Abstract
The blood-stage infection of the malaria parasite, Plasmodium falciparum, exhibits ts a48-hour developmental cycle that culminates in the synchronous release of parasites from red blood cells, triggering 48-hour fever cycles in the host. This cycle could be driven extrinsically by host circadian processes, or by a parasite-intrinsic oscillator. To distinguish between hypotheses, we examined the P. falciparum cycle in an in vitro culture system and show that the parasite has molecular signatures associated with circadian and cell-cycle oscillators. Each of four strains examined has a unique period, indicating strain-intrinsic period control. Finally, we demonstrate that parasites have low cell-to-cell variance in cycle period, on par with a circadian oscillator. We conclude that an intrinsic oscillator maintains Plasmodium’s rhythmic life cycle.
One Sentence Summary:
Periodicity of the malaria parasite does not require rhythmic cues from the host, but rather from an intrinsic parasite oscillator.
Malaria and its causal parasite, the Plasmodium genus, are fundamentally rhythmic entities. Patients infected with P. falciparum often exhibit 48-hour fever cycles, and these cycles coincide with the blood stage of the infection where the parasite progresses through the asexual intraerythrocytic cycle. After infection of the erythrocyte (red blood cell; RBC), parasites transit through three morphologically distinct developmental stages that can be visualized by light microscopy: rings, trophozoites, and schizonts. Parasites are in the ring stage immediately after RBC invasion and divide asexually multiple times during the schizont stage. At the end of the schizont stage the RBC bursts, releasing merozoites which quickly invade new host cells and begin the cycle anew. The infecting population of parasites in the host tend to undergo this cycle synchronously, and the subsequent release of merozoites is responsible for the characteristic periodic fevers seen in many patients (1). The human-infecting species of Plasmodium repeat this cycle every 24, 48, or 72 hours (depending on the species), suggesting that cycles could be driven by a host circadian cycle or a parasite-intrinsic oscillator with circadian periodicity (2). Interestingly, multiple animal studies have demonstrated that Plasmodium infections appear to synchronize with their host’s 24-hour circadian rhythms (2–5).
The source of the parasite’s rhythmic life cycle is a central, unsolved question. It is possible that the intraerythrocytic cycle periodicity is driven by extrinsic temporal cues in the host environment triggering the parasite’s developmental cascade (Fig. S1A). Several rhythmic host factors have been suggested to effect Plasmodium dynamics, including temperature (2), melatonin (6), glucose, and tumor necrosis factor α (TNFα) (7). In addition, recent studies have revealed the existence of an independent 24-hour oscillator in the redox state of peroxiredoxins, a highly conserved family of prominent cellular proteins; these oscillations continue even in isolated RBCs (8). Thus, it is possible that Plasmodium, even grown in culture, merely responds to an extrinsic oscillating program (Fig. S1A). An example can be found in plants, where several rhythmic biological processes are driven by light, rather than the plant’s innate rhythm (9).
Alternatively, Plasmodium may possess an intrinsic biological oscillator which generates its rhythms independently from the host. The best-known examples of endogenous oscillators are circadian biological clocks, found across a wide range of taxa and affecting a substantial array of functions. These 24-hour oscillators emerge from highly interconnected, auto-regulatory gene networks containing transcription-translation feedback loop motifs (10, 11). The oscillators themselves are free-running, but temporally align to cyclic entrainment signals, driven by the 24-hour cycle imposed by the Earth’s rotation. A wide variety of genes may be under circadian control, exhibiting 24-hour cyclic expression. The period length of the circadian rhythm is set by the core gene network underlying the clock (11); mutations may result in shorted, lengthened, or absent rhythms (12, 13). Similar programs of periodic gene expression are observed during cell division and have also been proposed to be driven by oscillating gene-regulatory networks (14, 15). Unlike circadian oscillators, cell-cycle oscillators do not necessarily exhibit 24-hour periods and are not necessarily aligned with or entrained by light-dark cycles.
The fact that Plasmodium spp. exhibit rhythms that are usually multiples of 24 hours suggests they may have an intrinsic oscillator network similar to a circadian oscillator. Although Plasmodium genomes do not appear to contain orthologs of canonical circadian clock genes (5), this does not rule out the possibility of an intrinsic network, similar in structure to either circadian or cell cycle networks, and capable of producing rhythms at multiples of 24 hours.
In this study, we investigate the rhythmic behavior of the P. falciparum intraerythrocytic cycle in an in vitro culture system where canonical circadian signals from the host were not present. Using high-density time-series transcriptomics and microscopy for four strains of P. falciparum, we compare several key molecular features of these cycles with molecular signatures produced by circadian networks and eukaryotic cell-cycle oscillators (14, 16). Our findings provide strong evidence for the existence of an intrinsic oscillator in Plasmodium and suggest parasites have evolved mechanisms to drive periodicity that may align with host circadian rhythms.
Plasmodium falciparum shows qualitative similarities to biological oscillator transcriptomes:
To assess the molecular features of the P. falciparum temporal transcriptome, we performed high-density time-series transcriptomic analyses for the unique strains 3D7 (17), FVO-NIH (18), SA250 (19), and D6 (20). These strains were chosen for their diverse geographic origins and cycle lengths, with some strains varying from the wild-type 48-hour cycle. Synchronized parasite populations were cultured in vitro for 60–70 hours, with time points sampled every three hours for transcriptomic analysis and microscopic observation. This sampling schedule allowed for the completion of 1–2 intraerythrocytic cycles per strain. in. RNA was isolated from each time point and subjected to RNA-seq analysis to quantitate the abundance of P. falciparum transcripts at each time point (Fig.1, File S1). For three of the four strains (D6, FVO-NIH, SA250), experiments were performed on the same days in the same conditions with blood from a single human donor to eliminate variability due to growth conditions (Materials and Methods).
Fig. 1.
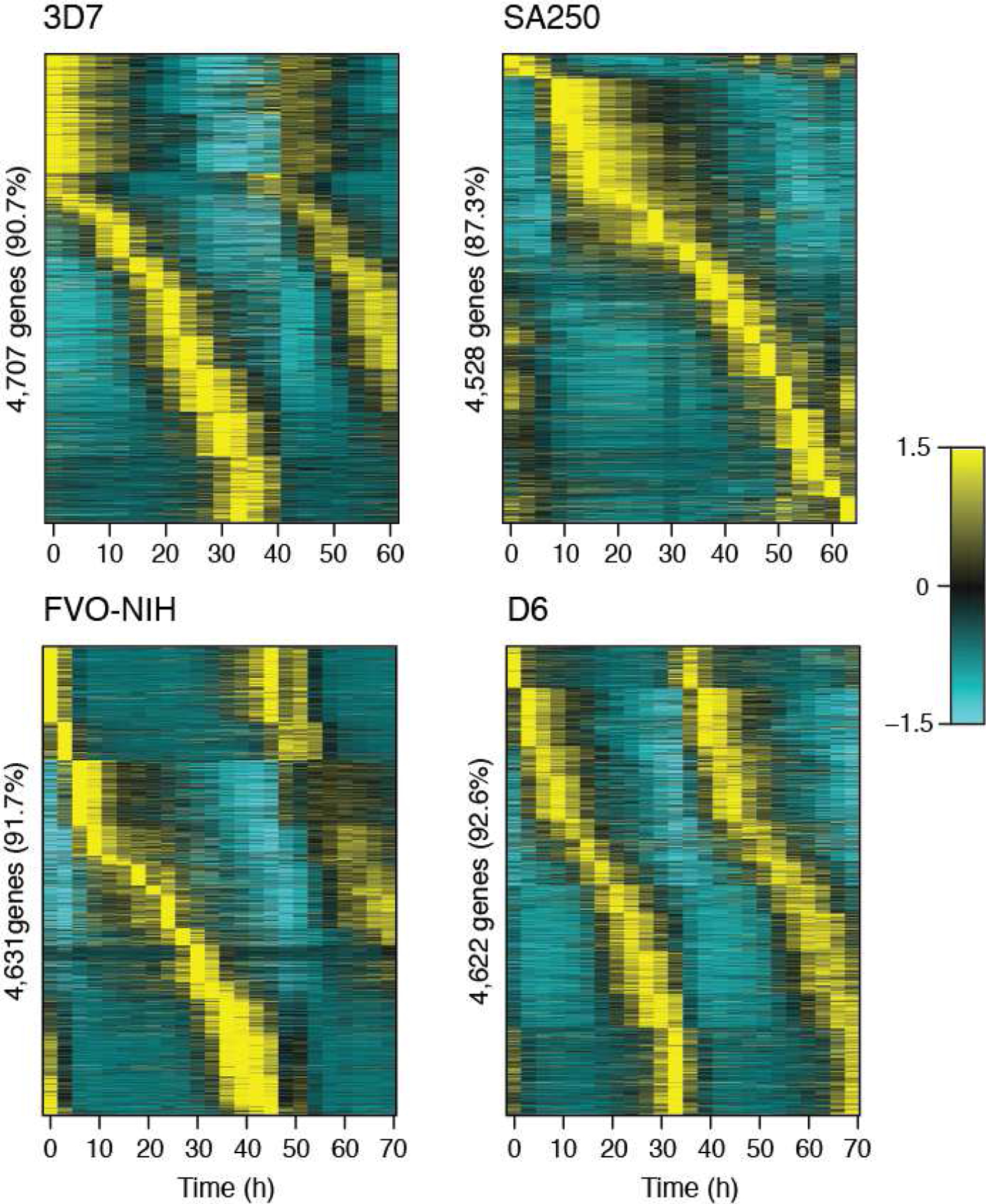
The majority of P. falciparum genes are periodically transcribed. Four strains of P. falciparum were cultured in vitro and transcriptionally profiled using time-series RNA-seq. Periodic genes were identified in each strain by JTK_CYCLE, and strain-intrinsic period length is evident. Each vertical line represents a time point, and gene expression is displayed horizontally. Expression values are mean-normalized for each gene and depicted as a z-score of standard deviations from the mean. Genes are ordered per strain by peak expression time.
A key characteristic of circadian or cell-cycle oscillators is a well-ordered program of periodic gene expression (14, 16). For each P. falciparum strain, we used the periodicity detecting algorithm JTK_CYCLE (21) to estimate the number of rhythmic genes (Table S1, File S1 and S2). Periodic genes are largely conserved among the four strains, with the majority of each strain’s periodic genes (79 – 82%) overlapping all other strains (Fig. S2), similar to previous observations (22, 23). The vast majority of the mapped transcriptome (between 87.3% and 92.5%) in P. falciparum appears periodically expressed (Fig. 1, Table S1, File S1–2) by visual inspection. These phase-specific, oscillating genes peak in expression across the entire cycle, forming a cascade of rhythmic genes, in a manner highly reminiscent of other oscillators.
Mammalian transcriptomic studies have noted that circadian genes tend to peak in phase clusters (“rush hours”) near dawn and dusk, and clustering is thought to represent activation of expression in anticipation of metabolic events (16, 24). To evaluate timing of gene peaks in P. falciparum, 3,703 periodic genes shared between all strains were mapped to a single representative cycle using a “wrapping” procedure that averaged overlapping measurements, similar in principle to phase dispersion minimization (25) (see Materials and Methods). This approach allowed comparison between parasites with different cycle times and, using microscopic assessments of intraerythrocytic-cycle phase (Fig. S3), mapping of the timing of gene peaks throughout the cycle (Fig. 2). Interestingly, we did not observe any evidence of “rush hours” in any of the phases of the intraerythrocytic cycle (Fig. 2).
Fig. 2.
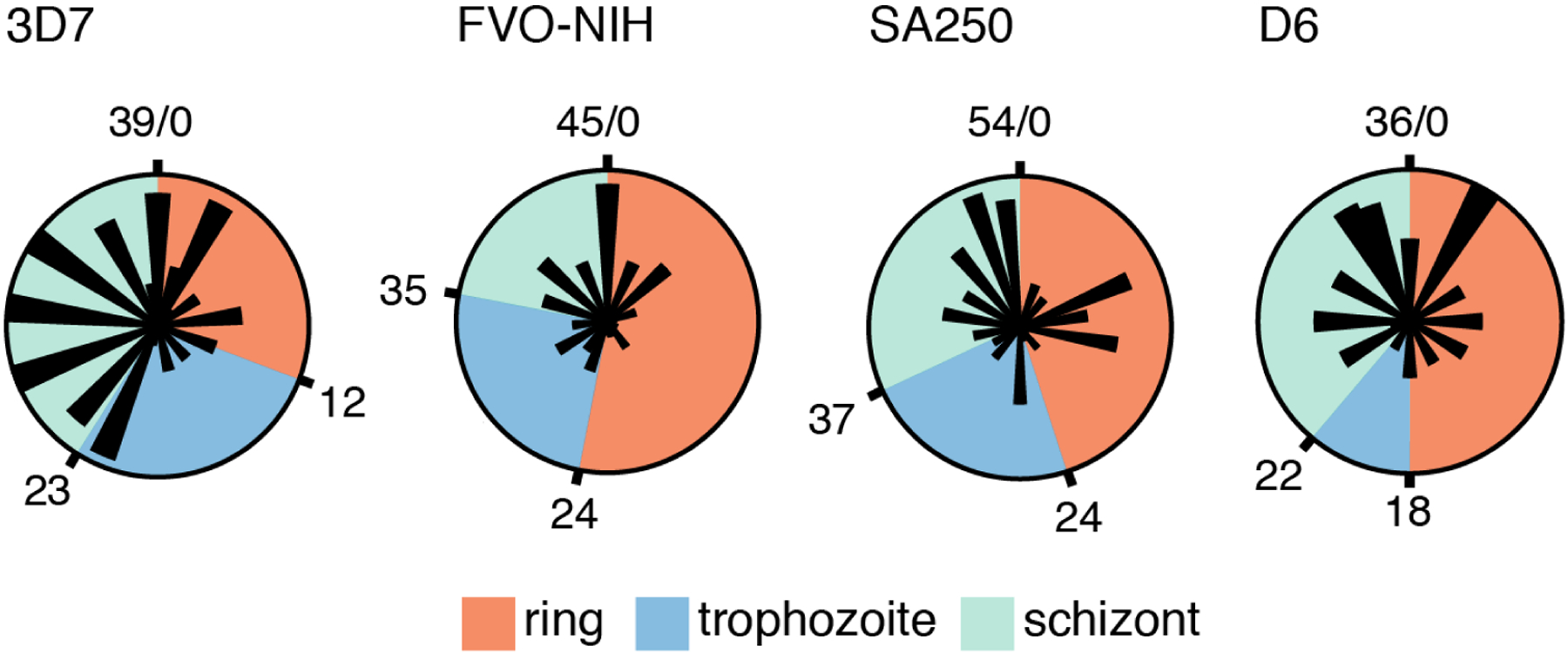
The periodic genes of P. falciparum are expressed in multiple phases of the intraerythrocytic cycle. Polar graphs depict the relative numbers of genes peaking per hour of each strain’s cycle. The boundaries between phases (ring, trophozoite, and schizont) were determined by microscopy (Fig. S3) and marked in hours (Table S5). Wrapped, interpolated expression data for the set of shared periodic genes (Materials and Methods) were used to assign peak times.
Multiple studies have shown that in both vertebrate and invertebrate circadian rhythms, a subset of genes oscillate at 8- and 12-hour periods (26, 27). Although the precise mechanisms of these “harmonic” rhythms have not been dissected, evidence from Clock gene mutants in mice indicate they arise from the core circadian clock mechanism (28). To search for such harmonics in P. falciparum, we took advantage of JTK_CYCLE’s period prediction features by running the algorithm with a period search range of 6 to 60 hours (6 to 54 hours in the case of D6). A minority of genes (between 3.5% and 4.3%) in each strain have a predicted period length roughly half that of the strain (Fig. 3, Table S2). Visualizing the expression of these genes confirms that they peak twice per cycle (Fig. 3, Fig. S4). Among this set of harmonic genes, most are unique to one strain; only three harmonic genes were identified in all four strains (Fig. S5).
Fig. 3.
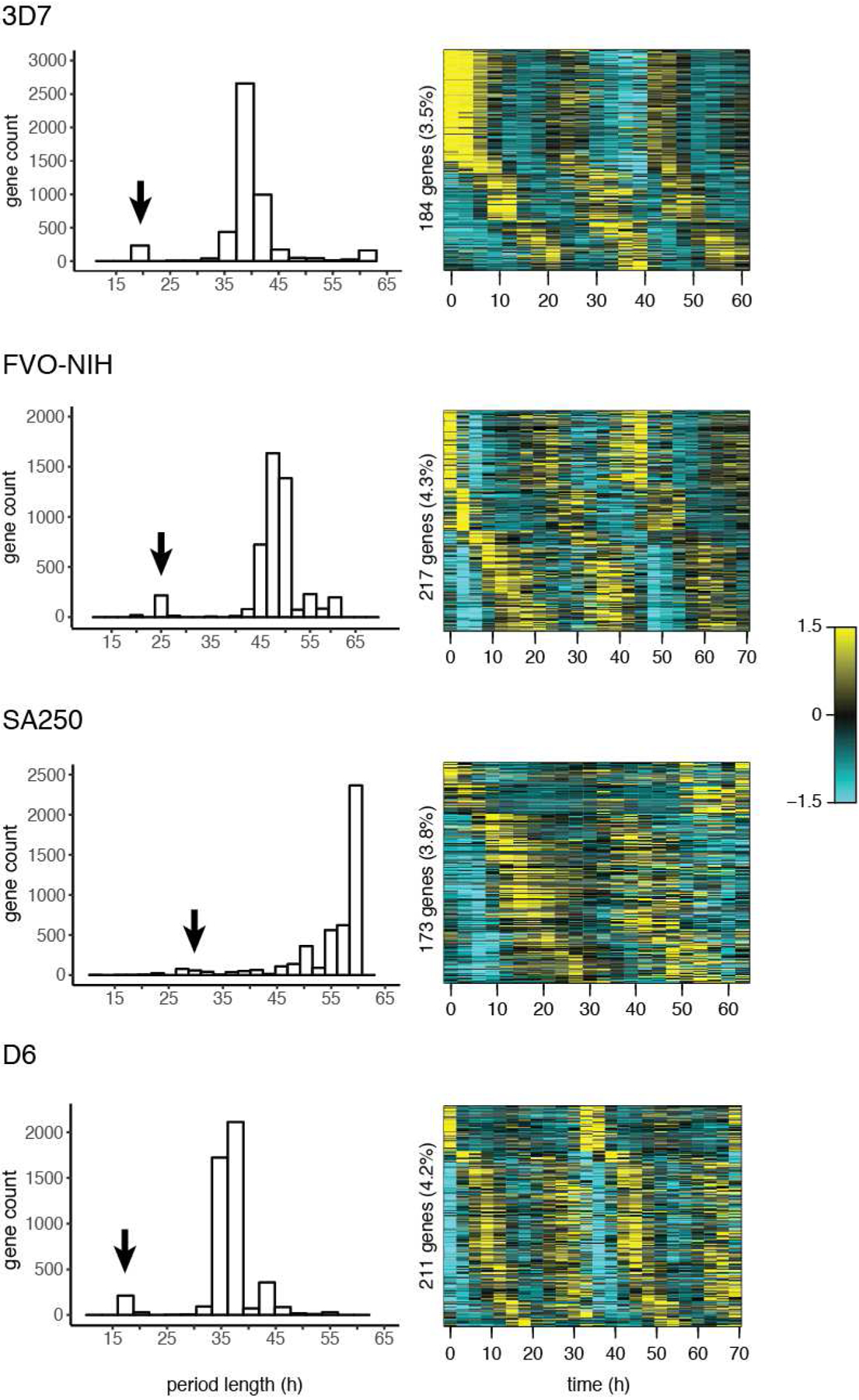
P. falciparum strains exhibit periodic genes at half the normal cycle length (harmonic expression). JTK_CYCLE was used to search for genes in a wide range of predicted period lengths; the distribution of predicted period lengths is shown for each strain. A minority of genes oscillate at approximately half the dominant period in each strain, indicated with an arrow; 21 hours in 3D7, 24 hours in FVO-NIH, 27 – 33 hours in SA250, and 18 hours in D6. These genes are plotted by heat map; each vertical line represents a time point, and gene expression is displayed horizontally. Expression values are mean-normalized for each gene and depicted as a z-score of standard deviations from the mean.
Quantitative characteristics of biological oscillators are found in P. falciparum:
Cell-cycle and circadian oscillators tend to produce well-ordered programs of transcription; however, substantial evolutionary divergence between species may yield novel ordering of orthologous periodic genes, correlating with substantial phenotypic change that may reflect rewired networks (29). We assessed the temporal ordering of the transcriptome between strains using a set of periodic genes shared between all strains. For each heatmap in Fig. 4A, the genes are plotted in the order of peak time of expression in the strain 3D7. Visual inspection indicates that the ordering of peak gene expression is globally well-conserved between all four strains (Fig. 4A). Similar qualitative levels of conservation were observed when the analysis was repeated using each of the remaining strains as the ordering standard (Fig. S6).
Fig. 4.
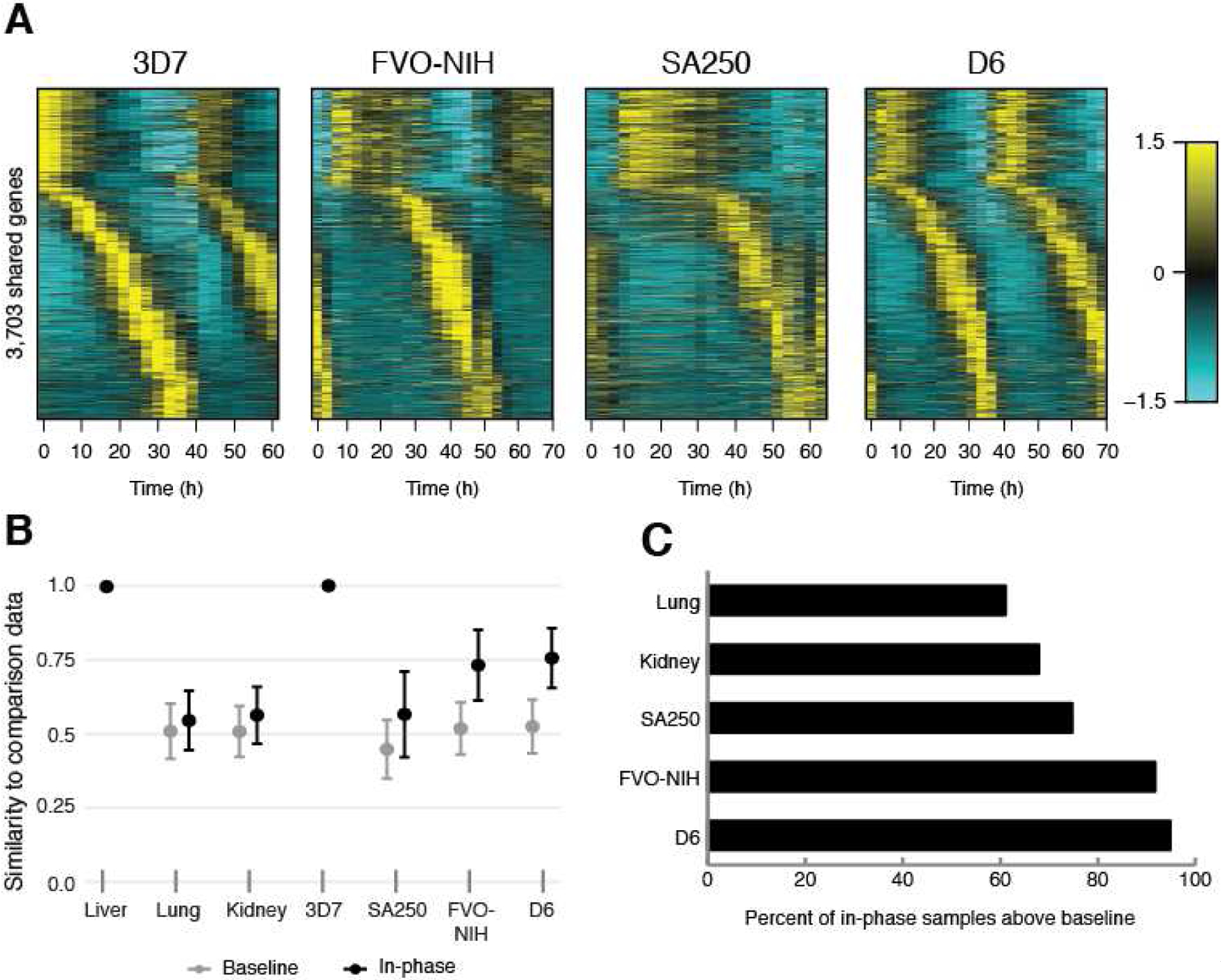
Ordering of periodic gene expression is broadly conserved among four strains of P. falciparum comparable to mouse circadian genes. (A) The ordering of genes for 3D7, as determined by peak expression time, was applied to the remaining three strains (FVO-NIH, SA250, D6). The resulting heat maps show highly conserved ordering (see Fig. 1 for comparison). Each vertical line represents a time point, and gene expression is displayed horizontally. Expression values are mean-normalized for each gene and depicted as a z-score of standard deviations from the mean. (B,C) A set of periodic genes from parasite (119 genes) and mouse circadian data (107 genes) were identified which peaked in very similar cycle phase to the reference strain (3D7) or tissue (liver). A null baseline distribution was created, and both the baseline and “in-phase” genes were sampled 5,000 times in sets of six genes to produce estimates of gene ordering similarity across strains or tissues. (B) Mean n and standard deviation of ordering similarity to reference strain/tissue, averaged across all samples and all applied levels of noise (6 – 10% ε.) (C) Percent of “in-phase” samples above baseline.
To make a quantitative determination of the similarity in ordering, a recently developed method was used to measure the conservation of gene ordering between data sets (30). In brief, extrema (peaks and troughs) for each gene’s normalized expression pattern were assigned a time interval and a partial order was computed. The use of partial orders allows a quantitative assessment while acknowledging that the order of some extrema cannot be distinguished because they both fall in the same time point. To account for stochastic behavior of gene expression and the relatively coarse sampling, partial orders were evaluated in the presence of 6 – 10% noise (ε) (Fig. 4B and C). A similarity score for each ε is computed between partial orders of the same subset of genes from two data sets (30). We calculated conservation of gene ordering among our strains of P. falciparum and compared this conservation to a collection of time-series data obtained from an established circadian oscillator. For the latter, we used the high-density time-series data of Zhang et al. (16), which profiled the circadian transcriptomes of 12 mouse organ tissues every two hours for 48 hours. In both species we used subsets of periodic genes that peaked at similar times across parasite strains or mouse tissues, which we call “in-phase” subsets, and computed a null baseline by randomizing the corresponding time series within each dataset (Materials and Methods).
We found that the ordering of similarity scores of the null baselines for circadian and parasite data were comparable, with in-phase similarity scores equivalent or higher in P. falciparum (Figs. 4B and 4C, Table S3). Furthermore, the percent of samples above the null baseline in all three parasite strains (75–95%) was greater than in both mouse circadian tissues (61% in lung and 68% in kidney, Fig. 4C, Table S3). Although it has been observed that circadian genes are not perfectly ordered across mouse tissues (16), the observation that P. falciparum strains exhibit ordering comparable to or better than circadian genes suggest that the mechanisms guiding the parasite transcriptional cycle are capable of maintaining high-fidelity ordering.
The analyses thus far indicate that P. falciparum’s transcriptome dynamics share features with known cell-intrinsic biological oscillators, yet the results could still be consistent with an extrinsic mechanism for the control of periodicity. While most conventional rhythmic host cues are absent in RBC culture conditions, a 24-hour peroxiredoxin clock identified in RBCs (8) could be sufficient to drive intraerythrocytic cycle periodicity by repeated cascades of gene expression.
To ascertain the intraerythrocytic cycle period length of each of our cultured strains, we used several distinct metrics to avoid bias from any one method. We interpolated and wrapped each strain’s transcriptome (Fig. S7) and microscopic culture progression curves for ring, trophozoite, and schizont stages (Fig. S8) to find the periods that minimized overlapping error for each strain (Materials and Methods, Table S4). We also measured the distances between recurring expression peaks and expression troughs for each rhythmic gene and identified the modal value in these peak-distance and trough-distance distributions, providing two more estimates of genetic period length (Fig. S9–10, Table S2). Due to some disagreement between metrics in some cases, we established a final estimated period by weighted average of all four metrics (Table S2). We observe that the strains differ in period regardless of the method of estimation and that the rank order of strains in terms of period length is the same.
While the typical in vivo P. falciparum infection exhibits a 48-hour periodicity, we observed that each cultured strain had a unique period length that sometimes varied substantially from 48 hours (Table S2). These variations are also apparent in visualization of each strain’s periodic transcriptomes (Fig. 1), and are not due to growth conditions, as three of four strains were cultured in parallel in blood from a single donor (Materials and Methods and File S3). Strain SA250 has the longest estimated cycle at 54 hours, while strain D6’s cycle is a mere 36 hours long. These results are incompatible with the hypothesis that the 24-hour peroxiredoxin clock is responsible for controlling the parasite’s rhythm; if such were the case, period lengths in all four strains would be roughly 48 hours long. The observations are, however, consistent with what is known about cell-intrinsic oscillators, in which period length is genetically controlled (12, 31, 32).
The significant diversity in cycle period lengths between P. falciparum strains raises the question of how such shortening and lengthening from the wild-type 48 hours has occurred. In the eukaryotic cell cycle, the length of G1 is the most flexible because factors such as cell size and the availability of nutrients or growth factors all affect the ability of the cells to move into S-phase. If P. falciparum’s intraerythrocytic cycle stages are analogous to the cell-cycle phases, as has been suggested (33), the bulk of the change in period length between strains may be confined to particular stages.
Using microscopy data re-wrapped to the final estimated cycle lengths (Table S4, Fig. S11) stages were labeled based on dominant parasite phase (>50% of parasites) and the length of each stage was calculated as a percent of the total period length. We are unable to detect a single stage or stages that show particular conservation or flexibility in length, whether measured in hours or in percent of the period length (Table S5). While the ring stage shows the most variability, and the trophozoite stage appears the most stable, there is no consistent correlation between total period lengths and stage lengths. The intraerythrocytic cycle appears to be plastic in terms of lengthening and shortening throughout all stages.
Variance in period length in P. falciparum culture is comparable to a circadian model system:
When Trager and Jensen published the first protocol for in vitro culture of P. falciparum in 1976, they noted that an initially synchronous parasite population from a clinical malaria sample eventually desynchronizes in culture, becoming a heterogenous mix of ring, trophozoite, and schizont stages (34). It has come to be broadly accepted in the field that synchronized parasites lose synchrony “rapidly” in culture, an observation that would appear to be largely inconsistent with a robust intrinsic oscillator. However, cells synchronized in the cell-cycle lose synchrony over time (35), and circadian oscillators also lose synchrony in cell-based systems in the absence of entrainment cues (36) due to variance in the individual cycle times.
To determine whether synchrony loss due to cell-to-cell variance observed in populations of P. falciparum is compatible with the variance in periods observed in cell-intrinsic circadian oscillations, we fit a simple phase-oscillator model (see Materials and Methods) to the microscopic staging data for the four strains along with an additional strain, HB3, from a prior study (23) (Fig. S1B and C, S12). We calculated the coefficient of variation (CoV) for all P. falciparum strains from this model and compared these values to the calculated CoV of single-cell traces of a circadian reporter gene in human fibroblast cells (37) (Table S6). The CoV of circadian cycles in the fibroblast population was estimated to be 0.0845, meaning that the period lengths exhibit a standard deviation of roughly 8.45% of the average period length. Strikingly, the estimated CoVs of the five P. falciparum strain cycles were similar or smaller, ranging between 0.23%–10.18% of estimated mean cycle period depending on the strain (Fig. 5, Table S6). Increasing or decreasing the mean cycle period by one hour does not appreciably change the estimated CoV (Fig. 5, Table S6).
Fig. 5.
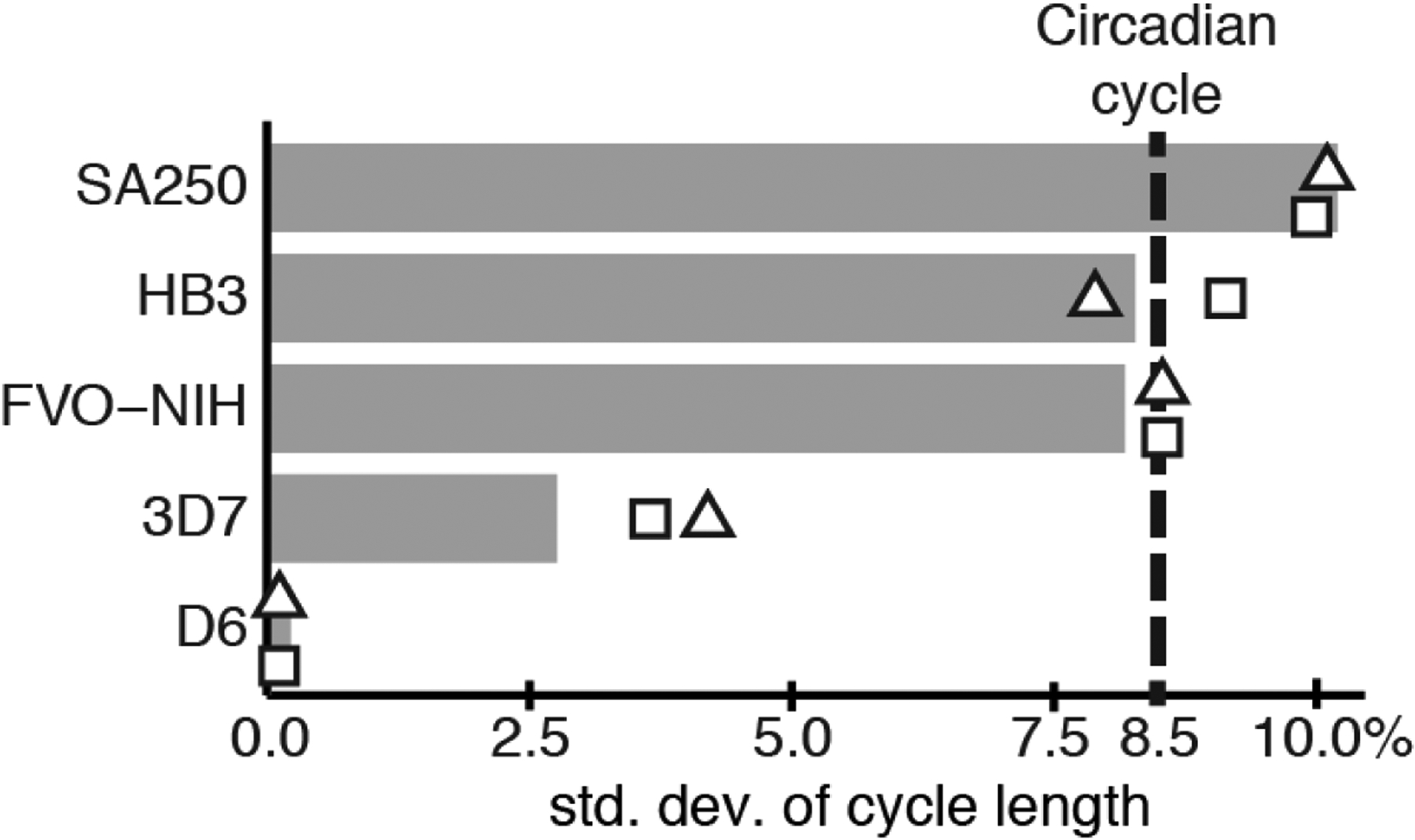
Variation of period lengths between cycles in P. falciparum is comparable to a circadian model. Microscopic times-series data from this study, along with additional data from strain HB3 (23), was fit to a phase-oscillator model, yielding an estimated standard deviation of cycle length as percent of the mean cycle length. The dashed line represents the empirical standard deviation of the circadian cycle derived from single-cell imaging of circadian reporters in fibroblast cells (37). Estimates are also shown if the mean cycle length is lengthened or shortened by one hour (Table S6).
These results were produced using best-fit estimates for parameters in the phase-oscillator model, such as strain-specific period length and initial population synchrony (Table S6). In order to ensure the model wasn’t artificially lowering estimates for period variability by adjusting other parameters, we recomputed optimal model parameters while enforcing a minimum allowed period variability that is larger than the empirically determined circadian oscillator (see Materials and Methods, Table S7). This alteration significantly decreases the model’s fit to the experimental data, particularly in the second round of replication where dampening due to synchrony loss becomes more apparent (Fig. S12).
Recently, it was suggested that microscopic staging curves over-estimate parasite population synchrony of P. falciparum in culture (38) because parasite replication during the schizont stage enforces synchrony on the schizont-ring transition. To make sure that our models were not unfairly biasing our estimates of variance, we explicitly added replication to the model at the schizont-ring transition, assuming a parasite multiplication rate of N = 4, 8, or 16 (Materials and Methods). We find that with the addition of replication, the dynamics of the experimental staging curves are better matched by modeling populations with smaller variance in period length than found in the simple oscillator model (Fig. S13). Thus, including replication in our model bolsters the finding that P. falciparum’s estimated period length variability, and thus the rate at which the population loses synchrony, is comparable to a circadian oscillator.
Discussion:
Periodic biological processes can arise from cell or organism-intrinsic oscillators or may be imposed by extrinsic rhythmic factors. Classic examples of cell-intrinsic biological oscillators include circadian rhythms and the eukaryotic cell cycle (14, 32). Here we investigated the origin of periodicity in the intraerythrocytic cycle of P. falciparum using an in vitro culture system where extrinsic rhythmic cues are largely eliminated. We find that P. falciparum shares many molecular signatures of well-characterized cellular oscillators, and that the data are most consistent with a model in which rhythmic behaviors are driven by a parasite-intrinsic oscillator.
Consistent with previous studies (22, 23, 39), we find that the majority (87.3% to 92.6%) of P. falciparum’s mapped genes are rhythmic in their expression during the cycle. This large periodic program of phase specific transcription is also observed in cell-cycle (~15–20% of the genome) (14) and circadian systems (~40–80% of the genome) (16, 24), although rhythmicity itself does not indicate an intrinsic oscillator. Circadian control of gene expression has been proposed to link various physiological processes with light/dark cycles (e.g. sleep/wake cycles) (11) or to temporally separate incompatible biochemical process (40), whereas the temporal program of expression in the cell-cycle is a foundational mechanism for ordering cell-cycle events (41). Unlike circadian gene expression that tends to cluster roughly near dawn and dusk, there does not appear to be a consistent pattern of phase clustering in P. falciparum. Given the ordered nature of P. falciparum development during the intraerythrocytic cycle, it is likely the temporal program of transcription serves a purpose similar to the cell-cycle transcriptional program, where just-in-time gene expression helps maintain temporal ordering. It has been suggested that these parasite stages may be analogous to the familiar G1, S, G2, and M stages of the eukaryotic cell cycle (33, 42), with the ring stage specifically suggested to be the equivalent of G1. In a typical cell cycle, the cell-cycle period is adjusted mostly by expanding or shortening in the G1 phase (43). However, in the four P. falciparum strains we examined, there was no discernible pattern of stage expansion/contraction to explain the diversity in strain cycle lengths.
Like circadian systems from several organisms (26–28), genes that oscillate at the first harmonic of the period length (i.e. twice as fast) are observed in all four P. falciparum strains. Importantly, the harmonic genes are half-period regardless of the different period length of the strains, indicating that like circadian oscillators, harmonic expression is driven by the core network and not by an alternative oscillator. The fact that strains have a unique period, despite three of the four strains growing in RBC cultures from the same donor, indicates that periodicity is not driven by the peroxiredoxin oscillations reported to function with a 24-hour period in RBCs. Moreover, the periods are not exact multiples of 24 hours, so they are unlikely to result from some alternative coupling to the peroxiredoxin cycle. These findings are supported by studies that indicate genetic variation or mutation of Plasmodium genes can lead to altered cycle period length (33, 42).
It has been suggested that synchronized populations of P. falciparum in in vitro cultures lose synchrony rapidly. Rapid synchrony loss argues that without input from the host, the parasite is unable to maintain a robust period. However, synchronized populations of cells under circadian- and cell-cycle oscillator controls have also been shown to lose periodicity due to variation in period (35, 36), raising the question of how variable the P. falciparum intraerythrocytic cycle is when compared to cell-cycle or circadian controlled systems. To examine this question, we used a phase-oscillator model to estimate the variation in cycle period lengths between parasites in culture, and found it is comparable to variation found in circadian cell lines. Thus, the degree of synchrony loss observed for synchronized P. falciparum is completely compatible with a model in which the intraerythrocytic cycle is driven by a robust intrinsic clock network.
Collectively, our findings are incompatible with a mechanism in which P. falciparum’s intraerythrocytic cycle is controlled only by extrinsic cues. Our findings point to a mechanism in which the cycle is driven by an intrinsic oscillator with molecular characteristics of circadian or cell-cycle oscillators. The control mechanisms for circadian and cell-cycle oscillators have been identified as gene regulatory networks comprised of transcriptional regulators, kinases, and ubiquitin ligases with negative feedback loops that drive oscillation. Within the genes we have identified as periodic, we found ApiAP2 transcription factor genes (44) along with genes annotated as kinases, suggesting the possibility that gene regulatory networks resembling circadian- or cell-cycle oscillators may be present. It is not surprising that orthologs to known circadian clock genes have not been identified in Plasmodium spp. (5), as even gene networks with similar oscillating functions often do not share genes. Genes in oscillating networks from different organism may not be highly conserved, but network motifs and topologies are (11, 45).
Farnert et al. found that in chronic infections with P. falciparum, 48-hour rhythms in parasitemia levels could be observed in patients that did not exhibit fever cycles, and led them to speculate that “mechanisms other than fever might be involved” (46). The existence of an intrinsic oscillator in P. falciparum is a likely mechanism, and reframes the models of periodicity in malarial disease as the coupling between a parasite oscillator with a 48-hour period and the host circadian oscillator with a 24-hour period. Indeed, there is evidence for phase alignment between Plasmodium parasites and animal models (2, 7). Plasmodium’s oscillator may entrain to circadian rhythms in the host, much as circadian clocks themselves ultimately entrain to light (47). The host signal(s) that the parasite uses for entrainment are as yet unsettled (6, 7). It is also possible that the parasite can manipulate the host circadian cycle to achieve better alignment. In mouse experiments, entrainment of the intraerythrocytic cycle to the host shows benefits for the parasite (4, 48). The benefit of phase alignment to the host circadian rhythm is a likely explanation for why mammalian-infecting Plasmodium species have cycle lengths that are multiples of 24 hours. The lack of this selective pressure in vitro (and/or genetic bottlenecking during culture establishment) may also explain why cultured P. falciparum strains, such as s those in this study, can vary substantially from their 48-hour wild-type period length.
Plasmodium is not the only parasite with a highly rhythmic life cycle as several parasites exhibit time-of-day elements in their life cycle (5). Moreover, an innate circadian rhythm was recently identified in the Trypanosoma brucei, the causal organism of sleeping sickness, using traditional metrics of circadian clocks (49). Recent reports indicate that nearly 80% of all genes in a primate genome are under circadian control in at least one tissue (24). Given that animal physiology is so broadly controlled by circadian rhythm it makes sense that pathogens have evolved to take advantage of the 24-hour periodicity of the host, and it is likely that many pathogens will display periodic behaviors.
Materials and Methods
Plasmodium falciparum strains, culturing and synchronization
P. falciparum parasites were synchronized by alanine treatment and temperature cycling as described (50). Details can be found in Supplemental Material.
RNA extraction, sequencing, and read processing
Total RNA from frozen, packed RBCs was extracted via a phenol-chloroform protocol as previously described (51). Library preparation and sequencing was performed by the Duke Sequencing Core. STAR(52) was used to create a genome index for read mapping (Table S8), using the P. falciparum 3D7 v3.0 genome as found on www.genedb.org, and the Sanger Institute’s 3D7 annotation downloaded on March 4th, 2016. Reads are presented as FPKM. Details can be found in Supplemental Material.
Periodic gene detection and ordering.
Genes were filtered for peak FPKM expression of at least 2 (or 3, with the addition of 1) to reduce noisy genes. We used JTK_CYCLE (21) to filter for periodic genes. After arriving at a more precise determination of period length for each strain (see “Estimation of period length per strain” and Table S2). Details can be found in the Supplementary Material.
Computing gene ordering similarities
A measure of similarity was developed (30) to compare the well-ordering of gene expression time-series data with regard to phase shift. This similarity measure is based on the timing of peaks and valleys for a collection of genes compared across data sets and is independent of amplitude due to time series normalization on the interval [−0.5,0.5]. Details can be found in the Supplementary Material.
Estimation of period length per strain
A consensus cycle length for each strain was determined on the basis of four measurements: (1) error-minimizing expression cycle length, (2) error-minimizing microscopy cycle length, (3) the distribution of distances in hours of the two largest expression maxima for all genes in each strain, and (4) the distribution of distances of the two smallest expression minima for all genes in each strain. Details can be found in the Supplementary Material.
Comparison of parasite stage lengths between strains
To compare the relative lengths of each parasite intraerythrocytic cycle stage (ring, trophozoite, and schizont,) we re-wrapped the microscopy data using the above described wrapping procedure, this time to the consensus period length determined d by the method explained above. Fig. S11 visualizes this procedure and Table S4 provides RMSE values for the wrap.
Supplementary Material
Acknowledgments:
Thanks to Dr. David K. Welsh (Department of Psychiatry, University of California, San Diego) for kindly providing us with data from the primary fibroblasts. We also thank Robert Moseley and Sophia Campione for critical reading of the manuscript, and S. Campione for technical help with figures. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
Funding: FM, SBH, JH, ARL and CMK were funded by the Defense Advanced Research Projects Agency, D12AP00025. SBH, LMS, TG, and BC were also funded by NIH 1R01GM126555-01. TG and BC were funded NSF DMS-1839299. RRN was funded by NIH R01 GM126555-01. SBH and JH are members of Mimetics, LLC. JH is CEO of Geometric Data Analytics, Inc. TG and BC are on the board of Kanto, Inc.
Footnotes
Modelling Synchrony Loss with a Population of Phase Oscillators
See Supplemental Materials for details of the model.
Competing interests: Authors declare no competing interests.
Data and materials availability: All data used in the paper are available in supplemental files. RNA sequences are deposited at the Gene Expression on Omnibus under series GSE141653. All code used in analyses are available in public repositories (53–55).
List of Supplementary Materials:
Data Files S1 to S4
References and Notes:
- 1.Kwiatkowski D, Nowak M, Periodic and chaotic host-parasite interactions in human malaria. Proc Natl Acad Sci U S A 88, 5111–5113 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawking F, The clock of the malaria parasite. Sci Am 222, 123–131 (1970). [DOI] [PubMed] [Google Scholar]
- 3.Gautret P, Deharo E, Tahar R, Chabaud AG, Landau I, The adjustment of the schizogonic cycle of Plasmodium chabaudi chabaudi in the blood to the circadian rhythm of the host. Parasite 2, 69–74 (1995). [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell AJ, Mideo N, Reece SE, Disrupting rhythms in Plasmodium chabaudi: costs accrue quickly and independently of how infections are initiated. Malar J 12, 372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reece SE, Prior KF, Mideo N, The Life and Times of Parasites: Rhythms in Strategies for Within-host Survival and Between-host Transmission. J Biol Rhythms 32, 516–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotta CT et al. , Calcium-dependent modulation by melatonin of the circadian rhythm in malarial parasites. Nat Cell Biol 2, 466–468 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Hirako IC et al. , Daily Rhythms of TNFalpha Expression and Food Intake Regulate Synchrony of Plasmodium Stages with the Host Circadian Cycle. Cell Host Microbe 23, 796–808 e796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill JS, Reddy AB, Circadian clocks in human red blood cells. Nature 469, 498–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nozue K, Maloof JN, Diurnal regulation of plant growth. Plant Cell Environ 29, 396–408 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Zhang EE, Kay SA, Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11, 764–776 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Takahashi JS, Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konopka RJ, Benzer S, Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millar AJ, Straume M, Chory J, Chua NH, Kay SA, The Regulation of Circadian Period by Phototransduction Pathways in Arabidopsis. Science 267, 1163–1166 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Orlando DA et al. , Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453, 944–947 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbison CT et al. , Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walliker D et al. , Genetic-Analysis of the Human Malaria Parasite Plasmodium-Falciparum. Science 236, 1661–1666 (1987). [DOI] [PubMed] [Google Scholar]
- 18.Jensen JB, Trager W, Plasmodium-Falciparum in Culture - Establishment of Additional Strains. Am J Trop Med Hyg 27, 743–746 (1978). [DOI] [PubMed] [Google Scholar]
- 19.Awandare GA et al. , Plasmodium falciparum field isolates use complement receptor 1 (CR1) as a receptor for invasion of erythrocytes. Mol Biochem Parasit 177, 57–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathod PK, McErlean T, Lee PC, Variations in frequencies of drug resistance in Plasmodium falciparum. P Natl Acad Sci USA 94, 9389–9393 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes ME, Hogenesch JB, Kornacker K, JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL, Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34, 1166–1173 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bozdech Z et al. , The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. Plos Biol 1, E5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mure LS et al. , Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, 1232–+ (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stellingwerf RF, Period Determination Using Phase Dispersion Minimization. Astrophys J 224, 953–960 (1978). [Google Scholar]
- 26.Hughes ME et al. , Harmonics of circadian gene transcription in mammals. PLoS Genet 5, e1000442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins BM, Das AK, Antunes L, Locke JC, Frequency doubling in the cyanobacterial circadian clock. Mol Syst Biol 12, 896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes ME et al. , Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet 8, e1002835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moseley RC et al. , Conservation and Diversification of Circadian Rhythmicity Between a Model Crassulacean Acid Metabolism Plant Kalanchoe fedtschenkoi and a Model C3 Photosynthesis Plant Arabidopsis thaliana. Front Plant Sci 9, 1757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry E et al. , Using extremal events to characterize noisy time series. J Math Biol 80, 1523–1557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons Kovacs LA et al. , Cyclin-dependent kinases are regulators and effectors of oscillations driven by a transcription factor network. Mol Cell 45, 669–679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janse CJ et al. , Malaria parasites lacking eef1a have a normal S/M phase yet grow more slowly due to a longer G1 phase. Molecular Microbiology 50, 1539–1551 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Trager W, Jensen JB, Human malaria parasites in continuous culture. Science 193, 673–675 (1976). [DOI] [PubMed] [Google Scholar]
- 34.Orlando DA et al. , A probabilistic model for cell cycle distributions in synchrony experiments. Cell Cycle 6, 478–488 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA, Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14, 2289–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leise TL, Wang CW, Gitis PJ, Welsh DK, Persistent cell-autonomous circadian oscillations in fibroblasts revealed by six-week single-cell imaging of PER2::LUC bioluminescence. PLoS One 7, e33334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greischar MA, Reece SE, Savill NJ, Mideo N, The Challenge of Quantifying Synchrony in Malaria Parasites. Trends Parasitol 35, 341–355 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Le Roch KG et al. , Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Boxall SF, Dever LV, Knerova J, Gould PD, Hartwell J, Phosphorylation of Phosphoenolpyruvate Carboxylase Is Essential for Maximal and Sustained Dark CO2 Fixation and Core Circadian Clock Operation in the Obligate Crassulacean Acid Metabolism Species Kalanchoe fedtschenkoi. Plant Cell 29, 2519–2536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haase SB, Wittenberg C, Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 196, 65–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reilly Ayala HB, Wacker MA, Siwo G, Ferdig MT, Quantitative trait loci mapping reveals candidate pathways regulating cell cycle duration in Plasmodium falciparum. BMC Genomics 11, 577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan DO, The cell cycle: principles of control. Primers in biology (Published by New Science Press in association with Oxford University Press; Distributed inside North America by Sinauer Associates, Publishers, LondonSunderland, MA, 2007), pp. xxvii, 297 p. [Google Scholar]
- 43.Painter HJ, Campbell TL, Llinas M, The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol 176, 1–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertoli C, Skotheim JM, de Bruin RA, Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 14, 518–528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farnert A, Snounou G, Rooth I, Bjorkman A, Daily dynamics of Plasmodium falciparum subpopulations in asymptomatic children in a holoendemic area. Am J Trop Med Hyg 56, 538–547 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Roenneberg T, Daan S, Merrow M, The art of entrainment. J Biol Rhythms 18, 183–194 (2003). [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell AJ, Schneider P, McWatters HG, Reece SE, Fitness costs of disrupting circadian rhythms in malaria parasites. Proc Biol Sci 278, 2429–2436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, Takahashi JS, Figueiredo LM, Trypanosoma brucei metabolism is under circadian control. Nat Microbiol 2, 17032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roche K, kimberlyroche/plasmodium_periodicity: First release, version v1.0.0, Zenodo (2020); 10.5281/zenodo.3710104. [DOI]
- 50.Cummins B, breecummins/2019-Intrinsic-Oscillator-Malaria: Version 0.1.0, version v0.1.0, Zenodo (2020); 10.5281/zenodo.3710436. [DOI]
- 51.Motta FC, P. falciparum synchrony loss, Zenodo (2020); 10.5281/zenodo.3712146. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


