Abstract
Colony-stimulating factor 1 receptor (CSF1R) inhibition has been proposed as a method for microglia depletion, with the assumption that it does not affect peripheral immune cells. Here, we show that CSF1R inhibition by PLX5622 indeed affects the myeloid and lymphoid compartments, causes long-term changes in bone marrow-derived macrophages by suppressing interleukin 1β, CD68, and phagocytosis but not CD208, following exposure to endotoxin, and also reduces the population of resident and interstitial macrophages of peritoneum, lung, and liver but not spleen. Thus, small-molecule CSF1R inhibition is not restricted to microglia, causing strong effects on circulating and tissue macrophages that perdure long after cessation of the treatment. Given that peripheral monocytes repopulate the central nervous system after CSF1R inhibition, these changes have practical implications for relevant experimental data.
Keywords: microglia, macrophages, CSF1R, hematopoiesis, CNS
Colony-stimulating factor 1 receptor (CSF1R) inhibition has been proposed as specific for central nervous system (CNS) microglia depletion without significant effect on peripheral immune cells (1–4). This has been based on rather crude assays and indirect assessment of peripheral cell subtypes and function. Given that peripheral monocytes have been shown to participate in CNS disease via both infiltration and repopulation of neuroglia following microglia depletion (4–10), it is important to determine if CSF1R inhibition causes functional changes in peripheral immune cells that can become part of the CNS (3, 4, 10–14).
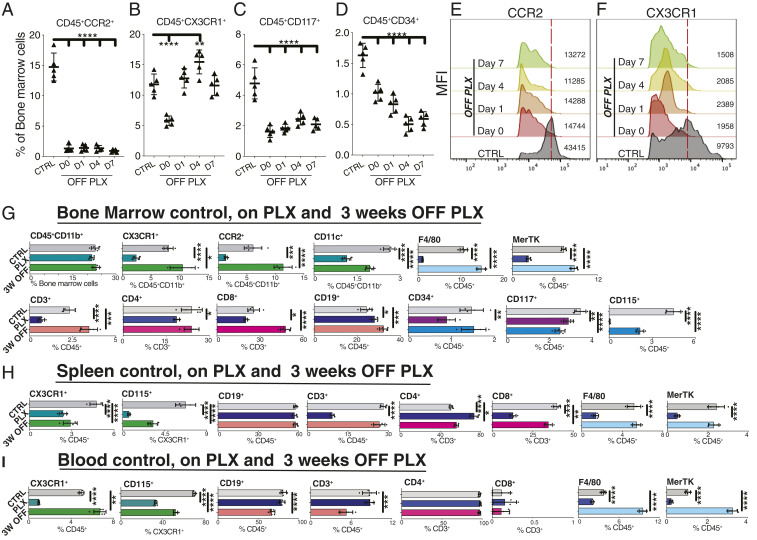
Here we show that, contrary to the accepted notion (1, 11), PLX5622, a commonly used CSF1R inhibitor (8, 10, 11), does not affect only microglia but also leads to long-term changes in the myeloid and lymphoid compartments of the bone marrow, spleen, and blood by suppressing CCR2+ monocyte progenitor cells, CX3CR1+ bone marrow-derived macrophages (BMDMs), CD117+ (C-KIT+) hematopoietic progenitor cells, F4/80+, MerTK+, and CD34+ hematopoietic stem cells (Fig. 1 A–G). Most importantly, these cell populations either do not recover or rebound after cessation of CSF1R inhibition, with the exception of CD45+ CD11b+ cells which remain unaffected (Fig. 1 A–G). CSF1R inhibition also alters the lymphoid compartment of the bone marrow by suppressing T cells (CD3+, CD4+, and CD8+) and up-regulating CD19+ B cells (Fig. 1G). Cessation of CSF1R inhibition causes rebound of some, but not all, lymphoid cells (Fig. 1G).
Fig. 1.
CSF1R inhibition affects the myeloid and lymphoid compartments of the bone marrow, spleen, and blood. Flow cytometric analysis of bone marrow cells isolated from CCR2+/RFP::CX3CR1+/GFP mice treated with PLX5622 for 3 wk, at different time points after inhibitor treatment cessation. (A–F) CSF1R inhibition suppresses CCR2+, CX3CR1+, CD117+, and CD34+ cells. One week after cessation of inhibitor, only macrophages recover in number, although with a lower expression of CX3CR1. (G) CSF1R inhibition does not affect CD45+, CD11b+, or Ly6C+ bone marrow myeloid cell populations but does suppress CD11c+ dendritic cells, CD4+ and CD8+ T lymphocytes, and CD115+, CD117+, and CD34+ hematopoietic subsets and up-regulates CD19+ B cells. Three weeks after cessation of CSF1R-inhibition, CX3CR1+, CCR2+, Ly6C+ CD3+, and CD8+ subpopulations rebound; Ly6G+ granulocytes, CD115+, and CD117+ cells remain suppressed; CD4+ T cells and CD34+ cells recover; and CD19+ B cells remain up-regulated. (H) Effects on spleen’s myeloid and lymphoid populations. Only CD19+ B cells remain unaffected. (I) CSF1R inhibition causes immediate suppression in the myeloid compartment and late suppression of the lymphoid compartment of the blood. n = 5 per group, mean ± SD, one-way analysis of variance with Dunnett’s correction for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
In addition to the effects on bone marrow cells, CSF1R inhibition also suppresses splenic CX3CR1+ cells that persist for at least 3 wk after cessation of treatment (Fig. 1H). Moreover, splenic CD3+ T cells (primarily CD8+) become suppressed, whereas CD19+ B cells are not affected (Fig. 1H).
Circulating CX3CR1+, CD115+, F4/80+, and MerTK+ blood cells are suppressed, but not the lymphoid CD3+ and CD4+ populations (Fig. 1I). Cessation of CSF1R inhibitor causes rebounding of the CX3CR1+, F4/80+, and MerTK+ but not CD115+ blood cells and leads to delayed suppression of CD19+, CD3+, and CD4+ lymphoid cell in the circulation (Fig. 1I).
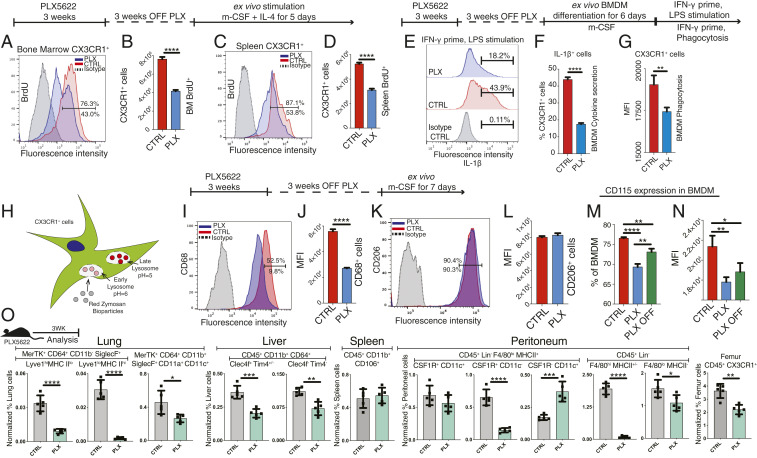
CSF1R inhibition also suppresses the proliferation of bone marrow and spleen macrophages (Fig. 2 A–D) and impairs the function of BMDMs for the long term. Three weeks after cessation of CSF1R inhibition, BMDMs display reduced interleukin (IL)-1β expression in response to endotoxin (Fig. 2 E and F), diminished phagocytosis (Fig. 2 E–G), reduced CD68 but not CD206 expression (Fig. 2 I–L), and suppressed CD115 labeling (Fig. 2 M and N). Moreover, CSF1R inhibition reduced the number of tissue-resident and interstitial macrophages of the lung (15, 16), liver (17), and peritoneum (18) and suppressed femur macrophages but did not affect CD45+ CD11b+ CD106+ spleen (19) and F4/80lo MHCII+ CSF1R+ CD11c+ peritoneum macrophage subsets (18) (Fig. 2O).
Fig. 2.
CSF1R inhibition affects function and survival of resident and interstitial macrophages. (A–D) Ex vivo evaluation of the function of BMDMs from CX3CR1+/GFP mice 3 wk after cessation of CSF1R inhibitor. Macrophages from the bone marrow or spleen exhibit reduced proliferation 3 wk after cessation of CSF1R inhibition. (E–L) CSF1R inhibition suppresses IL-1β, CD68 expression, and phagocytosis of BMDMs following exposure to LPS but does not affect CD206 expression. (H) Schematic representation of the phagocytosis assay. (M and N) CSF1R inhibition causes long-term suppression of CD115 macrophage marker. (O) CSF1R-inhibitor reduces the number of resident and interstitial macrophages of the lung, liver, peritoneum, and femur but does not affect CD45+ CD11b+ CD106+ spleen and F4/80lo MHCII+ CSF1R+ CD11c+ peritoneal macrophages. n = 5 per group, mean ± SD, Independent t test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Previous studies have suggested that microglia depletion by CSF1R inhibition can either promote or exacerbate neurodegeneration (4, 10). These puzzling and contradictory results may not only be due to the differing role of microglia in various disease models but also to the varying relative contribution of peripheral and circulating macrophages on disease phenotype. Considering that BMDMs permanently engraft into diseased CNS tissue (8, 10), this work suggests that small-molecule inhibition is not restricted to microglia but additionally affects the turnover and function of bone marrow-derived, circulating, and tissue-resident macrophages. These effects perdure long after cessation of the treatment and have implications for the interpretation of relevant experimental data.
Materials and Methods
Mouse Model.
Experiments were conducted in accordance with the Association for Research in Vision and Ophthalmology and the NIH guidance for use of laboratory animals and approved by the institutional Animal Care Committee of Massachusetts Eye and Ear - Harvard Medical School. Mice (6 to 12 mo old) used included C57BL/6J (000664), B6.129(Cg)-Ccr2tm2.1lfc/J (017586), and B6.129P- Cx3cr1tm1Litt/J (005582) from The Jackson Laboratory. CCR2RFP/+::CX3CR1EGFP/+ mice were generated by crossing B6.129(Cg)-Ccr2tm2.1lfc/J with B6.129P- Cx3cr1tm1Litt/J. CX3CR1EGFP/+ mice were generated by crossing male B6.129P- Cx3cr1tm1Litt/J with female C57BL/6J. Mice were bred in-house. For microglia depletion, PLX5622 chow (Plexxikon Inc.) was administered for 3 wk. Flow cytometry and ex vivo BMDM evaluation were performed as previously described (8, 10). Blood cells were collected by cardiac puncture and centrifugation. Peritoneum-, lung-, liver-, spleen-, and femur-resident macrophages were evaluated using appropriate markers (16, 19) in BMT CX3CR1+/EGFP reporter mice following 3-wk exposure to PLX5622.
Flow Cytometry Markers.
Bone marrow and spleen cells from CX3CR1+/GFP and CX3CR1+/EGFP::CCR2+/RFP reporter mice were blocked with CD16/32 (clone: 2.4G2), analyzed with IL-1β (clone: NJTEN3), Lyve1 (clone: ALY7) (eBiosciences); CD45 (clone: 104), CD11b (clone: M1/70), CD11c (clone: N418), CD3 (clone: 17A2), CD4 (clone: GK1.5), CD8 (clone: 53-5.8), CD19 (clone: 6D5), CD117 (clone: 2B8), CD34 (clone: HM34), CD115 (clone: AFS98), CD64 (clone X54-5/7.1), Clec4f (clone 3E3F9), Tim-4 (clone RMT4-54), Siglec F (clone S17007L), PE Lineage mixture (catalog no. 133303), CD68 (clone: FA-11), CD206 (clone: C068C2), CCR2 (clone: SA203G11), BrdU(clone: Bu20a), F4/80 (clone: BM8), MerTK (clone: 2B10C42), CD11a (clone: I21/7), CD102 (clone: 3C4), CD106 (clone: 429), and I-A/I/E (clone: M5/114.15.2) (BioLegend). Intracellular staining was performed by fixing cells in paraformaldehyde-based fixation buffer (BioLegend) followed by permeabilization with Perm/Wash buffer (BioLegend). Cells were analyzed on a BD LSR II cytometer (BD Biosciences) using FlowJo software (Tree Star).
Lipopolysaccharide Stimulation Assay.
BMDMs were primed with 150 U/mL interferon (IFN)-γ (6 h) followed by 10 ng/mL lipopolysaccharide (LPS) (Sigma-Aldrich) for 20 h. Brefeldin A (5 μg/mL) (BD Pharmingen) was added 4 h before flow cytometry.
Phagocytosis Assay.
The pHrodo Red BioParticles Conjugates for Phagocytosis (P35364; Molecular Probes) kit was used. Six days after cell plating with macrophage colony-stimulating factor, and 1 d prior to the assay, BMDMs were recovered from culture and seeded. Cells were stimulated with IFN-γ for 4 h and then culture medium was replaced with reconstituted red Zymosan A BioParticles. Cells were incubated at 37 °C for 2 h, trypsinized, and evaluated with flow cytometry.
Statistics.
Data were analyzed with GraphPad Prism version 2.8.1 using two-tailed unpaired t test and ordinary one-way ANOVA with Dunnett’s correction for multiple comparisons. Statistical significance was determined at P < 0.05.
Acknowledgments
This work was supported by the Boston Keratoprosthesis Research Fund, Research to Prevent Blindness, core grants P30EY003790 and EY014104, the Yeatts Family Foundation, and Monte J. Wallace Chair R21EY023079. PLX5622 was kindly provided by Plexxikon Inc.
Footnotes
The authors declare no competing interest.
Data Availability.
All study data are included in the paper. Materials are available commercially and are listed in Materials and Methods.
References
- 1.Elmore M. R. P. et al., Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mok S. et al., Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res. 74, 153–161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilla A. M., Diekmann H., Fischer D., Microglia are irrelevant for neuronal degeneration and axon regeneration after acute injury. J. Neurosci. 37, 6113–6124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellver-Landete V. et al., Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 10, 518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans T. A. et al., High-resolution intravital imaging reveals that blood-derived macrophages but not resident microglia facilitate secondary axonal dieback in traumatic spinal cord injury. Exp. Neurol. 254, 109–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynn T. A., Vannella K. M., Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlen S. J. et al., Monocyte infiltration rather than microglia proliferation dominates the early immune response to rapid photoreceptor degeneration. J. Neuroinflammation 15, 344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paschalis E. I. et al., Permanent neuroglial remodeling of the retina following infiltration of CSF1R inhibition-resistant peripheral monocytes. Proc. Natl. Acad. Sci. U.S.A. 115, E11359–E11368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschalis E. I. et al., The role of microglia and peripheral monocytes in retinal damage after corneal chemical injury. Am. J. Pathol. 188, 1580–1596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paschalis E. I. et al., Microglia regulate neuroglia remodeling in various ocular and retinal injuries. J. Immunol. 202, 539–549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagher N. N. et al., Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflammation 12, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kokona D., Ebneter A., Escher P., Zinkernagel M. S., Colony-stimulating factor 1 receptor inhibition prevents disruption of the blood-retina barrier during chronic inflammation. J. Neuroinflammation 15, 340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halder S. K., Milner R., A critical role for microglia in maintaining vascular integrity in the hypoxic spinal cord. Proc. Natl. Acad. Sci. U.S.A. 116, 26029–26037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson J. M., Isaacson L. G., Elimination of microglia in mouse spinal cord alters the retrograde CNS plasticity observed following peripheral axon injury. Brain Res. 1721, 146328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbings S. L. et al., Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 57, 66–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakarov S. et al., Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Scott C. L. et al., The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 49, 312–325.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bain C. C. et al., Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 7, ncomms11852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier E. L. et al.; Immunological Genome Consortium , Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the paper. Materials are available commercially and are listed in Materials and Methods.