Highlights
-
•
The network provided auto-segmentations with high overlap with ground truth volumes.
-
•
Evaluation of femoral heads/bladder auto-segmentations showed highest overlap.
-
•
Annotated structure sets from daily clinical practice is feasible as ground truth.
Abbreviations: AI, artificial intelligence; CNN, convolutional neural network; CT, computer tomography; CTVN, clinical target volume nodes; MSD, mean surface distance; OAR, organs at risk; PET-CT, positron emission tomography/computer tomography
Keywords: Cervical cancer radiotherapy, Organs-at-risk, Clinical Target Volume, Automatic segmentation, Convolutional neural network
Abstract
Background
It is time-consuming for oncologists to delineate volumes for radiotherapy treatment in computer tomography (CT) images. Automatic delineation based on image processing exists, but with varied accuracy and moderate time savings. Using convolutional neural network (CNN), delineations of volumes are faster and more accurate. We have used CTs with the annotated structure sets to train and evaluate a CNN.
Material and methods
The CNN is a standard segmentation network modified to minimize memory usage. We used CTs and structure sets from 75 cervical cancers and 191 anorectal cancers receiving radiation therapy at Skåne University Hospital 2014-2018. Five structures were investigated: left/right femoral heads, bladder, bowel bag, and clinical target volume of lymph nodes (CTVNs). Dice score and mean surface distance (MSD) (mm) evaluated accuracy, and one oncologist qualitatively evaluated auto-segmentations.
Results
Median Dice/MSD scores for anorectal cancer: 0.91–0.92/1.93–1.86 femoral heads, 0.94/2.07 bladder, and 0.83/6.80 bowel bag. Median Dice scores for cervical cancer were 0.93–0.94/1.42–1.49 femoral heads, 0.84/3.51 bladder, 0.88/5.80 bowel bag, and 0.82/3.89 CTVNs. With qualitative evaluation, performance on femoral heads and bladder auto-segmentations was mostly excellent, but CTVN auto-segmentations were not acceptable to a larger extent.
Discussion
It is possible to train a CNN with high overlap using structure sets as ground truth. Manually delineated pelvic volumes from structure sets do not always strictly follow volume boundaries and are sometimes inaccurately defined, which leads to similar inaccuracies in the CNN output. More data that is consistently annotated is needed to achieve higher CNN accuracy and to enable future clinical implementation.
1. Introduction
Positron emission tomography/computer tomography (PET-CT), is performed at our center for diagnostic and radiation therapy planning in all cases of cervical and anorectal cancer scheduled for radiotherapy. Using CT images, a radiation oncologist routinely performs the segmentation of the tumor with margin (clinical target volume (CTV)), clinical target volume of lymph nodes (CTVN)) and organs at risk (OAR) (for radiation toxicity).
Although guidelines exist, manual segmentations are subjective and time consuming, and an automated approach would make it possible to increase reproducibility, improve clinical workflow, and improve cancer care locally [1], [2] and globally [3]. According to reviews on radiation oncology and automated segmentation based on artificial intelligence (AI), there are indeed clinical benefits, but there are also challenges including inaccurate or incomplete auto segmentation due to software performance or unrecognized anatomical variations [1], [2], [4], [5].
In the last few years, interest in AI has increased dramatically in medical imaging and radiation oncology. More specifically, the interest is directed towards a specific family of models called convolutional neural networks (CNNs). The parameters of a convolutional neural network are updated through a general learning procedure using input data [6].
CNN-based methods have proven highly accurate for automated segmentation in magnetic resonance imaging (MRI) of the prostate, CT of the liver and bladder, and PET/CT images of skeletal structures [7], [8], [9], [10], [11]. Furthermore, in the field of radiation oncology, studies on head and neck cancer have adhered to the fact that correctly implemented AI techniques generates better efficiency and standardization of treatment for patients with head and neck cancer [12], [13], [14]. In the case of rectal cancer, a previous study evaluated the auto-segmentation of target volume and OAR, and showed varying segmentation accuracy based on the Dice similarity coefficient ranging from 61.8% (colon) to 93.4% (bladder) [15]. Recently on cervical cancer, Liu et al [16] described a method for OAR (but not CTVN) segmentation using CNNs, with structure sets as ground truth. Their method used a modified version of 2D U-Net, hence not fully utilizing the 3D nature of the data.
Two general but important concerns about the application of AI to segmentation in radiation oncology are the limited size of datasets available for research and the use of human evaluation of a network as a gold standard [2], [4]. To address these issues, this study takes advantage of existing segmented structure sets for comparison to the CNN segmentation [15], [16], thus eliminating the need for time-consuming renewed manual segmentation for research purposes [8] and allowing for larger sample populations. Furthermore, using an existing treatment plan gives an image of the general performance of manual segmentation in daily clinical practice (since it is performed by several radiation oncologists). This may be preferable to using the evaluations by a single radiation oncologist as a gold standard.
The aim of this project was to develop a CNN-based method for automated segmentation of OARs (left/right femoral heads, bladder, bowel bag), and CTVNs (cervical cancer only) for patients with anal, rectal, and cervical cancer, to test its overlap against manually segmented structure sets, and to evaluate the performance qualitatively.
2. Material and methods
2.1. Study design
The eligible cases comprised 75 cases of cervical cancer and 191 cases of anorectal cancer with available PET-CT as part of radiation therapy planning at the Department of Oncology, Skane University Hospital, Lund, Sweden, from 2014 to 2018. In this study, only CT-images was used for analyses. The study population was divided into one group for training and validation (65 cases and 161 cases for cervical cancer and anorectal cancer, respectively) and one test group (10 cases and 30 cases for cervical cancer and anorectal cancer, respectively). The test group was used for a final evaluation of the trained network and was not in any way used during training.
CNNs were trained to segment five volumes automatically: the left and the right femoral heads, the urinary bladder, the bowel bag [17], and CTVNs (cervical cancer only in the latter case). Three PET-CT scanners were used during the time frame: Philips Gemini TF, GE Discovery 690, and GE Discovery MI. CT images (supine treatment position) were obtained with intra-venous contrast for cervical cancer cases and without intra-venous contrast for anorectal cases according to clinical standards. The number of abdominal CT images per examination was approximately 150 images (+/- 10 images). The pixel size was 0.977 mm for all patients. Slice thickness was 2.5 mm or 3 mm. The study has received ethical approval (Dnr 2016/417 and 2018/753).
2.2. Automated segmentation
The network is a fully-convolutional 3D segmentation network described in [18]. The output from the network is one channel per organ class with softmax activation plus one channel for the background class.
As preprocessing, the HU values were truncated to [-800, 800] and divided by 800, resulting in input values between –1 and 1. Postprocessing is performed to remove noisy pixels; only the largest connected component (26-connected neighborhood) of each label is kept, and morphological hole filling (26-connected neighborhood) is applied to the resulting segmentation.
2.3. Training the network
A single CNN was trained on 226 manually segmented CT scans using a negative log-likelihood loss. Labels missing in the annotations are merged with the background label, resulting in the same loss if a pixel is classified as background or the missing label. The model was trained using CT patches of size 136 × 136 × 72 pixels and a batch size of 50. The patches were augmented using moderate rotations (−0.15 to 0.15 rad), scaling (−10% to +10%) and intensity shifts (−100 to +100 HU) to enrich the training data.
As usual in machine learning, the images were divided into a training set for direct parameter estimation (80% of the images) and a validation set (the remaining 20%). The optimization was performed using the Adam method [19] with Nesterov momentum. The learning rate was initialized at 0.0001 and when the validation loss reached a plateau. After 50 epochs (with 500 batches per epoch), the model was evaluated on the training group. Patches whose center points were classified as false positives were sampled more frequently (10% of the samples) when the training was restarted. This cycle was performed fifty times, varying the validation set to allow for all 226 images to be used for direct parameter estimation.
2.4. Manually segmented structure sets
As part of radiation therapy planning, manually segmented structure sets linked to CT images are created by a radiation oncologist and saved in Aria (ARIA Oncology Information System (Varian Medical Systems, Inc.)) which is the program where structure sets are performed and stored. All structure sets from clinical practice have delineations performed by senior radiation oncologists. The target volumes and OAR were outlined in accordance with national guidelines for the particular diagnoses, based on international consensus recommendations [20], [21], [22]. Due to the heterogeneously segmented structure sets in anorectal cancer (CTVNs are delineated differently depending on the level of the primary tumor), CTVNs were not eligible for auto-segmentation. Original contours used in the treatment of the patients were retrieved and used as the gold standard.
2.5. Qualitative evaluation
For the qualitative evaluation, a radiation oncologist rated the quality of the automated segmentations, using a slightly modified version of a previously described qualitative evaluation method [23]. The auto-segmentations were evaluated structure by structure and patient by patient. The rating was performed using four categories: excellent (almost no modification necessary), good (limited number of slices to be corrected), acceptable (automatic segmentations can be used but require modification of several slices), and not acceptable (useless). The radiation oncologist (MB) performing the qualitative evaluation have over twenty years of experience in radiation oncology.
2.6. Quantitative evaluation
The distribution of Mean Surface Distance (MSD) (mm) and Dice scores [24], [25] is illustrated descriptively per patient (separately for cervical cancer cases and anorectal cancer cases) and summarized with mean and median values. A Dice value of 1 indicates perfect overlap between manual segmentations and auto-segmentations. On MSD, for each surface voxel in the AI-segmented volume the nearest surface voxel in the manually segmented volume was found and an average Euclidean distance between all voxel-pairs was calculated, with the non-isotropic voxels taken into consideration. The same procedure was performed starting with the manually segmented volume and the MSD was calculated as the average of the two average distances. The qualitative evaluation is illustrated descriptively (n (%)) per group (excellent/good/acceptable/not acceptable) and shown separately for cervical and anorectal cancer cases.
3. Results
3.1. Comparison between CNN results and the structure sets
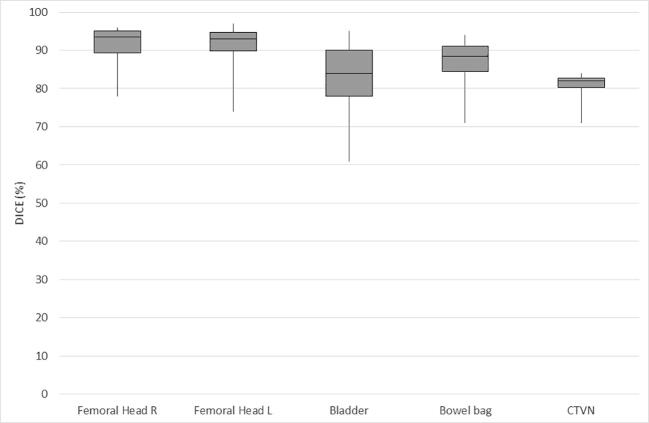
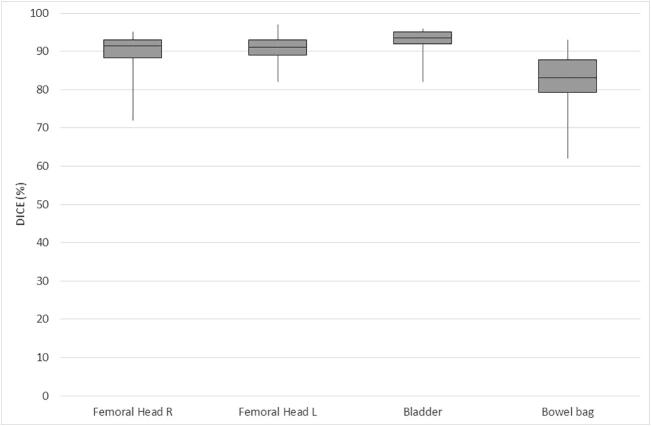
The distribution of Dice scores per case is illustrated in Table 1a, Table 1b and Fig. 1a, Fig. 1b with cervical and anorectal cancer cases presented separately. In the test set, there were 30 cases with anorectal cancer and 10 cases with cervical cancer. For anorectal cancer, the overall range of Dice scores was smaller. The urinary bladder had the highest median Dice scores (0.94), followed by the femoral heads (0.91 and 0.92 for the left and right femoral heads, respectively) and bowel bag (0.83). For cervical cancer cases, the femoral heads had the highest median Dice scores (0.93 and 0.94 for the left and right femoral heads, respectively). The median Dice score for the other evaluated organs at risk were 0.84 for the urinary bladder and 0.88 for the bowel bag. For the target volume (CTVN), the median Dice score was 0.82.
Table 1a.
Overlap (Dice values) for OAR and CTVN in cervical cancer.
| Cervical | Femoral Head | Femoral Head | Bladder | Bowel bag | CTVN |
|---|---|---|---|---|---|
| R | L | ||||
| 0.96 | 0.89 | 0.83 | 0.91 | 0.82 | |
| 0.78 | 0.74 | 0.87 | 0.92 | 0.82 | |
| 0.93 | 0.85 | 0.85 | 0.91 | 0.80 | |
| 0.90 | 0.95 | 0.94 | 0.88 | 0.78 | |
| 0.95 | 0.93 | 0.77 | 0.86 | 0.84 | |
| 0.89 | 0.92 | 0.95 | 0.89 | 0.83 | |
| 0.95 | 0.97 | 0.78 | 0.71 | 0.81 | |
| 0.96 | 0.97 | 0.78 | 0.84 | 0.71 | |
| 0.89 | 0.93 | 0.91 | 0.74 | 0.82 | |
| 0.94 | 0.94 | 0.61 | 0.94 | 0.84 | |
| Mean | 0.92 | 0.91 | 0.83 | 0.86 | 0.81 |
| Median | 0.94 | 0.93 | 0.84 | 0.88 | 0.82 |
Table 1b.
Overlap (Dice values) for OAR in anorectal cancer.
| Anorectal | Femoral Head | Femoral Head | Bladder | Bowel bag |
|---|---|---|---|---|
| R | L | |||
| 0.83 | 0.86 | 0.90 | 0.76 | |
| 0.91 | 0.91 | 0.95 | 0.89 | |
| 0.87 | 0.86 | 0.92 | 0.83 | |
| 0.95 | 0.95 | 0.90 | 0.79 | |
| 0.93 | 0.82 | 0.74 | ||
| 0.89 | 0.90 | 0.93 | 0.93 | |
| 0.92 | 0.88 | 0.95 | 0.75 | |
| 0.92 | 0.93 | 0.95 | 0.79 | |
| 0.94 | 0.92 | 0.94 | 0.89 | |
| 0.77 | 0.84 | 0.93 | 0.90 | |
| 0.94 | 0.91 | 0.94 | 0.88 | |
| 0.94 | 0.92 | 0.95 | 0.80 | |
| 0.93 | 0.92 | 0.93 | 0.84 | |
| 0.91 | 0.97 | 0.83 | 0.89 | |
| 0.91 | 0.95 | 0.95 | 0.88 | |
| 0.72 | 0.93 | 0.94 | 0.91 | |
| 0.88 | 0.91 | 0.95 | 0.82 | |
| 0.92 | 0.92 | 0.91 | 0.84 | |
| 0.94 | 0.91 | 0.89 | 0.83 | |
| 0.95 | 0.95 | 0.95 | 0.85 | |
| 0.91 | 0.90 | 0.93 | 0.84 | |
| 0.86 | 0.87 | 0.93 | 0.72 | |
| 0.90 | 0.88 | 0.94 | 0.71 | |
| 0.92 | 0.94 | 0.92 | 0.83 | |
| 0.93 | 0.94 | 0.96 | 0.87 | |
| 0.81 | 0.82 | 0.95 | 0.82 | |
| 0.92 | 0.90 | 0.93 | 0.86 | |
| 0.88 | 0.89 | 0.89 | 0.82 | |
| 0.92 | 0.95 | 0.96 | 0.81 | |
| 0.91 | 0.91 | 0.95 | 0.62 | |
| Mean | 0.90 | 0.91 | 0.93 | 0.82 |
| Median | 0.92 | 0.91 | 0.94 | 0.83 |
Fig. 1a.
Overlap (Dice values) for OAR and CTVN in cervical cancer (median/range).
Fig. 1b.
Overlap (Dice values) for OAR in anorectal cancer (median/range).
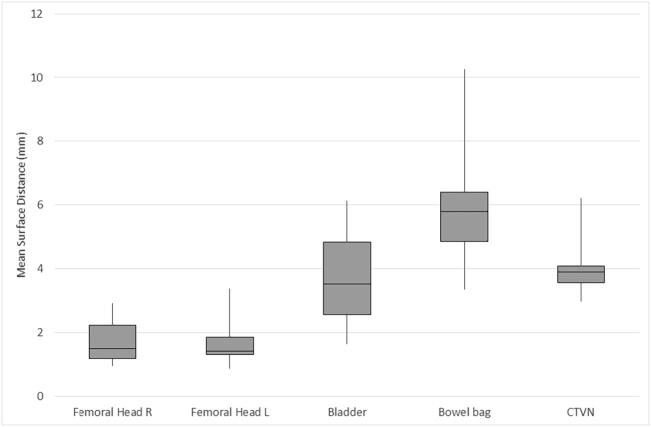
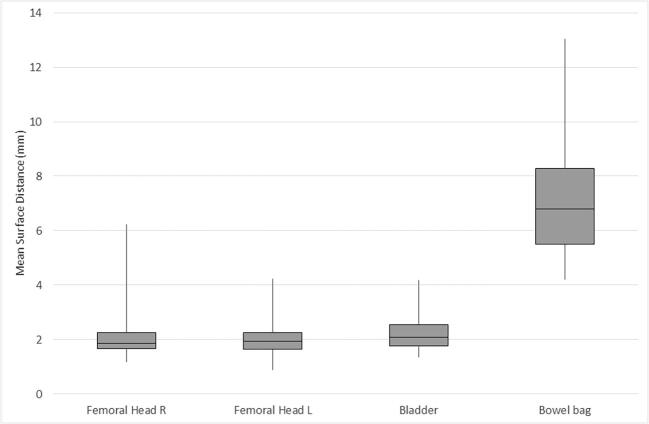
The distribution of MSD (mm) per case is illustrated in Table 2a, Table 2b and Fig. 2a, Fig. 2b with cervical and anorectal cancer cases presented separately. For anorectal cancer, the femoral heads had an MSD of 1.93 mm (left) and 1.86 mm (right), the urinary bladder 2.07 mm, and bowel bag 6.80 mm. For cervical cancer cases, the femoral heads had a median MSD of 1.42 mm (left) and 1.49 mm (right). The median MSD for the other evaluated organs at risk were 3.51 mm for the urinary bladder and 5.80 mm for the bowel bag. For the target volume (CTVN), the median MSD was 3.89 mm.
Table 2a.
Overlap (mean surface distance (mm)) for OAR and CTVN in cervical cancer.
| Cervical | Femoral Head | Femoral Head | Bladder | Bowel bag | CTVN |
|---|---|---|---|---|---|
| R | L | ||||
| 0.95 | 1.93 | 3.62 | 5.53 | 4.20 | |
| 2.92 | 3.39 | 2.53 | 4.77 | 3.57 | |
| 1.58 | 2.79 | 3.82 | 4.77 | 4.04 | |
| 2.31 | 1.29 | 2.35 | 6.42 | 3.95 | |
| 1.15 | 1.35 | 3.40 | 6.07 | 2.97 | |
| 2.36 | 1.41 | 1.64 | 5.09 | 3.42 | |
| 1.27 | 0.86 | 5.18 | 9.85 | 4.09 | |
| 1.10 | 0.92 | 6.12 | 10.26 | 6.22 | |
| 1.98 | 1.64 | 2.69 | 6.35 | 3.55 | |
| 1.40 | 1.42 | 5.82 | 3.34 | 3.83 | |
| Mean | 1.70 | 1.70 | 3.72 | 6.25 | 3.98 |
| Median | 1.49 | 1.42 | 3.51 | 5.80 | 3.89 |
Table 2b.
Overlap (mean surface distance (mm)) for OAR in anorectal cancer.
| Anorectal | Femoral Head | Femoral Head | Bladder | Bowel bag |
|---|---|---|---|---|
| R | L | |||
| 3.33 | 3.15 | 2.62 | 8.29 | |
| 2.09 | 2.04 | 2.07 | 5.42 | |
| 2.98 | 3.08 | 2.33 | 10.83 | |
| 1.17 | 1.33 | 3.16 | 10.41 | |
| 1.76 | 4.18 | 13.04 | ||
| 2.29 | 2.16 | 2.08 | 4.20 | |
| 1.98 | 2.88 | 1.43 | 9.23 | |
| 1.97 | 1.88 | 1.72 | 8.21 | |
| 1.57 | 1.92 | 2.58 | 6.36 | |
| 4.00 | 2.94 | 2.28 | 5.01 | |
| 1.35 | 1.93 | 1.76 | 5.05 | |
| 1.71 | 2.25 | 1.91 | 7.62 | |
| 1.65 | 1.99 | 2.38 | 6.27 | |
| 2.06 | 0.88 | 3.56 | 4.97 | |
| 1.94 | 1.47 | 1.70 | 5.02 | |
| 6.24 | 1.83 | 2.50 | 7.09 | |
| 1.80 | 1.40 | 1.73 | 6.51 | |
| 1.87 | 1.64 | 3.12 | 7.25 | |
| 1.25 | 1.82 | 2.34 | 5.92 | |
| 1.27 | 1.44 | 1.88 | 8.26 | |
| 1.85 | 2.50 | 2.03 | 8.00 | |
| 2.85 | 2.64 | 2.59 | 9.84 | |
| 2.14 | 2.12 | 1.99 | 9.07 | |
| 1.62 | 1.16 | 2.46 | 6.17 | |
| 1.74 | 1.64 | 1.57 | 5.41 | |
| 3.70 | 4.23 | 1.48 | 6.42 | |
| 1.77 | 2.09 | 1.84 | 5.67 | |
| 2.44 | 2.04 | 3.88 | 8.19 | |
| 1.77 | 0.97 | 1.35 | 5.45 | |
| 1.54 | 1.76 | 1.92 | 10.57 | |
| Mean | 2.19 | 2.04 | 2.28 | 7.32 |
| Median | 1.86 | 1.93 | 2.07 | 6.80 |
Fig. 2a.
Overlap (mean surface distance (mm)) for OAR and CTVN in cervical cancer (median/range).
Fig. 2b.
Overlap (mean surface distance (mm)) for OAR in anorectal cancer (median/range).
3.2. Qualitative evaluation of the auto-segmentations
The qualitative performance of the auto-segmented organs is illustrated in Table 3. For cervical cancer (n = 10) and anorectal cancer (n = 30), there was “excellent” performance for the bladder and femoral heads auto-segmentation in the majority of the cases according to the radiation oncologist. For CTVN (cervical cancer only, n = 10), the largest group (40%) was “not acceptable” and would have needed to be redone completely, but the second most common group was “good” (30%) with a limited number of slices to be corrected. For the bowel bag, the results were more mixed, with a more varying distribution between the groups of “good,” “acceptable,” and ”not acceptable.”
Table 3.
Qualitative evaluation of auto-segmentations, n (%).
| Excellent | Good | Acceptable | Not Acceptable | |
|---|---|---|---|---|
| Cervical cancer | ||||
| Bladder | 4(40) | 2(20) | 1(10) | 3(30) |
| Femoral head right | 9(90) | 1(10) | ||
| Femoral head left | 8(80) | 2(20) | ||
| Bowel bag | 1(10) | 4(40) | 4(40) | 1(10) |
| CTVN | 1(10) | 3(30) | 2(20) | 4(40) |
| Anorectal cancer | ||||
| Bladder | 15(50) | 9(30) | 3(10) | 3(10) |
| Femoral head right | 20(67) | 8(27) | 1(3) | 1(3) |
| Femoral head left* | 24(83) | 5(17) | ||
| Bowel bag | 1(3) | 8(27) | 11(37) | 10(33) |
*One patient with surgical implant left hip excluded.
4. Discussion
In this study, we have presented a CNN-based method for the automated segmentation of OAR in anorectal cancer, and OAR, and CTVNs in cervical cancer. Using a separate test set, we found that the CNN-based method performed well overall in the selected OAR and CTVNs, but the majority of organs/volumes would have needed to be manually corrected to some degree according to the qualitative evaluation.
A well-defined ground truth is needed to evaluate the CNN. This is a challenge considering that all human segmentations are subjective by nature [26], and an actual ground truth is difficult to establish. At our institution, radiation oncologists strive to adhere to international guidelines for manual segmentations, but the manually segmented organs and volumes from structure sets do not always strictly follow the organ and volume boundaries and are sometimes also inaccurately defined. This inevitably leads to similar inaccuracies in the CNN output, which is a problem for CNN performance.
More data, and more consistently annotated data, are needed to achieve higher CNN accuracy and to enable future clinical implementation. Overall, there will always be variations and even inaccuracies in manually segmented structure sets if and when they are used as the ground truth. However, we believe that the upsides of using available structure sets (based on the performance of several radiation oncologists at a large university hospital), which provide the possibility of scaling up study populations, will outweigh the downside of segmentation variations.
A quantitative evaluation of the CNN is needed, and the most commonly used in the literature is Dice making this overlap score particularly valuable for comparison with previous studies. In this study, all auto-segmented structures received a median Dice score ≥ 0.8. Previous studies on auto-segmentation, using different systems, have shown moderate to high levels of overlap (as measured with the Dice score). The segmentation of femoral heads is highly concordant in general according to previous literature, with a Dice score range of 90–95% [15], [16], [27], [28], [29]), which is in line with the results in this present study (median: 0.91–0.94). The reason for this high overlap is probably the femoral heads having good contrast with the surrounding tissue and often being regular with a well-defined shape. Further, the auto-segmentation of femoral heads in this study showed high performance not only with Dice, but also illustrated with MSD and with qualitative evaluation (for both cervical and anorectal cancer patients). Considering the bladder, which may be more varied in fill level and shape, showed a wider range of published accuracy (Dice range in previous studies: 67–93%[15], [16], [23], [27], [28], [29], [30], [31]), and our results were in the higher interval (0.84 and 0.94 for cervical and anorectal cancers, respectively). Furthermore, the intestine is even more difficult to delineate, and the difference from the surrounding tissue may be vague. This fact yielded a lower accuracy for auto-segmentation of the intestines, both in our study (bowel bag 0.83–0.88), Liu et al., (0.79–0.83), and lastly a study by Men et al., which had even lower accuracy (60–65%) [15]. However, the intestinal outlines are not directly comparable between the studies.
In our study, CTVN (cervical cancer group only) had the lowest Dice score (0.82). A CTVN has a small volume and occurs in an area with high anatomical variation. Therefore, it may be prone to variability in manual segmentation and more difficult for the CNN to master on a limited sample size [32]. In the anorectal cancer group, the manual segmentation of CTV for nodes was too variable to use for training because the volume is subdivided and segmented differently depending on the cancer’s subtype and pattern. In addition, the cervical and anorectal tumor volumes have even more variety (in size, boundaries, and localization) and are therefore challenging. They were not included in this study but could be research focus for future studies.
The Dice score reflects the overlap between auto-segmentation and manual segmentation. Notably, the distance between the auto-segmentation and manual segmentation will result in a low Dice score, but it may be that it is the auto-segmentation that more closely outlines the anatomical structure, which is a drawback of the Dice score method. To complement Dice, mean surface distance was analyzed as an additional quantitative measurement, in general showing the same performance (in terms of for which OAR/CTVN auto segmentations performed well or less well) as the Dice results. To evaluate the auto-segmentations further, we let one radiation oncologist qualitatively evaluate all the auto-segmented images patient by patient and structure by structure. As expected, some structures were more difficult for the network to delineate (the bowel bag and CTVNs) because of the more varying distribution in the abdomen and more indistinct border with surrounding structures (which is especially challenging with lean patients). The majority of these cases would have needed to be corrected for several image slices according to the qualitative evaluation.
Interestingly, the presence or absence of intravenous contrast did not seem to affect the performance of the auto-segmentations, given the quite similar qualitative evaluation in cervical and anorectal cancer (in the latter patient group, CT was performed without intravenous contrast according to clinical guidelines). Other automated structure delineations (femoral heads and bladder) fit very well with clinical segmentation standards, and the majority of cases needed only very few corrections.
Only one radiation oncologist evaluating the auto-segmentations may be a drawback. However, to the best of our knowledge, this approach of using structure sets as the ground truth has not been tried previously, and the qualitative evaluation is merely part of a first step to exploring the feasibility and gaining an overall view of the possibility of scaling up this approach. In future studies, potentially including more annotated cases, it would be interesting to let several radiation oncologists evaluate the auto-segmentations.
The approach of using already segmented structure sets as ground truth have been previously and successfully tried for OARs in cervical cancer [16] and rectal cancer [15]. Further, this approach is beneficial since it may be scaled up to larger populations and datasets, which is essential for successful AI training [1]. This study was conducted at a single institution, which is a drawback, but now that this approach has been successfully tested, collaborations with other sites are planned in upcoming studies. To take the evaluation one step further, for future studies, a highly interesting evaluation could be to create new alternative radiation treatment plans based on the CNN based segmentations. If only insignificant differences in the planned doses to the target volume and OAR were seen (as compared to fully manually annotated radiation treatment plans), a shift to an AI system for auto-segmentations could be clinically considered.
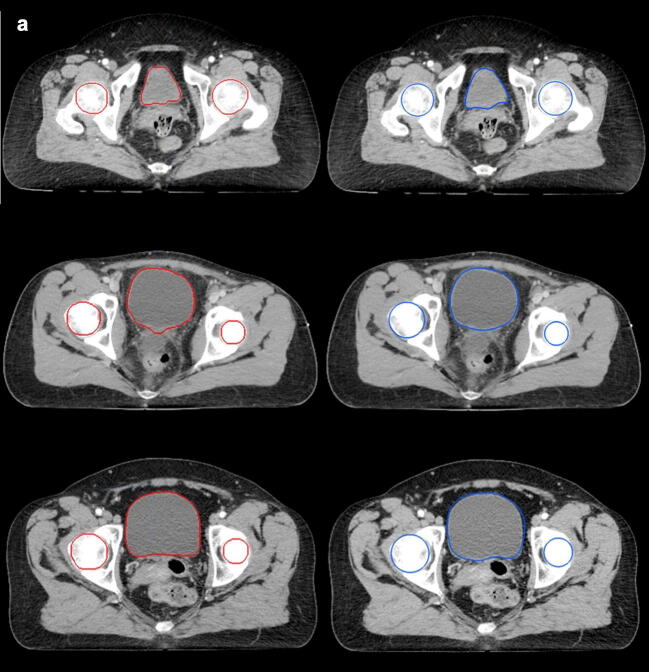
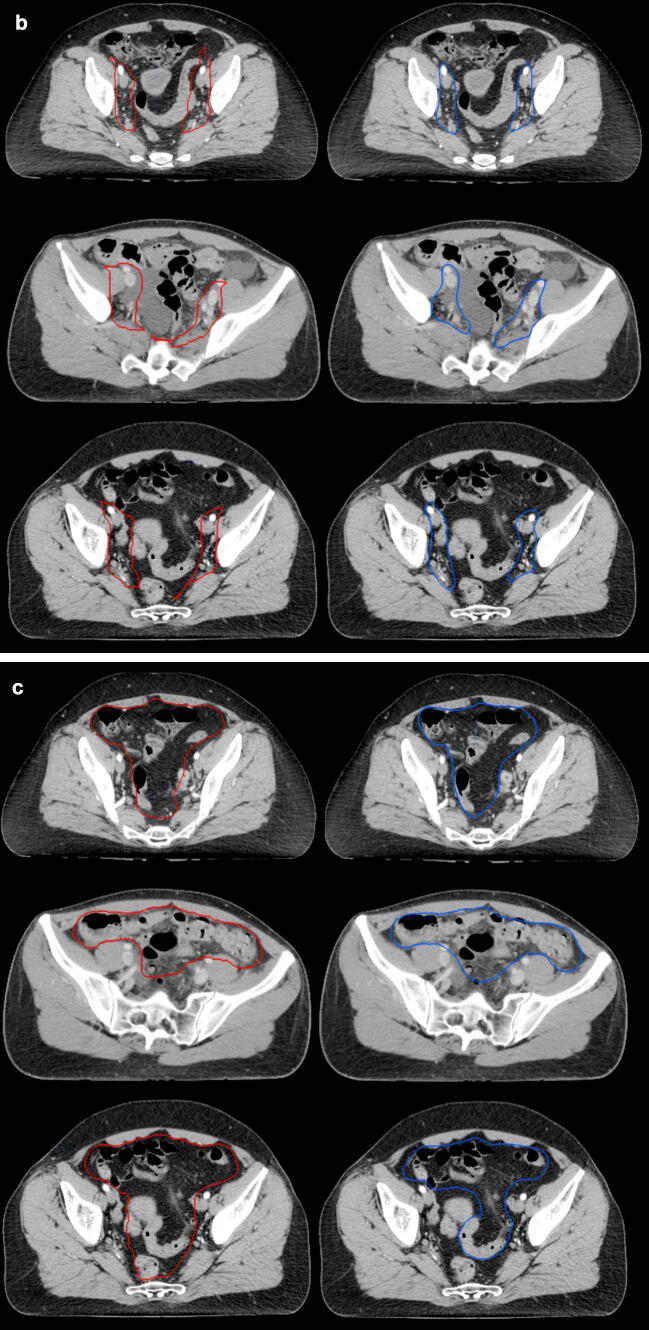
In conclusion, we have shown that it is possible to train a CNN in cervical and anorectal cancer with good overlap using clinically available structure sets as the ground truth. More data and consistently annotated data are most likely needed to achieve higher CNN overlap and enable future clinical implementation. Since one of the largest challenges for radiation oncologists is to prioritize and divide time correctly between different tasks, a feasible auto-segmentation model may aid in the clinical care of cancer patients (Fig. 3).
Fig. 3.
Auto-segmentations (left/red) and manual segmentations (right/blue) in three patients (a: femoral heads and urinary bladder, b: CTVN, c: bowel bag).
Declaration of Competing Interest
OE and JU are board members and stockholders of Eigenvision AB, which is a company working with research and development in automated image analysis, computer vision and machine learning. The other authors declare that they have no conflict of interest.
Acknowledgement
The study was financed by generous support from Knut and Alice Wallenberg foundation, Region Skåne and Lund University and by Governmental Funding of Clinical Research within National Health Services.
References
- 1.Feng M., Valdes G., Dixit N., Solberg T.D. Machine learning in radiation oncology: opportunities, requirements, and needs. Front Oncol. 2018;8 doi: 10.3389/fonc.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarrett D., Stride E., Vallis K., Gooding M.J. Applications and limitations of machine learning in radiation oncology. Br J Radiol. 2019;92:20190001. doi: 10.1259/bjr.20190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mak R.H., Endres M.G., Paik J.H., Sergeev R.A., Aerts H., Williams C.L. Use of crowd innovation to develop an artificial intelligence-based solution for radiation therapy targeting. JAMA Oncol. 2019;5:654–661. doi: 10.1001/jamaoncol.2019.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson R.F., Valdes G., Fuller C.D., Carpenter C.M., Morin O., Aneja S. Artificial intelligence in radiation oncology: a specialty-wide disruptive transformation? Radiother Oncol. 2018;129:421–426. doi: 10.1016/j.radonc.2018.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas C.E., Yang J., Anderson B.M., Court L.E., Brock K.B. Advances in auto-segmentation. Seminars Radiat Oncol. 2019;29:185–197. doi: 10.1016/j.semradonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Erickson B.J., Korfiatis P., Akkus Z., Kline T.L. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou Q., Yu L., Chen H., Jin Y., Yang X., Qin J. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal. 2017;41:40–54. doi: 10.1016/j.media.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Lindgren Belal S., Sadik M., Kaboteh R., Enqvist O., Ulén J., Poulsen M.H. Deep learning for segmentation of 49 selected bones in CT scans: first step in automated PET/CT-based 3D quantification of skeletal metastases. Eur J Radiol. 2019;113:89–95. doi: 10.1016/j.ejrad.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y., Gao Y., Shen D. Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans Med Imaging. 2016;35:1077–1089. doi: 10.1109/TMI.2015.2508280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Zhou F., Liu B. Automatic bladder segmentation from CT images using deep CNN and 3D fully connected CRF-RNN. Int J Comput Assist Radiol Surg. 2018;13:967–975. doi: 10.1007/s11548-018-1733-7. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi S., Nishio M., Yakami M., Nakagomi K., Togashi K. Bone segmentation on whole-body CT using convolutional neural network with novel data augmentation techniques. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103767. [DOI] [PubMed] [Google Scholar]
- 12.Kosmin M., Ledsam J., Romera-Paredes B., Mendes R., Moinuddin S., de Souza D. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radiother Oncol. 2019;135:130–140. doi: 10.1016/j.radonc.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 13.van der Veen J., Willems S., Deschuymer S., Robben D., Crijns W., Maes F. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol. 2019;138:68–74. doi: 10.1016/j.radonc.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Vrtovec T., Močnik D., Strojan P., Pernuš F., Ibragimov B. Auto-segmentation of organs at risk for head and neck radiotherapy planning: from atlas-based to deep learning methods. Med Phys. 2020 doi: 10.1002/mp.14320. [DOI] [PubMed] [Google Scholar]
- 15.Men K., Dai J., Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377–6389. doi: 10.1002/mp.12602. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Liu X., Xiao B., Wang S., Miao Z., Sun Y. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys Med. 2020;69:184–191. doi: 10.1016/j.ejmp.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Kavanagh B.D., Pan C.C., Dawson L.A., Das S.K., Li X.A., Ten Haken R.K. Radiation dose-volume effects in the stomach and small bowel. Int J Radiat Oncol Biol Phys. 2010;76:S101–S107. doi: 10.1016/j.ijrobp.2009.05.071. [DOI] [PubMed] [Google Scholar]
- 18.Trägårdh E., Borrelli P., Kaboteh R., Gillberg T., Ulén J., Enqvist O. RECOMIA—a cloud-based platform for artificial intelligence research in nuclear medicine and radiology. EJNMMI Phys. 2020;7:51. doi: 10.1186/s40658-020-00316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
- 20.Nijkamp J., Doodeman B., Marijnen C., Vincent A., van Vliet-Vroegindeweij C. Bowel exposure in rectal cancer IMRT using prone, supine, or a belly board. Radiother Oncol. 2012;102:22–29. doi: 10.1016/j.radonc.2011.05.076. [DOI] [PubMed] [Google Scholar]
- 21.Lim K., Small W., Jr., Portelance L., Creutzberg C., Jurgenliemk-Schulz I.M., Mundt A. Consensus guidelines for delineation of clinical target volume for intensity-modulated pelvic radiotherapy for the definitive treatment of cervix cancer. Int J Radiat Oncol Biol Phys. 2011;79:348–355. doi: 10.1016/j.ijrobp.2009.10.075. [DOI] [PubMed] [Google Scholar]
- 22.Ng M., Leong T., Chander S., Chu J., Kneebone A., Carroll S. Australasian Gastrointestinal Trials Group (AGITG) contouring atlas and planning guidelines for intensity-modulated radiotherapy in anal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1455–1462. doi: 10.1016/j.ijrobp.2011.12.058. [DOI] [PubMed] [Google Scholar]
- 23.Huyskens D.P., Maingon P., Vanuytsel L., Remouchamps V., Roques T., Dubray B. A qualitative and a quantitative analysis of an auto-segmentation module for prostate cancer. Radiother Oncol. 2009;90:337–345. doi: 10.1016/j.radonc.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 25.Yeghiazaryan V., Voiculescu I. Department of Computer Science, University of Oxford; 2015. An overview of current evaluation methods used in medical image segmentation. [Google Scholar]
- 26.Roques T.W. Patient selection and radiotherapy volume definition — can we improve the weakest links in the treatment chain? Clin Oncol. 2014;26:353–355. doi: 10.1016/j.clon.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Delpon G., Escande A., Ruef T., Darréon J., Fontaine J., Noblet C. Comparison of automated atlas-based segmentation software for postoperative prostate cancer radiotherapy. Front Oncol. 2016;6 doi: 10.3389/fonc.2016.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwee J., Louie A.V., Gaede S., Bauman G., D'Souza D., Sexton T. Technology assessment of automated atlas based segmentation in prostate bed contouring. Radiat Oncol. 2011;6:110. doi: 10.1186/1748-717X-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong WKH, Leung LHT, Kwong DLW. Evaluation and optimization of the parameters used in multiple-atlas-based segmentation of prostate cancers in radiation therapy. Br J Radiol. 2016;89:20140732. [DOI] [PMC free article] [PubMed]
- 30.Li D., Zang P., Chai X., Cui Y., Li R., Xing L. Automatic multiorgan segmentation in CT images of the male pelvis using region-specific hierarchical appearance cluster models. Med Phys. 2016;43:5426. doi: 10.1118/1.4962468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Schoot A.J., Schooneveldt G., Wognum S., Hoogeman M.S., Chai X., Stalpers L.J. Generic method for automatic bladder segmentation on cone beam CT using a patient-specific bladder shape model. Med Phys. 2014;41 doi: 10.1118/1.4865762. [DOI] [PubMed] [Google Scholar]
- 32.Anders L.C., Stieler F., Siebenlist K., Schafer J., Lohr F., Wenz F. Performance of an atlas-based autosegmentation software for delineation of target volumes for radiotherapy of breast and anorectal cancer. Radiother Oncol. 2012;102:68–73. doi: 10.1016/j.radonc.2011.08.043. [DOI] [PubMed] [Google Scholar]