Abstract
OBJECTIVE
Data engineering is the foundation of effective machine learning model development and research. The accuracy and clinical utility of machine learning models fundamentally depend on the quality of the data used for model development. This article aims to provide radiologists and radiology researchers with an understanding of the core elements of data preparation for machine learning research. We cover key concepts from an engineering perspective, including databases, data integrity, and characteristics of data suitable for machine learning projects, and from a clinical perspective, including the HIPAA, patient consent, avoidance of bias, and ethical concerns related to the potential to magnify health disparities. The focus of this article is women’s imaging; nonetheless, the principles described apply to all domains of medical imaging.
CONCLUSION
Machine learning research is inherently interdisciplinary: effective collaboration is critical for success. In medical imaging, radiologists possess knowledge essential for data engineers to develop useful datasets for machine learning model development.
Keywords: artificial intelligence, breast imaging, data engineering, machine learning, women’s imaging
Radiology and pathology have been at the forefront of machine learning (ML) medical research [1–4] primarily because vast stores of rich information are encoded in medical images and, in the case of radiology, routinely stored in health care system archives. Numerous ML models, especially those based on deep learning networks, have been developed to extract patterns in data from different women’s imaging modalities. For example, computer-aided mammographic breast cancer detection technologies use a wide range of ML methods, including support vector machines [5, 6], pixel-based ML [7], artificial neural networks [8], and the latest deep learning methods [9, 10]. Several ML techniques have been used to identify ovarian masses on ultrasound images [11], and a preliminary model of automated ovarian cancer classification has been developed by applying a deep learning architecture to cytologic images [12].
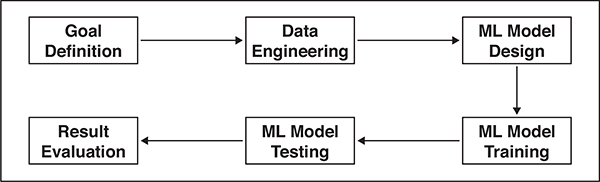
A typical ML project can be conceptualized as a pipeline or flowchart (Fig. 1). The first phase is to define a goal: what we wish to predict. In biomedical research, goal definition requires close collaboration between clinicians and data scientists to establish two critical parameters: clinical relevance and technical feasibility. The second phase is to prepare relevant data for building ML models—that is, data engineering—which is the foundation of effective ML model development and research. Data engineering refers to a set of processes required to determine which data are relevant and how the data should be accessed, securely stored, and modified in a manner that meets often-complex scientific, engineering, institutional, and regulatory requirements. ML systems have numerous parameters that require optimization. These parameters vary across different datasets and different hypotheses, even when using the same data. ML model development is therefore an iterative process of model design, training, testing, and validation, until system performance is deemed satisfactory.
Fig. 1—
Flowchart illustrating pipeline of machine learning (ML) project.
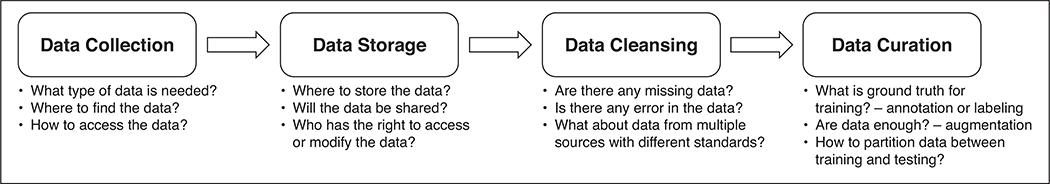
Data engineering is a central requirement in every ML project. It lays the foundation for the subsequent steps of model design, training, and testing. The accuracy and clinical utility of ML models fundamentally depend on the quality of the data used for model development. This article aims to provide radiologists and radiology researchers with an understanding of the core elements of data preparation for ML research. We will present key concepts and steps in data engineering (Fig. 2) with a focus on women’s imaging. Clinical, ethical, and legal considerations associated with data engineering will also be discussed.
Fig. 2—
Flowchart illustrating key components of data engineering.
Key Concepts in Data Engineering
In medical imaging, most data are acquired for clinical purposes and not for ML research or model development. For this reason, clinical data, including imaging data, are generally unstructured or only partially structured and are often stored in a variety of disparate formats and locations. The field of data engineering covers the entire process of data acquisition, curation, secure storage, and retrieval for ML model development. As shown in Figure 2, data engineering can be divided into four key components: data collection, data storage, data cleansing, and data curation. We will discuss each component in the context of developing clinically relevant ML models.
Clinical Considerations in Data Engineering
How to Identify Key Data and Data Sources
Data to be collected and curated in the data engineering process can vary on the basis of the clinical question and the chosen ML method [13, 14]. Supervised learning requires labeled data. Unsupervised learning uses unlabeled data to identify patterns and generate meaningful labels to improve the classification or organization of the data. These labels can then be used for supervised training approaches, a hybrid method known as semisupervised learning [13, 14]. Labeling medical data requires time and domain expertise. Although the vast majority of data in the world are unlabeled, relatively few medical ML studies use unlabeled data [15, 16]. The main reason for this is the large amount of training data required to recognize meaningful patterns in unlabeled data. As such, most of the published ML studies in women’s imaging and breast imaging rely on labeled data with supervised learning approaches [9, 10, 17–28] (Table 1).
TABLE 1:
Examples of Machine Learning Research Studies in Breast Imaging
| Study | Year | Data Source | Data Type | Image or Text | IRB Approval or Consent | Specialty | Subspecialty |
|---|---|---|---|---|---|---|---|
| Al-Antari et al. [18] | 2018 | Public database (INbreast) | Labeled, annotated | Image | None | Radiology | Breast |
| Al-Masni et al. [19] | 2018 | Public database (DDSM) | Labeled, annotated | Image | None | Radiology | Breast |
| Arevalo et al. [20] | 2016 | Public database (BCDR) | Labeled, annotated | Image | None | Radiology | Breast |
| Bahl et al. [21] | 2018 | Single center | Labeled | Text | IRB, consent waived | Radiology | Breast |
| Bandeira Diniz et al. [22] | 2018 | Public database (DDSM) | Labeled, annotated | Image | None | Radiology | Breast |
| Becker et al. [9] | 2017 | Single center for training and public database (BCDR) for testing | Labeled, annotated | Image | Local ethics committee, consent waived | Radiology | Breast |
| Cheng et al. [17] | 2016 | Single center and public database (LIDC) | Labeled, annotated | Image | IRB, consent waived | Radiology | Breast and chest |
| Chougrad et al. [23] | 2018 | Three public databases (DDSM, BCDR, INbreast) | Labeled, annotated | Image | None | Radiology | Breast |
| Dhungel et al. [24] | 2017 | Public database (INbreast) | Labeled, annotated | Image | None | Radiology | Breast |
| Ehteshami Bejnordi et al. [25] | 2017 | Two hospitals | Labeled, annotated | Image | IRB, consent waived | Radiology | Breast |
| Kooi et al. [26] | 2017 | Large-scale screening program in The Netherlands | Labeled | Image | None | Radiology | Breast |
| Ribli et al. [27] | 2018 | Single center and two public databases (DDSM, INbreast) | Labeled, annotated | Image | IRB, consent waived for single center | Radiology | Breast |
| Samala et al. [28] | 2016 | Two centers and public database (DDSM) | Labeled | Image | IRB, consent waived | Radiology | Breast |
| Wang et al. [10] | 2016 | Two centers | Labeled | Image | Ethics committees, consent obtained | Radiology | Breast |
Note—IRB = institutional review board, DDSM = Digital Database for Screening Mammography, BCDR = Breast Cancer Digital Repository, LIDC = Lung Image Database Consortium.
Expert data annotation and curation and the creation of massive datasets are resource intensive. Careful project selection and definition of the clinical question are essential for generating outcomes that justify the substantial investment required to successfully execute and deploy ML models in clinical care. It is critical that project selection be informed by both radiologists and ML scientists. Radiologists play a lead role in defining high-yield use cases (i.e., clinical questions and goals), constructing datasets, and establishing ground truth, whereas data scientists assess technical feasibility and approach.
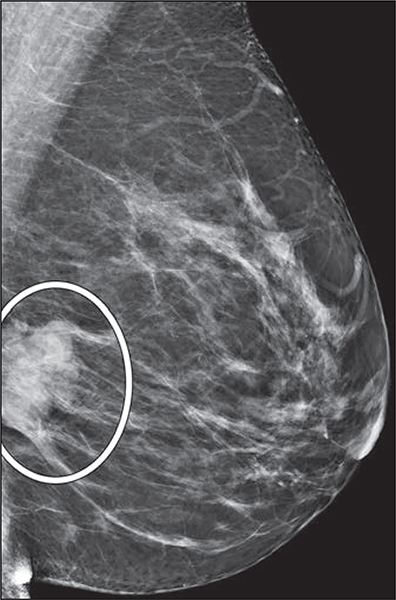
Once the use case for an ML research project is defined, the clinical question dictates the labels, or ground truth, required for supervised training and downstream algorithm testing and validation. For instance, if the outcome of interest is the detection of malignancies, radiologists could provide screening mammograms annotated with findings (Fig. 3) that are correlated with the final pathologic results and imaging and clinical follow-up as labels. Another example is risk prediction based on breast density: radiologists could provide screening mammograms without image annotation but with a breast density assessment, whether based on consensus or individual expert readers versus volumetric quantification by software, and use subsequent breast cancer diagnosis within a predefined time frame as labels. Again, radiologists play a crucial role in defining ground truth to ensure that model outputs augment the radiologists’ perception and interpretation without sacrificing the specificity of the imaging tests [29].
Fig. 3—
47-year-old woman. Screening mediolateral-oblique mammographic image shows annotation of recalled finding (oval).
The clinical question also dictates whether a text-based, image-based, or combined text-and image-based approach would be most informative, which then guides the process of identifying data sources and collecting key data. Table 1 summarizes practical examples of key data in text- or image-based ML studies in breast imaging [9, 10, 17–28]. Bahl et al. [21] applied text-based ML models to predict the upgrade rate of high-risk breast lesions using clinical information from the electronic health records (EHRs), mammography reports, and core needle biopsy reports as input, with surgical pathologic reports and follow-up outcomes as labels. To accurately detect and localize cancers on mammograms, Ribli et al. [27] applied an image-based convolutional neural network trained with screening mammograms, with pixel-level ground truth annotation of recalled lesions and histologic proof of cancer or benignity. Key data commonly include known or suspected factors associated with the clinical question or any additional information that might have potential clinical, social, biologic, or scientific relevance to the clinical question.
In its guidance document regarding premarket notification (510(k)) submissions for computer-assisted detection devices applied to radiologic images and radiologic data, the U.S. Food and Drug Administration provides nonbinding recommendations for the types of key data and reference standards to be included in databases for training, testing, and validation [30]. Public databases serve as useful prototype references for the design and construction of useful databases [31–33]. However, although these databases provide a useful template, they may not contain all the elements required for specific ML research projects.
Once radiologists, data scientists, and data engineers have identified the key data needed for specific ML applications, the team will usually collaboratively determine the data sources. The data source is of paramount importance, because incorporating the correct data stream can facilitate model development and improve model predictive accuracy [34]. Common data sources are public databases, private databases from single centers or partnering institutions, and a combination of both private and public databases [9, 10, 17–28] (Table 1). Public databases range from single-center datasets (e.g., INbreast) to collaborative datasets from multiple centers and industries (e.g., Digital Database for Screening Mammography and Lung Image Database Consortium image collection) [31, 32, 35, 36]. As an example, Ribli et al. [27] trained their models on the public Digital Database for Screening Mammography and a private institutional dataset and tested their models on the INbreast database, before becoming the second place winner of the Digital Mammography DREAM challenge, which consisted of 86,000 examinations without annotation, except for a binary label of positive or negative breast cancer diagnosis within 12 months [37]. Developed as solutions to image sharing for improved clinical processes, the Integrating the Healthcare Enterprise and the Radiological Society of North America’s Image Share Network provide image exchange platforms across centers that could serve as models to facilitate multisite sharing and database construction of deidentified images specific for research [38, 39].
Multisite data sources provide more-comprehensive and diverse data across health care settings, practice types, geography, and patient demographics for ML algorithm training, which is likely to improve the generalizability of developed models. Because data labeling is costly and time-consuming, preexisting databases commonly do not contain labels appropriate for the specific research question. Crowdsourcing presents a solution to collate ground-truth annotations via an online open call to a large group of individual participants of varying experiences and knowledge to voluntarily undertake the task [40]. Unfortunately, efforts to create large labeled databases using multisite data or labeling sources introduce data standardization challenges [31, 40].
In May 2017, the American College of Radiology (ACR) launched the Data Science Institute with the goals of guiding and facilitating appropriate development and implementation of artificial intelligence tools to help improve medical imaging care [41, 42]. To lead the definition of use cases and data elements, the ACR Data Science Institute recently released a group of use cases for industry feedback [43]. In addition, it is working to provide tools for institutions interested in developing annotated image datasets around the specific use cases defined by the ACR Data Science Institute. Ultimately, the success of the ACR Data Science Institute and partnering institutions would lead to availability of standardized multisite datasets applicable to specific high-yield use cases, creating a framework for the development and deployment of clinically relevant ML models. Before such multisite datasets with standardized data elements become readily available and accessible, radiology researchers aiming to use their own institutional data and construct their own databases should set up processes of obtaining, curating, annotating, and storing their own datasets.
How to Obtain and Store Data
Commonly within a single institution, clinical data are fragmented across imaging systems, departmental servers, EHR systems, and other health care information systems. The increasing trend toward health care organization consolidation with the absorption of multiple hospitals and practices into conglomerate enterprises has contributed to the growing complexity of clinical data consolidation. The Health Information Technology for Economic and Clinical Health Act of 2009 has led to significant increases in hospitals’ adoption of EHR systems and contributed to the advent of big clinical data [44]. Transition to a unified EHR system, combined with other data management and health information technology infrastructure efforts, required an investment of more than $1 billion at our institution [45]. Although initially targeted at improved clinical care, such efforts have created the context for eventual large-scale ML technology development and adoption success. Provided that the research activity is approved by an appropriate institutional review board, research investigators at our institution can request relevant clinical data and download imaging studies via a platform supported by the Research Patient Data Registry, a centralized data repository that consolidates and houses around 7 million patients and 3 billion rows of clinical data within the health care enterprise.
Many academic institutions have similar infrastructures with different levels of clinical data granularity or patient pool size for research purposes. If institution-wide resources do not exist, many radiology departments have internal radiology information systems, which can be queried to generate relevant radiology reports, examination requisition information, and patient demographic information. Availability of these technologic tools and personnel support is vital to the collection of necessary clinical data for ML algorithm development.
Text-based data
Most EHR data can be formatted into text-based data, which can be stored, organized, managed, and retrieved by widely used desktop programs, such as Microsoft Excel and Microsoft Access. These tools allow clinical experts, data engineers, and computer scientists share the same data relatively easily. Meticulous attention to the integrity and security of these data is essential in the prevention of unauthorized and undesirable data disclosure at a massive scale. Depending on the research aims, the research team may decide to deidentify the data. Password encoding is recommended as a layer of protection and security against unauthorized use. The prevention of unintended or unauthorized disclosure of protected health information (PHI) requires advanced measures that are governed by the HIPAA. Data security requirements vary by institution and governing authority. However, several principles are likely to apply generally: Transmission and storage of datasets should stay within the confines of secured servers and network, data transfer should take place using encrypted and secured file transfer protocols, and data management policies should be defined by appropriate experts within the medical or academic institution. These processes are particularly important if data are not deidentified, but remain important and relevant, even when data are robustly deidentified.
Imaging data
In most large health care organizations, imaging data are stored on a PACS and are typically also archived on hospital clinical servers for clinical use. Medical imaging data are typically stored in DICOM format to facilitate system interoperability. Besides image data, the DICOM standards contain related metadata for each imaging examination as a wrapper within each data file. Several programming languages (e.g., Python and Matlab) provide a DICOM library that allows us to access both the metadata and the image data. For example, a 2D ultrasound breast image in a DICOM file can be read into a 2D matrix in either Python or Matlab. Each element of the 2D matrix represents the intensity value of the corresponding pixel in the image. We can then apply different image-processing techniques to the 2D matrix. For medical imaging data to be organized, managed, retrieved, and manipulated for ML purposes, while minimizing the risk of disruption to clinical services, imaging or annotation data should be securely stored in alternate storage, such as secured research servers or offline research environments separate from the PACS and clinical servers. The research team, particularly the radiologists, must decide whether image annotation should take place in the PACS to create a markup database before image export or later in the research environment during data curation and image preprocessing. Creating markups in the PACS may save time and allow image review on U.S. Food and Drug Administration–approved display devices, which is particularly important to high-resolution images such as mammography. However, archiving of annotations within the PACS as part of the clinical record may have medicolegal implications and may affect subsequent interpretations when present on comparison studies. Creation of a separate annotation-only database is one strategy to leverage the availability of images from the clinical PACS for ML model development. However, this approach introduces additional network data transfer demands onto the clinical PACS, potentially disrupting clinical care operations. This risk can be mitigated by secure storage of select deidentified images from the PACS along with their annotations in a system separate from the PACS, but able to query the PACS and retrieve related data as required.
In general, deidentification of medical images is desirable for risk mitigation and will optimally occur at the time of image export from the PACS before storage in a secure data environment for research purposes. Medical imaging data poses unique challenges to the deidentification process; data engineers should familiarize themselves with the specific intricacies of the DICOM format, particularly PHI contained in the data [46]. Resources and tools to deidentify DICOM exist to help researchers [46–48]. Data engineers must take care when using existing deidentification toolkits to prevent the risk of disclosing PHI [47]. Patient identification mapping can be performed for linkage with text-based EHR data during the deidentification process.
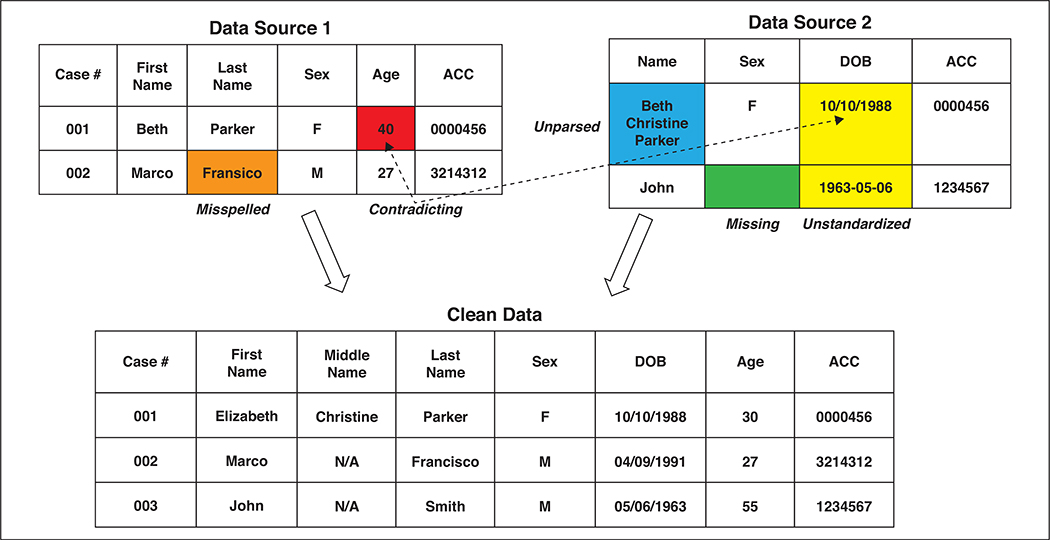
As shown in Figure 4, data transfer and storage vary depending on the sources of data and the levels of data usage. A secure local drive may be preferred if data usage is within a specific research group. Institutional network storage (and computation) has been growing rapidly over the past decade. The Big Data to Knowledge initiative, launched by the National Institutes of Health in 2012, aims to build six to eight investigator-initiated big data centers to improve data sharing and accessibility and knowledge discovery [49]. Institutional centers, such as The Center for Clinical Data Science, a joint effort of Massachusetts General Hospital and Brigham & Women’s Hospital in Boston, provide a platform for sharing data and ML expertise [50]. Cloud-based data storage is also growing rapidly with products such as MongoDB Atlas, Microsoft Azure for health, Amazon Web Service in Healthcare, and Google Cloud for Healthcare, all of which offer massive data storage systems and ML tools to health care organizations.
Fig. 4—
Example of data cleansing (applied to information of fictitious persons). Specifically, cleansing process includes correcting misspelling, filling missing entry, standardizing date-of-birth (DOB) format, correcting contradicting age information, and merging two data sources. ACC = Accession.
Each of these data storage solutions has advantages and disadvantages. Local storage has the merits of easy setup, convenient data access, and direct control by the research group. However, storage capacity may be limited when compared with nonlocal storage providers. Moreover, data security, backup, and access permissions require active management, which requires expertise and personnel. The initial capital costs of personnel and equipment may be prohibitive for many research groups. Nonlocal storage and computational infrastructure can be shared among multiple research groups. Shared capital costs, particularly when underwritten by institutional or departmental support, mitigate these challenges and provide a pathway to self-sufficiency for research groups starting out in medical imaging ML. Institutional data centers and commercial cloud offerings are also able to support ML analysis tools and provide new research groups with standardized frameworks and tools to integrate ML model generation into their research. Data backup and restoration of cloud storage may be slower compared with local storage because of limited Internet bandwidth. Although cloud storage offers greater accessibility, data access is completely blocked during Internet or service outage. Concerns with data security of cloud storage cause the most unease at many institutions.
Data Cleansing
Data cleansing is the process of identifying incomplete, incorrect, inaccurate, and irrelevant data and modifying, deleting, improving, or replacing those data. Data cleansing is a routine part of almost every ML project. Some of the key characteristics of well-conditioned data include a wide variety of the conditions being considered, a balanced representation of classes, high-quality labels (i.e., high interrater agreement), missing data filled in with meaningful values, and consistent data format that can be represented mathematically.
For text-based medical data, common problems are spelling, grammar, and punctuation errors; missing entries in patient records; contradictory data; nonunique identifiers, such as one patient with two forms of the same name; and data integration problems for more than one EHR system. Data cleansing typically involves single-file cleansing processes, such as parsing text, correcting mistakes, and adding missing data; and multiple-file cleansing processes, such as format standardization, entry matching, and consolidation of multiple files. Figure 4 is an example of text-based data cleansing.
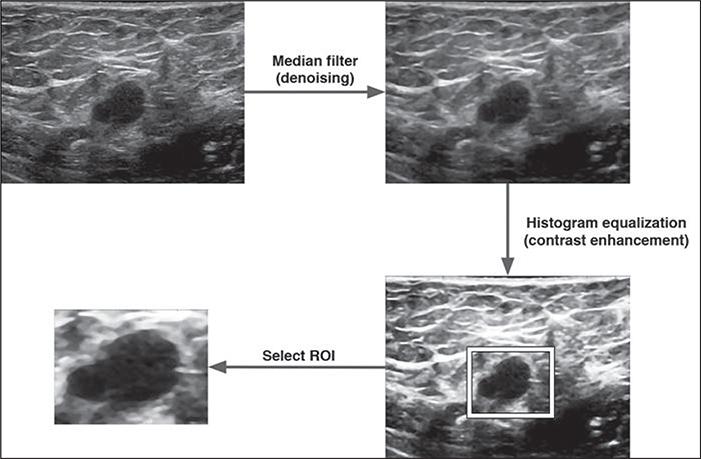
Medical images have specific characteristics that may require preprocessing before the ML model development. For example, ultrasound images encounter greater challenges in deidentification when PHI is embedded in the images. Preprocessing using special software is required to remove PHI from the images; even then, removing the metadata with potential PHI from certain images remains difficult and may be incomplete. Ultrasound images also tend to be highly variable owing to operator-dependent acquisition. Objects such as cysts on breast sonography may exhibit variation in echogenicity and artifactual internal echoes owing to different device settings or transducer positions. Ultrasound machines from different system integrators typically use different gray scales and may also have different default FOVs. To mitigate this variability, conventional image-processing techniques, such as denoising, contrast enhancement, normalization, and morphologic transformation (down- or up-sampling), are typically performed. Figure 5 is an example of image-processing techniques typically applied to breast ultrasound data. This preprocessing is analogous to data cleansing and is, in our experience, a routine part of ML model development with sonographic image data. More complicated preprocessing schemas, typically comprising a sequence of techniques, have been applied to several domains in women’s imaging. For example, Khazendar et al. [51] preprocessed ultrasound images of ovarian tumors in three steps, including a nonlocal mean filter for denoising, a negative transformation of the denoised image, and the absolute difference between the results from the first two steps, to generate data suitable for the ML method used (a support vector machine). In summary, whether text or image based, data generally have to be processed or cleansed in a robust and standardized manner before the ML model development.
Fig. 5—
50-year-old woman. Example of breast ultrasound image preprocessing for machine learning is shown.
How to Curate and Annotate Data
Data curation comprises organizing, integrating, and processing data collected from various sources into usable data. It should allow continued active management, maintenance, and reuse of data over time. Example curation processes of multiple public imaging databases, such as the Curated Breast Imaging Subset of Digital Database for Screening Mammography and collections in The Cancer Imaging Archive, are available as references [32, 33, 46, 52]. Data curation may be manual, automated, or semiautomated. For example, semantic parsing of DICOM metadata, EHR data, and radiology and pathology reports can extract useful features and compile these into usable sortable forms or spreadsheets [52]. ML models may be used to curate data, followed by human review and additional annotation, depending on the specific requirements of the project.
Accurate data annotation and labeling are vital to the success of algorithm development. As clinical experts, appropriately qualified radiologists should generally oversee or conduct the labeling process in a manner that preserves accuracy while minimizing variability. Prior radiology and pathology studies have relied on independent experts to label the findings on images; agreements between both were adopted as ground truths [31, 53]. Discordant labels are presented to both experts for discussion or to additional expert adjudicators, with the final decisions based on consensus [31, 53].
Data Augmentation and Synthetic Data
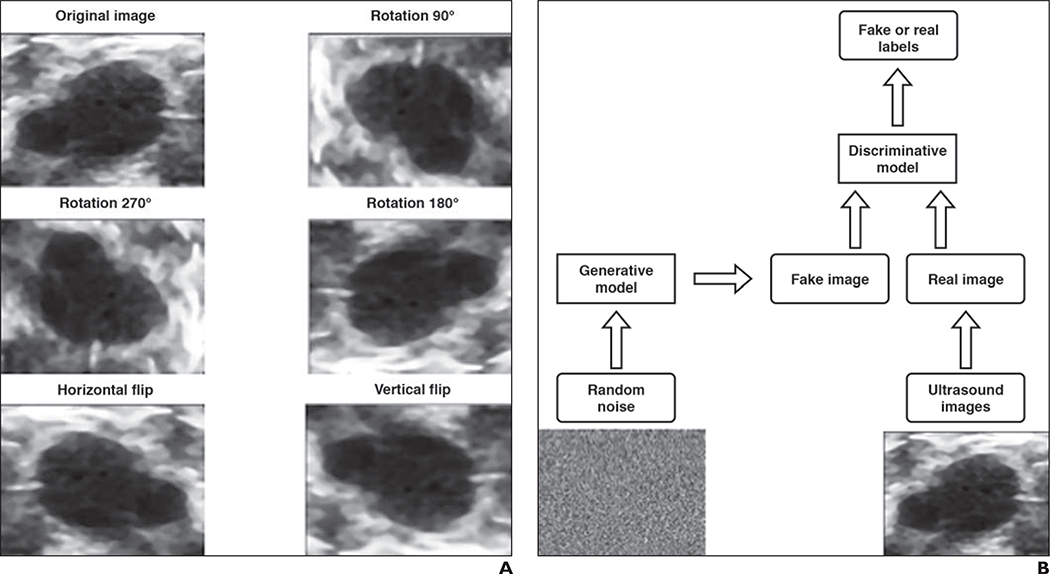
The scarcity of accurately annotated medical data has been a critical challenge for studies intended to apply ML methods to clinical questions. Data augmentation has proven to be an effective way to enlarge training dataset size. The early idea of augmenting data generated a large public-domain image database of numeric and alphabetic characters by learning the parameters of image defects [54]. This is the essence of conventional data augmentation, also called data warping. For example, new images with geometric (or color) transformations are created by performing image-processing techniques on original images. Figure 6A shows a few examples of commonly used data augmentation techniques: rotation and flipping. Vasconcelos et al. [55] describe a number of data-warping techniques before applying a convolutional neural network to classify skin lesions in their study. These techniques include geometric augmentation (rotation and flipping), color augmentation, and distortion by the lesion’s main axis size. The benefits of using data augmentation in different ML models have also been discussed previously [56].
Fig. 6—
50-year-old woman (same patient as Fig. 5). Examples of data augmentation and simplified framework for generative adversarial network are shown.
A, Data augmentation of ultrasound images is shown.
B, Flowchart shows simplified framework for generative adversarial network.
Although standard data augmentation directly manipulates existing data, generative adversarial networks [57], a recent technical advance in deep learning, can create synthetic data through learning from data. A simplified example of a generative adversarial network is shown in Figure 6B. There are two neural network–based models, a generative model and a discriminative model, to be trained. The purpose of the generative model is to generate better fake images from random noise to fool the discriminative model, whereas the discriminative model is trained to better distinguish real images from fake ones. The discriminative model and generative model are analogous to two players playing a game in which the desired outcome—correct image classification—is known. Generative adversarial networks have been effective in many applications, especially those with small datasets [58]. More details of generative adversarial networks can be found elsewhere [57–60].
Ethical Considerations in Data Engineering
The Belmont report, published in 1979 [61], establishes the basic ethical principles for research involving human subjects, including respect for persons, beneficence, and justice. Medical ML model development routinely uses large amounts of human subject data and, therefore, must even more dutifully and cautiously comply with these basic ethical principles. The black-box tendency of certain ML methods, compounded by the power imbalance and knowledge gap between individual subjects included in these datasets and the data controllers, implies that governance by these ethical principles should occur deliberately at the outset of the data engineering process [62, 63] and that ethical considerations should be carefully considered when providing or restricting access to data.
Ethical considerations of data engineering begin at goal setting. Developing clear objectives in medical ML research can be challenging when exploratory studies rely on algorithms to identify correlations without underlying hypotheses [63]. Clinical experts and data engineers should have the common goal of protecting stakeholder interests, especially those of subjects whose data have been incorporated into the data-engineering process, individuals who may be subjected to subsequent model application (e.g., risk prediction or profiling) by the models generated, and society at large [63]. Explicit, conscientious, and thorough vetting of collected data helps identify deficiencies in the dataset during the data engineering process. This awareness may also prevent the use of classification models in inappropriate populations during downstream applications.
How to Avoid Bias
In traditional clinical studies, researchers strive to carefully construct their cohorts or control groups in well-designed retrospective or prospective studies to avoid biases and confounding. ML research commonly uses real-world data from the EHR, insurance claims, and personal devices acquired for nonresearch purposes. Patient self-selection, which is confounding by indication, and inconsistent outcome availability are likely inadvertent consequences given these uncontrolled data sources [34]. Social constructs and economic and political systems shaping the social determinants of health extend to the types of data available; at the simplest level, it is intuitive that underserviced populations will have fewer data, and, therefore, data-derived models may not generalize well to underserviced populations. However, bias, both recognized and unrecognized, distorts the delivery of medical care in more complex ways. Data derived from the clinical work stream reflects these biases. Moreover, the high cost of health care information technology and the shortage of ML expertise create distortions. For example, hospitals with the infrastructure to manage and analyze large data may also service the wealthy, resulting in a widening gap between those who have or lack resources [62, 64]. Health care delivery is known to vary by race. Few, if any, outcome or genetic studies are available in nonwhite populations, with barriers related to distrust from historical and potential discrimination and challenges in establishing contact [65–67]. Medical treatments and guidelines are commonly extrapolated from research data derived from largely white populations. Research has shown that breast cancer age distribution and prognosis differ by race, with peak diagnoses at younger age in nonwhite women and poorer survival in black women compared with white women [68–70]. Stapleton et al. [70] state that the U.S. Preventive Services Task Force’s “age-based screening guidelines that do not account for race may adversely affect nonwhite populations.” Similarly, ML research using clinical data based on unconsciously biased medical decisions may mirror these biases in its models.
Recognition of and vigilance for potential biases in the data sources are the first steps to mitigating bias in data engineering. Purpose limitation to collecting data for specific and well-thought-out research aims, prohibiting arbitrary data reuse, and minimizing collection to high-quality relevant data are ideal [63], but may not be practical at the time of database construction.
At the project level, radiologists and data engineers should deliberately construct a balanced and diverse dataset, to ensure sound clinical evaluation of ML model performance and to avoid engineering and human bias. Overfitting, which refers to a learning model overly customized to its training data [71], should also be avoided.
At the policy level, researchers can use ML models to help resolve disparities and identify areas of needed research [72]. Fairness, accountability, governance, and respect for stakeholder and public interests should guide ethical considerations in data engineering.
Legal Considerations in Data Engineering
Currently and in the foreseeable near future, radiologists are likely to approve final reports and retain primary responsibility for the acquisition and interpretation of imaging studies, with or without the support of ML applications [13]. Therefore, it is vital for radiologists to recognize potential legal implications throughout the development of an ML algorithm, along each step of data collection and engineering, algorithm training, testing, and validation, and ultimately algorithm implementation in clinical care.
HIPAA and Informed Consent
Privacy is both an ethical and legal real-world concern in ML and big data research. Data protection is of paramount importance in the era of advanced data-mining and cloud-computing technologies. The HIPAA Privacy Rule governs strict PHI policy in the United States. The U.S. Department of Health and Human Services provides guidance on two deidentification methods—Expert Determination and Safe Harbor—to satisfy HIPAA’s standard [73]. Health care systems have been vulnerable to security breaches, similar to other industries and organizations. Imaging data remain insecure, even within the confines of hospital radiology servers [74]. Researchers must safeguard identifiable data in the research environment and remain compliant with both institutional policies and governmental regulations.
HIPAA requires written authorization from patients for the disclosure and use of PHI for purposes other than treatment, payment, and operations. This authorization is in the form of an informed consent for researchers to obtain PHI. The institutional review boards have typically deemed radiology ML studies as posing minimal risk to patients and have waived the requirement for informed consent. Neither institutional review board approval nor informed consent is required for researchers using public databases (Table 1).
Compared with HIPAA, however, privacy regulations are more stringent with broader coverage under the European Union’s General Data Protection Regulation (GDPR). Put into effect in May 2018 and introduced by the U.K. Information Commissioner’s Office, the GDPR applies to all research involving personal data of any resident located in the European Union [75]. Coded data are treated as identifiable data under the GDPR. Processing of personal health, genetic, and biometric data is prohibited; exceptions include explicit consent provided by the data subject and processing for reasons of substantial public interest [75]. Consent poses a great challenge in ML and big data research, given that the purposes of data collection may be unclear, reuse of the collected data may not have been anticipated, and the opaque nature of ML algorithms [63]. To address this difficulty, the Information Commissioner’s Office suggests a graduated consent model with just-in-time notifications to foster trust as part of an ongoing relationship with the individual [75]. Radiologists must familiarize themselves with the basic GDPR requirements before receiving personal data from the European Union.
Data Ownership, Intellectual Property, and Data Sharing
As part of the consent disclosures under GDPR, data subjects have the rights to request access, rectification, erasure, or restriction of processing, or to object to processing of their personal data [75]. Enabling patient access to and control of personal health care data appears to encourage active patient engagement in improving their health [76]. Besides patients, health care providers and hospital systems are also stakeholders with vested responsibilities and interests in the health care data [62]. Ownership of subsequently developed intellectual property from the use of health care data are equally unclear. A data use agreement signed by the patient could specify data quality, security, and use with more comprehensive rules for data reuse and intellectual property [64, 76].
The contribution of health care data to larger shared databases is useful, particularly in the context of larger organizational efforts where a data-sharing framework has been established. In such cases, it is important to ensure that patient privacy is protected and government regulations are followed. Trends and requirements in data sharing have increased in recent years, particularly regarding clinical trial research data. The National Institutes of Health and the International Committee of Medical Journal Editors issued requirements for researchers to address data sharing in a data-sharing statement [77, 78]. Nonetheless, research data generated or collected as part of a research study or clinical trial are different from clinical data, which are the primary data source of ML research. Some large health care institutions have established committee oversight of data-sharing requests with third parties. Compliance with HIPAA and GDPR, where applicable, should be enforced in data sharing; violation of privacy laws can incur enormous penalties [62, 75].
Conclusion
Data engineering represents one of the most important and challenging tasks in ML research. It greatly influences downstream training, testing, validation, and application of ML models with significant implications for patient care and ethical practice, including the perpetuation of bias and systematic unfair discrimination. Radiologists are well suited to the role of clinical information manager, gatekeeper of data, and collaborative definer of use cases [79]. Because radiologists are intimately familiar with imaging and other sources of clinical information required for the development of ML models, their role should expand to the interpretation of the output of ML models, by interpreting and contextualizing imaging with the patient’s overall clinical picture, with the aid of augmented intelligence. Interdisciplinary collaboration is paramount to the success of ML research. Radiologists possess the critical knowledge to help data engineers curate the most useful and unbiased data for ML.
Acknowledgments
Supported in part by Air Force Contract No. FA8702-15-D-001 from the Assistant Secretary of Defense for Research and Engineering. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Assistant Secretary of Defense for Research and Engineering. This work is also supported by award numbers K23 EB020710 from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health.
S. H. S. Chou is a coinvestigator on a GE Healthcare research agreement. C. D. Lehman is a member of a GE Healthcare advisory board and receives research support from GE Healthcare. A. E. Samir is a consultant for Astra Zeneca, Bracco Diagnostics, Bristol Myers Squibb, Founder Collective, General Electric, Gerson Lehman Group, Supersonic Imagine, Novartis, Pfizer, Philips Healthcare, Parexel Informatics, and WorldCare Clinical; serves on the advisory or scientific boards of General Electric and Remedy Medical; has received research support from Analogic Corporation, General Electric, Hitachi, Philips Healthcare, Supersonic Imagine, and Toshiba Medical Systesm; and has received research funding from Allergan, Analogic Corporation, Boehringer Ingelheim, Department of Defense, Echosens, Foundations for the National Institutes of Health, Genentech, General Electric, Gilead, Intercept, Medimmune, Novo Nordisk, the National Institutes of Health, Partners Healthcare, Philips Healthcare, Supersonic Imagine, Takeda, Toshiba Medical Systems, and Siemens Healthcare.
References
- 1.Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012; 16:933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal 2016; 33:170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kononenko I Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001; 23:89–109 [DOI] [PubMed] [Google Scholar]
- 4.Obermeyer Z, Emanuel EJ. Predicting the future: big data, machine learning, and clinical medicine. N Engl J Med 2016; 375:1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Shi L, Ann Heng P. Automatic detection of breast cancers in mammograms using structured support vector machines. Neurocomputing 2009; 72:3296–3302 [Google Scholar]
- 6.Muthu Rama Krishnan M, Banerjee S, Chakraborty C, Chakraborty C, Ray AK. Statistical analysis of mammographic features and its classification using support vector machine. Expert Syst Appl 2010; 37:470–478 [Google Scholar]
- 7.Suzuki K Pixel-based machine learning in medical imaging. Int J Biomed Imaging 2012; 2012:792079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayer T, Alagoz O, Chhatwal J, Shavlik JW, Kahn CE Jr, Burnside ES. Breast cancer risk estimation with artificial neural networks revisited: discrimination and calibration. Cancer 2010; 116:3310–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 2017; 52:434–440 [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Yang X, Cai H, Tan W, Jin C, Li L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci Rep 2016; 6:27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pathak H, Kulkarni V. Identification of ovarian mass through ultrasound images using machine learning techniques. IEEE website. ieeexplore.ieee.org/document/7434224. Published 2015. Accessed November 20, 2018 [Google Scholar]
- 12.Wu M, Yan C, Liu H, Liu Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci Rep 2018; 38:BSR20180289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. AJR 2017; 208:754–760 [DOI] [PubMed] [Google Scholar]
- 14.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017; 37:2113–2131 [DOI] [PubMed] [Google Scholar]
- 15.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zech J, Pain M, Titano J, et al. Natural language-based machine learning models for the annotation of clinical radiology reports. Radiology 2018; 287:570–580 [DOI] [PubMed] [Google Scholar]
- 17.Cheng JZ, Ni D, Chou YH, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in CT scans. Sci Rep 2016; 6:24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Antari MA, Al-Masni MA, Choi MT, Han SM, Kim TS. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int J Med Inform 2018; 117:44–54 [DOI] [PubMed] [Google Scholar]
- 19.Al-Masni MA, Al-Antari MA, Park JM, et al. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput Methods Programs Biomed 2018; 157:85–94 [DOI] [PubMed] [Google Scholar]
- 20.Arevalo J, Gonzalez FA, Ramos-Pollan R, Oliveira JL, Guevara Lopez MA. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed 2016; 127:248–257 [DOI] [PubMed] [Google Scholar]
- 21.Bahl M, Barzilay R, Yedidia AB, Locascio NJ, Yu L, Lehman CD. High-risk breast lesions: a machine learning model to predict pathologic upgrade and reduce unnecessary surgical excision. Radiology 2018; 286:810–818 [DOI] [PubMed] [Google Scholar]
- 22.Bandeira Diniz JO, Bandeira Diniz PH, Azevedo Valente TL, Correa Silva A, de Paiva AC, Gattass M. Detection of mass regions in mammograms by bilateral analysis adapted to breast density using similarity indexes and convolutional neural networks. Comput Methods Programs Biomed 2018; 156:191–207 [DOI] [PubMed] [Google Scholar]
- 23.Chougrad H, Zouaki H, Alheyane O. Deep convolutional neural networks for breast cancer screening. Comput Methods Programs Biomed 2018; 157:19–30 [DOI] [PubMed] [Google Scholar]
- 24.Dhungel N, Carneiro G, Bradley AP. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Anal 2017; 37:114–128 [DOI] [PubMed] [Google Scholar]
- 25.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318:2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kooi T, Litjens G, van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 2017; 35:303–312 [DOI] [PubMed] [Google Scholar]
- 27.Ribli D, Horvath A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep 2018; 8:4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys 2016; 43:6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohli A, Jha S. Why CAD failed in mammography. J Am Coll Radiol 2018; 15:535–537 [DOI] [PubMed] [Google Scholar]
- 30.U.S. Department of Health & Human Services; U.S. Food & Drug Administration (FDA). Computer-assisted detection devices applied to radiology images and radiology device data: premarket notification [510(k)] submissions—guidance for industry and food and drug administration staff. FDA website. www.fda.gov/RegulatoryInformation/Guidances/ucm187249.htm. Published July 3, 2012. Accessed July 15, 2018
- 31.Moreira IC, Amaral I, Domingues I, Cardoso A, Cardoso MJ, Cardoso JS. INbreast: toward a fullfield digital mammographic database. Acad Radiol 2012; 19:236–248 [DOI] [PubMed] [Google Scholar]
- 32.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013; 26:1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prior F, Smith K, Sharma A, et al. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci Data 2017; 4:170124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen JH, Asch SM. Machine learning and prediction in medicine: beyond the peak of inflated expectations. N Engl J Med 2017; 376:2507–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armato SG 3rd, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011; 38:915–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heath MBK, Kopans D, Moore R, Kegelmeyer WP. The digital database for screening mammography In: Yaffe MJ, ed. Proceedings of the Fifth International Workshop on Digital Mammography. Madison, WI: Medical Physics Publishing, 2001:212–218 [Google Scholar]
- 37.[No authors listed] The digital mammography DREAM challenge. DREAM challenge website. www.synapse.org/Digital_Mammography_DREAM_challenge. Published June 28, 2016. Revised March 20, 2018. Accessed October 1, 2018 [Google Scholar]
- 38.Mendelson DS, Bak PR, Menschik E, Siegel E. Informatics in radiology: image exchange—IHE and the evolution of image sharing. RadioGraphics 2008; 28:1817–1833 [DOI] [PubMed] [Google Scholar]
- 39.Greco G, Patel AS, Lewis SC, et al. Patient-directed internet-based medical image exchange: experience from an initial multicenter implementation. Acad Radiol 2016; 23:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging 2016; 35:1313–1321 [DOI] [PubMed] [Google Scholar]
- 41.Allen B, Dreyer K. The artificial intelligence ecosystem for the radiological sciences: ideas to clinical practice. J Am Coll Radiol 2018; 15:1455–1457 [DOI] [PubMed] [Google Scholar]
- 42.McGinty GB, Allen B Jr. The ACR Data Science Institute and AI Advisory Group: harnessing the power of artificial intelligence to improve patient care. J Am Coll Radiol 2018; 15:577–579 [DOI] [PubMed] [Google Scholar]
- 43.American College of Radiology (ACR). ACR DSI releases initial use cases for industry feedback. ACR website. www.acr.org/Media-Center/ACR-News-Releases/2018/ACR-DSI-Releases-Initial-Use-Cases-for-Industry-Feedback. Published July 5, 2018. Accessed July 11, 2018 [Google Scholar]
- 44.Adler-Milstein J, Jha AK. HITECH act drove large gains in hospital electronic health record adoption. Health Aff (Millwood) 2017; 36:1416–1422 [DOI] [PubMed] [Google Scholar]
- 45.Dreyer KJ, Geis JR. When machines think: radiology’s next frontier. Radiology 2017; 285:713–718 [DOI] [PubMed] [Google Scholar]
- 46.Moore SM, Maffitt DR, Smith KE, et al. De-identification of medical images with retention of scientific research value. RadioGraphics 2015; 35:727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aryanto KY, Oudkerk M, van Ooijen PM. Free DICOM de-identification tools in clinical research: functioning and safety of patient privacy. Eur Radiol 2015; 25:3685–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ju K De-identification knowledge base. Cancer Imaging Archive website. wiki.cancerimagingarchive.net/display/Public/De-identification+Knowledge+Base. Published July 5, 2017. Accessed July 19, 2018 [Google Scholar]
- 49.U.S. Department of Health & Human Services; National Institutes of Health (NIH). NIH commits $24 million annually for Big Data Centers of Excellence. NIH website. www.nih.gov/news-events/news-releases/nih-commits-24-million-annually-big-data-centers-excellence. Published July 22, 2013. Accessed July 20, 2018 [Google Scholar]
- 50.Davenport T, Bean R. Revolutionizing radiology with deep learning at partners healthcare–and many others. Forbes website. www.forbes.com/sites/tomdavenport/2017/11/05/revolutionizingradiology-with-deep-learning-at-partners-healthcare-and-many-others/. Published November 5, 2017. Accessed July 20, 2018 [Google Scholar]
- 51.Khazendar S, Sayasneh A, Al-Assam H, et al. Automated characterisation of ultrasound images of ovarian tumours: the diagnostic accuracy of a support vector machine and image processing with a local binary pattern operator. Facts Views Vis ObGyn 2015; 7:7–15 [PMC free article] [PubMed] [Google Scholar]
- 52.Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data 2017; 4:170177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veta M, van Diest PJ, Willems SM, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal 2015; 20:237–248 [DOI] [PubMed] [Google Scholar]
- 54.Bunke H, Wang PSP, Baird H. Document image defect models In: O’Gorman L, Kasturi R, eds. Document image analysis. Los Alamitos, CA: IEEE Computer Society Press, 1994:315–325 [Google Scholar]
- 55.Vasconcelos C, Nader Vasconcelos B. Increasing deep learning melanoma classification by classical and expert knowledge based image transforms. ResearchGate website. www.researchgate.net/publication/313910523_Increasing_Deep_Learning_Melanoma_Classification_by_Classical_And_Expert_Knowledge_Based_Image_Transforms. Published February 2017. Accessed November 20, 2018 [Google Scholar]
- 56.Wong SC, Gatt A, Stamatescu V, McDonnell MD. Understanding data augmentation for classification: when to warp? arXiv website. arxiv.org/pdf/1609.08764.pdf. Published 2016. Accessed November 20, 2018 [Google Scholar]
- 57.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. arXiv website. arxiv. org/abs/1406.2661. Published June 10, 2014. Accessed November 20, 2018 [Google Scholar]
- 58.Gurumurthy S, Sarvadevabhatla RK, Radhakrishnan VB. DeLiGAN: generative adversarial networks for diverse and limited data. arXiv website. arxiv.org/abs/1706.02071. Published June 7, 2017. Accessed November 20, 2018 [Google Scholar]
- 59.Goodfellow I NIPS 2016 tutorial: generative adversarial networks. arXiv website. arxiv.org/abs/1701.00160. Published December 31, 2016. Accessed November 20, 2018 [Google Scholar]
- 60.Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. arXiv website. arxiv.org/abs/1712.04621. Published December 13, 2017. Accessed November 20, 2018 [Google Scholar]
- 61.National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research. The Belmont report: ethical principles and guidelines for the protection of human subjects of research. Bethesda, MD: National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research, 1978:172–173 [Google Scholar]
- 62.Balthazar P, Harri P, Prater A, Safdar NM. Protecting your patients’ interests in the era of big data, artificial intelligence, and predictive analytics J Am Coll Radiol 2018; 15:580–586 [DOI] [PubMed] [Google Scholar]
- 63.Butterworth M The ICO and artificial intelligence: the role of fairness in the GDPR framework. Comput Law Secur Rep 2018; 34:257–268 [Google Scholar]
- 64.Kohli M, Geis R. Ethics, artificial intelligence, and radiology. J Am Coll Radiol 2018; 15:1317–1319 [DOI] [PubMed] [Google Scholar]
- 65.Hartz SM, Johnson EO, Saccone NL, Hatsukami D, Breslau N, Bierut LJ. Inclusion of African Americans in genetic studies: what is the barrier? Am J Epidemiol 2011; 174:336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katz RV, Wang MQ, Green BL, et al. Participation in biomedical research studies and cancer screenings: perceptions of risks to minorities compared with whites. Cancer Control 2008; 15:344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald JA, Barg FK, Weathers B, et al. Understanding participation by African Americans in cancer genetics research. J Natl Med Assoc 2012; 104:324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017; 67:439–448 [DOI] [PubMed] [Google Scholar]
- 69.Jemal A, Robbins AS, Lin CC, et al. Factors that contributed to black-white disparities in survival among nonelderly women with breast cancer between 2004 and 2013. J Clin Oncol 2018; 36:14–24 [DOI] [PubMed] [Google Scholar]
- 70.Stapleton SM, Oseni TO, Bababekov YJ, Hung YC, Chang DC. Race/ethnicity and age distribution of breast cancer diagnosis in the United States. JAMA Surg 2018; 153:594–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018; 286:800–809 [DOI] [PubMed] [Google Scholar]
- 72.Char DS, Shah NH, Magnus D. Implementing machine learning in health care: addressing ethical challenges. N Engl J Med 2018; 378:981–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.U.S. Department of Health & Human Services (HHS). Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. HHS website. www.hhs.gov/hipaa/for-professionals/privacy/specialtopics/de-identification/index.html. Published November 26, 2012. Updated November 6, 2015. Accessed November 20, 2018 [Google Scholar]
- 74.Stites M, Pianykh OS. How secure is your radiology department? Mapping digital radiology adoption and security worldwide. AJR 2016; 206:797–804 [DOI] [PubMed] [Google Scholar]
- 75.Information Commission’s Office (ICO). Data Protection Act and General Data Protection Regulation: big data, artificial intelligence, machine learning and data protection. ICO website. ico.org.uk/media/for-organisations/documents/2013559/big-data-ai-ml-and-data-protection.pdf. Published 2017. Accessed July 3, 2018 [Google Scholar]
- 76.Mikk KA, Sleeper HA, Topol EJ. The pathway to patient data ownership and better health. JAMA 2017; 318:1433–1434 [DOI] [PubMed] [Google Scholar]
- 77.National Institutes of Health (NIH). NIH data sharing policy. NIH website. grants.nih.gov/grants/policy/data_sharing/. Published February 26, 2003. Updated April 17, 2007. Accessed June 25, 2018 [Google Scholar]
- 78.Taichman DB, Sahni P, Pinborg A, et al. Data sharing statements for clinical trials: a requirement of the International Committee of Medical Journal Editors. N Engl J Med 2017; 376:2277–2279 [DOI] [PubMed] [Google Scholar]
- 79.Jha S, Topol EJ. Information and artificial intelligence. J Am Coll Radiol 2018; 15:509–511 [DOI] [PubMed] [Google Scholar]