Kim SJ, et al. Docetaxel induced-JNK2/PHD1 signaling pathway increases degradation of HIF-1alpha and causes cancer cell death under hypoxia. Sci Rep. 2016;6:27382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: an epigenetic mark frequently deregulated in cancer. Biochim Biophys Acta. 2015;1855(2):144–154. [DOI] [PubMed] [Google Scholar]
- [40].Wen L, Li X, Yan L, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15(3):R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Taylor SE, Li YH, Wong WH, et al. Genome-wide mapping of DNA hydroxymethylation in osteoarthritic chondrocytes. Arthritis Rheumatol. 2015;67(8):2129–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sepulveda W, Sebire NJ, Hughes K, et al. The lambda sign at 10-14 weeks of gestation as a predictor of chorionicity in twin pregnancies. Ultrasound Obstet Gynecol. 1996;7(6):421–423. [DOI] [PubMed] [Google Scholar]
- [43].Gao Y, He Z, Wang Z, et al. Increased expression and altered methylation of HERVWE1 in the human placentas of smaller fetuses from monozygotic, dichorionic, discordant twins. PLoS One. 2012;7(3):e33503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Groene SG, Tollenaar L, Slaghekke F, et al. Placental characteristics in monochorionic twins with selective intrauterine growth restriction in relation to the umbilical artery Doppler classification. Placenta. 2018;71:1–5. [DOI] [PubMed] [Google Scholar]
- [45].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- [46].Chen C, Khaleel SS, Huang H, et al. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med. 2014;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Frazee AC, Pertea G, Jaffe AE, et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(3):243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. [DOI] [PubMed] [Google Scholar]
- [51].Li XC, Jin F, Wang BY, et al. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9(13):3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Huang N, Tan L, Xue Z, et al. Reduction of DNA hydroxymethylation in the mouse kidney insulted by ischemia reperfusion. Biochem Biophys Res Commun. 2012;422(4):697–702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


