Abstract
Intrahepatic cholangiocarcinoma (ICC) is a rare but fatal tumor. The isocitrate dehydrogenase 1 and 2 (IDH1/2) genes are known to be mutated in ICC. IDH1/2 mutations tend to be accompanied by enhanced hypermethylation at a subset of genomic loci. We sought to clarify the clinicopathological features, including prognostic value, of ICCs with IDH1/2 mutation-associated hypermethylation at a subset of genes. The mutation status of IDH1/2 and methylation status of 30 gene CpG island loci were analyzed in 172 cases of ICC using pyrosequencing and the MethyLight assay, respectively. The mutation status of IDH1/2 was correlated with clinicopathological features and the DNA methylation status at 30 gene loci. Then, the clinicopathological characteristics were analyzed regarding three-tiered methylation statuses in genes showing IDH1/2 mutation-associated methylation. IDH1/2 mutations were found in 9.3% of ICCs, and IDH1/2-mutated tumors were associated with the histological subtype, including the bile ductular type and small duct type, and poor differentiation. Eight DNA methylation markers showed associations with IDH1/2 mutations, and ICCs with > 5/8 methylated markers were associated with the bile ductular type or small duct type, absence of mucin production, absence of biliary intraepithelial neoplasia, and presence of chronic liver disease. > 5/8 methylated markers were an independent prognostic marker associated with better survival in both cancer-specific survival and recurrence-free survival. In summary, by analyzing the association between IDH1/2 mutations and DNA methylation in individual genes, we developed a panel of DNA methylation markers that were significantly associated with IDH1/2 mutations and were able to identify a subset of ICC with better clinical outcomes.
Subject terms: DNA methylation, Bile duct cancer
Introduction
Approximately 2.8 × 107 5-methyl CpG dinucleotides are present in the genome, and the enzymes that generate and maintain methylation at the CpG dinucleotides are DNMT3A/B and DNMT1, respectively. The enzyme that demethylates 5-methyl CpG dinucleotides was first identified in 2009 by Rao and colleagues, who demonstrated that TET1 catalyzes 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC)1. In 2011, hydroxyglutarate produced by mutant IDH1/2 was demonstrated to inhibit the catalytic activity of the TET family of 5-mC hydroxylases, which have ketoglutarate- and oxygen-dependent demethylation activity2. Zhang and colleagues further demonstrated that not only TET1 but also TET2 or 3 generates 5-hmC, 5-formyl cytosine, and 5-carboxyl cytosine through iterative oxidation of 5-mC3. IDH1/2 mutations are mainly found in gliomas, acute myelogenous leukemia, and chondroid tumors but rarely found in epithelial malignancies except for cholangiocarcinomas4. Of the cholangiocarcinomas, IDH1/2 mutations are found in intrahepatic cholangiocarcinomas (ICCs) and rarely, if ever, in perihilar or extrahepatic cholangiocarcinomas5,6.
ICC is a heterogeneous disease entity in terms of histology and is further classified into three histological subtypes, including the bile ductular (BD) type, small duct (SD) type, and large duct (LD) type7–9. The putative cell of origin of ICC varies according to the histological subtype10–13. ICC of the BD type might originate from hepatic progenitor cells at the canal of Herring or from biliary epithelial cells lining bile ductules, whereas ICC of the SD type might arise from biliary epithelial cells lining bile ductules or interlobular bile ducts, and ICC of the LD type might originate from epithelial cells lining the intrahepatic large bile ducts or peribiliary glands. According to the recently updated 2019 World Health Organization (WHO) classification, BD type was classified as a subtype of SD type14. Mutant IDHs produce 2-hydroxyglutarate and reduced nicotinamide adenine dinucleotide phosphate, both of which exert an effect on blocking the differentiation of hepatic progenitor cells toward hepatocytes but have no effect on bile duct differentiation15. The application of 2-hydroxyglutarate or induction of mutant IDHs in hepatic progenitor cells blocks differentiation toward hepatocytes16. The action of mutant IDHs in an analogous fashion has been suggested in leukemogenesis in which mutant IDHs disrupt hematopoietic stem cell differentiation17.
Although ICC has a relatively high frequency of IDH1/2 mutations, the histomorphological features of ICC with IDH1/2 mutations have not been well characterized. Kipp and colleagues first identified IDH1/2 mutations in cholangiocarcinomas and a high frequency of IDH1/2 mutations in ICCs compared with a low frequency of IDH1/2 mutations in extrahepatic cholangiocarcinomas (28% vs. 7%, P = 0.030); they characterized the histological features of IDH1/2-mutated tumors as poor differentiation, clear cytoplasm, organoid arrangement of tumor cells, and relatively little desmoplasia5. However, it remains to be clarified whether ICCs with IDH1/2 mutations show specific histological subtypes. Furthermore, controversy has been raised regarding the prognostic implications of IDH1/2 mutations in patients.
In a previous study, we analyzed the methylation statuses of ICCs in 30 genes and found that frequently methylated genes were different between ICCs of the BD or SD type and ICCs of the LD type18. Of the 30 genes, six genes, including DLEC1, RASSF1A, RIP3, SOCS3, PTGS2, and TNFRSF10C, were more frequently methylated in ICCs of the BD type or SD type than in ICCs of the LD type. Of the six genes, five genes except for TNFRSF10C were more frequently methylated in hepatocellular carcinoma than in extrahepatic cholangiocarcinoma. Because it is known that DNA methylation profiles signifying the cell of origin are maintained during carcinogenesis, DNA methylation in the five genes might be postulated to signify a hepatic progenitor cell of origin in ICCs of the BD or SD type. In the present study, we analyzed 172 cases of ICC for their mutation statuses of IDH1/2 and methylation statuses of 30 gene CpG island loci using the pyrosequencing assay and MethyLight assay, respectively. We sought to elucidate the clinicopathological features of ICCs with IDH1/2 mutations, including the histological subtype and prognosis. By analyzing the association between IDH1/2 mutations and DNA methylation at individual genes, we found that 8 DNA methylation markers, including RIP3, DLEC1, PTGS2, MINT2, TNFRSF10C, RASSF1A, SOCS3, and ITF2, were significantly associated with IDH1/2 mutations. DNA methylation markers associated with IDH1/2 mutations may be a signature of the IDH pathway. Because IDH1 and 2 are not the only genes involved in the IDH pathway, the analysis of IDH1/2 mutations will miss the identification of ICC cases that are not mutated at IDH1/2 but are disturbed by alterations in other genes involved in the IDH pathway and disposed to hypermethylation at a specific set of the genes. We speculated that a panel of these eight DNA methylation markers were able to identify a subset of ICCs which are enriched in IDH1/2 mutations. We found that the panel might help to identify a subset of ICC with characteristic clinicopathological features, including frequent IDH1/2 mutations and better clinical outcomes.
Material and methods
Specimens
We collected archival tissue material from a consecutive series of ICC patients (n = 172) operated on at Seoul National University Hospital between January 2005 and December 2012. Pathological staging was determined according to the 7th version of the American Joint Committee on Cancer/Union for International Cancer Control (AJCC/UICC) tumor, node, metastasis (TNM) staging system. The determination of gross types, histological subtypes, and tumor grades were conducted as described previously18. Although the recently updated 2019 WHO classification classified ICCs into two main types, LD and SD, in a previous study, ICCs showed different clinical outcomes of a specific methylation marker between the BD type and SD type18. Thus, the present study retained three histologic types, BD, SD, and LD, rather than two main types, SD and LD. Through microscopic examination, we evaluated tissue slides for perineural invasion, lymphatic embolus, venous invasion, and the accompaniment of biliary disease, biliary intraepithelial neoplasia (BilIN), or chronic liver disease. This study was approved by the Institutional Review of Board of Seoul National University Hospital (IRB No. 1804-168-942). This study was conducted in compliance with the principles of the Declaration of Helsinki and its later amendments.
DNA extraction
Tumor areas with the highest tumor purity and the most representative histology in the individual cases were marked through microscopic examination of the available tissue slides. The corresponding areas on unstained tissue slides were manually scraped with single edge-razor blades and collected into the microtubes. The collected tissues were incubated in lysis buffer containing 50 mM Tris, 1 mM EDTA (pH 8.0), 1% Tween-20, and proteinase K (3 mg/ml) at 55 °C for 24 h. The tissue lysates were subjected to heating at 95 °C for 30 min to inactivate proteinase K and denature genomic DNA. After centrifugation, the supernatants were transferred into newly labeled, clean microtubes. We did not purify DNA from the supernatants of tissue lysates but used tissue lysates directly for polymerase chain reaction (PCR) or bisulfite modification.
Pyrosequencing assay
DNA from the tissue lysates was amplified by PCR with oligonucleotide primers specific to IDH1 codon 132 and IDH2 codon 172 (Supplementary Table 1). PCR was conducted in a total volume of 25 µL with the PyroMark PCR kit (Qiagen, Hilden, Germany) and final concentrations of 1X PyroMark PCR Mastermix, 1 × CollaLoad Concentrate, and 0.2 μM of each primer. Thermal cycling consisted of 45 cycles with denaturing (95 °C, 30 s), annealing (53 °C, 30 s), and elongation (72 °C, 30 s) steps, preceded by an initial denaturation step (95 °C, 10 min) and succeeded by a final extension step (72 °C, 10 min). Pyrosequencing for the detection of IDH1 (codon 132) and IDH2 (codon 172) mutations was performed using PyroGold reagents on the PyroMark Q24 pyrosequencer (Qiagen) according to the manufacturer’s instructions. Pyrogram outputs (Supplementary Fig. 1) were analyzed by PyroMark Q24 software (Qiagen).
Bisulfite modification and the MethyLight assay
For the bisulfite modification of DNA samples, the EZ DNA Methylation Kit was used (Zymo Research, Orange, CA, USA). The modified DNA samples were subjected to Alu-based MethyLight control reaction to measure the amount of the modified DNA and were diluted with distilled water if the cross-threshold value went below 18. MethyLight assays were performed for the 30 CpG island loci as described previously19.
Statistical analysis
IBM SPSS statistics (version 25) was used for the statistical analysis. Two-sided chi-square test was used for 2 × 2 contingency tables with minimal sample size > 5, whereas Fisher’s exact test (bidirectional) was used for 2 × 2 contingency tables with minimal sample size ≤ 5. Wilcoxon’s rank sum test and Kruskal–Wallis test were performed for ordinal variables and non-ordinal variables (contingency tables greater than 2 × 2), respectively. To identify whether the number of methylated genes was normally distributed in ICC tissue samples, normalization test was performed using the Shapiro–Wilk test, which revealed that the number of methylated genes were not normally distributed. Mean values of the number of methylated genes across two groups or across three or more groups were compared using the Mann–Whitney test and Kruskal–Wallis test, respectively. The cancer-specific survival (CSS) time was calculated from the date of surgery to the date of the death of the patient due to ICC. The recurrence-free survival (RFS) time was calculated from the date of surgery to the date of recurrence or death, whichever came first. CSS and RFS were compared between groups using the Kaplan–Meier method and log rank test. A Cox proportional hazards model with backward regression was used to investigate the association between the survival time of patients and one or more predictor variables.
Ethical approval
This study was approved by the institutional review board of Seoul National University Hospital (IRB No. 1804-168-942). Under the condition of retrospective archival tissue collection and patient data anonymization, our study was exempted from the acquisition of informed consent from patients by the institutional review board of Seoul National University Hospital.
Results
The overall study population included 172 patients who underwent surgical resection for ICC (Table 1). The median age at admission for operation was 63 years (range 38–80 years), and 70.3% of the patients were male. Of the 172 ICCs, the tumor growth patterns were the mass-forming type in 82.0%, periductal infiltrative type in 4.7%, intraductal growth type in 10.5%, and mixed type in 2.9%. The histological subtypes were BD type in 12.8%, SD type in 39.5%, and LD type in 47.7%. Chronic liver diseases and chronic biliary disease were found in the background liver of 24.4% and 7.6% of ICCs, respectively.
Table 1.
Clinicopathological features of intrahepatic cholangiocarcinoma with IDH1/2 mutations.
| n | IDH1/2 | P-value | ||
|---|---|---|---|---|
| No mutation | mutation | |||
| Sex | ||||
| M | 121 | 110 (90.9%) | 11 (9.1%) | 0.999** |
| F | 51 | 46 (90.2%) | 5 (9.8%) | |
| Age | ||||
| <64 years | 87 | 78 (89.7%) | 9 (10.3%) | 0.794 |
| 64 years | 85 | 78 (91.8%) | 7 (8.2%) | |
| Gross type | ||||
| Mass forming | 141 | 126 (89.4%) | 15 (10.6%) | 0.589^ |
| Periductal infiltrative | 8 | 8 (100%) | 0 | |
| Intraductal growing | 18 | 17 (94.4%) | 1 (5.6%) | |
| Mixed | 5 | 5 (100%) | 0 | |
| Differentiation | ||||
| Well, moderate | 117 | 110 (94.0%) | 7 (6.0%) | 0.031# |
| Poor | 55 | 46 (83.6%) | 9 (16.4%) | |
| Histologic subtype | ||||
| Bile ductular type | 22 | 18 (81.8%) | 4 (18.2%) | 0.012^ |
| Small duct type | 68 | 58 (85.3%) | 10 (14.7%) | |
| Large duct type | 82 | 80 (97.6%) | 2 (2.4%) | |
| Intraglandular and/or extraglandular mucin production | ||||
| Absent | 78 | 68 (87.2%) | 10 (12.8%) | 0.190 |
| Present | 94 | 88 (93.6%) | 6 (6.4%) | |
| Lymphatic emboli | ||||
| Absent | 102 | 91 (89.2%) | 11 (10.8%) | 0.594 |
| Present | 70 | 65 (92.9%) | 5 (7.1%) | |
| Venous invasion | ||||
| Absent | 95 | 87 (91.6%) | 8 (8.4%) | 0.793 |
| Present | 77 | 69 (89.6%) | 8 (10.4%) | |
| Perineural invasion | ||||
| Absent | 118 | 104 (88.1%) | 14 (11.9%) | 0.098** |
| Present | 54 | 52 (96.3%) | 2 (3.7%) | |
| Chronic liver disease | ||||
| Absent | 130 | 118 (90.8%) | 12 (9.2%) | 1.000** |
| Present | 42 | 38 (90.5%) | 4 (9.5%) | |
| BilIN* | ||||
| Absent | 102 | 88 (86.3%) | 14 (13.7%) | 0.010** |
| Present | 65 | 64 (98.5%) | 1 (1.5%) | |
| T category | ||||
| T1 | 48 | 44 (91.7%) | 4 (8.3%) | 0.887# |
| T2 | 52 | 47 (90.4%) | 5 (9.6%) | |
| T3 | 47 | 42 (89.4%) | 5 (10.6%) | |
| T4 | 25 | 23 (92.0%) | 2 (8.0%) | |
| N category | ||||
| pN0 | 133 | 119 (89.5%) | 14 (10.5%) | 0.312# |
| pN1 | 39 | 37 (94.9%) | 2 (5.1%) | |
| M category | ||||
| pM0 | 161 | 145 (90.1%) | 16 (8.9%) | 0.278# |
| pM1 | 11 | 11 (100%) | 0 | |
| TNM staging | ||||
| Stage I | 40 | 36 (90.0%) | 4 (10.0%) | 0.280# |
| Stage II | 37 | 33 (89.2%) | 4 (10.8%) | |
| Stage III | 30 | 25 (83.3%) | 5 (16.7%) | |
| Stage IVA | 54 | 51 (94.4%) | 3 (5.6%) | |
| Stage IVB | 11 | 11 (100%) | 0 | |
*Biliary intraepithelial neoplasia; 5 cases could not be assessed for biliary intraepithelial neoplasia.
**Fisher’s exact test,
^Kruskal–Wallis test,
#Wilcoxon’s rank-sum test.
IDH1/2 mutations and their association with clinicopathological features
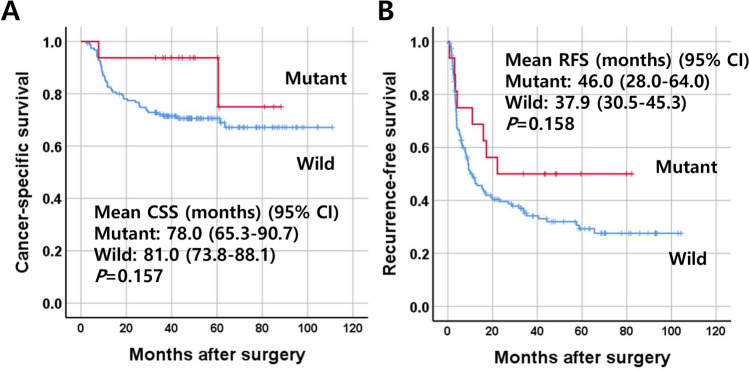
A total of 16 patients (9.3%) showed either IDH1 mutation (n = 12, 7.0%) or IDH2 mutation (n = 4, 2.3%). IDH1/2 mutations were associated with the histological subtypes: IDH1/2 mutations were more frequent in the BD type and SD type (18.2% and 14.7%, respectively) than in the LD type (2.4%) (P = 0.003) (Table 1). IDH1/2 mutations were associated with poor differentiation (P = 0.046) (Fig. 1). IDH1/2 mutations were less frequent in ICCs with BilIN than in those without BilIN (1.5% vs. 13.7%, P = 0.010). Although marginally significant, ICCs with IDH1/2 mutations tended to show less frequent perineural invasion (P = 0.098). In the survival analysis, ICCs with IDH1/2 mutations tended to show better clinical outcomes in both CSS and RFS, which were statistically insignificant (Fig. 2).
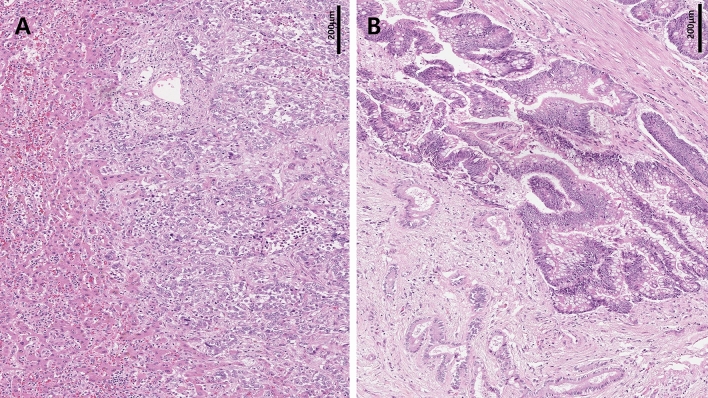
Figure 1.
Histological features of IDH1/2-mutated intrahepatic cholangiocarcinomas (ICCs). ICCs with IDH1/2 mutations were associated with a higher frequency of poor differentiation compared with ICCs without IDH1/2 mutations. A case of ICC with IDH1/2 mutations (A) and a case of ICC with no IDH1/2 mutations (B).
Figure 2.
Survival curves of patients with intrahepatic cholangiocarcinoma according to the mutation status of IDH1/2. Cancer-specific survival (A) and recurrence-free survival (B).
IDH1/2 mutation-associated methylation markers and their usefulness for the identification of a subset of ICC with characteristic clinicopathological features
In a previous study, we analyzed the methylation statuses of ICC and normal duct tissues in the CpG island loci of 105 genes using the MethyLight assay and found 30 individual genes to be methylated in ICC tissues by more than 15% over normal duct tissues; these were regarded as genes showing cancer-related methylation. In the present study, we analyzed the relationship between IDH1/2 mutations and the methylation of the 30 genes and found that 8 genes showed increased methylation frequencies in association with IDH1/2 mutations, including RIP3, DLEC1, PTGS2, MINT2, TNFRSF10C, RASSF1A, SOCS3, and ITF2 (in an order of decreasing P-values) (Supplementary Fig. 2 and Supplementary Table 2). When the relationships between the number of methylated genes and the clinicopathological parameters of ICC were examined among these 8 genes, an increased number of methylated genes was associated with male sex, lower T stage, BD or SD type, the absence of intraglandular and/or extraglandular mucin production, the absence of perineural invasion, the absence of BilIN, the presence of chronic liver disease, the absence of nodal metastasis, and lower TNM stage (Table 2). When we analyzed the relationship between the number of methylated genes and IDH1/2 mutation (Supplementary Table 3), ICCs with > and ≤ 5/8 methylated genes showed IDH1/2 mutation at a frequency of 52.2% and 2.7%, respectively, Compared with low-methylated ICCs (≤ 5/8 methylated genes), high-methylated ICCs (> 5/8 methylated genes) were associated with histological subtype of the BD or SD type, the absence of intraglandular and/or extraglandular mucin production, the absence of BilIN, and venous invasion (Supplementary Table 4).
Table 2.
Comparison of the number of methylated markers in the panel in relation to clinicopathological features.
| n | No. of methylated markers | Standard deviation | P-value* | |
|---|---|---|---|---|
| Sex | ||||
| M | 121 | 3.6 | 1.82 | 0.008 |
| F | 51 | 2.8 | 1.84 | |
| Age (years) | ||||
| < 64 | 87 | 3.3 | 1.86 | 0.775 |
| ≥ 64 | 85 | 3.4 | 1.86 | |
| Gross type | ||||
| Mass forming type | 141 | 3.5 | 1.91 | 0.161 |
| Periductal infiltrative type | 8 | 2.1 | 1.25 | |
| Intraductal growing type | 18 | 3.3 | 1.53 | |
| Mixed type | 5 | 2.60 | 1.52 | |
| Subtype | ||||
| Bile ductular type | 22 | 4.5 | 1.68 | <0.001 |
| Small duct type | 68 | 3.8 | 2.13 | |
| Large duct type | 82 | 2.8 | 1.41 | |
| Grade | ||||
| Well to moderately differentiated | 117 | 3.2 | 1.67 | 0.074 |
| Poorly differentiated | 55 | 3.8 | 2.14 | |
| Intraglandular and/or extraglandular mucin | ||||
| Absent | 78 | 3.8 | 2.05 | 0.012 |
| Present | 94 | 3.0 | 1. 60 | |
| Lymphatic invasion | ||||
| Absent | 102 | 3.5 | 1.88 | 0.178 |
| Present | 70 | 3.2 | 1.90 | |
| Vascular invasion | ||||
| Absent | 95 | 3.5 | 1.68 | 0.232 |
| Present | 77 | 3.3 | 2.06 | |
| Perineural invasion | ||||
| Absent | 118 | 3.7 | 1.87 | 0.001 |
| Present | 54 | 2.7 | 1.63 | |
| T stage | ||||
| T1 | 48 | 3.8 | 1.60 | 0.013 |
| T2 | 52 | 3.7 | 2.08 | |
| T3 | 47 | 2.7 | 1.66 | |
| T4 | 25 | 3.3 | 1.90 | |
| N stage | ||||
| pN0 | 133 | 3.6 | 1.96 | 0.010 |
| pN1 | 39 | 2.7 | 1.79 | |
| M stage | ||||
| pM0 | 161 | 3.5 | 1.87 | 0.074 |
| pM1 | 11 | 2.4 | 1.29 | |
| TNM stage | ||||
| I | 40 | 3.8 | 0.260 | 0.005 |
| II | 37 | 4.1 | 0.344 | |
| III | 30 | 3.0 | 0.294 | |
| IVA | 54 | 2.9 | 0.253 | |
| IVB | 11 | 2.4 | 0.388 | |
| BilIN | ||||
| Absent | 102 | 3.7 | 2.11 | 0.031 |
| Present | 60 | 2.9 | 1.24 | |
| Chronic liver disease | ||||
| Absent | 130 | 3.2 | 1.79 | 0.068 |
| Present | 42 | 3.9 | 1.98 | |
| Chronic biliary disease | ||||
| Absent | 159 | 3.4 | 1.86 | 0.391 |
| Present | 13 | 2.9 | 1.82 | |
Survival analysis according to the methylation statuses of eight methylation markers
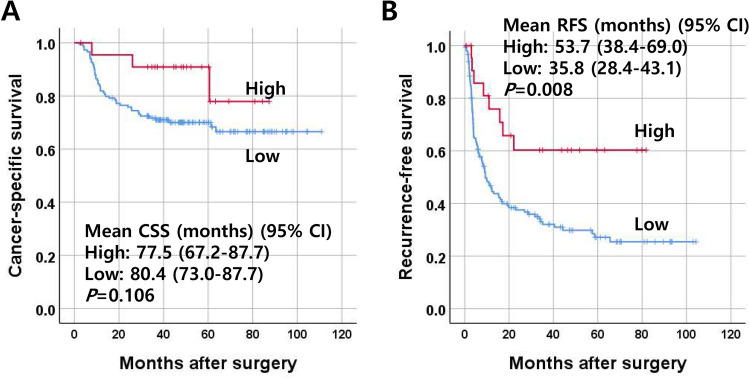
Kaplan–Meier survival curve analysis revealed that the recurrence-free survival rate of ICCs with high-methylation status (> 5/8 methylated markers in the marker panel) was significantly higher than that of ICCs with low- methylation status. No significant difference was noted in the cancer-specific survival rate of ICCs according to the methylation status in the marker panel (Fig. 3A, B). When we performed multivariate analysis, high methylation status (> 5/8 methylated genes) was found to be an independent prognostic parameter for better survival in terms of CSS and RFS (Table 3). When we performed subgroup analysis, the prognostic implications of the high-methylation status (> 5/8 methylated markers) in the marker panel were different depending on IDH1/2 mutation status, a significant association with survival was found in ICCs with no IDH1/2 mutation. For RFS, ICCs with high methylation showed better survival than ICCs with low methylation (Supplementary Fig. 3A–D).
Figure 3.
Survival curves of patients with intrahepatic cholangiocarcinoma (n = 172) according to the methylation status of the eight DNA methylation markers. ICCs were classified into two subgroups according to the number of methylated markers, including low-(≤ 5/8 methylated markers), and high-(> 5/8 methylated markers) methylation. Cancer-specific survival (A) and recurrence-free survival (B).
Table 3.
Multivariate survival analysis.
| HR (95% CI) | P-value | ||
|---|---|---|---|
| Cancer-specific survivala | |||
| Methylation status of the eight methylation markers | |||
| Low methylation | 149 | Ref | |
| High methylation | 23 | 0.30 (0.09–0.97) | 0.040 |
| Recurrence-free survivala | |||
| Methylation status of the eight methylation markers | |||
| Low methylation | 149 | Ref | |
| High methylation | 23 | 0.35 (0.17–0.72) | 0.005 |
aAdjusted for gross type, histological type, differentiation grade, lymphatic emboli, venous invasion, perineural invasion, T stage, N stage, M stage, and IDH1/2 mutation status.
Discussion
In the present study, we analyzed the mutation status of ICCs in codon 132 of IDH1 and codon 172 of IDH2 using a pyrosequencing assay and found that IDH1 or 2 was mutated in 9.3% of ICCs. When we explored cBioPortal for IDH1/2 mutations in biliary tract cancers (n = 182), including gallbladder cancer, IDH1 and IDH2 mutations were found to be restricted to codon 132 and codon 172, respectively20,21. The frequency of IDH1/2 mutations was lower in our study than in Western studies of ICCs. In Jiao et al.’s study in which a series of ICC specimens (n = 32) were sequenced for their exomes, 6 ICCs (19%) showed IDH1/2 mutations22. In Kwong et al.’s study in which 32 ICC specimens were sequenced, IDH1/2 mutations were found in 7 ICCs (22%)23. In a study using the SNaPshot genotyping assay, 40 of 200 ICCs (20%) showed IDH1/2 mutations24. However, in a study that performed a comparative analysis to ICCs from Asian patients and compared the mutation frequency of IDH1/2 between liver fluke-related and nonrelated ICCs, a significant difference was found in the mutation frequency of IDH1/2 between liver fluke-related and nonrelated ICCs; the mutation frequency of IDH1/2 was significantly higher in Opisthorchis viverrini-nonrelated ICCs (n = 27) than in O. viverrini-related ICCs (n = 62) (22.2% vs. 3.2%, P = 0.009)25. The mutation frequency of IDH1/2 in Asian individuals with O. viverrini-nonrelated ICC was in agreement with the mutation frequency of IDH1/2 in Western individuals with O. viverrini-nonrelated ICC. Although we could not obtain information regarding the infection status of Clonorchis sinensis in patients with ICC, Korea has geographical variation in terms of C. sinensis prevalence ranging from 2.1 to 31.3%26, and 9.5% of cholangiocarcinomas are estimated to be caused by chronic infection with C. sinensis27. In the endemic areas of Korea, 22.6% of cholangiocarcinoma cases were attributable to C. sinensis infection27. Thus, the lower rate of IDH1/2 mutations in our ICC cases might be partly attributable to C. sinensis infection. Another attributing factor is related to the high proportion of the LD subtype in ICCs, and the LD subtype was less likely to contain IDH1/2 mutations compared with the other subtypes of ICCs.
When we explored the relationship between IDH1/2 mutations and clinicopathological features, we found that the histological subtype was significantly associated with IDH1/2 mutations. IDH1/2 mutations were mainly found in ICCs of the BD or SD subtype but rarely found in ICCs of the LD type. ICCs with IDH1/2 mutations showed more frequent poor differentiation compared with ICCs with no IDH1/2 mutations, which is in line with the findings of Kipp et al.’s study5. Kipp et al. analyzed the mutation statuses in IDH1/2 of 94 cholangiocarcinomas and correlated them with clinicopathological features, revealing that ICCs with IDH1/2 mutations were poorly differentiated. In Kipp et al.’s study, IDH1/2-mutated tumors were characterized by an organoid arrangement of tumor cells featured by an amphophilic cytoplasm and discrete cell borders or a compact arrangement of small tumor glands in stroma with relatively little desmoplasia. Although it is unclear whether such histological descriptions are compatible with the BD or SD histological subtypes, it is clear that such descriptions counter those of the LD subtype, which is characterized by mucin production, relatively large gland lumina, and desmoplastic stroma. However, in Borger et al.’s study, which found IDH1/2 mutations in 9 of 40 ICCs, IDH1/2-mutated ICCs were all well to moderately differentiated, with tumor glands embedded within moderate to abundant stromal tissue6.
In the present study, we explored the relationship between IDH1/2 mutations and the methylation of 30 genes and found that 8 genes showed associations between the methylation of individual genes and IDH1/2 mutations. However, when we compared the number of methylated genes between ICCs with and without IDH1/2 mutations, no significant difference was identified in the 30 examined genes (18.6 vs. 16.1, P = 0.136; Mann–Whitney U-test). These results indicate that IDH1/2 mutations did not lead to the diffuse enhancement of promoter CpG island loci but instead to the selective enhancement of specific CpG island loci, which was evidenced in Kwong et al.’s study, which analyzed the genome-wide methylation statuses of 38 cases of cholangiocarcinoma using the Illumina 450 K Infinium bead assay23. Kwong et al. performed unsupervised clustering of cholangiocarcinoma cases using CpG sites that showed cancer-specific methylation changes, which generated four clusters. They found two distinct hypermethylation clusters in which IDH1/2-mutated tumors belonged to one cluster only. The cluster containing IDH1/2-mutated tumors showed a lower number of methylated CpG sites compared with the other hypermethylated cluster. Furthermore, the CpG sites that were frequently methylated in the cluster containing IDH1/2-mutated tumors were less frequently methylated in the other hypermethylated cluster.
In the present study, a panel of IDH1/2 mutation-associated DNA methylation markers could identify a subset of highly methylated ICCs that is characterized by the BD or SD histological subtype, the absence of mucin production, the absence of BilIN, and better survival. Such clinicopathological features remind us of a molecular subclass enriched with IDH1/2 mutations that was defined by integrated multiplatform clustering analysis (genome, epigenome, and transcriptome)23,28. Such a molecular subclass has also been suggested in the review paper of Sia et al., who proposed two distinct molecular classes of ICCs, including the proliferation and inflammation classes. The proliferation class contains a subtype with stem cell–like features, hypermethylation, and IDH1/2 mutations29. However, integrated multiplatform clustering analysis is less likely to be adopted in clinical practice because of the requirement of fresh tissue samples and the high expense for the examination, which warrants the development of a more practical method to identify a subclass of ICCs enriched with IDH1/2 mutations. Further study is needed to determine how much highly methylated ICC defined by the eight-marker panel of the present study coincides with the IDH1/2 mutation-enriched molecular subtype defined by multiplatform analysis.
To validate our finding, on a different cohort, that the methylation status of eight genes could identify a subset of ICC with better clinical outcomes, we conducted Kaplan–Meier survival analysis in the ICC cohort (n = 35) of pancancer methylation data listed in the UCSC Xena browser30. Cut-off was set at beta-value of 0.15 and 0.2 to define methylation (Supplementary Fig. 4). Regardless of the cut-off value, no significant difference was identified in the survival rate between ICCs with > 5/8 methylated genes and ICCs with ≤ 5/8 methylated genes. The reason for which survival analysis result was not reproduced in the TCGA cohort might be related to the following: (1) the small sample size of subgroups in ICC cohort of the TCGA, which undermines the power of survival analysis. (2) The collection of ICC cases from a single high-volume hospital or from multiple hospitals over the world, including Brazil, Italy, Canada and USA, might induce bias in survival analysis because of the heterogeneity of clinical methods and surgical strategies in a multi-center study23. Clinical outcomes are more favorable in high-volume centers than in less experienced centers31. And, (3) Analytical technology was different between TCGA and the present study to assess methylation levels at eight genes. The Infinium technology determines methylation percentage at single CpGs, whereas the MethyLight technology assess the relative prevalence of a particular pattern of DNA methylation at multiple CpGs. A large-scaled study from a high-volume expert center is needed to validate our finding that the methylation status at eight genes determined by the MethyLight technology could identify a subset of ICCs with better clinical outcomes.
In conclusion, IDH1/2 mutations were observed in 9.3% of ICCs, and most of them were observed in the mass-forming gross type, bile ductular or small duct histological types. Mutations in IDH1/2 were not significantly associated with the prognosis of the patients. Although the IDH1/2 mutations did not show prognostic value, ICCs with IDH1/2 mutations exhibited a propensity toward better clinical outcomes. However, 8 gene CpG island loci were found to be significantly enhanced in methylation in association with IDH1/2 mutations, and methylation at > 5/8 of these genes was found to be able to identify a subset of ICCs with distinct clinicopathological features and good prognosis. Future studies are needed to investigate whether the eight-methylation marker panel can identify IDH1/2 mutation-enriched molecular subtypes defined by multiplatform analysis.
Supplementary information
Author contributions
K.L. collected the data, contributed data, conceived the study, and performed the analysis. Y.S.S. performed the analysis. Y.S. performed the analysis. X.W. performed the analysis. Y.K. collected the data and contributed to the interpretation of the data. N.Y.C. performed the analysis, collected the data, and contributed to the interpretation of the data. J.M.B. conducted statistical analyses. G.H.K. conceived the study, wrote the draft, and interpreted the data. All the authors participated in the final approval of the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was financially supported by a grant from the National Research Foundation (NRF) funded by the Korean Ministry of Science and ICT (2016M3A9B6026921), an NRF grant funded by the Korean government (the Ministry of Science and ICT) (2019R1F1A1061227), and a grant (no 0320190010) from the SNUH Research Fund.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72810-0.
References
- 1.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Niu L, Xu X, Zhang S, Gao F. A comprehensive theoretical investigation of intramolecular proton transfer in the excited states for some newly-designed diphenylethylene derivatives bearing 2-(2-hydroxy-phenyl)-benzotriazole part. J. Fluoresc. 2011;21:1721–1728. doi: 10.1007/s10895-011-0867-6. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark O, Yen K, Mellinghoff IK. Molecular pathways: isocitrate dehydrogenase mutations in cancer. Clin. Cancer Res. 2016;22:1837–1842. doi: 10.1158/1078-0432.CCR-13-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, Zhang L, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum. Pathol. 2012;43:1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aishima S, Oda Y. Pathogenesis and classification of intrahepatic cholangiocarcinoma: different characters of perihilar large duct type versus peripheral small duct type. J. Hepatobiliary Pancreat. Sci. 2015;22:94–100. doi: 10.1002/jhbp.154. [DOI] [PubMed] [Google Scholar]
- 8.Bridgewater J, Galle PR, Khan SA, Llovet JM, Park JW, Patel T, et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Nakanuma Y, Kakuda Y. Pathologic classification of cholangiocarcinoma: new concepts. Best Pract. Res. Clin. Gastroenterol. 2015;29:277–293. doi: 10.1016/j.bpg.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Bormann F, Rodriguez-Paredes M, Lasitschka F, Edelmann D, Musch T, Benner A, et al. Cell-of-origin DNA methylation signatures are maintained during colorectal carcinogenesis. Cell Rep. 2018;23:3407–3418. doi: 10.1016/j.celrep.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 11.Cardinale V, Carpino G, Reid L, Gaudio E, Alvaro D. Multiple cells of origin in cholangiocarcinoma underlie biological, epidemiological and clinical heterogeneity. World J. Gastrointest. Oncol. 2012;4:94–102. doi: 10.4251/wjgo.v4.i5.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bragazzi MC, Ridola L, Safarikia S, Matteo SD, Costantini D, Nevi L, et al. New insights into cholangiocarcinoma: multiple stems and related cell lineages of origin. Ann. Gastroenterol. 2018;31:42–55. doi: 10.20524/aog.2017.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182–188. doi: 10.1111/his.13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha SK, Parachoniak CA, Bardeesy N. IDH mutations in liver cell plasticity and biliary cancer. Cell Cycle. 2014;13:3176–3182. doi: 10.4161/15384101.2014.965054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, Lee K, Jeong S, Wen X, Cho NY, Kang GH. DLEC1 methylation is associated with a better clinical outcome in patients with intrahepatic cholangiocarcinoma of the small duct subtype. Virchows Arch. 2019;475:49–58. doi: 10.1007/s00428-018-02511-7. [DOI] [PubMed] [Google Scholar]
- 19.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J. Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017;19:2878–2880. doi: 10.1016/j.celrep.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, et al. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Ann. Surg. Oncol. 2014;21:3827–3834. doi: 10.1245/s10434-014-3828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 26.Lim MK, Ju YH, Franceschi S, Oh JK, Kong HJ, Hwang SS, et al. Clonorchis sinensis infection and increasing risk of cholangiocarcinoma in the Republic of Korea. Am. J. Trop. Med. Hyg. 2006;75:93–96. doi: 10.4269/ajtmh.2006.75.93. [DOI] [PubMed] [Google Scholar]
- 27.Shin HR, Oh JK, Lim MK, Shin A, Kong HJ, Jung KW, et al. Descriptive epidemiology of cholangiocarcinoma and clonorchiasis in Korea. J. Korean Med. Sci. 2010;25:1011–1016. doi: 10.3346/jkms.2010.25.7.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 30.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagino M, Ebata T, Yokoyama Y, Igami T, Sugawara G, Takahashi Y, et al. Evolution of surgical treatment for perihilar cholangiocarcinoma: a single-center 34-year review of 574 consecutive resections. Ann. Surg. 2013;258:129–140. doi: 10.1097/SLA.0b013e3182708b57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.