ABSTRACT
EAR motif-containing proteins are able to repress gene expression, therefore play important roles in regulating plants growth and development, plant response to environmental stimuli, as well as plant hormone signal transduction. ABA is a plant hormone that regulates abiotic stress tolerance in plants via signal transduction. ABA signaling via the PYR1/PYLs/RCARs receptors, the PP2Cs phosphatases, and SnRK2s protein kinases activates the ABF/AREB/ABI5-type bZIP transcription factors, resulting in the activation/repression of ABA response genes. However, functions of many ABA response genes remained largely unknown. We report here the identification of the ABA-responsive gene SlEAD1 (Solanum lycopersicum EAR motif-containing ABA down-regulated 1) as a novel EAR motif-containing transcription repressor gene in tomato. We found that the expression of SlEAD1 was down-regulated by ABA treatment, and SlEAD1 repressed reporter gene expression in transfected protoplasts. By using CRISPR gene editing, we generated transgene-free slead1 mutants and found that the mutants produced short roots. By using seed germination and root elongation assays, we examined ABA response of the slead1 mutants and found that ABA sensitivity in the mutants was increased. By using qRT-PCR, we further show that the expression of some of the ABA biosynthesis and signaling component genes were increased in the slead1 mutants. Taken together, our results suggest that SlEAD1 is an ABA response gene, that SlEAD1 is a novel EAR motif-containing transcription repressor, and that SlEAD1 negatively regulates ABA responses in tomato possibly by repressing the expression of some ABA biosynthesis and signaling genes.
KEYWORDS: EAR motif-containing protein, ABA, SlEAD1, transcription factor, tomato
Introduction
The EAR (ERF-associated amphiphilic repression) motifs, which contain a conserved sequence pattern of (L/F)DLN(L/F)xP, were initially identified in the class II ERFs (Ethylene-responsive factors) and some C2H2 family proteins, and have been shown to confer transcriptional repression activities.1 Based on sequence comparison of the EAR motifs in class II ERFs, C2H2 family proteins, and some other EAR motif-containing proteins, the consen sus sequence patterns of EAR motifs were further refined as LxLxL and DLNxxP.2 EAR motif-proteins mediated transcriptional repression has been considered to be the main form of transcriptional repression in plants,3 and a genome wide searching of LxLxL or DLNxxP sequence-containing proteins have identified more than 20,000 EAR motif-containing proteins from 71 different plant species.4
EAR motif-containing proteins are able to function as transcription repressors or recruit co-repressors to repress gene expression, therefore play important roles in regulating plants growth and development, as well as plant response to environmental stimuli.4 For example, KIX8 (KINASE-INDUCIBLE DOMAIN INTERACTING 8) and KIX9 repress leaf growth via function as adaptor proteins for the co-repressor TOPLESS,5 OFP1 (Ovate family protein 1) regulates cell elongation via repressing the expression of GA20ox1 (Gibberellin 20-oxidase 1),6 TOE1 (TARGET OF EARLY ACTIVATION TAGGED (EAT) 1) and TOE2 regulate flowering via repressing the expression of FT (FLOWERING LOCUS T),7 whereas several repressor ERFs such as ERF4 and ERF7 are involved in the regulation of plant responses to stresses.1,8,9 Several tomato ERFs have also been found to regulate plant growth and development including stomatal density, photosynthesis and fruit ripening, and plant resistance pathogen infections.10–12
EAR motif-containing proteins are also involved in the regulation of hormone signaling, for example, ERFs regulate ethylene signaling,1 JAZ (Jasmonate ZIM domain) proteins regulate jasmonic acid signaling, Aux/IAA proteins regulate auxin signaling, and D53 (DWARF 53) and SMXL7 (SMAX1-LIKE 7) regulate strigolactone signaling.2,13-16 At least in some cases, changes in hormone singling caused by EAR motif-containing proteins lead to alternation in plants growth and development, as well as plant response to environmental stimuli. For example, D53 and SMXL7 are involved in the regulation of shoot development.15,16 Whereas AITRs (ABA-induced transcription repressors), a novel family of transcription repressors involved in the regulation of ABA (Abscisic acid) signaling, are able to regulate plant response to abiotic stresses.17
ABA is one of the five classic hormones in plants and plays a key role in regulating plant responses to abiotic stresses such as drought, heat, cold, and salinity.18–20 ABA regulates plant abiotic stress responses via signal transduction.18–22 ABA signaling is started by the recognition of ABA molecules by the PYR1 (Pyrabactin resistance 1)/PYLs (PYR1-likes)/RCARs (Regulatory component of ABA receptors) receptors.23–25 Binding of ABA by the PYR/PYLs/RCARs receptors enables their interaction with the A-group PP2Cs (PROTEIN PHOSPHATASE 2Cs) phosphatases,26,27 who are interacted, at the absence of ABA, with the SnRK2s (NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASES 2s) protein kinases and inhibiting their activities.28 Interaction of PYR/PYLs/RCARs with PP2Cs led to the release of SnRK2s, which in turn is able to activate the downstream ABF/AREB/ABI5-type bZIP (basic region leucine zipper) transcription factors,29,30 resulting in the activation/repression of ABA response genes, and therefore the changes of plant responses to abiotic stresses.22,26-28,31
Among the ABA response genes, some have been identified as transcription factor genes from several different families such as the R2R3 MYB family, the NAC (NAM, ATAF1/2, and CUC) family, the bHLH (basic Helix-Loop-Helix) family, the GARP (Golden2, ARR-B, Psr1) family, and the WDR (WD40-repeat) family.31–39 As mentioned above, we have previously identified a novel family of transcription factors AITRs, we found that AITRs are conserved in angiosperms, and function as negative regulators of ABA signaling.17 However, even though some of the AITRs contain a full conserved LxLxL EAR motif,17 AITRs were not identified in the previously genomes wide searching for EAR motif-containing proteins,4 possibly due to the amino acid sequence diversity of the EAR motif-containing proteins.4 This suggests that there may still be more EAR motif-containing proteins remained unidentified.
To identify novel EAR motif-containing regulators in ABA signaling in tomato, we performed data mining by using available transcriptome datasets to identify ABA-responsive genes with unknown functions, and then searched for the presence of EAR motif in the candidates. We found that the expression of SlEAD1 (Solanum lycopersicum EAR motif-containing ABA down-regulated 1) was down-regulated by ABA treatment, and SlEAD1 is a novel EAR motif-containing transcription repressor in tomato. By generating and characterizing gene edited slead1 mutants, we found that SlEAD1 is involved in the regulation of root elongation, and functions as negative regulator in regulating ABA responses in tomato.
Materials and Methods
Plant Materials and Growth Conditions
The Columbia-0 (Col) wild type Arabidopsis (Arabidopsis thaliana) was used for protoplast isolation. The Micro-Tom wild type tomato (Solanum lycopersicum) was used for protoplast isolation and plant transformation, and as a control for seed germination and root elongation assays. The slead1 mutants were generated by using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) gene editing in the Micro-Tom wild type background.
To generate seedlings for plant transformation and protoplast isolation, seeds of Micro-Tom wild type tomato were surface-sterilized and germinated as described previously.40 For seed germination and root elongation assays, surface-sterilized seeds of the Micro-Tom wild type and the slead1 mutants were plated on 1/2 MS plates with 0.8% agar and 1% sucrose. The plates were kept at 4°C in darkness for 2 days and then transferred into a plant growth chamber.
For protoplast isolation from Arabidopsis, seeds of the Col wild type were sown directly into soil pots and grew in a growth chamber. The growth conditions in the growth chamber were set at 22°C for Arabidopsis and 24°C for tomato, with a light density of ~120 μmol m−2 s−1, and a 16 h/8 h light/dark cycle.
Bioinformatics Analysis of EADs
Homologues of SlEAD1 in other plants were identified by using “Protein Homologs” on phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#), and check on PlantEAR (http://structuralbiology.cau.edu.cn/plantEAR) to see if they have been identified as EAR motif-containing proteins. The full-length amino acid sequences of EADs (Supplemental file 1) were used for phylogenetic analysis by using “One Click” mode on phylogeny (http://www.phylogeny.fr/simple_phylogeny.cgi) with default settings, and for sequence alignment by using BioEdit.
ABA Treatment, RNA Isolation and Quantitative RT-PCR (QRT-PCR)
To examine the expression of SlEAD1 in response to ABA, 14-day-old Micro-Tom wild type seedlings were cut into pieces and treated with 50 μM ABA or solvent methanol as a control for 4 h in darkness on a shaker at 40 rpm. Samples were frozen in liquid N2 and used for RNA isolation. To examine the expression of ABA signaling and biosynthesis genes in the Micro-Tom wild type and the slead1 mutants, 14-day-old Micro-Tom wild type and slead1 mutant seedlings were frozen in liquid N2 and used for RNA isolation.
Total RNA from tomato seedlings were isolated by using the Plant RNA Kit (OMEGA), cDNA was synthesized as described previously,37 and used as templates for qRT-PCR to examine the expression of SlEAD1, and ABA signaling and biosynthesis genes. The primers used for qRT-PCR analysis of SlEAD1 are 5ʹ-CATCGTGCTAGTGGTTCCCC-3ʹ and 5ʹ-ATCATCACCACCAAAGAGCGA-3ʹ. The primers used for qRT-PCR analysis of SlACT2 (Solyc11g005330) are 5ʹ-TGGATCTTGCTGGTCGTGATTTA-3ʹ and 5ʹ-AATTTCCCGTTCAGCAGTGGT-3ʹ. The primers used for qRT-PCR analysis of SlSnRK2.1 (Solyc5g056550), SlSnRK2.4 (Solyc02g090390), and SlNCED2 (Solyc08g016720) have been described previously.41,42
The EazyScript First-Strand DNA Synthesis Super Mix (TransGen Biotech) used for cDNA synthesis are able to remove DNA in the RNA sample, and synthesized cDNA were further examined by PCR amplification of SlACT2 by using intron-spanning primers as described previously,43 to further ensure there is no DNA contamination. TB Green® Premix EX TaqTM (Takara) were used for qRT-PCR analysis on StepOnePlus (Thermo Fisher Scientific) following a procedure described previously .44
Constructs
The construct NLS-RFP was used as a nuclear indicator.45 The reporter construct LexA-Gal4:GUS, the activator construct LD-VP and the control effector construct GD were used for protoplast transfection to examine transcription repressor activities.6,46
To generate GD-SlEAD1 and GFP-SlEAD1 constructs for protoplast transient transfection, the full length open reading frame (ORF) sequence of SlEAD1 was amplified by RT-PCR using RNA isolated from 14-day-old Micro-Tom wild type seedlings, double digested with NdeI and SacI enzymes, and then cloned into digested pUC19 vector with an N-terminal GD and GFP tag, respectively, under the control of the CaMV 35S promoter.6,46
To generate gene editing CRISPR/Cas9 constructs of SlEAD1, genomic sequence of SlEAD1 (contains a single exon) was subjected to CRISPRscan (http://www.crisprscan.org) for potential target sequences identification, and selected target sequences were evaluated on Cas-OFFinder (http://www.rgenome.net/cas-offinder/) for potential off-targets. The specific target sequences selected were 5ʹ-CGCGGCCTTATTAGCGGTGG(TGG)-3ʹ and 5ʹ-GCCACACGCGGCCTTATTAG(CGG)-3ʹ. The first sequence was used to generate CRISPR/Cas9 construct by using the pHDE vector and following the procedure described by Gao et al.,47 and both sequences were used to generate CRISPR/Cas9 construct by using the pHEE401E vector and following the procedure described by Wang et al.48 The primers used to generate pHDE-SlEAD1 construct were 5ʹ-CGCGGCCTTATTAGCGGTGGGTTTTAGAGCTAGAAATAGCAAGTTA-3ʹ and 5ʹ-CCACCGCTAATAAGGCCGCGAATCACTACTTCGACTCTAGC-3ʹ. The primers used to generate pHEE-SlEAD1 construct were SlEAD1-DT1-BsF, 5ʹ-ATATATGGTCTCGATTGGCGGCCTTATTAGCGGTGGGTT-3ʹ, SlEAD1-DT1-F0, 5ʹ- TGGCGGCCTTATTAGCGGTGGGTTTTAGAGCTAGAAATAGC-3ʹ, SlEAD1-DT2-R0, 5ʹ-AACCTAATAAGGCCGCGTGTGGCAATCTCTTAGTCGACTCTAC-3ʹ and SlEAD1-DT2-BsR, 5ʹ-ATTATTGGTCTCGAAACCTAATAAGGCCGCGTGTGGC-3ʹ. The primers pER8-U95-F and E9-U29-R, and U626-IDF and U629-IDR described previously 47-49) were used for colony PCR and sequencing of pHDE-SlEAD1 and pHEE401E-SlEAD1 constructs, respectively.
Plant Transformation, Transgenic Plant Selection and Cas9-free Mutant Isolation
Cotyledons and hypocotyls were collected from ~10-day-old Micro-Tom wild type seedlings and transformed with the CRISPR/Cas9 constructs via agrobacterium-mediated transformation, by using the plant tissue culture method as described previously.40 Transgenic T1 plants were identified by examining the present of Cas9 in the regenerated plants. Gene editing status in confirmed T1 transgenic plants was examined by amplifying and sequencing the genomic sequence of SlEAD1. T2 seeds were collected from gene edited T1 plants, germinated and grew in soil pots, and used to identify homozygous Cas9-free mutant by PCR amplification and sequencing.
DNA Isolation and PCR
Genomic DNA was isolated as described previously.50 To identify T1 transgenic plants, DNA was isolated from leaves of regenerated plants and used for PCR amplification of Cas9 fragment. To identify gene edited mutants for SlEAD1, DNA was isolated from leaves of the confirmed T1 transgenic plants and used for PCR amplification and sequencing of SlEAD1. To isolate homozygous Cas9-free mutants, DNA was isolated from leaves of T2 plants germinated from the confirmed mutants in T1 generation, and used for PCR amplification of Cas9 fragment, as well as amplification and sequencing of SlEAD1. The primers for amplifying Cas9 in pHDE-SlEAD1 transformed plants are 5ʹ-GACAAGAAGTACTCCATTGGG-3ʹ and 5ʹ-CAAACAGGCCGTTCTTCTTC-3ʹ, and in pHEE-SlEAD1 transformed plants are as described previously.49
Plasmid DNA Isolation, Protoplast Isolation and Transfection
Plasmid DNA of the reporter and effector were isolated by using Endo-Free Plasmid Maxi Kit (OMEGA). Arabidopsis protoplasts were isolated from rosette leaves of 3- to 4-week-old Col wild type plants as described previously.6,17,51 Tomato protoplasts were isolated by using the procedure for Arabidopsis protoplast isolation as described previously,51 except that cotyledons of 8- to 10-day-old Micro-Tom wild type plants were used for protoplast isolation, and the time for enzyme digestion increased from ~3 h to 6–8 h.
For subcellular location assay in Arabidopsis and tomato protoplasts, plasmids of GFP-SlEAD1 and NLS-RFP were co-transfected into protoplasts. After 20–22 h incubation in darkness, GFP and RFP florescence in the transfected protoplasts were observed under a fluorescence microscope.
For transcriptional activity assay in Arabidopsis and tomato protoplasts, plasmids of the reporter gene LexA-Gal4:GUS, the activator gene LD-VP and the effector gene GD-SlEAD1 were co-transfected into protoplasts, the effector gene GD was co-transfected as a control. After 20–22 h incubation in darkness, GUS activities in the transfected protoplasts were measured by using a microplate reader.
ABA Sensitivity Assays
ABA inhibited seed germination and root elongation assays were performed as described previously with some modifications.52–54
For ABA inhibited seed germination, seeds of the Micro-Tom wild type and the slead1 mutants were surface-sterilized and plated on 1/2 MS plates containing 2.5 μM ABA or without ABA as a control. Pictures were taken 4 d after the plated were transferred into the growth chamber, the number of germination seeds was counted, and the germination rate was calculated. Thirty seeds for each genotype were used for the experiment, and the experiment was repeated three times.
For primary root length and ABA inhibited root elongation assays, seeds of the Micro-Tom and the slead1 mutants were surface-sterilized and plated on 1/2 MS plates. After 3 d, germinated seeds were chosen and transferred to 1/2 MS plates with 0, 5, and 10 μM ABA and grown vertically. Then, pictures were taken 6 d after the transfer, root length was measured, and percentage of inhibition was calculated. At least 10 seedlings for each genotype were used for the experiment, and the experiment was repeated three times.
Results
Expression of SlEAD1 Is Down-regulated by ABA Treatment
In an attempt to identify novel players in ABA signaling in tomato, we performed data mining by using available transcriptome datasets, we found that an unknown function gene SlEAD1 (Solyc12g099500) was identified as a differential expressed gene in response to ABA during tomato fruit ripening,55 but it is expression level remained largely unchanged in tomato leaves 24 h after the spray of ABA.56
Considering that ABA response genes response to ABA in hours or even minutes, we examined the expression of SlEAD1 in tomato seedlings treated with ABA for 4 h. Quantitative RT-PCR analysis shows that the expression level of SlEAD1 reduced ~2 folds in the ABA treated seedlings (Fig. 1) suggest that SlEAD1 is ABA response gene.
Figure 1.
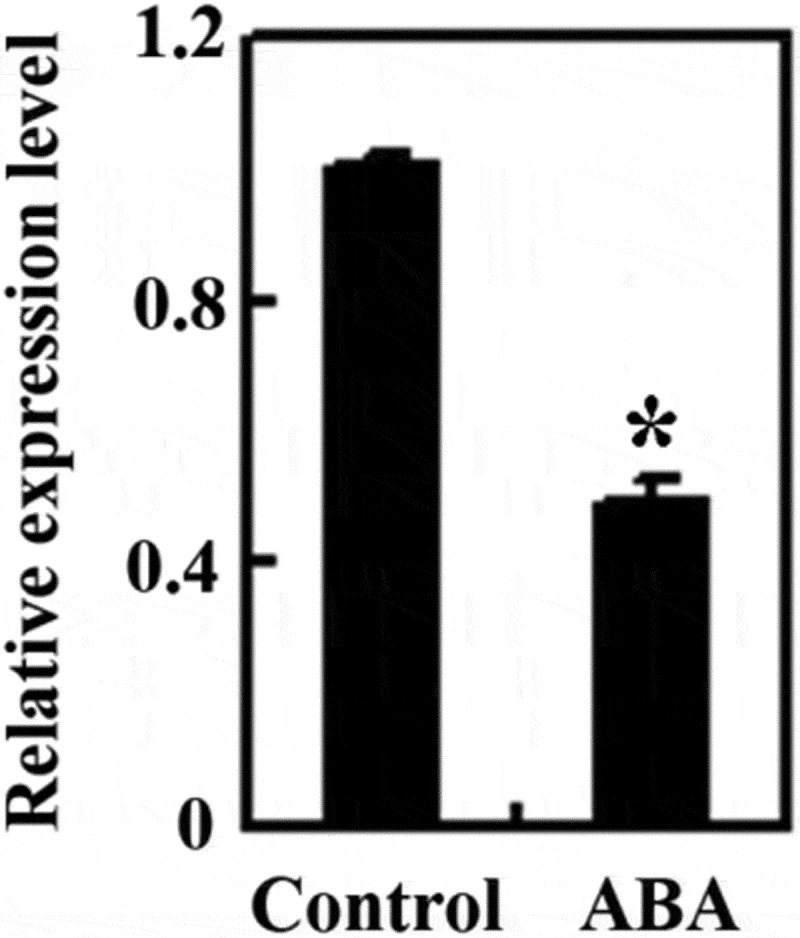
Expression of SlEAD1 in response to ABA treatment. The expression of SlEAD1 was down-regulated by ABA treatment. Fourteen-day-old Micro-Tom wild type seedlings were cut into pieces and treated in darkness for 4 h with 50 µM ABA or solvent methanol as a control. Samples were frozen in liquid N2, and total RNA was isolated and used for cDNA synthesis. The synthesized cDNA was used as template for qRT-PCR to examine the expression of SlEAD1, and the expression of SlACT2 was examined and used as an inner control. The expression level of SlEAD1 in mock-treated samples was set as 1, and its relative expression level in ABA treated samples was calculated. Data represent the mean ± SD of three replicates. *Significantly different from that in the mock-treated samples (p < .001).
Amino acid sequence BLASTing on NCBI (https://blast.ncbi.nlm.nih.gov) indicates that EADs are plant specific proteins, and are likely presented only in the flowering plants. However, considering that some genome databases are very preliminary, we could not rule out the possibility that EADs may also present in other plants. Phylogenetic analysis of SlEAD1 and some representative EADs from several selected plants including the model plant Arabidopsis, the tree poplar, the crops soybean, rice, sorghum and corn, and the grass Panicum hallii indicates that EADs from dicots formed a clade, whereas that from monocots formed another clade (Fig. 2a). Yet amino acids in the representative EADs are highly conserved (Fig. 2b). In addition, an LxLxL type EAR repressor motif initial found in repressor ERFs was presented in SlEAD1 (Fig. 2b); therefore, it was named Solanum lycopersicum EAR motif-containing ABA down-regulated 1. We also found that there are actually two overlapped LxLxL type EAR motifs in all the EADs examined (Fig. 2b), similar to that in the chimeric repressor SRDX.57
Figure 2.
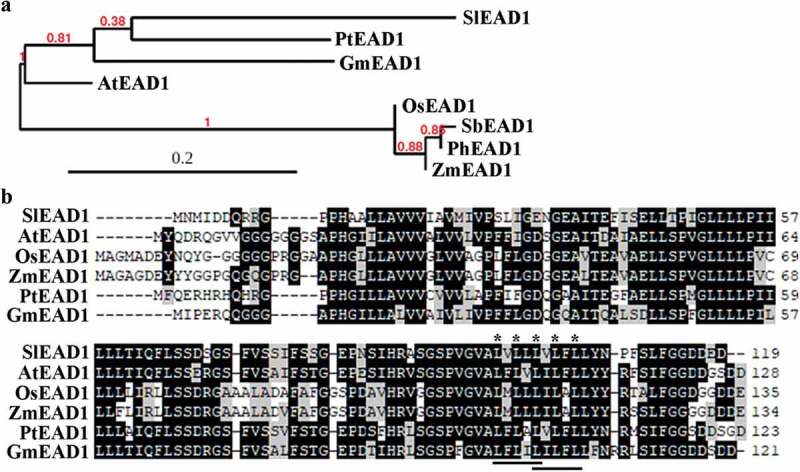
Phylogenetic analysis and amino acid alignment of EADs in different plants. (a) Phylogenetic analysis of SlEAD1 and EADs in several other plant species. EADs in other plants were identified on phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#), and their full-length amino acid sequence were used for phylogenetic analysis by using “One Click” mode on phylogeny (http://www.phylogeny.fr/simple_phylogeny.cgi) with default settings. (b) Amino acid sequence alignment of EADs. Full-length amino acid sequences of EADs were used for sequence alignment by using BioEdit. The identical amino acids were shaded in black, and the similar ones in gray. Underlines indicate the overlapped LxLxL EAR motifs, and stars indicate the conserved L residues.
SlEAD1 Is a Transcription Repressor
Considering that the EAR motif is found in, and is required, at least partially for the repression function of transcription repressors including the repressor ERFs, the Aux/IAA proteins, the OFPs, and the AITRs,1,6,14,17 we assume that SlEAD1 may also functions as a transcription repressor.
To examine if that is the case, we first observed the subcellular localization of SlEAD1 by using protoplast transit transfection assays. Plasmid DNA of GFP-SlEAD1 was transfected into protoplasts isolated from Arabidopsis leaves, and GFP fluorescence was observed by using a fluorescence microscope. We found that GFP fluorescence was predominantly observed in the nucleus (Fig. 3a). We also transfected plasmid DNA of GFP-SlEAD1 into protoplasts isolated from tomato cotyledons and found that SlEAD1 was predominantly localized in the nucleus (Fig. 3a).
Figure 3.
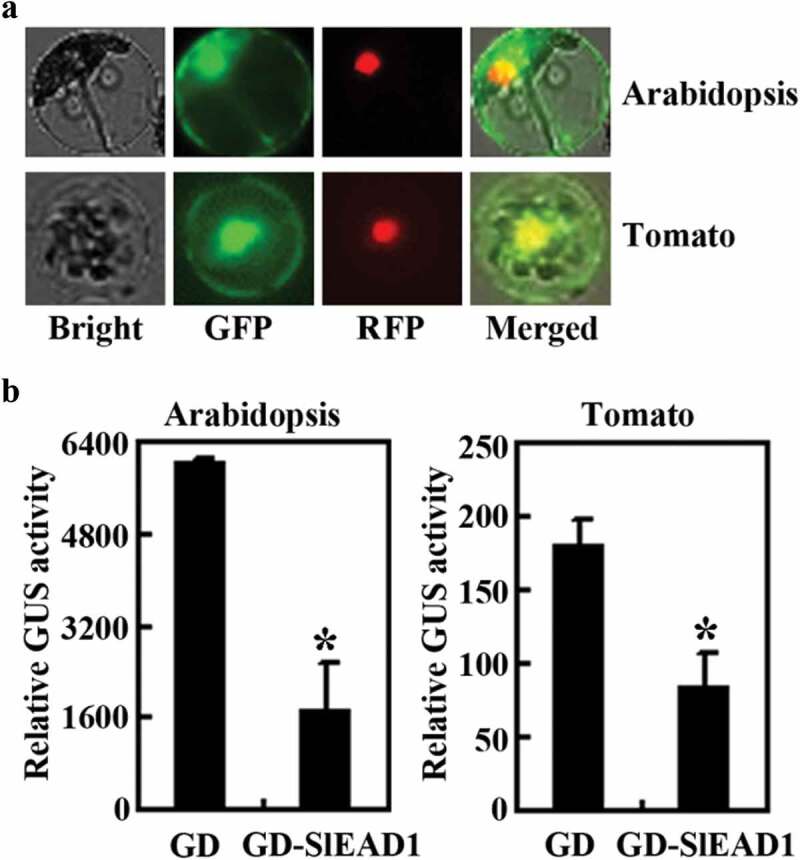
SlEAD1 is a transcription repressor. (a) Subcellular localization of SlEAD1. Plasmids of the effector gene GFP-SlEAD1 and NLS-RFP were co-transfected into protoplasts isolated from Arabidopsis leaves and tomato cotyledons, respectively. After transfection, the protoplasts were incubated at room temperature in darkness for 20~22 h, then the GFP and RFP fluorescence was observed under a confocal microscope. (b) Transcriptional activities of SlEAD1. Plasmids of the reporter gene LexA-Gal4:GUS, the transcription activator gene LD-VP and the effector gene GD-SlEAD1 were co-transfected into protoplasts isolated from Arabidopsis leaves and tomato cotyledons, respectively. Co-transfection of the effector gene GD was used as a control. After transfection, the protoplasts were incubated at room temperature in darkness for 20~22 h, then GUS activities were measured by using amicroplate reader. Data represent the mean ± SD of three replicates. *Significantly different from that of the GD control (p < .001).
We then examined transcriptional activities of SlEAD1 in transfected protoplasts. Plasmids of the reporter gene LexA-Gal4:GUS, the activator gene LD-VP and the effect gene GD-SlEAD1 or the control gene GD were co-transfected into protoplasts isolated from Arabidopsis leaves, and GUS activities in the protoplasts were measured by using a microplate reader. As shown in Fig. 3b, GUS activity activated by the activator LD-VP was repressed by the co-transfection of the effector gene GD-SlEAD1. Similarly, Co-transfection of the effector gene GD-SlEAD1 in protoplasts isolated from tomato cotyledons also resulted in repression of the reporter gene (Fig. 3b). These results indicate that SlEAD1 functions as a transcription repressor.
The Gene Editing Slead1 Mutants are Hypersensitive to ABA
Having shown that the expression of SlEAD1 was suppressed by ABA and SlEAD1 is as a transcription repressor, we wanted to further examine the function of SlEAD1 in regulating ABA response in tomato.
To do that, we decided to generate mutants by using CRISPR/Cas9 gene editing, since it has been able to generate transgene-free mutants in the model plant Arabidopsis as well as crops.47,58-60 Two CRISPR/Cas9 constructs, pHDE-SlEAD1 and pHEE-SlEAD1 targeting one and two sites, respectively (Fig. 4a,b), were generated and used for tomato plant transformation. Gene editing was observed in T1 plants generated with both constructs, and transgene-free mutants were obtained in T2 generations. The slead1-c1 mutant was generated with pHDE-SlEAD1 construct, and has a single nucleotide insertion in the target site of SlEAD1, whereas the slead1-c2 mutant was generated with pHEE-SlEAD1 construct, and has a single nucleotide insertion in one target site, and 5 bp deletion in another target site of SlEAD1 (Fig. 4c). In both mutants, the nucleotide indels led to a few amino acids substitution and a premature stop in SlEAD1 (Fig. 4d).
Figure 4.
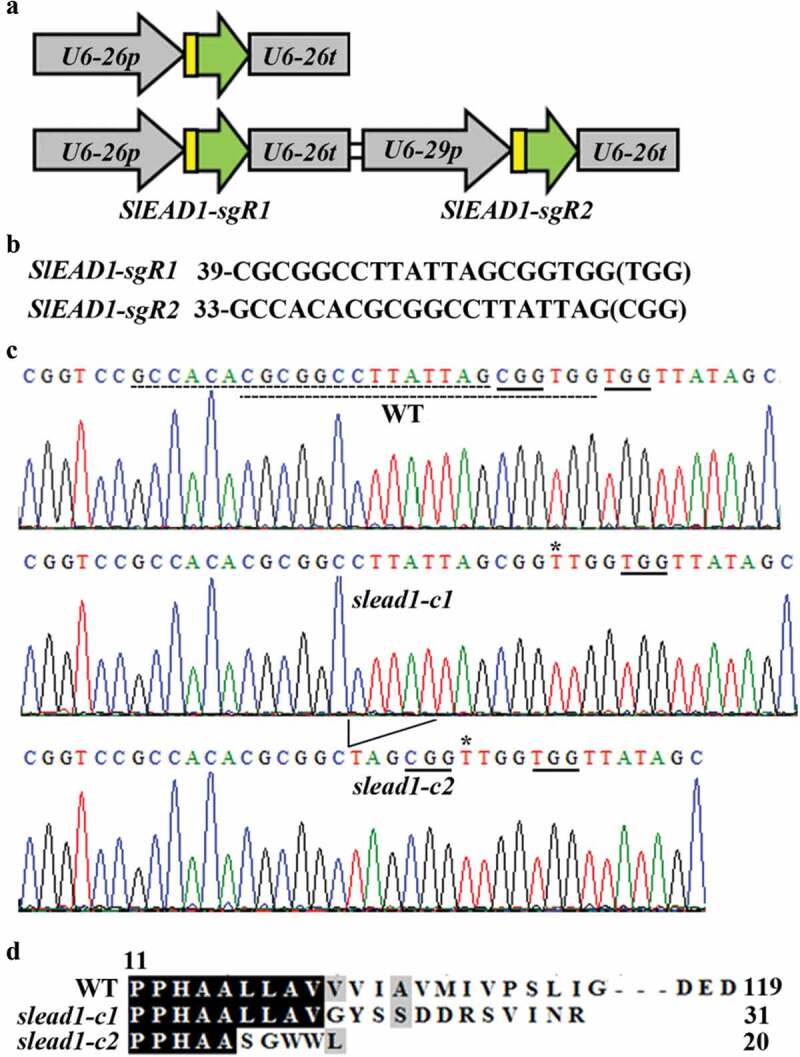
Generation of transgene-free slead1 mutants. (a) Diagram of the sgRNA expression cassettes cloned into the pHDE-SlEAD1 (up panel) and the pHEE-SlEAD1 (low panel) vectors, respectively. (b) Target sequences of SlEAD1. The numbers indicate the positions of the first nucleotide in the target sequences relative to the first nucleotide in the coding sequence of SlEAD1. The NGG PAM sites immediately after the target sequences are in brackets. (c) Editing status of SlEAD1 in the slead1 mutants. The slead1-c1 and slead1-c2 mutants were obtained by transforming Micro-Tom wild type plants with the pHDE-SlEAD1 and pHEE-SlEAD1 constructs, respectively. Editing status of SlEAD1 in T1 plants was examined, and transgene-free homozygous mutant plants were isolated from T2 generations. Stars indicate the single T nucleotide insertion in the slead1-c1 and slead1-c2 mutants, and arrow head indicates the 5 bp deletion in the slead1-c2 mutant. Solid underlines indicate the PAM sites, and dash underlines indicated the target sequences. (d) Amino acid alignment of SlEAD1 in the Micro-Tom wild type and the slead1 mutants. ORFs of SlEAD1 in the slead1 mutants were identified by using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/), and predicted amino acid sequences were used for alignment with the amino acid sequence of SlEAD1. Number at the N-terminal indicates the amino acid position relative to the first M amino acid, and the numbers at the C-terminal indicate total amino acid numbers of SlDAE1 in the Micro-Tom wild type and the slead1 mutants.
By using the mutants obtained, we examined if SlEAD1 may regulate ABA response in tomato plants. In seed germination assays, we found that ABA inhibited seed germination in both the Micro-Tom wild type and the slead1 mutants (Fig. 5a). However, no different was observed for both the Micro-Tom wild type and the slead1 mutants in the control plates, they all reached an ~60% generation rate (~20 out of 30) 4 d after the transfer, but that in ABA-contained plates were ~50% (~15 out of 30) and 30% (~9 out of 30), respectively, for the Micro-Tom wild type and the slead1 mutants (Fig. 5b), indicating that the slead1 mutants are more sensitivity to ABA treatment.
Figure 5.
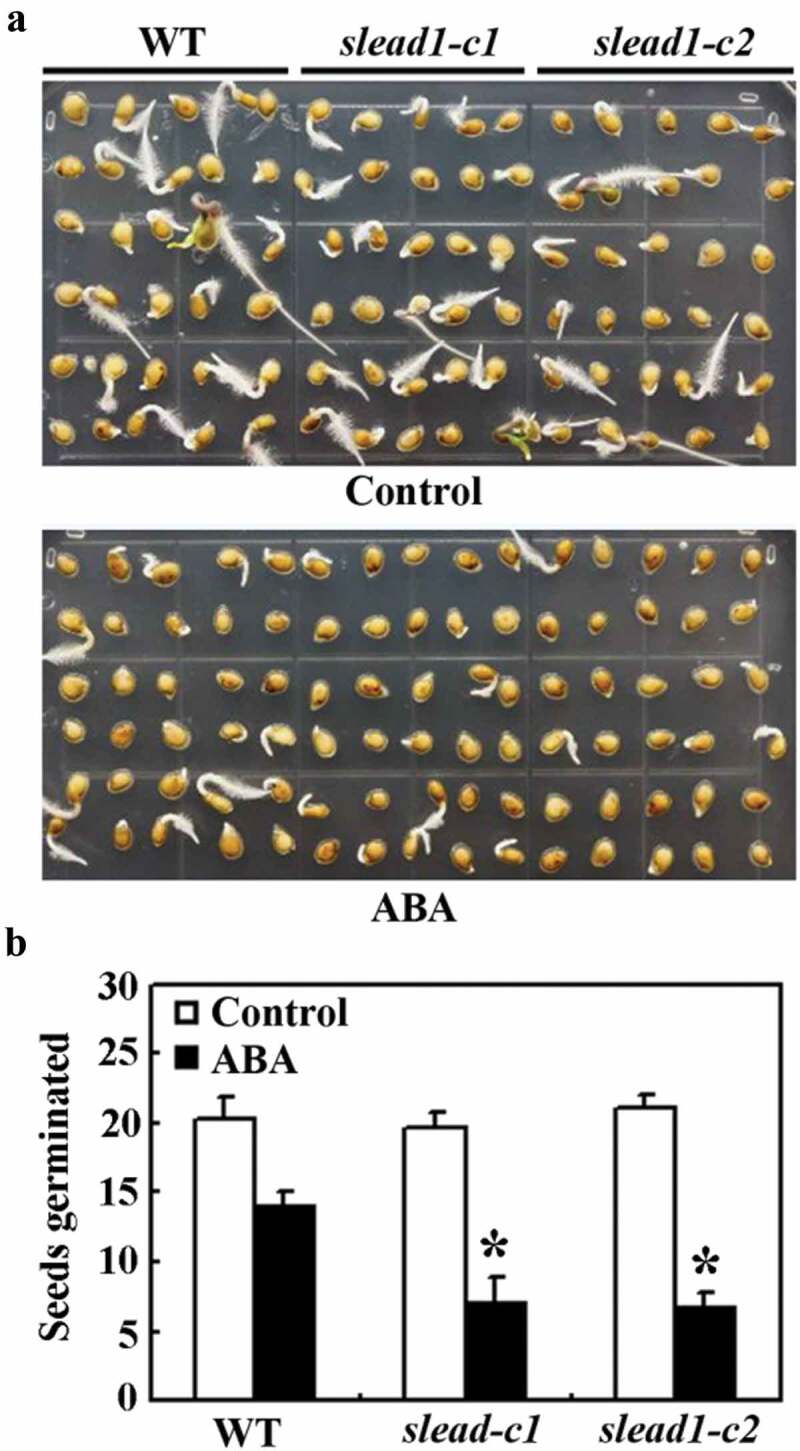
Effects of ABA on seed germination of the Micro-Tom wild type and the slead1 mutants. (a) Seed germination of the Micro-Tom wild type and the slead1 mutants in response to ABA treatment. Seeds were sterilized and plated on 1/2 MS plates or plates with 2.5 µM ABA. The plates were kept at 4°C in darkness for 2 d, and then transferred to a growth room. Pictures were taken 4 d after the transfer. (b) Number of seed germinated of the Micro-Tom wild type and the slead1 mutants in response to ABA treatment. Seeds germinated were counted 4 d after the transfer, and average number of seed germinated was calculated. Data represent the mean ± SD of three replicates. *Significantly different from that of the wild type (p < .01).
Similarly, the slead1 mutants also showed an increased sensitivity to ABA treatments in root elongation assays (Fig. 6). We also noted that the primary root length in the slead1 mutant seedlings was shorter when compared with the Micro-Tom wild type seedlings (Fig. 6a). Quantitative results show that the root length for 9-day-old Micro-Tom wild type seedlings was ~9 cm, but that for the slead1 mutants was about ~8 cm (Fig. 6b). Root elongation in the Micro-Tom wild type seedlings was inhibited about ~50% and ~60%, respectively, by 5 μM and 10 μM ABA, where as that for the slead1 mutant seedlings was ~60% and ~70%, respectively (Fig. 6c).
Figure 6.
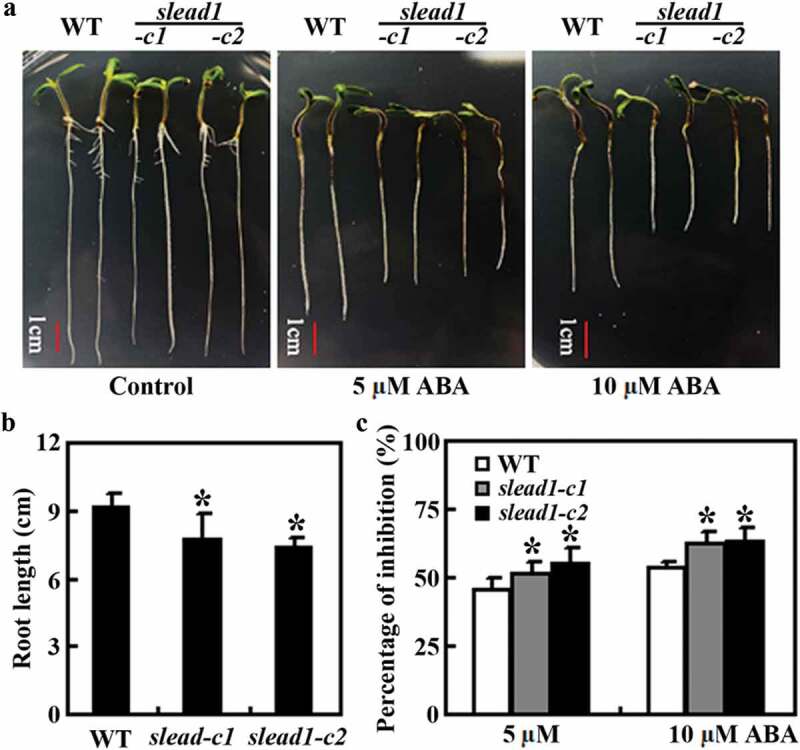
Effects of ABA on root elongation of the Micro-Tom wild type and the slead1 mutants. (a) Root elongation of the Micro-Tom wild type and the slead1 mutants in response to ABA treatment. Seeds were sterilized and plated on 1/2 MS plates. The plates were kept at 4°C in darkness for 2 d, and then transferred to a growth room. After 3 d, germinated seeds were transferred to control plates, or plates containing 5 and 10 μM ABA, respectively, and grown vertically for 6 d before the pictures were taken. (b) Root length of Micro-Tom wild type and the slead1 mutants. Root length of the seedling was measured 6 d after the transfer. Data represent means ± SD of at least 10 seedlings. *Significantly different from that of the wild type (p < .05) (c) Percentage of inhibition on root elongation by ABA. Root length of the seedling was measured 6 d after the transfer, and the percentage of inhibition on root elongation was calculated. Data represent means ± SD of at least 10 seedlings. *Significantly different from that of the wild type (p < .01).
Expression of ABA Signaling and Biosynthesis Genes Was Affected in the Slead1 Mutants
Changes in ABA sensitivity may caused by alternation in ABA signaling and/or ABA biosynthesis, to examine why ABA sensitivity was increased in the slead1 mutants, we examined the expression levels of the core ABA signaling regulator genes and ABA biosynthesis genes in the slead1 mutants. We found that the expression levels of the core ABA signaling regulator genes SlSnRK2.1 and SlSnRK2.4, and the ABA biosynthesis gene SlNCED2 were increased in the slead1 mutants (Fig. 7a). These results suggest that SlEAD1 may regulate ABA response in tomato by negatively regulating the expression of ABA signaling genes and ABA biosynthesis genes (Fig. 7b).
Figure 7.
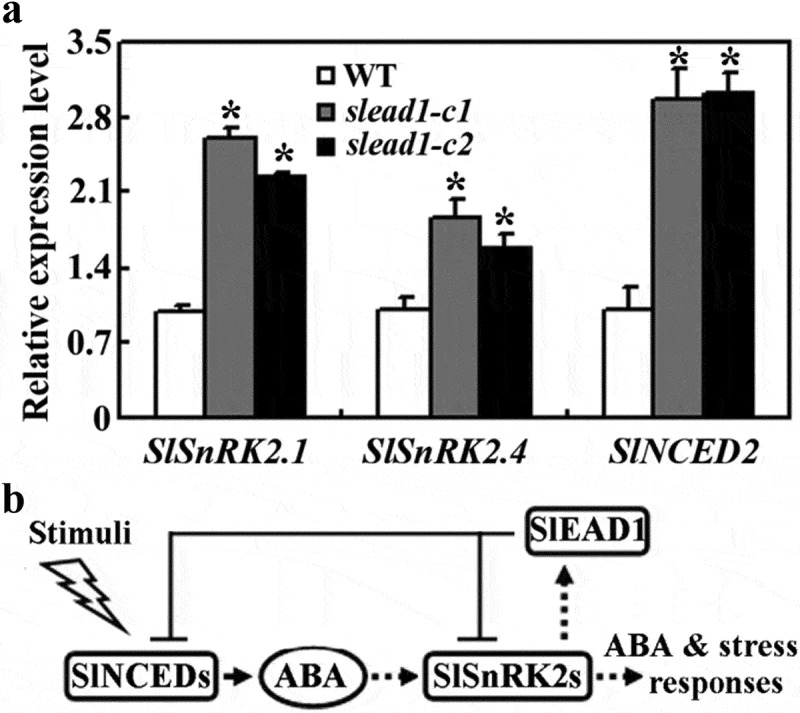
Expression of ABA signaling and biosynthesis genes in the Micro-Tom wild type and the slead1 mutants. (a) Expression of SlSnRKs and SlNCED2 in the wild type and slead1 mutant tomato seedlings. Total RNA was isolated from 14-day-old Micro-Tom wild type and slead1 mutant seedlings and used for cDNA synthesis. The synthesized cDNA was used as template for qRT-PCR to examine the expression of ABA signaling and biosynthesis genes, and the expression of SlACT2 was examined and used as an inner control. The expression levels of the corresponding genes in the Micro-Tom wild type seedlings were set as 1, and their relative expression levels in slead1 mutant seedlings were calculated. Data represent the mean ± SD of three replicates. *Significantly different from that in the wild type (p < .01). (b) A diagram showing the regulation and roles of SlEAD1 in ABA signaling in tomato.
Discussion
EAR motif-containing proteins are involved in the regulation of hormone signaling, including auxin signaling, ethylene signaling, jasmonic acid signaling, strigolactone signaling.1,2,13-16 We previously identified AITRs as a novel family of transcription repressors that are involved in the regulation of ABA signaling, and at least some of the AITRs contain a full conserved LxLxL EAR motif.17 We identified here SlEAD1 as a novel EAR motif-containing protein that plays a role in regulating ABA response in tomato, and EADs may represent a novel family of transcription repressors in plants.
First, SlEAD1 contains two overlapped LxLxL EAR motifs(Fig. 2b), similar to that in SRDX, a chimeric activate repressor that can convert a transcription activator to a repressor,57 and have been used to study the functions of transcription factors from different families, such as the ERF family 4 Huang et al. 61], the R2R3 MYB family,62–64 the WRKY family,65–67 and the NAC family.68,69 Most importantly, the overlapped LxLxL EAR motifs are conserved on EADs from other plant species such as Arabidopsis, rice, and poplar (Fig. 2). Second, SlEAD1 functions as a transcription repressor, SlEAD1 was found to be predominately localized in nucleus in transfected protoplasts, and consistent with the presence of EAR motifs, SlEAD1 repressed reported gene expression in both Arabidopsis and tomato protoplasts (Fig. 3). Third, ABA sensitivities in slead1 mutants were increased (Fig. 5, Fig. 6), suggesting that SlEAD1 may play a negative role in regulating plant response to ABA.
EAR motif-containing proteins mediated transcription repression can be achieved by at least two different ways. One is epigenetic modification3 by recruit a histone deacetylase (HDAC) to form a HDAC complex via interactions with co-suppressors. For example, ERF7 can interact with SIN3, whereas ERF3 and ERF4 can interact with SAP18 (SIN3 ASSOCIATED POLYPEPTIDE P18), to recruit HDA19 to form an HDAC complex.8,9,70,71 Another is to interfere with the activities of other transcription factors via directly or indirectly binding to them. For example, the Aux/IAA proteins interact with activator ARFs to repress their activities, the JAZ (JASMONATE ZIM) domain proteins interact with MYC activators to repress their activities, whereas OFP1 and OFP4 interact with KNAT7 to enhance its repression activities.2,13,14,72-75 We found that SlEAD1 is an EAR motif-containing protein (Fig. 2), and functions as transcription repressors (Fig. 3), however, it is unclear how SlEAD1 may mediate transcription repression, identification of proteins that can interact with SlEAD1 may enable to figure this out.
ABA signaling via the PYR/PYLs/RCARs receptors, the PP2Cs phosphatases and SnRK2s protein kinases activates the downstream ABF/AREB/ABI5-type bZIP transcription factors, leading to the activation/repression of ABA response genes and plant responses to abiotic stresses.22,26-28,31 We show that the expression of SlEAD1 was down-regulated by ABA; therefore, it will be of interest to examine if ABF/AREB/ABI5-type bZIP transcription factors may directly regulate the expression of SlEAD1.
The function mechanisms of SlEAD1 are also needed to be further studied. Our data show that ABA sensitivities were increased in the slead1 mutants (Fig. 5, Fig. 6), and consistent with SlEAD1’s transcription repression activities, the expression levels of ABA biosynthesis gene SlNCED2 and ABA signaling key component genes SlSnRK2.1 and SlSnRK2.4 were increased in the slead1 mutants, indicating that SlEAD1 may play a feedback regulating role in ABA signaling (Fig. 7), it is worthwhile to examine if SlEAD1 is involved in the regulation of plant abiotic stress responses, to examine if these genes are directly targets of SlEAD1, and to examine how SlEAD1 may regulate the expression of these genes, therefore to uncover the functional mechanism of SlEAD1 in regulating ABA signaling and plant response to abiotic stresses.
In summary, we found that SlEAD1 is a novel EAR motif-containing transcription repressor, the expression of SlEAD1 was down-regulated by ABA, and SlEAD1 negatively regulates ABA responses in tomato.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFD0101900) and a start up funding from Linyi University (LYDX2019BS039). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding Statement
This work was supported by the National Key R&D Program of China [2016YFD0101900], and a start up fund from Linyi University [LYDX2019BS039].
Author Contributions
SW conceived the study. SW, WW, and TW designed the experiments. WW, XW, YW, GZ, CW, SH, A, and RL performed the experiments. WW and SW analyzed the data. SW and WW drafted the manuscript. All the authors participated in the revision of the manuscript.
Declaration Of Interest Statement
The authors declared no conflict of interest.
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M.. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13(8):1959–68. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kagale S, Links MG, Rozwadowski K.. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in arabidopsis. Plant Physiol. 2010;152(3):1109–34. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6(2):141–46. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Liu Y, Yan H, Tian T, You Q, Zhang L, Xu W, Su Z. PlantEAR: functional analysis platform for plant EAR motif-containing proteins. Front Genet. 2018;9; 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez N, Pauwels L, Baekelandt A, De Milde L, Van Leene J, Besbrugge N, Heyndrickx KS, Cuéllar Pérez A, Durand AN, De Clercq R, et al. A repressor protein complex regulates leaf growth in arabidopsis. Plant Cell. 2015;27(8):2273–87. doi: 10.1105/tpc.15.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Chang Y, Guo J, Chen JG. Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007;50:858–72. doi: 10.1111/j.1365-313X.2007.03096.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhai QZ, Zhang X, Wu FM, Feng HL, Deng L, Xu L, Zhang M, Wang Q, Li C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in arabidopsis. Plant Cell. 2015;27:2814–28. doi: 10.1105/tpc.15.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139:949–59. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu J. Role of an arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–96. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Gomes BL, Mila I, Purgatto E, Peres LE, Frasse P, Maza E, Zouine M, Roustan JP, Bouzayen M, et al. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016;170(3):1732–44. doi: 10.1104/pp.15.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang Z, Liu S, Huang L, Hong Y, Li X, Huang L, Zhang Y, Zhang H, Li D, Song F. Tomato SlERF.A1, SlERF.B4, SlERF.C3 and SlERF.A3, members of B3 group of ERF family, are required for resistance to Botrytis cinerea. Front Plant Sci. 2016;7:1964. doi: 10.3389/fpls.2016.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upadhyay RK, Soni DK, Singh R, Dwivedi UN, Pathre UV, Nath P, Sane AP. SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J Exp Bot. 2013;64(11):3237–47. doi: 10.1093/jxb/ert162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nat. 2007;448(7154):666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 14.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–60. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nat. 2013;504(7480):401–05. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Ward S, Li P, Bennett T, Leyser O. SMAX1-LIKE7 signals from the nucleus to regulate shoot development in arabidopsis via partially EAR motif-independent mechanisms. Plant Cell. 2016;28:1581–601. doi: 10.1105/tpc.16.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian H, Chen S, Yang W, Wang T, Zheng K, Wang Y, Cheng Y, Zhang N, Liu S, Li D, et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017;40(12):2958–71. doi: 10.1111/pce.13058. [DOI] [PubMed] [Google Scholar]
- 18.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–85. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 2012;10(1):2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–39. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du S, Jiang T, et al. The Mg-chelatase H subunit of arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–35. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in aba responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–39. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Sci. 2009;324:1064–68. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 24.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Sci. 2009;324:1068–71. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–88. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 26.Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez PL, Leube MP, Grill E. Molecular cloning in arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol. 1998;38(5):879–83. doi: 10.1023/A:1006012218704. [DOI] [PubMed] [Google Scholar]
- 28.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in arabidopsis. Plant Cell. 2007;19:485–94. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YT, Liu HX, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013;75:965–76. doi: 10.1111/tpj.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Stone SL. Abscisic acid increases arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010;22(8):2630–41. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR. A transcription factor hierarchy defines an environmental stress response network. Sci. 2016;354(6312):aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad R, Liu Y, Wang TJ, Meng Q, Yin H, Wang X, Wu Y, Nan N, Liu B, Xu ZY. GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 2019;179:1844–60. doi: 10.1104/pp.18.01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran L-SP, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–76. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 34.Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426:183–96. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 35.Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–35. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell. 2010;22:1716–32. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian H, Guo H, Dai X, Cheng Y, Zheng K, Wang X, Wang S. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci Rep. 2015;5(1):17587. doi: 10.1038/srep17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng K, Wang Y, Wang S. The non-DNA binding bHLH transcription factor PACLOBUTRAZOL RESISTANCES are involved in the regulation of ABA and salt responses in arabidopsis. Plant Physiol Biochem. 2019;139:239–45. doi: 10.1016/j.plaphy.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 2003;15:1689–703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kai W, Fu Y, Wang J, Liang B, Li Q, Leng P. Functional analysis of SlNCED1 in pistil development and fruit set in tomato (Solanum lycopersicum L. Sci Rep. 2019;9(1):16943. doi: 10.1038/s41598-019-52948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Wu N, Zhang L, Ahammed GJ, Chen X, Xiang X, Zhou J, Xia X, Shi K, Yu J, et al. Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol. 2018;176(2):1311–26. doi: 10.1104/pp.17.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yáñez M, Cáceres S, Orellana S, Bastías A, Verdugo I, Ruiz-Lara S, Casaretto JA. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep. 2009;28:1497–507. doi: 10.1007/s00299-009-0749-4. [DOI] [PubMed] [Google Scholar]
- 44.Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. bHLH122 is important for drought and osmotic stress resistance in arabidopsis and in the repression of ABA catabolism. New Phytol. 2014;201(4):1192–204. doi: 10.1111/nph.12607. [DOI] [PubMed] [Google Scholar]
- 45.Lee, Y.J., Kim, D.H., Kim, Y.-W., Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell. 2001;13:2175––2190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiwari SB, Hagen G, Guilfoyle TJ. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–43. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Chen J, Dai X, Zhang D, Zhao Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by RISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, Zhang N, Zhang Q, Zhou G, Tian H, Hussain S, Ahmed S, Wang T, Wang S. Genome editing to integrate seed size and abiotic stress tolerance traits in arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int J Mol Sci. 2019;20(11):2695. doi: 10.3390/ijms20112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiwari S, Wang S, Hagen G, Guilfoyle TJ. Transfection assays with protoplasts containing integrated reporter genes. Methods Mol Biol. 2006;323:237–44. doi: 10.1385/1-59745-003-0:237. [DOI] [PubMed] [Google Scholar]
- 52.Bi L, Weng L, Jiang Z, Xiao H. The tomato IQD gene SUN24 regulates seed germination through ABA signaling pathway. Planta. 2018;248(4):919–31. doi: 10.1007/s00425-018-2950-6. [DOI] [PubMed] [Google Scholar]
- 53.Ijaz R, Ejaz J, Gao S, Liu T, Imtiza M, Ye Z, Wang T. Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci Rep. 2017;7:12087. doi: 10.1038/s41598-017-11168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren Z, Wang X. SlTIR1 is involved in crosstalk of phytohormones, regulates auxin-induced root growth and stimulates stenospermocarpic fruit formation in tomato. Plant Sci. 2016;253:13–20. doi: 10.1016/j.plantsci.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Mou W, Li D, Luo Z, Mao L, Ying T. Transcriptomic analysis reveals possible influences of ABA on secondary metabolism of pigments, flavonoids and antioxidants in tomato fruit during ripening. PLoS One. 2015;10:e0129598. doi: 10.1371/journal.pone.0129598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Tao X, Tang XM, Xiao L, Sun JL, Yan XF, Li D, Deng HY, Ma XR. Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genomics. 2013;14:841. doi: 10.1186/1471-2164-14-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in arabidopsis. Plant J. 2003;34(5):733–39. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 58.He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant. 2018;11(9):1210–13. doi: 10.1016/j.molp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Lu HP, Liu SM, Xu SL, Chen WY, Zhou X, Tan YY, Huang JZ, Shu QY. CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol J. 2017;15:1371–73. doi: 10.1111/pbi.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–84. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 61.Huang, P-Y., Zhang, J., Jiang, B., Chan, C., Yu, J H., Lu, Y-P.., Chung, K., Zimmerli, L. (2018). NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. Journal of Experimental Botany. 70, 1033-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS. An arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013;587(12):1773–78. doi: 10.1016/j.febslet.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 63.Liang G, He H, Li Y, Ai Q, Yu D. MYB82 functions in regulation of trichome development in arabidopsis. J Exp Bot. 2014;65(12):3215–23. doi: 10.1093/jxb/eru179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in arabidopsis and torenia fournieri. Plant Cell. 2013;25(5):1609–24. doi: 10.1105/tpc.113.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grunewald W, De Smet I, De Rybel B, Robert HS, van de Cotte B, Willemsen V, Gheysen G, Weijers D, Friml J, Beeckman T. Tightly controlled WRKY23 expression mediates arabidopsis embryo development. EMBO Rep. 2013;14:1136–42. doi: 10.1038/embor.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in catharanthus roseus. Plant Physiol. 2011;1570(4):2081–93. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016;85:96–106. doi: 10.1111/tpj.13092. [DOI] [PubMed] [Google Scholar]
- 68.Guo S, Dai S, Singh PK, Wang H, Wang Y, Tan JLH, Wee W, Ito T. A membrane-bound NAC-like transcription factor OsNTL5 represses the flowering in Oryza Sativ . Front Plant Sci. 2018;9: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shih CF, Hsu WH, Peng YJ, Yang CH. The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J Exp Bot. 2014;65:621–39. doi: 10.1093/jxb/ert412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–96. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- 72.Goossens J, Swinnen G, Vanden Bossche R, Pauwels L, Goossens A. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 2015;206(4):1229–37. doi: 10.1111/nph.13398. [DOI] [PubMed] [Google Scholar]
- 73.Li E, Wang S, Liu Y, Chen JG, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in arabidopsis thaliana. Plant J. 2011;67(2):328–41. doi: 10.1111/j.1365-313X.2011.04595.x. [DOI] [PubMed] [Google Scholar]
- 74.Song S, Huang H, Wang J, Liu B, Qi T, Xie D. MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses. Plant Cell Physiol. 2017;58(10):1752–63. doi: 10.1093/pcp/pcx112. [DOI] [PubMed] [Google Scholar]
- 75.Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, Howe GA. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015;82(4):669–79. doi: 10.1111/tpj.12841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


