Abstract
The mammalian Target of Rapamycin complex 1 (mTORC1) nutrient-sensing pathway is a central regulator of cell growth and metabolism and is dysregulated in diabetes. The eukaryotic translation initiation factor 4E (EIF-4E) protein, a key regulator of gene translation and protein function, is controlled by mTORC1 and EIF-4E Binding Proteins (EIF4EBPs). Both EIF4EBPs and ribosomal protein S6K kinase (RP-S6K) are downstream effectors regulated by mTORC1 but converge to regulate two independent pathways. We investigated whether the risk of type 2 diabetes varied with genetically predicted EIF-4E, EIF-4A, EIF-4G, EIF4EBP, and RP-S6K circulating levels using Mendelian Randomization. We estimated the causal role of EIF-4F complex, EIF4EBP, and S6K in the circulation on type 2 diabetes, based on independent single nucleotide polymorphisms strongly associated (p = 5 × 10–6) with EIF-4E (16 SNPs), EIF-4A (11 SNPs), EIF-4G (6 SNPs), EIF4EBP2 (12 SNPs), and RP-S6K (16 SNPs). The exposure data were obtained from the INTERVAL study. We applied these SNPs for each exposure to publically available genetic associations with diabetes from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) case (n = 26,676) and control (n = 132,532) study (mean age 57.4 years). We meta-analyzed SNP-specific Wald-estimates using inverse variance weighting with multiplicative random effects and conducted sensitivity analysis. Mendelian Randomization (MR-Base) R package was used in the analysis. The PhenoScanner curated database was used to identify disease associations with SNP gene variants. EIF-4E is associated with a lowered risk of type 2 diabetes with an odds ratio (OR) 0.94, 95% confidence interval (0.88, 0.99, p = 0.03) with similar estimates from the weighted median and MR-Egger. Similarly, EIF-4A was associated with lower risk of type 2 diabetes with odds ratio (OR) 0.90, 95% confidence interval (0.85, 0.97, p = 0.0003). Sensitivity analysis using MR-Egger and weighed median analysis does not indicate that there is a pleiotropic effect. This unbiased Mendelian Randomization estimate is consistent with a protective causal association of EIF-4E and EIF-4A on type 2 diabetes. EIF-4E and EIF-4A may be targeted for intervention by repurposing existing therapeutics to reduce the risk of type 2 diabetes.
Subject terms: Diseases, Risk factors
Introduction
Diabetes is one of the most prevalent chronic diseases globally and in the US and is associated with several co-morbidities. In 2015, it was estimated that 9.4% of the US population, or approximately 30.3 million people have diabetes1. Among those, only 23.1 million people are diagnosed, and about 7.2 million are undiagnosed. Type 2 diabetes represents approximately 90 to 95% of all diabetes cases. The estimated direct medical costs of diagnosed diabetes in 2017 was $237 billion in the US with an average medical expenditure of diagnosed person of $13,700 per year2. It is further estimated that 33.9% of US adults have prediabetes. To date, the causes of type 2 diabetes are not fully understood. Diabetes is also associated with multiple co-morbidities and complications, including obesity. Thus, there is an urgent need to determine causal associations and develop new strategies for prevention, early detection, diagnosis, and treatment of type 2 diabetes.
The mechanistic Target of Rapamycin (mTOR) is a highly conserved serine/threonine kinase and a key regulator of cell growth and metabolism3–5. As such, mTOR protein is a central metabolic integrator and is dysregulated in type 2 diabetes and diabetes-associated co-morbidities6–8. Importantly, mTOR is a druggable protein and therefore is a potential target for type 2 diabetes interventions. It nucleates two functionally-distinct and mutually-exclusive complexes, namely mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2). Both mTORC1 and mTORC2 regulate cellular metabolism, survival, proliferation, and growth. mTORC1 is a central hub for nutrient-sensing and energy metabolism and, as such, coordinates anabolic protein and nucleotide synthesis and catabolic autophagy9–16. On the other hand, mTORC2 drives insulin signaling by phosphorylating Akt (Ser473)/PKB downstream of the phosphoinositide 3-kinase (PI3K)/insulin pathway17–20.
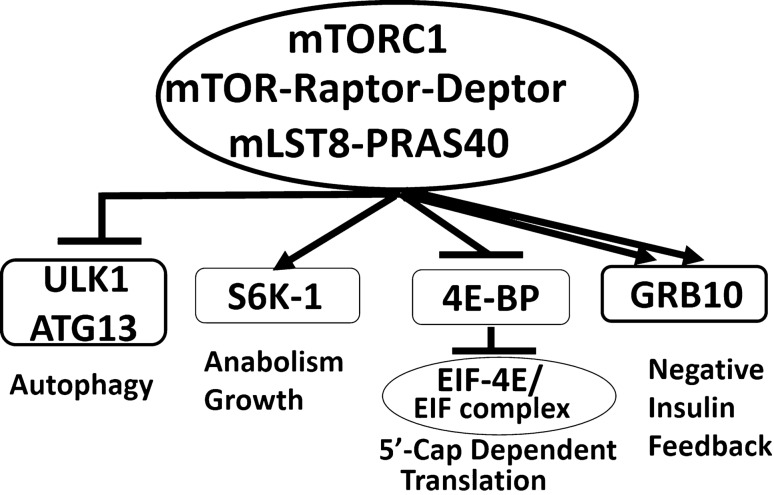
The mTORC1, which binds exclusively to Raptor protein and other partners (Fig. 1), regulates two downstream effectors, namely, Ribosomal Protein-S6 kinase 1 (RP-S6K 1) and eukaryotic translation initiation factor 4E-binding protein (EIF4EBP). The first mTORC1 target, RP-S6K1 phosphorylates eukaryotic translation initiation factor 4B (EIF-4B), which is a positive regulator of EIF-4F complex21. In addition, RP-S6K also phosphorylates the programmed cell death protein 4 (PDCD4), which is a negative regulator of EIF-4A and targets it to degradation via the ubiquitin pathway22. The second mTORC1 downstream target, EIF4EBP, is a repressor of the translation initiation complex (EIF-4F), which is required for 5′cap-dependent translation of mRNA (Fig. 1). Once phosphorylated by mTORC1, EIF4EBP dissociates from EIF-4E, allowing for the assembly of the EIF-4F complex and initiation of 5′cap-dependent translation. The EIF-4F complex consists of EIF-4E, EIF-4G, and EIF-4A. EIF-4E binds to 7-methylguanosine cap at the 5′-UTR of eukaryotic mRNA and mediates the recruitment of mRNA on ribosomes to start protein translation. As mentioned earlier, the mTOR kinase attracts different protein partners to generate two functionally distinct complexes, namely, mTORC1 and mTORC2. While mTORC1 integrates inputs from nutrients and growth factors and coordinates cellular growth and metabolism23–25; mTORC2 is activated by growth factor signals only and responds by phosphorylating the C-terminal hydrophobic motif of Akt/PKB on serine 47317,26–29. As such, mTOR complexes and their downstream targets are actionable proteins and metabolic targets in managing type 2 diabetes and mediate mTOR central role in glucose and energy metabolism and in pancreatic progenitor cell growth. Dysregulation of mTORC1 and mTORC2 complexes is linked to insulin resistance and type 2 diabetes via several reciprocal mechanisms in animal models and cell culture30–41. However, the causal association of mTOR pathways with diabetes is difficult to establish using observational studies in humans, and randomized controlled trials (RCTs) would be expensive. The Mendelian Randomization (MR), instrumental variable analysis using genetic variants, provides unbiased and unconfounded estimates by taking advantage of the randomization during meiosis and conception42. Studies in animal models and cell culture document a biphasic response of mTOR on glucose and energy metabolism16,40,41,43,44. Recently, studies in the conditional knockout mice found that loss of mTORC1 impairs pancreatic beta-cell mass and functions as well as the cap-dependent protein translation45,46. Therefore, we hypothesized that the elevated levels of circulating proteins of the downstream signals of mTORC1 could be protective from type 2 diabetes mellitus.
Figure 1.
Diagrammatic representation of the mTORC1 complex. mTOR protein kinase nucleates two distinct and mutually exclusive complexes, namely mTORC1 and mTORC2. mTORC1 receives inputs from nutrients, insulin, growth factors, cytokines, and environmental cues; and transmits signals to downstream targets including S6K1, 4E-BP, and EIF complex, GRB10, and ULK1/ATG13.
In this study, we investigated whether the risk of type 2 diabetes varied with EIF4EBP2, EIF-4E, EIF-4G, EIF-4A, and RP-S6K levels using the Mendelian Randomization approach. We hypothesized that mTOR downstream effectors are metabolic targets in managing type 2 diabetes due to the mTOR central role in glucose and energy metabolism and in the pancreatic progenitors' cell growth.
Subjects and methods
Study design and participant flowchart
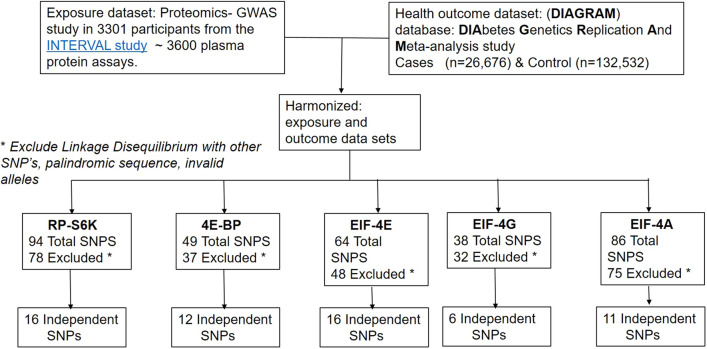
We employed a two-sample MR design, where we used aggregate summary statistics from two Genome-wide Association Studies (GWAS) (Fig. 2). For the exposures, we used proteomics- GWAS INTERVAL study47. For diabetes mellitus health outcomes, we leveraged the publically available DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) GWAS (Supplemental Table 1). We applied the genetic predictors of mTOR downstream targets, namely RP-S6K, EIF4EBP2, and EIF complexes in the circulation, EIF-4E, EIF-4A, and EIF-4G, to the DIAGRAM genotyped case–control study to determine the causal association between mTOR-downstream targets and type 2 diabetes. The flowchart and the selection process of the data included in the study design are summarized in Fig. 2.
Figure 2.
Flow diagram of the selection of SNPs genetic variants as instrumental variables related to exposure harmonized with the dataset for diabetes outcome. We employed a two-sample MR, using summary statistics from two GWAS genetic datasets. For exposure, we used Proteomics-GWAS study39. For the health effect outcome, we leveraged the publically available DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) case (n = 26,676), and control (n = 132,532) study (mean age 57.4 years). We applied the genetic predictors of mTOR downstream targets, namely RP-S6K, EIF4E-BP2, EIF-4E, EIF-4A, and EIF-4G, to DIAGRAM genotyped case–control study to determine the causal association between mTOR-targets and diabetes mellitus. Mendelian Randomization (MR-Base) R package was used in the analysis.
Approach
Genetic prediction of the exposure
Data about mTOR-related gene exposure were obtained from the publically available INTERVAL study Proteomics- GWAS data in 3,301 participants from the INTERVAL study, which included ~ 3,600 plasma protein assay https://www.phpc.cam.ac.uk/ceu/proteins/47. The protein concentrations were measured using the SomaLogic method. This SomaScan proteomic assay utilizes a protein-capture Slow Off-rate Modified Aptamer (SOMAmer). The SOMAmer reagents have dual characteristics as protein-affinity binding agents and unique nucleotide sequences recognized by DNA probes for quantification, which enhances the sensitivity of the proteomic assay48. Protein concentrations were quantified as relative fluorescent units47. The initial INTERVAL study recruited 45,264 healthy blood donors who were 18 years or older and were eligible to donate blood within 25 National Health Service (NHS) Blood and Transplant Centers in England49,50. The trial was registered under # ISRCTN24760606, and recruitments were conducted from June 2012–June 2014. We used the publically available datasets.
As mentioned above, in this study, we obtained single nucleotide polymorphisms (SNPs) which strongly (p-value 5 × 10–6), and independently (r2 < 0.05) predicted RP-S6K, EIF4EBP2, EIF-4E, EIF-4A, and EIF-4G from a GWAS of 3,301 participants in the INTERVAL study47. Genetic associations were obtained using linear regression of the natural log-transformed protein abundances adjusted for age, gender, the time between blood collections, processed for biochemical analysis, and scaled to account for population stratification47,51. To ensure the SNPs for each exposure were independent (i.e., not in linkage disequilibrium), we used MR Base function "clump data" to select and remove SNPs correlated SNPs.
Genetic prediction of health outcomes
Data on diabetes health outcomes were obtained from publically available genetic associations with diabetes from the DIAbestes Genetics Replication And Meta-analysis (DIAGRAM) case–control study https://www.diagram-consortium.org/download. We applied the SNPs predicting the exposures mentioned above to publically available genetic associations with diabetes from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) case (n = 26,676), and control (n = 132,532) study (mean age 57.4 years).
PhenoScanner curated database
The phenoScanner database contains curated large scale GWAS data and facilitates phenome scans, and genotype–phenotype associations across databases to link SNPs with disease biology and mechanistic pathways51,52. We used the PhenoScanner to link the identified SNPs in the current study with possible mechanistic pathways of type 2 diabetes mellitus. We inputted the specific SNPs associated with circulating protein levels of interest to determine the associations with disease traits.
Statistical analysis
We assessed the strength of the genetic instruments from the F-statistic using an approximation, an F-statistics > 10 makes weak instrument bias unlikely (Supplemental Table 2)53. To obtain the MR estimates, we used a meta-analysis of SNP-specific Wald estimates from inverse variance weighting (IVW) with multiplicative random effects54–57. The SNPs were aligned based on the allele letter and allele frequency. The Wald estimate was calculated as the ratio of the estimate for SNPs on diabetes divided by the estimate for SNPs on exposure for each exposure, i.e., RP-S6K, EIF4EBP2, EIF-4E, EIF-4A, and EIF-4G. SNPs were excluded if they were not in the 1,000 Genomes catalog. We corrected for multiple comparisons by using Bonferroni correction. Mendelian Randomization (MR-Base) R package version 3.6.1 was used in the analysis58.
Sensitivity testing
For the IVW estimate to be valid, all the SNPs should be valid instruments. To establish the validity of the SNPs as genetic instrumental variables (IV), we used the following tests: a weighted median and MR-Egger estimates59. The weighted median provides correct estimates when SNPs accounting for > 50% of the weight are valid60. MR-Egger adapts the IVW analysis by allowing a non-zero intercept. This balance would allow for the detection of an unbiased causal effect, even if the IV2 assumption was violated. To test for heterogeneity, we used the Cochrane Q statistic, as well as I2, which is calculated as 100% × (Q − df)/Q, where Q is Cochran's Heterogeneity statistics, and df is the degrees of freedom61.
MR-Egger can give an accurate estimate when the assumption of independent effects are satisfied and that there is little evidence of the pleiotropic effects of SNPs on the outcome (Supplemental Figures 1–3). In addition, if MR-Egger does not correct for unknown pleiotropy, we also used MR-PRESSO, which detects and corrects for outliers62,63.
We used the R package "clump_data" to obtain independent SNPs, and the R Package "Mendelian Randomization" and MR-PRESSO to obtain MR estimates. MR-Base analytical platform web application and MR-Base R package were used64–66. MR-Base web applications were used to generate some of the graphs, URL: https://www.mrbase.org/. We leveraged R (version 3.6.2). R language and environment for statistical analysis were used for all analyses (URL: https://www.R-project.og/) Foundation for Statistical Computing, Vienna, Austria. Mendelian Randomization requires several stringent assumptions to be fulfilled (Fig. 3). First, no confounders are associated with the genetic instrument; second, the genetic proxy of exposure (SNP) should not be independently associated with the disease outcome, but only mediates its effect via the relevant exposure. The SNPs used in the study had no association with the confounding variables (U), and no independent association with diabetes.
Figure 3.
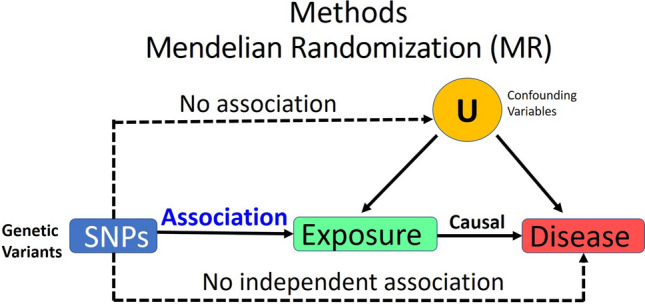
Mendelian Randomization unbiased causal associations and assumptions. Mendelian Randomization (MR) is an application of the instrumental variable using genetic variants Genome-Wide Association Studies (GWAS), and Single Nucleotide Polymorphisms (SNPs). MR requires several stringent assumptions to be fulfilled. First, no confounders are associated with the genetic instrument; and second, the genetic proxy of exposure (SNP) should not be independently associated with the disease outcome, but only mediates its effect via the relevant exposure. The SNPs used in the study had no association with the confounding variables (U) and no independent association with diabetes.
Ethics
We conducted secondary analysis from publically available aggregate summary data with no involvement of the participants in the primary studies. No original data were generated from this manuscript. Ethical approval of each of the studies used is available in the original publications. There is no required IRB for the secondary analysis of summary data. This study follows the ethical guidelines of the Declaration of Helsinki 1975.
Results
As mentioned earlier, for genetic prediction of exposure, we obtained SNPs that strongly and independently predicted RP-S6K, EIF4EBP2, EIF-4E, EIF-4A, and EIF-4G circulating plasma levels from a GWAS of 3,301 participants in the INTERVAL study47. For the genetic prediction of diabetes outcomes, we applied these SNPs predicting the exposures to publically available genetic associations with diabetes from the DIAGRAM case (n = 26,676) and control (n = 132,532) study (mean age 57.4 years). We used the R packages "clump_data" to obtain independent SNPs.
Of the 94 SNPs predicting RP-S6K, 78 correlated SNPs were excluded, with the remaining 16 SNPs that are uncorrelated were included in the analysis. Of the 49 SNPs predicting EIF4EBP2, 37 correlated SNPs were excluded, giving 12 independent SNPs. Of the 64 SNPs predicting EIF-4E, 48 correlated SNPs were excluded, leaving 16 independent SNPs; of the 38 SNPs predicting EIF-4G, 32 correlated SNPs were excluded, resulting in 6 independent SNPs. Finally, of the 86 SNPs predicting EIF-4A, 75 correlated SNPs were excluded, yielding 11 independent SNPs (Fig. 2).
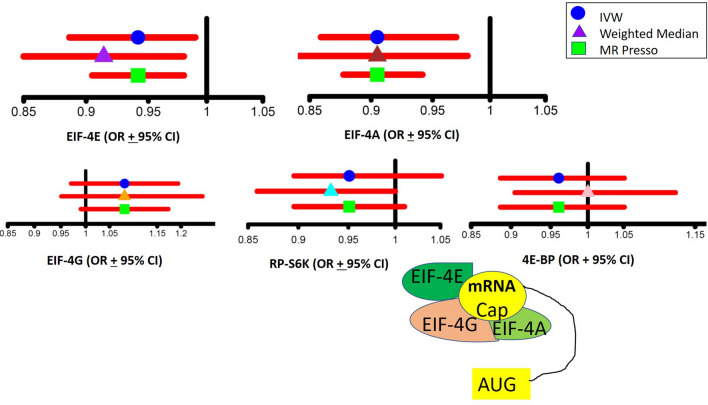
Table 1 shows that EIF-4E cap-dependent translation factor circulating level was associated with a lower risk of type 2 diabetes with an odds ratio (OR) 0.94, 95% confidence interval (0.88, 0.99, p = 0.03) with similar estimates from the weighted median and MR-Egger. Protein concentrations were measured using the SOMAscan assay47. Protein concentrations were quantified as relative fluorescent units. Similarly, EIF-4A cap-dependent translation factor was associated with a lower risk of type 2 diabetes with an odds ratio (OR) 0.90, 95% confidence interval (0.85, 0.97, p = 0.0003). The estimates were very similar in sensitivity analysis, although the MR-Egger estimate had wide confidence intervals.
Table 1.
Mendelian randomization estimates for the association of EIF-4E (based on 16 independent SNPs with the p-value of 5 × 10–6), EIF-4A (based on 11 independent SNPs with the p-value of 5 × 10–6), EIF-4G (based on 6 independent SNP's with the p-value of 5 × 10–6), EIF4E BP2(based on 12 independent SNPs with the p-value of 5 × 10–6), and RP-S6K (based on 16 independent SNPs, with the p-value of 5 × 10–6) with diabetes, using public summary data from DIAGRAM meta-analysis, http://www.cardiogramplusc4d.org/media/cardiogramplusc4d-consortium/data-downloads/UKBB.GWAS1KG.EXOME.CAD.SOFT.META.PublicRelease.300517.txt.gz, and human plasma proteome (Sun BB et al. Nature 201847). Protein concentrations were quantified as relative fluorescent units.
| Exposure | Mendelian randomization method | Odds ratio | 95% confidence interval | p-value | Cochran's Q statistic (p-value) | MR-Egger | |
|---|---|---|---|---|---|---|---|
| Intercept p-value | I2 | ||||||
|
EIF-4E Effect size |
Inverse variance weighted | 0.94 | 0.88, 0.99 | 0.027 | 8.397 (0.906) | – | – |
| Weighted median | 0.91 | 0.84, 0.98 | 0.018 | – | – | – | |
| MR-Egger | 0.93 | 0.83, 1.05 | 0.265 | 8.392 (0.867) | 0.944 | 83.4% | |
| MR PRESSO (uncorrected) | 0.94 | 0.90, 0.98 | 0.009 | – | – | – | |
|
EIF-4A Effect size |
Inverse variance weighted | 0.90 | 0.85, 0.97 | 0.003 | 2.662 (0.988) | – | – |
| Weighted median | 0.90 | 0.82, 0.98 | 0.017 | – | – | – | |
| MR-Egger | 0.91 | 0.80, 1.05 | 0.211 | 2.628 (0.977) | 0.853 | 88.3% | |
| MR PRESSO (uncorrected) | 0.90 | 0.87, 0.94 | 0.0002 | – | – | – | |
|
EIF-4G3 Effect size |
Inverse variance weighted | 1.08 | 0.97, 1.19 | 0.169 | – | – | |
| Weighted median | 1.08 | 0.95, 1.24 | 0.252 | – | – | – | |
| MR-Egger | 0.92 | 0.70, 1.20 | 0.540 | 1.819 (0.7) | 0.213 | 51.3% | |
| MR PRESSO (uncorrected) | 1.08 | 0.99, 1.17 | 0.154 | – | – | – | |
|
EIF4EBP2 Effect size |
Inverse variance weighted | 0.96 | 0.88, 1.05 | 0.372 | 16.75 (0.115) | – | – |
| Weighted median | 1.00 | 0.90, 1.12 | 0.973 | – | – | – | |
| MR-Egger | 1.20 | 0.98, 1.48 | 0.080 | 10.917 (0.364) | 0.021 | 0.6% | |
| MR PRESSO (uncorrected) | 0.96 | 0.88, 1.05 | 0.391 | – | – | – | |
| RP-S6K | Inverse variance weighted | 0.95 | 0.89, 1.01 | 0.077 | 18.747 (0.225) | – | – |
| Weighted median | 0.93 | 0.85, 1.00 | 0.061 | – | – | – | |
| MR-Egger | 0.92 | 0.82, 1.04 | 0.194 | 18.492 (0.185) | 0.660 | 89.5% | |
| MR PRESSO (uncorrected) | 0.95 | 0.89, 1.01 | 0.097 | – | – | – | |
The data shows that EIF-4E cap-dependent translation factor was associated with a lower risk of type 2 diabetes with an odds ratio (OR) 0.94, 95% confidence interval (0.88, 0.99, p = 0.03) with similar estimates from the weighted median and MR-Egger. Similarly, EIF-4A cap-dependent translation factor was associated with a lower risk of type 2 diabetes with an odds ratio (OR) 0.90, 95% confidence interval (0.85, 0.97, p = 0.0003). The estimates were very similar in sensitivity analysis, although the MR-Egger estimate had wide confidence intervals. Mendelian Randomization (MR-Base) R package version 3.6.1 was used in the analysis.
EIF-4G3 and EIF4EBP2 were not associated with diabetes with odds ratio (OR) 1.08, 95% confidence interval (0.97, 1.19, p = 0.169), and odds ratio (OR) 0.99, 95% confidence interval (0.86, 1.03, p = 0.09), respectively. Similarly, ribosomal protein S6K was not associated with diabetes, OR 0.95, 95% confidence interval (0.89, 1.01, p = 0.08). Sensitivity analysis gave consistent estimates.
The following 16 Independent SNPs were associated with EIF-4E circulating protein level (Table 2) and the harmonized outcomes were characterized, as shown in Fig. 4A. Similarly, we identified 11 independent SNPs associated with EIF-4A circulating levels and associated with protective effects from the risk of diabetes (Table 2). We leveraged the use of the PhenoScanner v2 curated database of human genotype–phenotype associations based on cataloging GWAS publically available data from over 150 million unique SNPs and 65 billion associations https://www.phenoscanner.medschl.cam.ac.uk51,52. Out of the 16 SNPs variants genetically associated with EIF-4E, twelve SNPs were located in the intron region of the following genes, RP11-266A241, AGBL1, LINCO2133, NLRP12 (3 SNPs), MARCO, ALK, PR11-138117.1, SPOCK3, RP11-434D9.1, CNGB3; also, three SNPs were located in the intergenic region of MIR4675, ACOO77851, and RLP7L1P4; and one SNP was located upstream of IZUMO3 gene. Most of the SNPs had no known disease associations. However, only three SNPs were associated with NLRP12, a marker of monocytes, and macrophages mediated inflammasome.
Table 2.
Association of genetically predicted mTOR-downstream targets exposures with diabetes outcome SNPS.
| Chr:position outcome | Beta. outcome | SE outcome | Effect allele | Other allele | p-value outcome | SNP | Beta exposure | SE exposure | Effect allele | Other allele | p-value exposure | Wald | Waldvar | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EIF4E | ||||||||||||||
| 1 | 10:20905192 | 0.017 | 0.021 | T | C | 0.41 | rs2209485 | 0.1637 | 0.034 | T | c | 1.51E−06 | 0.103849 | 0.016457 |
| 2 | 11:23,204,504 | 0.032 | 0.075 | T | G | 0.67 | rs74842834 | 0.4443 | 0.0947 | T | g | 2.69E−06 | 0.072023 | 0.028495 |
| 3 | 15:87118467 | 0.044 | 0.081 | T | G | 0.59 | rs192206210 | − 0.5846 | 0.1211 | T | g | 1.41E−06 | − 0.07527 | 0.019198 |
| 4 | 16:47761066 | − 0.014 | 0.042 | A | G | 0.73 | rs138236097 | − 0.2673 | 0.0583 | A | g | 4.47E−06 | 0.052376 | 0.024689 |
| 5 | 19:29350614 | 0.025 | 0.018 | T | C | 0.16 | rs741454 | − 0.1426 | 0.031 | T | c | 4.17E−06 | − 0.17532 | 0.015933 |
| 6 | 19:54311090 | − 0.019 | 0.021 | T | C | 0.36 | rs11084300 | 0.1585 | 0.0272 | T | c | 5.75E−09 | − 0.11987 | 0.017554 |
| 7 | 19:54317794 | 0.0077 | 0.032 | A | G | 0.81 | rs3848582 | − 0.1776 | 0.0364 | A | g | 1.07E−06 | − 0.04336 | 0.032465 |
| 8 | 19:54320939 | − 0.05 | 0.026 | A | G | 0.055 | rs62143198 | 0.4669 | 0.0296 | A | g | 6.76E−56 | − 0.10709 | 0.003101 |
| 9 | 2:119720418 | 0.024 | 0.075 | T | C | 0.75 | rs149036167 | 0.5168 | 0.108 | T | c | 1.70E−06 | 0.04644 | 0.021061 |
| 10 | 2:29505824 | − 0.065 | 0.051 | T | G | 0.2 | rs62130614 | 0.4606 | 0.0999 | T | g | 3.98E−06 | − 0.14112 | 0.01226 |
| 11 | 4:138007589 | 0.04 | 0.044 | A | G | 0.36 | rs113679228 | 0.3464 | 0.0732 | A | g | 2.24E−06 | 0.115473 | 0.016134 |
| 12 | 4:167757172 | − 0.01 | 0.019 | A | C | 0.6 | rs12640699 | 0.1574 | 0.0341 | A | c | 4.07E−06 | − 0.06353 | 0.014571 |
| 13 | 5:116455931 | − 0.017 | 0.027 | T | G | 0.52 | rs142569846 | 0.2303 | 0.0468 | T | g | 8.71E−07 | − 0.07382 | 0.013745 |
| 14 | 5:66,685,975 | − 0.098 | 0.074 | A | G | 0.19 | rs192028145 | 0.5295 | 0.1025 | A | g | 2.40E−07 | − 0.18508 | 0.019531 |
| 15 | 8:87691869 | 0.011 | 0.064 | T | C | 0.87 | rs116934738 | − 0.3758 | 0.077 | T | c | 1.05E−06 | − 0.02927 | 0.029003 |
| 16 | 9:24547232 | − 0.013 | 0.018 | A | G | 0.46 | rs2772577 | 0.1361 | 0.0292 | A | g | 3.02E−06 | − 0.09552 | 0.017492 |
| EIF-4G3 | ||||||||||||||
| 1 | 12:116387486 | 0.026 | 0.015 | A | G | 0.089 | rs7955609 | 0.1251 | 0.0263 | A | g | 2.00E−06 | 0.207834 | 0.014377 |
| 2 | 12:127417047 | − 0.089 | 0.1 | T | C | 0.38 | rs112309230 | 0.5745 | 0.1158 | T | c | 6.92E−07 | − 0.15492 | 0.030298 |
| 3 | 12:39641451 | − 3.00E−04 | 0.027 | T | C | 0.99 | rs143862167 | − 0.204 | 0.044 | T | c | 3.55E−06 | 0.001471 | 0.017517 |
| 4 | 17:26694861 | 0.013 | 0.015 | A | G | 0.37 | rs704 | 0.1618 | 0.0245 | A | g | 4.27E−11 | 0.080346 | 0.008595 |
| 5 | 3:53892603 | 0.054 | 0.08 | T | C | 0.5 | rs142978915 | 0.4055 | 0.0885 | T | c | 4.57E−06 | 0.133169 | 0.038922 |
| 6 | 9:3762150 | 0.029 | 0.057 | A | G | 0.61 | rs1411879 | 0.4092 | 0.089 | A | g | 4.27E−06 | 0.07087 | 0.019403 |
| EIF-4A | ||||||||||||||
| 1 | 12:95350637 | − 0.016 | 0.023 | A | G | 0.49 | rs74512707 | 0.1785 | 0.0385 | A | g | 3.55E−06 | − 0.08964 | 0.016603 |
| 2 | 15:86409988 | − 0.053 | 0.074 | A | G | 0.48 | rs151270869 | 0.4543 | 0.0938 | A | g | 1.29E−06 | − 0.11666 | 0.026532 |
| 3 | 19:54295370 | 0.0014 | 0.025 | T | G | 0.96 | rs3859507 | 0.1777 | 0.0327 | T | g | 5.75E−08 | 0.007878 | 0.019793 |
| 4 | 19:54311090 | − 0.019 | 0.021 | T | C | 0.36 | rs11084300 | 0.1431 | 0.0272 | T | c | 1.48E−07 | − 0.13277 | 0.021536 |
| 5 | 19:54327313 | − 0.052 | 0.026 | A | C | 0.042 | rs34436714 | 0.4687 | 0.0291 | A | c | 2.00E−58 | − 0.11095 | 0.003077 |
| 6 | 19:54329672 | 0.033 | 0.036 | A | G | 0.35 | rs146117463 | − 0.2062 | 0.042 | A | g | 9.33E−07 | − 0.16004 | 0.030481 |
| 7 | 2:29058128 | − 0.0015 | 0.036 | A | G | 0.97 | rs34131899 | − 0.2829 | 0.0591 | A | g | 1.70E−06 | 0.005302 | 0.016193 |
| 8 | 3:27245225 | 0.013 | 0.017 | A | G | 0.45 | rs6792693 | − 0.134 | 0.0286 | A | g | 2.82E−06 | − 0.09701 | 0.016095 |
| 9 | 3:74143047 | 0.024 | 0.015 | T | C | 0.11 | rs1447676 | − 0.1182 | 0.0257 | T | c | 4.37E−06 | − 0.20305 | 0.016105 |
| 10 | 7:85099898 | − 0.018 | 0.015 | T | G | 0.2 | rs2462049 | 0.1159 | 0.0249 | T | g | 3.31E−06 | − 0.15531 | 0.01675 |
| 11 | 9:87844548 | − 0.0031 | 0.015 | A | G | 0.83 | rs1931094 | 0.1163 | 0.0253 | A | g | 4.27E−06 | − 0.02666 | 0.016635 |
| EIF4EBP2 | ||||||||||||||
| 1 | 1:9425722 | 0.035 | 0.015 | A | G | 0.023 | rs10864412 | − 0.1334 | 0.0254 | A | g | 1.58E−07 | − 0.26237 | 0.012644 |
| 2 | 10:64948684 | 0.011 | 0.015 | T | C | 0.5 | rs10733789 | − 0.1254 | 0.0272 | T | c | 4.07E−06 | − 0.08772 | 0.014308 |
| 3 | 15:58881788 | − 0.028 | 0.054 | A | G | 0.61 | rs72743058 | − 0.3839 | 0.0827 | A | g | 3.47E−06 | 0.072936 | 0.019786 |
| 4 | 15:90454175 | 0.032 | 0.065 | T | C | 0.62 | rs79943794 | − 0.357 | 0.0766 | T | c | 3.16E−06 | − 0.08964 | 0.033151 |
| 5 | 16:1547477 | − 0.012 | 0.026 | T | C | 0.65 | rs2745108 | 0.1927 | 0.0402 | T | c | 1.58E−06 | − 0.06227 | 0.018205 |
| 6 | 16:1846089 | − 0.047 | 0.02 | T | C | 0.017 | rs2575348 | 0.1527 | 0.0333 | T | c | 4.47E−06 | − 0.30779 | 0.017155 |
| 7 | 2:167837496 | − 0.035 | 0.04 | T | C | 0.39 | rs79613514 | − 0.3407 | 0.0738 | T | c | 3.89E−06 | 0.10273 | 0.013784 |
| 8 | 21:21934726 | − 0.049 | 0.074 | T | C | 0.51 | rs17003636 | − 0.4607 | 0.0984 | T | c | 2.88E−06 | 0.10636 | 0.0258 |
| 9 | 3:14547897 | 0.013 | 0.053 | T | C | 0.8 | rs113664570 | 0.2666 | 0.0584 | T | c | 4.90E−06 | 0.048762 | 0.039521 |
| 10 | 5:152342669 | 0.026 | 0.017 | T | C | 0.13 | rs72806713 | − 0.1366 | 0.0296 | T | c | 3.89E−06 | − 0.19034 | 0.015488 |
| 11 | 7:113007324 | − 0.021 | 0.027 | T | C | 0.43 | rs76802510 | − 0.2684 | 0.0496 | T | c | 6.46E−08 | 0.078241 | 0.01012 |
| 12 | 8:106583124 | 0.021 | 0.017 | A | G | 0.2 | rs4734879 | 0.1343 | 0.028 | A | g | 1.62E−06 | 0.156366 | 0.016023 |
| RP-S6K | ||||||||||||||
| 1 | 11:11949472 | − 0.0032 | 0.015 | T | C | 0.84 | rs1355191 | − 0.125 | 0.0263 | T | c | 1.95E−06 | 0.0256 | 0.0144 |
| 2 | 15:83650787 | − 0.016 | 0.068 | A | C | 0.82 | rs77394885 | − 0.5291 | 0.1122 | A | c | 2.40E−06 | 0.03024 | 0.016517 |
| 3 | 19:54294400 | 0.003 | 0.025 | A | C | 0.91 | rs3859503 | − 0.1892 | 0.0333 | A | c | 1.35E−08 | − 0.01586 | 0.01746 |
| 4 | 19:54305398 | − 0.073 | 0.025 | T | G | 0.0037 | rs148800371 | 0.1637 | 0.0327 | T | g | 5.50E−07 | − 0.44594 | 0.023323 |
| 5 | 19:54320716 | − 0.047 | 0.026 | A | G | 0.075 | rs62143197 | 0.5347 | 0.029 | A | g | 8.13E−76 | − 0.0879 | 0.002364 |
| 6 | 19:54333199 | − 0.035 | 0.036 | T | C | 0.34 | rs117021160 | 0.2834 | 0.0433 | T | c | 6.17E−11 | − 0.1235 | 0.016136 |
| 7 | 20:58296885 | − 0.081 | 0.065 | T | C | 0.22 | rs75688971 | − 0.4179 | 0.0836 | T | c | 5.75E−07 | 0.193826 | 0.024193 |
| 8 | 3:22584610 | − 0.073 | 0.057 | T | C | 0.2 | rs58565824 | 0.4607 | 0.0937 | T | c | 8.91E−07 | − 0.15845 | 0.015308 |
| 9 | 3:27587457 | 0.053 | 0.036 | T | C | 0.14 | rs9833044 | − 0.319 | 0.0635 | T | c | 5.13E−07 | − 0.16614 | 0.012736 |
| 10 | 4:145934089 | − 0.043 | 0.059 | T | C | 0.46 | rs35747952 | 0.3736 | 0.0809 | T | c | 3.89E−06 | − 0.1151 | 0.02494 |
| 11 | 5:101423444 | 0.031 | 0.046 | T | C | 0.51 | rs1381968 | 0.3462 | 0.0746 | T | c | 3.55E−06 | 0.089544 | 0.017655 |
| 12 | 5:152488248 | − 0.013 | 0.014 | A | G | 0.38 | rs62398809 | 0.1129 | 0.0247 | A | g | 4.90E−06 | − 0.11515 | 0.015377 |
| 13 | 5:39170760 | 0.042 | 0.076 | A | G | 0.58 | rs148897689 | 0.4496 | 0.0943 | A | g | 1.91E−06 | 0.093416 | 0.028574 |
| 14 | 6:68218559 | − 0.053 | 0.074 | A | G | 0.48 | rs72881486 | 0.3633 | 0.0745 | A | g | 1.07E−06 | − 0.14588 | 0.041489 |
| 15 | 7:58384 | 0.025 | 0.02 | A | C | 0.21 | rs79777011 | 0.1634 | 0.0346 | A | c | 2.29E−06 | 0.152999 | 0.014982 |
| 16 | 8:56752146 | 0.0069 | 0.015 | A | G | 0.65 | rs7017005 | 0.1213 | 0.0256 | A | g | 2.14E−06 | 0.056884 | 0.015292 |
Association between genetically determined EIF-4E, EIF-4A, EIF-4G, EIF4EBP2, and RP-S6K, mTOR downstream targets with Diabetes health outcome. The data source for exposure is the human plasma proteomics–GWAS INTERVAL study (n = 3,301) participants from publically available aggregate summary data47. The source for diabetes health outcome is DIAGRAM database (DIAbetes Genetics Replication And Metaanalysis case (n = 26,676), and control (n = 132,532); https://www.diagram-consortium.org/. For sensitivity analysis, we used uncorrelated SNPs for EIF-4E, IF-4A, EIF-4G, EIF4EBP2, and RP-S6K. Mendelian Randomization (MR-Base) R package was used in the analysis.
Figure 4.
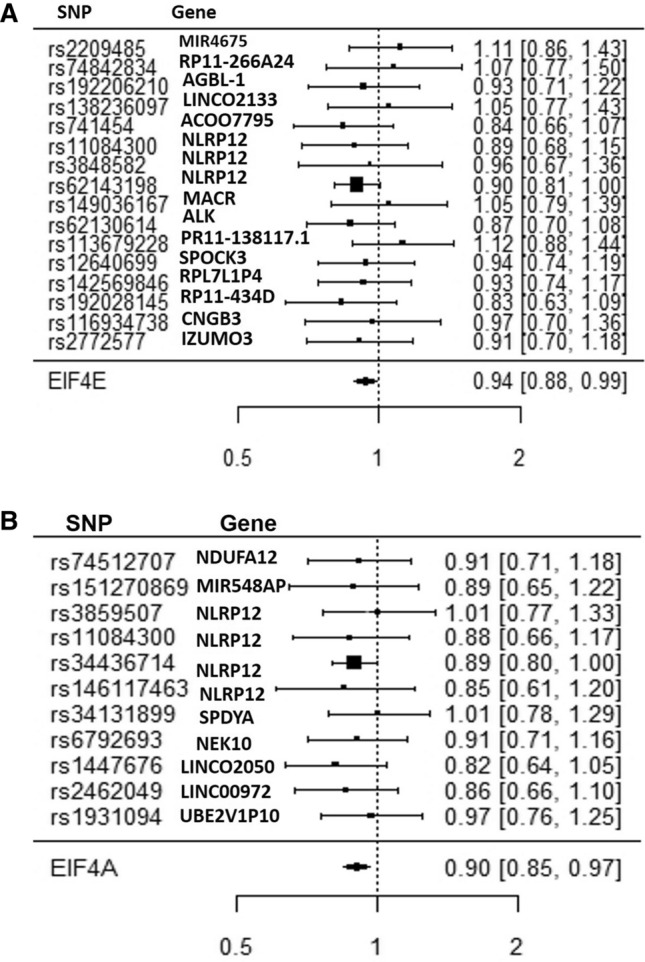
(A) SNP variants associated with EIF-4E and the risk of type 2 diabetes. Odds ratio ± 95% confidence interval of the EIf-4E causal estimates on types 2 diabetes are shown. Inverse Variance Weighted (IVW) analysis of individual SNPs and the composite odds ratio of all SNPs shows the independent SNPs that are strongly associated with EIF-4E. Using phenoScanner v2 database of human genotype–phenotype association based on the UKBB data https://www.phenoscanner.medschl.cam.ac.uk, the following 16 Independent SNPs associated with EIF-4E merged with diabetes health outcome. Out of the 16 SNPs, 10 were located in the intron region of the following genes, RP11-266A241, AGBL1, LINCO2133, NLRP12 (3 SNPs), MARCO, ALK, PR11-138117.1, SPOCK3, RP11-434D9.1, CNGB3; 3 SNPs were located intergenic region to the nearest genes MIR4675, ACOO77851, and RLP7L1P4. One SNP was located upstream of the IZUMO3 gene. Mendelian Randomization (MR-Base) R package was used in the analysis58. (B) SNP variants associated with EIF-4A and the risk of type 2 diabetes. Odds Ratio ± 95% Confidence Interval of Individual SNP causality on type 2 diabetes are shown. Inverse Variance Weighted (IVW) analysis of individual SNPs and the composite odds ratio of all SNPs shows the independent SNPs that are strongly associated with EIF-4A. Using phenoScanner v2 database of human genotype–phenotype association based on publically available databases https://www.phenoscanner.medschl.cam.ac.uk, the following 11 Independent SNPs associated with EIF-4A were harmonized with diabetes health outcome. Out of the 11 SNPs, 5 were located in the intron region of the following genes NDUFA12, SPDYA, NEK10, LINCOO972, and NLPR12; 3 SNPs were located intergenic region to the nearest genes, MIR548AP, LINCO2050, and UBE2VIP10. 1SNP was located upstream of gene NLPR12, 1 downstream NLPR12 gene, and 1 missense on gene NLPR12. Mendelian Randomization (MR-Base) R package was used in the analysis58.
Discussion
To our knowledge, this is the first study to leverage MR to determine the causal association of EIF-4E and EIF-4A circulating levels with diabetes mellitus in humans. The data suggest that higher circulating levels of EIF-4E (OR 0.94, CI [0.88, 0.99]) and EIF-4A (OR 0.90, CI [0.85, 0.97]) translation initiation proteins may be causally associated with a lower risk of type 2 diabetes mellitus. As shown in the leave-one-out sensitivity IVW analysis, the effect size for EIF-4E is − 0.06 (OR 0.94), and for EIF-4A is − 0.10 (OR 0.90) (Supplemental Figure 2). Per our study objectives, five mTORC1 downstream targets (RP-S6K, EIF4EBP2, EIF-4G3, EIF-4E cap-dependent translation, and EIF-4A cap-dependent translation factor), circulating levels were instrumented by strongly associated SNPs (Fig. 5). This unbiased MR approach found that EIF-4E and EIF-4A, which are essential for 5′-cap-dependent mRNA translation, could have a causal protective association with type 2 diabetes. The directionality of the MR association between circulating levels of EIF-4E and EIF-4A, mTORC1-dependent proteins, and type-2 diabetes mellitus was protection, i.e., higher circulating levels of EIF-4E and EIF-4A may be causally associated with decreased risk of type 2 diabetes mellitus. This data is consistent with experimental data in mTORC1 knockout mice showing an essential role of mTORC1 in beta cell functions and insulin processing45. Our data indicated that the risk of diabetes varied with mTOR downstream target EIF-4E and EIF-4A, but not with RP-S6K or EIF4EBP2. The association for both EIF-4E cap-dependent translation factor and EIF-4A were robust to Bonferroni correction.
Figure 5.
Comparisons of the Odds Ratio values and the 95% Upper Confidence Level and Lower Confidence Level between different MR Models. Comparison of the overall estimates of odds ratio values obtained by different MR models, including Inverse Variance Weighted (IVW), Weighted Median (WM), and MR PRESSO analyses of genetically determined EIF-4E, EIF-4A, EIF4G, EIF4EBP, and RP-S6K. Mendelian Randomization (MR-Base) R package and MR Base web applications were used in the analysis58.
In this study, we used the two-sample MR, which increases the sample size and provides unconfounded estimates. However, MR requires several stringent assumptions to be fulfilled. First, the genetic predictors should be strongly associated with the exposures (IV1); to satisfy this assumption, we selected only the SNPs strongly associated with EIF-4E, EIF-4A, EIF-4G, EIF4EBP2, and RP-S6K circulating plasma levels. Second, no confounders of the genetic instrument on outcome should exist, which we assume from genetic randomization at conception (IV2). The underlying genetic studies were controlled for population stratification when necessary. Third, genetic predictors of exposure should not be independently associated with diabetes (IV3), and only mediate their effects via the relevant exposure. In other words, there is little evidence of pleiotropy. Sensitivity analyses with different assumptions about pleiotropy gave estimates with the IVW (Fig. 5). Since this study only used aggregate summary statistics, we were not able to assess differences by age, gender, or other factors. However, causal effects are generally expected to act consistently.
In contrast to RCTs, MR studies inform the role of a specific causal pathway but do not reveal whether an intervention will be efficient. Furthermore, RCTs only report the effects of short-time exposures, while MR studies usually identify the lifetime exposures. The limitations of the MR study are those cohort populations recruited for the exposure (INTERVAL Study), and outcomes (DIAGRAM Study) were from European descent, which may limit the generalizability of the results to other populations.
Cell culture and animal studies have documented an integral role of mTORC1 in glucose metabolism and diabetes24,25,67–70. Blandino-Rosano and colleagues have shown in Raptor, a mTORC1 specific partner, in a conditional knockout mouse model led to pancreatic β-cell failure due to defects in insulin secretion and cell proliferation, and thus furthered the development of diabetes44–46,71,72. Ardestani and colleagues reported that mTORC1 positively regulates beta-cell growth and proliferation, thus promoting insulin secretion, while the hyperactivation of mTORC1 led to beta-cell failure in type 2 diabetes16. Similarly, Tuo and Xiang reported a paradoxical role of mTORC1 on insulin secretion and β-cell metabolism31. The biphasic response of pancreatic β cells to mTORC1 activation has also been reported in mice43. Shigeyama and colleagues found that ablated Tuberous Sclerosis Complex 2 (TSC2), an upstream inhibitor of mTORC1m in mice (βTSC−/− mice), led to an initial increase in β cell mass and size in an mTOR-dependent manner. Furthermore, the mice were hyperinsulinemic and hypoglycemic. However, after 40 weeks of age, these mice became hypoinsulinemic and hyperglycemic due to a reduction in β cell mass and number43. Taken together, these two observations suggest that mTOR-mediated a biphasic response on insulin secretion.
Similarly, a biphasic insulin sensitivity to mTOR inhibitor, rapamycin, has been reported in C2C12 myocytes37. Reciprocal regulation and cross-talk between mTORC1 and mTORC2 have been suggested35,73. Further, a distinctive role of mTORC2 in regulating insulin secretion and pancreatic β cell functions has also been reported18,35. Blandino-Rosano and colleagues found that the deletion of EIF-4EBP2 induced feedback and elevated glucose tolerance by increasing pancreatic beta-cell mass in knockout mice74. It is possible that EIF-4E and EIF-4A may regulate the mRNA translation of insulin protein, insulin receptor, or the enzymes involved in the regulation of glucose metabolism such as those regulating glycolysis and TCA cycle. We leveraged the use of the PhenoScanner curated database to explore possible causal mechanistic associations between the circulating levels of EIF-4E and EIF-4A and type 2 diabetes mellitus. We found 3 EIF-4E proxy SNPs namely, rs11084300, rs3848582, rs62143198, and 4 EIF-4A predicting SNPs, namely, rs3859507, rs11084300, rs34436714, rs146117463, that are associated with the NLRP12 gene. The NLPR12 is an inhibitory innate immune sensor and a marker of monocytes and macrophage inflammasomes. Recently, NLPR12 has been implicated in protecting against obesity by regulating the gut microbiota homeostasis, insulin-tolerance, and inflammation in mice75. Thus, we propose that EIF-4E and EIF-4A translation initiation proteins may have a protective effect on type 2 diabetes that is possibly mediated via NLPR12 inflammasome, which regulates the gut microbiome. Experimental laboratory data is warranted to explore the mechanistic pathway for this effect.
Conclusion
This unbiased MR study is consistent with a causal protective association of EIF-4E and EIF-4A with type 2 diabetes. EIF-4E and EIF-4A circulating levels, critical regulators of gene translation and protein functions, may be targets for intervention by repurposing existing therapeutics to reduce the risk of type 2 diabetes.
Supplementary information
Abbreviations
- SNPs
Single nucleotide polymorphism
- mTOR
Mammalian target of rapamycin
- EIF-4E
Eukaryotic translation initiation factor 4E
- EIF4EBP2
Eukaryotic translation initiation factor 4E binding protein 2 (4E-BP)
- mTORC1
Mammalian target of rapamycin complex 1
- mTORC2
Mammalian target of rapamycin complex 2
- GWAS
Genome-wide association studies
- IVW
Inverse variance weighted
- WM
Weighted median
- MR
Mendelian randomization
- RP-S6K
Ribosomal protein S6K kinase
Author contributions
The authors contributions to the manuscript and responsibilities are as follows: G.S. and C.M.S. designed the study, G.S. analyzed the data, interpreted the results, and drafted the paper, and C.M.S. critically revised the paper. The author's work is partially funded by the PSC-CUNY award. All authors had read and approved the final version of the manuscript.
Funding
Funding provided in part by a Professional Staff Council (PSC-CUNY) Grant # 63154-00 51.
Data availability
Data on diabetes health outcomes were obtained publically available genetic associations with diabetes from the DIAbestes Genetics Replication And Meta-analysis (DIAGRAM) case–control study https://www.diagram-consortium.org/download. Data about mTOR-related gene exposure were obtained from the publically available INTERVAL study Proteomics-GWAS data in 3,301 participants from the INTERVAL study, which included ~ 3,600 plasma protein assay https://www.phpc.cam.ac.uk/ceu/proteins/ (Sun et al., Nature, 201839). The authors would like to thank all investigators for sharing the data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71987-8.
References
- 1.1CDC. National Diabetes Statistics Report, 2017. Estimates of Diabetes and Its Burden in the United States. Center for Disease Control, National Center for Chronic Disease Prevention and Health Promotion, Division of Diabetes Translation. (2017).
- 2.American Diabetes, A Economic costs of diabetes in the U.S. in 2017. Diabetes Care. 2018;41:917–928. doi: 10.2337/dci18-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentges KE, et al. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc. Natl. Acad. Sci. USA. 2001;98:13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatini DM, Pierchala BA, Barrow RK, Schell MJ, Snyder SH. The rapamycin and FKBP12 target (RAFT) displays phosphatidylinositol 4-kinase activity. J. Biol. Chem. 1995;270:20875–20878. doi: 10.1074/jbc.270.36.20875. [DOI] [PubMed] [Google Scholar]
- 5.Soliman GA. The mammalian target of rapamycin signaling network and gene regulation. Curr. Opin. Lipidol. 2005;16:317–323. doi: 10.1097/01.mol.0000169352.35642.06. [DOI] [PubMed] [Google Scholar]
- 6.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efeyan A, Sabatini DM. mTOR and cancer: Many loops in one pathway. Curr. Opin. Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: From lysosomes to disease. Trends Mol. Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikani CK, et al. Activation of PASK by mTORC1 is required for the onset of the terminal differentiation program. Proc. Natl. Acad. Sci. USA. 2019;116:10382–10391. doi: 10.1073/pnas.1804013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatini DM. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA. 2017;114:11818–11825. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 12.Son S, et al. Cooperative nutrient accumulation sustains growth of mammalian cells. Sci. Rep. 2015;5:17401. doi: 10.1038/srep17401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamming DW, Sabatini DM. A central role for mTOR in lipid homeostasis. Cell Metab. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb. Perspect. Biol. 2012;4:1–4. doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardestani A, Lupse B, Kido Y, Leibowitz G, Maedler K. mTORC1 signaling: A double-edged sword in diabetic beta cells. Cell Metab. 2018;27:314–331. doi: 10.1016/j.cmet.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Jhanwar-Uniyal M, et al. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv. Biol. Regul. 2019;72:51–62. doi: 10.1016/j.jbior.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Yuan T, Lupse B, Maedler K, Ardestani A. mTORC2 signaling: A path for pancreatic beta cell's growth and function. J. Mol. Biol. 2018;430:904–918. doi: 10.1016/j.jmb.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Le Bacquer O, et al. mTORC1 and mTORC2 regulate insulin secretion through Akt in INS-1 cells. J. Endocrinol.0. 2013;216:21–29. doi: 10.1530/JOE-12-0351. [DOI] [PubMed] [Google Scholar]
- 20.Guertin DA, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Liu P, et al. Author correction: Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 2019;21:662–663. doi: 10.1038/s41556-019-0280-y. [DOI] [PubMed] [Google Scholar]
- 23.Soliman GA, et al. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J. Biol. Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol. Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JY, et al. mTORC1 and mTORC2 are differentially engaged in the development of laser-induced CNV. Cell Commun. Signal. 2019;17:64. doi: 10.1186/s12964-019-0380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Fan W, et al. mTORC1 and mTORC2 play different roles in the functional survival of transplanted adipose-derived stromal cells in hind limb ischemic mice via regulating inflammation in vivo. Stem Cells. 2013;31:203–214. doi: 10.1002/stem.1265. [DOI] [PubMed] [Google Scholar]
- 29.Rosner M, Hengstschlager M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: Rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum. Mol. Genet. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- 30.Cantley J, et al. Disruption of beta cell acetyl-CoA carboxylase-1 in mice impairs insulin secretion and beta cell mass. Diabetologia. 2019;62:99–111. doi: 10.1007/s00125-018-4743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuo Y, Xiang M. mTOR: A double-edged sword for diabetes. J. Leukoc. Biol. 2018;106(2):385–395. doi: 10.1189/JLB.3MR0317-095RR. [DOI] [PubMed] [Google Scholar]
- 32.Verges B. mTOR and cardiovascular diseases: Diabetes mellitus. Transplantation. 2018;102:S47–S49. doi: 10.1097/TP.0000000000001722. [DOI] [PubMed] [Google Scholar]
- 33.Rice KM, Manne N, Arvapalli R, Ginjupalli GK, Blough ER. Diabetes alters vascular mechanotransduction data: Pressure-induced regulation of mTor and associated signaling in the rat inferior vena cava. Data Brief. 2017;15:63–71. doi: 10.1016/j.dib.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin X, et al. Association of PI3K/AKT/mTOR pathway genetic variants with type 2 diabetes mellitus in Chinese. Diabetes Res. Clin. Pract. 2017;128:127–135. doi: 10.1016/j.diabres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Yuan T, et al. Reciprocal regulation of mTOR complexes in pancreatic islets from humans with type 2 diabetes. Diabetologia. 2017;60:668–678. doi: 10.1007/s00125-016-4188-9. [DOI] [PubMed] [Google Scholar]
- 36.Verges B, Cariou B. mTOR inhibitors and diabetes. Diabetes Res. Clin. Pract. 2015;110:101–108. doi: 10.1016/j.diabres.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Ye L, Varamini B, Lamming DW, Sabatini DM, Baur JA. Rapamycin has a biphasic effect on insulin sensitivity in C2C12 myotubes due to sequential disruption of mTORC1 and mTORC2. Front. Genet. 2012;3:177. doi: 10.3389/fgene.2012.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suhara T, Baba Y, Shimada BK, Higa JK, Matsui T. The mTOR signaling pathway in myocardial dysfunction in type 2 diabetes mellitus. Curr. Diab. Rep. 2017;17:38. doi: 10.1007/s11892-017-0865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie J, Herbert TP. The role of mammalian target of rapamycin (mTOR) in the regulation of pancreatic beta-cell mass: Implications in the development of type-2 diabetes. Cell Mol. Life Sci. 2012;69:1289–1304. doi: 10.1007/s00018-011-0874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leibowitz G, Cerasi E, Ketzinel-Gilad M. The role of mTOR in the adaptation and failure of beta-cells in type 2 diabetes. Diabetes Obes. Metab. 2008;10(Suppl 4):157–169. doi: 10.1111/j.1463-1326.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- 41.Fraenkel M, et al. mTOR inhibition by rapamycin prevents beta-cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes. 2008;57:945–957. doi: 10.2337/db07-0922. [DOI] [PubMed] [Google Scholar]
- 42.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 43.Shigeyama Y, et al. Biphasic response of pancreatic beta-cell mass to ablation of tuberous sclerosis complex 2 in mice. Mol. Cell Biol. 2008;28:2971–2979. doi: 10.1128/MCB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riahi Y, et al. Inhibition of mTORC1 by ER stress impairs neonatal beta-cell expansion and predisposes to diabetes in the Akita mouse. Elife. 2018;7:1–25. doi: 10.7554/eLife.38472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blandino-Rosano M, et al. Loss of mTORC1 signalling impairs beta-cell homeostasis and insulin processing. Nat. Commun. 2017;8:16014. doi: 10.1038/ncomms16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blandino-Rosano M, et al. mTORC1 signaling and regulation of pancreatic beta-cell mass. Cell Cycle. 2012;11:1892–1902. doi: 10.4161/cc.20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies DR, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA. 2012;109:19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore C, et al. The INTERVAL trial to determine whether intervals between blood donations can be safely and acceptably decreased to optimise blood supply: Study protocol for a randomised controlled trial. Trials. 2014;15:363. doi: 10.1186/1745-6215-15-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Angelantonio E, et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): A randomised trial of 45 000 donors. Lancet. 2017;390:2360–2371. doi: 10.1016/S0140-6736(17)31928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staley JR, et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamat MA, et al. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowden J, et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao JV, Kwok MK, Schooling CM. Effect of glutamate and aspartate on ischemic heart disease, blood pressure, and diabetes: A Mendelian randomization study. Am. J. Clin. Nutr. 2019;109:1197–1206. doi: 10.1093/ajcn/nqy362. [DOI] [PubMed] [Google Scholar]
- 55.Zhao JV, Schooling CM. Effect of linoleic acid on ischemic heart disease and its risk factors: A Mendelian randomization study. BMC Med. 2019;17:61. doi: 10.1186/s12916-019-1293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo S, Au Yeung SL, Zhao JV, Burgess S, Schooling CM. Association of genetically predicted testosterone with thromboembolism, heart failure, and myocardial infarction: Mendelian randomisation study in UK Biobank. BMJ. 2019;364:l476. doi: 10.1136/bmj.l476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Au Yeung SL, Luo S, Schooling CM. The impact of glycated hemoglobin (HbA1c) on cardiovascular disease risk: A mendelian randomization study using UK Biobank. Diabetes Care. 2018;41:1991–1997. doi: 10.2337/dc18-0289. [DOI] [PubMed] [Google Scholar]
- 58.Team, R. C. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, 2019) https://www.R-project.org.
- 59.Schooling CM, Ng JCM. Reproduction and longevity: A Mendelian randomization study of gonadotropin-releasing hormone and ischemic heart disease. SSM Popul. Health. 2019;8:100411. doi: 10.1016/j.ssmph.2019.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schooling CM, Kodali H, Li S, Borrell LN. ET (Endothelin)-1 and ischemic heart disease: A Mendelian randomization study. Circ. Genom Precis. Med. 2018;11:e002026. doi: 10.1161/CIRCGENETICS.117.002026. [DOI] [PubMed] [Google Scholar]
- 61.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corbin LJ, et al. BMI as a modifiable risk factor for type 2 diabetes: Refining and understanding causal estimates using Mendelian randomization. Diabetes. 2016;65:3002–3007. doi: 10.2337/db16-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker VM, et al. Using the MR-Base platform to investigate risk factors and drug targets for thousands of phenotypes. Wellcome Open Res. 2019;4:113. doi: 10.12688/wellcomeopenres.15334.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng J, et al. Recent developments in Mendelian randomization studies. Curr. Epidemiol. Rep. 2017;4:330–345. doi: 10.1007/s40471-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:1–29. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoxhaj G, et al. The mTORC1 signaling network senses changes in cellular purine nucleotide levels. Cell Rep. 2017;21:1331–1346. doi: 10.1016/j.celrep.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem. Soc. Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 69.Byles V, et al. The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 71.Bozadjieva N, et al. Loss of mTORC1 signaling alters pancreatic alpha cell mass and impairs glucagon secretion. J. Clin. Investig. 2017;127:4379–4393. doi: 10.1172/JCI90004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balcazar N, et al. mTORC1 activation regulates beta-cell mass and proliferation by modulation of cyclin D2 synthesis and stability. J. Biol. Chem. 2009;284:7832–7842. doi: 10.1074/jbc.M807458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F, et al. Crosstalks between mTORC1 and mTORC2 variagate cytokine signaling to control NK maturation and effector function. Nat.. Commun. 2018;9:4874. doi: 10.1038/s41467-018-07277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blandino-Rosano M, et al. 4E-BP2/SH2B1/IRS2 are part of a novel feedback loop that controls beta-cell mass. Diabetes. 2016;65:2235–2248. doi: 10.2337/db15-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Truax AD, et al. The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe. 2018;24:364–378. doi: 10.1016/j.chom.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data on diabetes health outcomes were obtained publically available genetic associations with diabetes from the DIAbestes Genetics Replication And Meta-analysis (DIAGRAM) case–control study https://www.diagram-consortium.org/download. Data about mTOR-related gene exposure were obtained from the publically available INTERVAL study Proteomics-GWAS data in 3,301 participants from the INTERVAL study, which included ~ 3,600 plasma protein assay https://www.phpc.cam.ac.uk/ceu/proteins/ (Sun et al., Nature, 201839). The authors would like to thank all investigators for sharing the data.