Significance
Developing a male-sterility system that is effective in multiple species is essential for hybrid seed production in different plants, especially for plants without cloned male-sterility genes. Here, we identified the transcriptional regulation mechanism for maize male-sterility gene ZmMs7 and thereby developed a dominant male-sterility system that was proved to be effective in maize, rice, and Arabidopsis. Compared with current male-sterility systems, this system has potential advantages, e.g., utilization of a single transgene cassette, high stability of male sterility under different genetic backgrounds, and producing fluorescent transgenic and normal color nontransgenic F1 hybrid seeds which can be used flexibly in different countries where transgenic crop cultivation is prohibited or allowed. Therefore, it is a simple, cost-effective, and multiple-crop-applicable biotechnology.
Keywords: ZmMs7, PHD finger, protein-protein interaction, dominant male-sterility system
Abstract
Understanding the molecular basis of male sterility and developing practical male-sterility systems are essential for heterosis utilization and commercial hybrid seed production in crops. Here, we report molecular regulation by genic male-sterility gene maize male sterility 7 (ZmMs7) and its application for developing a dominant male-sterility system in multiple species. ZmMs7 is specifically expressed in maize anthers, encodes a plant homeodomain (PHD) finger protein that functions as a transcriptional activator, and plays a key role in tapetal development and pollen exine formation. ZmMs7 can interact with maize nuclear factor Y (NF-Y) subunits to form ZmMs7-NF-YA6-YB2-YC9/12/15 protein complexes that activate target genes by directly binding to CCAAT box in their promoter regions. Premature expression of ZmMs7 in maize by an anther-specific promoter p5126 results in dominant and complete male sterility but normal vegetative growth and female fertility. Early expression of ZmMs7 downstream genes induced by prematurely expressed ZmMs7 leads to abnormal tapetal development and pollen exine formation in p5126-ZmMs7 maize lines. The p5126-ZmMs7 transgenic rice and Arabidopsis plants display similar dominant male sterility. Meanwhile, the mCherry gene coupled with p5126-ZmMs7 facilitates the sorting of dominant sterility seeds based on fluorescent selection. In addition, both the ms7-6007 recessive male-sterility line and p5126-ZmMs7M dominant male-sterility line are highly stable under different genetic germplasms and thus applicable for hybrid maize breeding. Together, our work provides insight into the mechanisms of anther and pollen development and a promising technology for hybrid seed production in crops.
Heterosis is a phenomenon in which heterozygous hybrid progeny are superior to both homozygous parents and can offer 20% to over 50% yield increases in various crops (1). Maize is one of the most successful crops of heterosis utilization. Manual or mechanical detasseling has been widely used for maize hybrid seed production. However, detasseling is not only time consuming, labor intensive, and expensive, but also detrimental to plant growth, and thus reduces the yield of maize hybrid seed (2). Therefore, the male-sterility line is critical for the commercial hybrid seed production in maize.
Male sterility mainly includes three types, i.e., cytoplasmic male sterility (CMS) caused by both mitochondrial and nuclear genes, genic male sterility (GMS) caused by nuclear genes alone, and photoperiod- and/or temperature-sensitive genic male sterility (2–4). Among them, the CMS-based three-line system has been successfully used for hybrid seed production in many crops (4). For example, the CMS hybrid rice has accounted for ∼40% of the total rice planting area in China since the late 1980s (5). However, the CMS system has suffered from several intrinsic problems, such as the limited genetic resources of the restorer lines, the low genetic diversity between the CMS lines and the restorer lines in rice (6), and the potentially increased disease susceptibility and unreliable restoration of CMS lines in maize (7). The photoperiod- and/or temperature-sensitive genic male sterility-based two-line system has been initially used for hybrid rice production in China since the 1990s (8). It eliminates the requirement for crossing to propagate the male-sterility female line, and almost any fertile line with a good combining ability can be used as a male parent, thus it can enhance the usage efficiency of genetic resources and further increase the yield of hybrid rice (3, 8). However, purity of hybrid seeds produced by using this system may be influenced by the undesired climate or environmental conditions (4).
The male gamete development is a well-orchestrated process; any disturbance may thus lead to male sterility (9). Up to now, more than 100 GMS genes have been identified in plants (2, 10, 11), and a great deal of effort has been made to develop biotechnology-based male-sterility systems by using these GMS genes and their corresponding mutants to maintain and propagate GMS lines as female parents for hybrid seed production (2, 4). For instance, a system called seed production technology (SPT) was developed to produce recessive GMS lines by DuPont Pioneer using the maize male sterility 45 (ZmMs45) gene, coupled with an α-amylase gene and a red fluorescence gene DsRed (12). Recently, we have updated the SPT system and developed the multi-control sterility (MCS) systems by using maize GMS genes ZmMs7, ZmMs30, and ZmMs33, respectively (13–15). A similar system was also constructed in rice (16). In addition, a dominant male-sterility system was developed to produce dominant GMS lines using the dominant GMS gene ms44 in maize (17). Although these biotechnology-based male-sterility systems have several advantages (18), they rely on the combination of the cloned GMS genes and their corresponding mutants. Therefore, developing a male-sterility system would be a valuable application for hybrid seed production in various plant species, especially for plants without cloned GMS genes.
ZmMs7 encodes a PHD-finger transcription factor (TF) and is a key regulator of postmeiotic anther development in maize (13). Its orthologs have been identified in several plant species, such as Arabidopsis male sterility 1 (AtMS1) (19–22), rice male sterility 1/Persistent Tapetal Cell 1 (OsMs1/OsPTC1) (23, 24), and barley male sterility 1 (HvMs1) (25). Mutations in AtMs1 and OsMs1/OsPTC1 result in abnormal tapetum programmed cell death (PCD) and defective pollen exine formation (22–24, 26). Both AtMs1 and OsMs1/OsPTC1 function as transcriptional activators (20, 23), and OsMs1/OsPTC1 interacts with tapetal regulatory factors, such as OsMADS15 and TDR Interacting Protein2 (TIP2), to regulate tapetal cell PCD and pollen exine formation (23). Both HvMs1 and OsPTC1 can functionally complement the Arabidopsis ms1 mutation and rescue male fertility, indicating functions of AtMs1 orthologs in higher plants may be conserved (24, 25). Nevertheless, the precise molecular mechanism of transcriptional regulation by AtMs1 orthologs remains largely unknown.
Here, we investigate molecular mechanism of ZmMs7 for regulating anther and pollen development, and find that ZmMs7 interacts with maize NF-Y subunits to form multiprotein complexes that directly activate target genes. Premature expression of ZmMs7 driven by p5126 results in the dominant and complete male sterility through dramatically altering the gene expression networks responsible for anther and pollen development. By using the p5126-ZmMs7 transgenic element, we construct a dominant male-sterility (DMS) system that is proved to be effective in maize, rice, and Arabidopsis.
Results
ZmMs7 Is Required for Tapetum Development and Pollen Exine Formation.
ZmMs7 was cloned and verified by using functional complementation in our laboratory (13). To further confirm its function in controlling male fertility, four ZmMs7 knockout lines were generated via the CRISPR-Cas9 system. Compared with wild type (WT), all of the Cas9-ZmMs7 knockout lines displayed complete male sterility with no exerted anthers, and the shriveled anthers lacked pollen grains at mature stage, which is similar to the ms7-6007 mutant (Fig. 1A and SI Appendix, Fig. S1), indicating that ZmMs7 is required for male fertility in maize.
Fig. 1.
Phenotypic and cytological comparison of WT, ms7-6007 mutant, and ZmMs7 knockout line generated via the CRISPR-Cas9 method. (A) Comparison of tassels (A1), anthers (A2), and pollen grains stained with I2-KI (A3) among WT, ms7-6007 mutant, and the Cas9-ZmMs7-01 line. (Scale bars, 1 mm in A2 and 200 μm in A3.) (B) Transverse section analysis of anthers in WT (B1 to B3) and ms7-6007 mutant (B4 to B6) from stages 9 to 11 during maize anther development. (Scale bar, 50 μm.) (C) SEM analysis of pollen grain development in WT (C1 to C3) and ms7-6007 mutant (C4 to C6). (Scale bar, 20 μm.) (D) TEM analysis of Ubisch body in WT (D1 to D3) and ms7-6007 mutant (D4 to D6). (Scale bar, 0.5 μm.) (E) TEM analysis of pollen exine in WT (E1 to E3) and ms7-6007 mutant (E4 to E6) (Scale bar, 0.5 μm.) Ba, bacula; CMsp, collapsed microspore; E, epidermis; En, endothecium; F, foot layer; ML, middle layer; Msp, microspore; Ta, tapetum; Te, tectum; and Ub, Ubisch body.
Maize anther development can be divided into 14 stages (stage 1 to stage 14) (2). To characterize the cytological defects in ms7-6007 anthers, we performed scanning electron microscopy (SEM), transverse section, and transmission electron microscopy (TEM) analyses and found that anther and microspore development progressed similarly between WT and ms7-6007 until stage 9 (Fig. 1 B–D and SI Appendix, Figs. S2–S4). The SEM analysis showed only transverse strips covered the outer surface of WT anthers until stage 10, and then the three-dimensional (3D) knitting cuticle formed at stage 11 and fully grew at stage 13. However, the 3D knitting cuticle appeared at stage 10 but gradually reduced thereafter in ms7-6007 anthers. On the inner surface, Ubisch bodies emerged at stage 9 in both WT and ms7-6007 anthers, and they enlarged subsequently in WT but gradually disappeared in ms7-6007 (SI Appendix, Fig. S2). The transverse section analysis showed developmental differences between WT and ms7-6007 anthers occurred after stage 9. For example, at stage 11, the vacuoles disappeared in the microspore and tapetal cells almost completely degenerated in WT anthers, while the mutant locules started to shrink and the microspores broke (Fig. 1B and SI Appendix, Fig. S3). Similarly, the SEM observation of pollen showed that microspore mother cells, dyads, tetrads, and microspores were successively generated from stages 7 to 9 in both WT and ms7-6007 anthers. After formation of the vacuolated microspores at stage 10, starch granules were gradually accumulated, and then the regular round-shaped mature pollen grains formed in WT anther locules at stage 13. By contrast, the ms7-6007 vacuolated microspores severely sank at stage 10 and disappeared at stage 12 (Fig. 1C and SI Appendix, Fig. S3). The TEM observation showed that Ubisch bodies appeared on the inner surface of WT and mutant anthers at stage 9, enlarged thereafter in WT, but not enlarged in ms7-6007. The WT microspores were enveloped with evident exine at stage 9, and then their distinctive layers (i.e., tectum, bacula, and foot layer) formed and thickened from stages 10 to 13. However, the ms7-6007 exine was much thinner than that of WT (Fig. 1 D and E and SI Appendix, Fig. S4). Taken together, loss of function of ZmMs7 results in delayed tapetal degeneration, abnormal Ubisch body development, and pollen wall formation, and these developmental defects ultimately cause complete male sterility.
ZmMs7 Functions as a Transcriptional Activator and Regulates Genes Involved in Tapetum Development and Pollen Exine Formation.
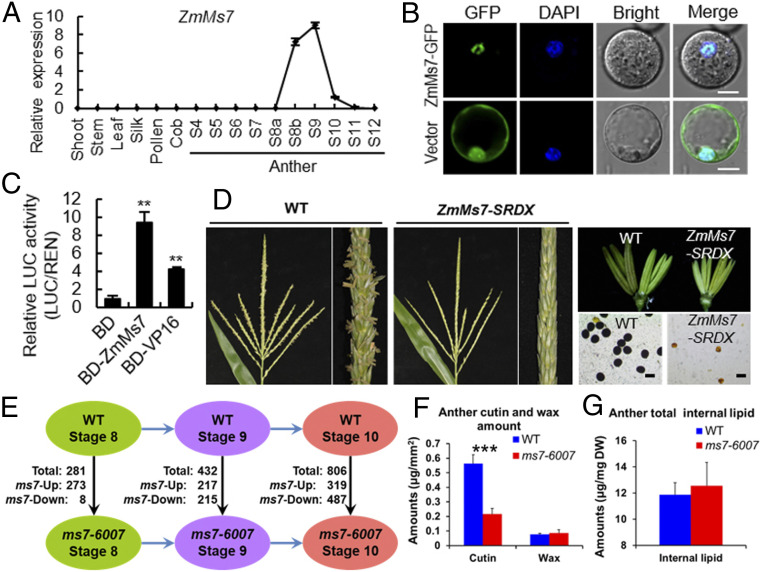
The spatiotemporal expression patterns of ZmMs7 in different tissues were detected by using real-time quantitative PCR (RT-qPCR). ZmMs7 transcript was only detected in WT anthers from stages 8b to 10, and peaked at stage 9, indicating that ZmMs7 is specifically expressed in maize anthers at tetrad and free haploid microspore stages (Fig. 2A). ZmMs7 encodes a PHD-finger protein (13) which is localized in the nucleus, supporting that it functions as a TF (Fig. 2B). ZmMs7 dramatically enhanced the reporter expression in a transient dual-luciferase reporter assay performed in maize protoplasts, indicating that ZmMs7 has a transcriptional activation activity (Fig. 2C). To further test the transcriptional activity in planta, ZmMs7 was fused with a conserved suppressing motif, SRDX (LDLDLELRLGFA), which can convert transcriptional activators into dominant repressors (27). Transformation of pZmMs7:ZmMS7-SRDX into maize plants inhibited anther and pollen development, and led to complete male sterility (Fig. 2D). These results strongly suggest that ZmMs7 acts as a transcriptional activator in maize anthers, being consistent with its homologs AtMs1 (20) and OsMs1/OsPTC1 (23), implying that ZmMs7-mediated transcriptional regulatory mechanism may be conserved among different plant species.
Fig. 2.
Gene expression, transcriptional activity, and transcriptome and lipidome analyses of ZmMs7. (A) RT-qPCR analysis of ZmMs7 expression in different organs of maize. Data are means ± SD, n = 3. (B) Subcellular localization of ZmMs7 in maize protoplasts. DAPI staining was used as a nuclear marker. (Scale bar, 10 μm.) (C) Transcriptional activation assay of ZmMs7 using a dual-luciferase system in maize protoplasts. GAL4 DNA binding domain (BD) and transcriptional activator VP16 were used as negative and positive controls, respectively. Data are means ± SD, n = 4. Asterisks indicate significant difference compared to BD (**P < 0.01, Student’s t test). (D) Male sterile phenotype of pZmMs7:ZmMS7-SRDX transgenic maize plants. Comparison of tassels, anthers, and pollen grains stained with I2-KI between WT and transgenic plants. (Scale bar, 100 μm.) (E) The numbers of DEGs between WT and ms7-6007 mutant anthers at stages 8, 9, and 10, respectively. (F) Total amount of anther cutin and wax per unit surface area and (G) total amount of anther internal lipid per dry weight (DW) in WT and ms7-6007 mutant. Data are means ± SD, n = 5. Asterisks indicate significant difference in comparison (***P < 0.001, Student’s t test).
To identify the downstream genes potentially regulated by ZmMs7 during anther development, RNA sequencing (RNA-seq) analysis was performed using WT and ms7-6007 anthers at stages 8 to 10 (SI Appendix, Fig. S5A). A total of 1,143 differentially expressed genes (DEGs) between WT and ms7-6007 were detected (SI Appendix, Fig. S5 B–F). Among these DEGs, 710 were down-regulated and 809 were up-regulated in ms7-6007 anthers during at least one stage (Fig. 2E and SI Appendix, Table S1). Gene ontology analysis of the 1,143 DEGs showed that ZmMs7 modulates biological processes such as pollen exine formation, polysaccharide catabolism, phenypropanoid biosynthesis, and cell differentiation processes, most of which are important for anther and pollen development (SI Appendix, Fig. S5G). Particularly, a set of genes putatively related to tapetum and pollen wall development showed altered expression patterns in ms7-6007 anthers, including ZmMs45 (28), Indeterminate Gametophyte1 (IG1) (29), IG1/AS2-like1 (IAL1) (29), maize basic Helix–loop–helix 122 (ZmbHLH122) (30), maize Metallothionein 2C (ZmMT2C) (31), maize Aspartyl Protease 37 (ZmAP37) (32), ZmMs6021, maize Dihydroflavonol 4-Reductase-Like 1/2 (ZmDRL1/2), maize Less Adhesive Pollen 5 (ZmLAP5), maize Acyl-COA-Synthetase 5 (ZmACOS5), and maize Quartet3 (ZmQRT3) (2, 11) (SI Appendix, Tables S1–S4).
Since a set of lipid metabolism-related genes displayed altered expression patterns in ms7-6007 anthers (SI Appendix, Table S1), the components of cutin, wax, and internal fatty acids in WT and ms7-6007 anthers were detected. The results showed a dramatic reduction for the total cutin content and most of the cutin monomers in ms7-6007 anthers (Fig. 2F and SI Appendix, Fig. S6A and Table S2). No significant difference was detected for the total wax and internal fatty acid contents between WT and ms7-6007 anthers (Fig. 2 F and G), although most of the individual components of wax and internal fatty acids displayed significant changes (SI Appendix, Fig. S6 B and C). These results suggested that ms7-6007 mutation impedes cutin biosynthesis and alters the constituents of wax and internal fatty acids, thereby affecting anther wall development and pollen exine formation.
ZmMs7 Forms Multiprotein Complexes with ZmNF-Y Subunits.
Like many PHD finger proteins, ZmMs7 has no DNA binding domain as analyzed by the InterPro online program (https://www.ebi.ac.uk/interpro/). To understand how ZmMs7 regulates its target genes, we used yeast two-hybrid (Y2H) assay to screen its interactors. Because the C-terminal domain of ZmMs7 possesses self-activation activity, the N-terminal region (amino acids 1 to 358) was used as bait for the Y2H assay. A total of 130 positive clones were obtained, and four NF-Y proteins were confirmed to interact with ZmMs7, including ZmNF-YA6, ZmNF-YC9, ZmNF-YC12, and ZmNF-YC15 (Fig. 3 A, Left and SI Appendix, Fig. S7 A and C). To validate these protein interactions, coimmunoprecipitation (Co-IP) assay was performed in maize protoplasts. The four NF-Y proteins were strongly coimmunoprecipitated by ZmMs7-cYFP-Myc, suggesting that ZmMs7 physically links the NF-YA6 and NF-YC9/12/15 subunits in plant cells (Fig. 3 A, Right). Bimolecular fluorescence complementation (BiFC) assay in maize protoplasts further conformed the results (Fig. 3D). Since NF-Y TFs are reported to form heterotrimeric complexes composed of NF-YA, NF-YB, and NF-YC subunits (33), the interactions between NF-YA and NF-YC subunits were tested by using Y2H, Co-IP, and BiFC assays. The results showed that NF-YA6 can interact with all of the three NF-YC9/12/15 subunits in both yeast and plant cells (Fig. 3 B and D). Next, to find out the NF-YB counterparts of the NF-Y complexes, we conducted Y2H screening between candidate NF-YBs and ZmMs7, NF-YA6 and NF-YC9/12/15. Among 18 NF-YB members of maize (34), NF-YB2 was found to interact with NF-YC9/12/15, but not with NF-YA6 and ZmMs7, which was further confirmed by Co-IP and BiFC assays (Fig. 3 C and D). Besides NF-YB2, NF-YB10 was found to interact with NF-YC15, but not with NF-YC9/12 (SI Appendix, Fig. S8). Therefore, we selected NF-YB2 for further studies. Meanwhile, the RT-qPCR analysis showed that NF-YA6, NF-YB2, and NF-YC9/12/15 genes were all expressed in maize anthers during stages 6 to 10 (SI Appendix, Fig. S7D). The coexpression patterns of ZmMs7 and NF-Y genes provided a prerequisite for the protein interactions between ZmMs7 with NF-Y subunits in maize anthers. Together, ZmMs7 can form protein complexes with NF-YA6, NF-YB2, and NF-YC9/12/15 (Fig. 4F).
Fig. 3.
Interaction of ZmMs7 with ZmNF-Y subunits. (A) Y2H and Co-IP assays of ZmMs7 interaction with NF-YA6 and NF-YC9/12/15. For Y2H assay, LAM and P53 were used as negative and positive controls, respectively. DDO, double dropout medium (SD-Trp-Leu); QDO, quadruple dropout medium (SD-Trp-Leu-His-Ade). Co-IP assays were performed using a transient expression system in maize protoplasts; the nYFP-FLAG was used as a negative control. The asterisk indicates a nonspecific band. (B) Y2H and Co-IP assays of NF-YA6 interaction with NF-YC9/12/15. IG1-nYFP-FLAG was used as a negative control. Others are as in A. (C) Y2H and Co-IP assays of NF-YB2 interaction with ZmMs7N, NF-YA6, and NF-YC9/12/15. Others are as in A. (D) BiFC analysis of in vivo interaction between ZmMs7, NF-YA6, NF-YB2, and NY-YC9/12/15 in maize protoplasts. Protein name-n indicates nYFP fusions. DAPI staining was used as a nuclear marker. The fluorescence was detected using confocal microscopy. (Scale bar, 10 μm.)
Fig. 4.
ZmMs7-NF-YA/YB/YC complexes directly activate target gene expression. (A) Transient dual-luciferase assay of ZmMT2C promoter activity activated by ZmMs7-NF-Y complexes in maize protoplasts. Data are means ± SD, n = 3. Different letters above each column indicate significant difference (P < 0.01, Student’s t test). (B) EMSA assay of ZmNF-YA6 binding to the CCAAT box in ZmMT2C promoter region. (C) The enrichments of ZmMT2C promoter analyzed by ChIP-qPCR with the primer sets (P1, P2, and P3), using the anther samples of ZmMs7-3x Myc transgenic maize plants. Data are means ± SD, n = 3. +Ab, presence of anti-c-Myc antibody; −Ab, absence of the antibody. The asterisks indicate significant difference between +Ab and −Ab (P < 0.01, Student’s t test). (D) Detection of DNA fragmentation by TUNEL assay in WT and ms7-6007 anthers. (Scale bar, 50 μm.) (E) TEM of tapetum degeneration in WT and ms7-6007 anthers. Ta, tapetum. (Scale bar, 10 μm.) (F) Model of ZmMs7-NF-YA/YB/YC complexes controlling maize male fertility through directly activating ZmMT2C expression and indirectly regulating cutin and sporopollenin biosynthesis-related genes such as ZmMs6021, ZmLAP5, ZmDRL1/2, and ZmACOS5.
The ZmMs7-NF-Y Complexes Directly Activate Target Gene Expression.
To identify target genes of ZmMs7, we selected a set of genes with down-regulated expressions in ms7-6007 anther, including ZmMs6021, ZmIAL1, ZmMT2C, ZmAP37, and ZmQRT3 that are homologous to the reported GMS genes in rice and Arabidopsis. Among these genes, the ZmMT2C promoter was significantly activated by the ZmMs7-NF-Y complexes in the transactivation assay, suggesting that ZmMs7 in cooperation with ZmNF-YA6-YB2-YC9/12/15 trimers activates the ZmMT2C promoter in maize protoplasts (Fig. 4A). Considering that ZmMT2C is potentially involved in tapetum development, we selected this gene for further testing. It was reported that NF-Y complex binds to the consensus motif CCAAT in the promoters of target genes (33). Electrophoretic mobility shift assays (EMSA) showed that MBP-NF-YA6 can specifically bind to the CCAAT box in the promoter region of ZmMT2C (Fig. 4B). Next, to examine whether ZmMs7 complexes can bind to the ZmMT2C promoter in vivo, we performed chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) analysis using the anther samples of ZmMs7-3xMyc transgenic maize plants. Compared with the control probes (P2 and P3), the P1 fragment containing the CCAAT box was significantly enriched by using anti-c-Myc antibody (Fig. 4C), suggesting that ZmMs7 binds to the ZmMT2C promoter in vivo through association with the CCAAT element. Collectively, ZmMs7-NF-Y complexes can directly activate expression of the target gene ZmMT2C.
Since ZmMT2C is a homolog of OsMT2b which is involved in tapetal cell PCD of rice anthers (31), we examined DNA fragmentation in WT and ms7-6007 anthers using a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay. TUNEL signals began to appear at stage 9 in WT anthers, but later appeared at stage 10 in ms7-6007 anthers, indicating that tapetal cell PCD is delayed in ms7-6007 anthers (Fig. 4D). Consistent with the TUNEL results, the TEM analysis showed delayed tapetum degeneration in ms7-6007 anthers compared with that in WT (Fig. 4E). Collectively, ZmMs7 may regulate tapetal cell PCD via directly activating the expression of its target gene ZmMT2C and modulate anther cuticle and pollen wall formation by indirectly regulating cutin and sporopollenin biosynthesis-related genes such as ZmMs6021, ZmDRL1/2, ZmLAP5, and ZmACOS5 (Fig. 4F).
ms7-6007 Mutation Is Genetically Stable and Applicable for Hybrid Maize Breeding and Seed Production.
A total of 403 maize inbred lines with broad genetic diversity were used to test whether male sterility caused by ms7-6007 mutation was stable under different maize germplasms (SI Appendix, Fig. S9A and Table S3). The 403 lines were pollinated by pollen of heterozygous plants (ZmMs7/ms7-6007) and then 403 F2 populations were generated from the self-pollination of F1 plants with the genotype of ZmMs7/ms7-6007 based on marker-assisted selection. Among the 403 F2 populations, 376 (93.3%) type II populations (0.05 < P values ≤ 1.0 and 1.6 < ratios ≤ 7.0) fitted to the 3:1 ratio of fertile plants to sterile plants, while six (1.5%) type I (0 ≤ P values < 0.05 and 1.2 < ratios ≤ 1.6) and 21 (5.2%) type III populations (0 ≤ P values < 0.05 and 7.0 < ratios ≤ 42.0) showed segregation deviated from the expected 3:1 ratio (SI Appendix, Fig. S9B and Table S3). To justify whether the deviated segregation from 3:1 in the 27 type I and III populations may result from environmental conditions or the exogenetic fertile pollen unexpectedly participating in the self-pollination of the 27 F1 plants, we randomly chose 4 from the 27 type I and III populations and performed molecular marker analysis using a ms7-6007 mutation marker. Interestingly, we found that the male-sterility genotypes of 4 F2 populations matched well with their corresponding sterile phenotypes (SI Appendix, Fig. S9C), indicating that male sterility of the ms7-6007 mutation is stable in the 27 populations with deviated segregation from 3:1. Additionally, the tassels of sterile F2 individuals were collected from 8 F2 populations with different tassel shapes from those of WT and ms7-6007 and showed complete male sterility and no anther shedding. Moreover, no pollen grain was observed in the anthers of these sterile individuals under different genetic backgrounds, which resembled the ms7-6007 mutant (SI Appendix, Fig. S9D). Therefore, male sterility of the ms7-6007 mutation is stable under diverse maize genetic backgrounds.
To test whether ms7-6007 mutation affects grain yield and other agronomic traits in hybrid maize production, we randomly selected 31 elite inbred lines and took them as male parents to cross with the ZmMs7/ZmMs7 and ms7-6007/ms7-6007 homozygous plants, respectively. The harvested F1 hybrids and their corresponding parental lines were grown in three locations in triplicate. Seventeen agronomic traits were investigated to compare the differences of heterosis and field production performance between each of the 31 pairs of hybrid combinations (SI Appendix, Table S4). The hybrids derived from ms7-6007 and its WT as female parents showed similar field performance in the three locations (SI Appendix, Fig. S10). The plot yields of 13 representative pairs of hybrid combinations, and ear morphologies of 4 pairs of hybrid combinations were shown in SI Appendix, Figs. S11 and S12A, as examples. All of the hybrid combinations greatly outperformed their parents, whereas plot yields showed almost no statistical difference between pairs of the 31 hybrid combinations (SI Appendix, Table S4). For other important traits, such as 100-seed weight, grain number per ear, plant height, and mature period, significant differences between a few pairs of hybrid combinations were detected under only one environmental condition. Nevertheless, the change trends showed complete randomness between pairs of the 31 hybrid combinations (SI Appendix, Fig. S12 B–E and Table S4). These results demonstrated that the ms7-6007 mutation has no obvious negative effects on maize heterosis and field production, suggesting that the ZmMs7 gene and its mutant ms7-6007 are applicable for hybrid maize breeding and seed production.
Premature Expression of ZmMs7 Driven by an Anther-Specific Promoter p5126 Disrupts Tapetum and Pollen Development and Results in Dominant Male Sterility in Maize.
The maize anther-specific promoter p5126 confers tapetal-specific expression at stages 7 to S8a (28) which is earlier than the expression of ZmMs7 at stages 8b to 10. Overexpression of ZmMs7 homologous genes, AtMs1 and HvMS1, results in dominant male sterility in Arabidopsis and barley, respectively, indicating precise control of AtMs1 and HvMS1 expression is critical for male fertility in plants (22, 25). Therefore, to develop a dominant male-sterility system, we constructed the p5126-ZmMs7M vector containing three functional modules, i.e., ZmMs7 expression cassette driven by p5126 to obtain a dominant male-sterility trait, the red fluorescence protein gene mCherry driven by the aleurone-specific LTP2 promoter to mark the color of transgenic seeds, and the herbicide-resistant gene Bar driven by the CaMV35S promoter to select transgenic plants (SI Appendix, Fig. S13A). As a result, five p5126-ZmMs7M transgenic events (p5126-ZmMs7M-01 to -05) showed dominant male sterility with smaller anthers and no pollen grain, whereas their vegetative development and female fertility were normal (Fig. 5A and SI Appendix, Fig. S13B). Due to the dominant male sterility, the p5126-ZmMs7M element can be genetically transmitted only through female gametes. Thus all of the transgenic lines produced nearly 50% transgenic fluorescent seeds and 50% nontransgenic normal color seeds as observed under green excitation light with different red fluorescence filters (Fig. 5B and SI Appendix, Fig. S13B).
Fig. 5.
Phenotypic and cytological comparison of WT and the p5126-ZmMs7M-01 dominant male-sterility line. (A) Comparison of tassels (A1), anthers (A2), and pollen grains stained with I2-KI (A3) between WT and the p5126-ZmMs7M-01 line. (Scale bars, 1 mm in A2 and 200 μm in A3.) (B) Comparison of ear phenotypes between WT and the p5126-ZmMs7M-01 line under bright light (B1, B4, and B5) and green excitation light with red fluorescence filter I (GREEN.L, China) (B2 and B6) and red fluorescence filter II (NIGHTSEA, United States) (B3 and B7). (C) Transverse section analysis of anthers in WT (C1 to C3) and the p5126-ZmMs7M-01 line (C4 to C6) from stage 8b to 10 during maize anther development. (Scale bar, 50 μm.) (D) SEM analysis of microspores in WT (D1 to D3) and the p5126-ZmMs7M-01 line (D4 to D6). (Scale bar, 20 μm.) (E) TEM analysis of Ubisch body in WT (E1 to E3) and the p5126-ZmMs7M-01 line (E4 to E6). (Scale bar, 0.5 μm.) (F) TEM analysis of pollen exine in WT (F1 to F3) and the p5126-ZmMs7M01 line (F4 to F6). (Scale bar, 0.5 μm.) Ba, bacula; CMsp, collapsed microspore; E, epidermis; En, endothecium; F, foot layer; LD, lipid droplet; ML, middle layer; Msp, microspore; Ta, tapetum; Tds, tetrads; Te, tectum; and Ub, Ubisch body.
Microscopic analyses were conducted to explore the morphological defects of anther and pollen in the p5126-ZmMs7M-01 line. The SEM analysis showed that the 3D knitting cuticle formed at stage 11 in both WT and p5126-ZmMs7M-01 anthers. But it was obviously tighter and thinner in p5126-ZmMs7M-01 anthers. On the inner surface of the p5126-ZmMs7M-01 anther wall, lots of lipid droplets emerged evidently at stage 9, but no typical Ubisch body was observed when compared with that of WT (SI Appendix, Fig. S2). The transverse section analysis showed that obvious differences between WT and p5126-ZmMs7M-01 anthers started to appear at stage 9. At this stage, abnormal tapetal cell proliferation and swelling occurred evidently in p5126-ZmMs7M-01 anthers. At stage 10, WT tapetum showed the hill-like structures indicating the degeneration of tapetal cells, while p5126-ZmMs7M-01 tapetum maintained a regular shape without degeneration. Anther locule of p5126-ZmMs7M-01 line started to shrink at stage 11, and ultimately formed a similar shape with that of ms7-6007 at stage 13 (Fig. 5C and SI Appendix, Fig. S3). Meanwhile, the SEM analysis of pollen showed that microspore mother cells, dyads, tetrads, and microspores were successively generated in p5126-ZmMs7M-01 anthers. However, the haploid microspores sank severely in the tetrads at stage 8b, the integrity of the microspore cell wall was disrupted at stage 10, and only cell debris remained within the locule of p5126-ZmMs7M-01 anthers from stages 11 to 13 (Fig. 5D and SI Appendix, Fig. S3). The TEM analysis showed that the obvious Ubisch bodies appeared at stage 9 and gradually enlarged from stages 10 to 13 on the inner surface of WT tapetum. However, lots of lipid droplets instead of Ubisch bodies accumulated on the inner surface of p5126-ZmMs7M-01 tapetum from stages 9 to 11 (Fig. 5E and SI Appendix, Fig. S4). Similarly, lipid droplets were observed on the p5126-ZmMs7M-01 microspore wall from stages 9 to 13, and no obvious exine was found when compared with that in WT (Fig. 5F and SI Appendix, Fig. S4). Taken together, premature expression of ZmMs7 driven by p5126 disrupts tapetum and pollen wall development, which accounts for the dominant male sterility of the p5126-ZmMs7M-01 line.
Premature Expression of ZmMs7 Alters Expression Patterns of Genes Required for Tapetum and Pollen Development in Maize.
To investigate the molecular basis underlying the dominant male sterility, anther transcriptomes of WT and the p5126-ZmMs7M-01 line were analyzed and compared during six developmental stages (S6 to S10) based on RNA-seq. At the six investigated stages, 62.3% (5,390/8,654) of DEGs were found to be up-regulated in p5126-ZmMs7M-01 anthers, being consistent with the activator function of ZmMs7. Among the 4,474 individual DEGs identified in p5126-ZmMs7M-01 anthers from stages 6 to 10, 492 genes were overlapped with the 1,143 DEGs identified in ms7-6007 anthers (SI Appendix, Fig. S14 A–C). The 492 shared DEGs were functionally enriched in pollen development and the exine formation process, which is consistent with the defective pollen wall phenotypes of the p5126-ZmMs7M-01 line (Fig. 5 C–F and SI Appendix, Fig. S14D). Because ZmMs7 acts as a transcriptional activator, to examine the direct effects of ZmMs7 premature expression on its downstream targeted or regulated genes, we identified 126 shared genes that were down-regulated in ms7-6007 anthers at all three stages (stages 8 to 10), but up-regulated in p5126-ZmMs7M-01 anthers from stages 7 to S8b (Fig. 6A and SI Appendix, Fig. S16 and Tables S5 and S6). The 126 genes were functionally enriched in biological processes such as cellular development, cell differentiation, lipid biosynthesis, and transmembrane transport, which are critical for development of the anther, tapetum, and pollen wall (Fig. 6B). To further confirm the transcriptomic results, we performed RT-qPCR analysis on six genes including ZmMs7, one target gene (ZmMT2C) regulated by ZmMs7-NF-Y complexes, and four GMS homologous genes (ZmAP37, ZmQRT3, ZmIAL1, and ZmMs6021) (Fig. 6C). ZmMs7 expression peaked at stage 9 in WT anther and at stage 8a in p5126-ZmMs7M-01 anther, showing that ZmMs7 expression was advanced by two stages in the p5126-ZmMs7M-01 line. Consistently, the expression peaks of ZmMT2C, ZmAP37, ZmQRT3, and ZmMs6021 were advanced by one to two stages in the p5126-ZmMs7M-01 line. The transcript level of ZmIAL1 was obviously elevated from stages 8b to 10 in the p5126-ZmMs7M-01 line (Fig. 6C). Collectively, the premature expression of ZmMs7 driven by p5126 altered expression patterns of genes involved in tapetum and pollen development, which likely led to the dominant male sterility of p5126-ZmMs7M lines.
Fig. 6.
p5126 promoter drives premature expression of ZmMs7-activated genes and results in the dominant male-sterility phenotype of the p5126-ZmMs7M-01 line. (A) The 126 shared DEGs between 144 down-regulated DEGs in the ms7-6007 mutant and 244 up-regulated DEGs in the p5126-ZmMs7M-01 line (A1). The expression patterns of the 50 representative genes in anther transcriptomes of ms7-6007 mutant at stages 8 to 9 (A2) and in the p5126-ZmMs7M-01 line at stages 6 to 10 (A3) are shown, respectively. (B) Gene ontology enrichment analysis of the 126 shared genes. (C) RT-qPCR analysis of ZmMs7 and five putative activated genes in WT, ms7-6007, and the p5126-ZmMs7M-01 line anthers at stages 6 to 10.
Construction of a Dominant Male-Sterility System Based on the Conserved Function of p5126-ZmMs7 in Maize, Rice, and Arabidopsis.
Functions of ZmMs7 and its orthologs have been found to be conserved in Arabidopsis, maize, rice, and barley (13, 19, 24, 25). This prompted us to further test whether premature expression of ZmMs7 can induce similar dominant male sterility in other plant species. Firstly, the plasmid of p5126-ZmMs7M was transformed into rice. The obtained four T0 transgenic rice lines exhibited normal vegetative growth and spikelet morphology, but their anthers were smaller and whitish, lacking viable pollen grains (Fig. 7A), indicating that the p5126-ZmMs7M element causes dominant male sterility in rice. The transgenic element can be transmitted through the female gamete when pollinated by WT pollen, and the harvested panicles contained about 50% of T1 fluorescent seeds with a hemizygous transgenic genotype (p5126-ZmMs7M/-) and 50% T1 normal color seeds without the transgene (Fig. 7A). Secondly, the plasmid p5126-ZmMs7-myc was introduced into Arabidopsis. The obtained three T1 transgenic events developed as WT except that their siliques were stunted and failed to set seeds (Fig. 7B). Abundant pollen grains were observed on WT stigma after anther dehiscence. In contrast, the anthers of the three transgenic Arabidopsis plants became brown and had few pollen grains without germination ability (Fig. 7B). Taken together, the dominant male sterility induced by premature expression of ZmMs7 is conserved in both monocot (e.g., maize and rice) and dicot (e.g., Arabidopsis). This type of biotechnology-based male sterility is thus proposed as a DMS system in plants. The detailed strategy is shown in SI Appendix, Fig. S13C using maize as an example.
Fig. 7.
Dominant male-sterility phenotype of p5126-ZmMs7 transgenic rice and Arabidopsis plants. (A) Four rice transgenic plants expressing p5126-ZmMs7-mCherry show complete male-sterility phenotypes. Comparison of whole plants after heading, panicles at anthesis, anthers (Scale bars, 1 mm.), pollen grains with I2-KI staining (Scale bars, 100 μm.), and seeds under bright field and a red fluorescence filter (Scale bars, 0.5 cm.) among WT and four p5126-ZmMs7 transgenic lines. (B) Three Arabidopsis plants expressing p5126-ZmMs7-myc show complete male sterility of pollen grains. Comparison of siliques, the surface of stigmas and anthers, pollen grains stained with I2-KI and pollen germination among WT and three transgenic lines. (Scale bars, 100 μm.) The numbers in A and B indicate different independent transgenic events.
Additionally, the stability of dominant male sterility caused by premature expression of ZmMs7 was evaluated by crossing the p5126-ZmMs7M-01 line with 392 maize inbred lines with broad genetic diversity (SI Appendix, Fig. S15A and Table S7). Then the fluorescent and normal color seeds in each of 392 F1 populations were manually sorted out and grown separately. All plants exhibited normal vegetative growth and female fertility. Plants from normal color seeds in all of the 392 F1 populations were male fertile, while plants from fluorescent seeds of 391 F1 populations were male sterile without exerted anthers and pollen grain (SI Appendix, Fig. S15 B–D). One exception was that plants from fluorescent seeds of the C460 F1 population showed male fertility, and the reason may be that the transgenic element p5126-ZmMs7M was disrupted or silenced in the C460 line. Nevertheless, the dominant male sterility induced by p5126-ZmMs7M is relatively stable under different maize genetic backgrounds.
Discussion
In this study, we investigated molecular regulation by ZmMs7 required for maize male fertility and hereby developed a DMS system. ZmMs7 acts as a transcriptional activator and interacts with ZmNF-Y subunits to form multiprotein complexes which are capable of activating downstream genes directly. Most interestingly, premature expression of ZmMs7 driven by p5126 altered the expression patterns of a series of genes involved in tapetum and pollen development and thus led to dominant male sterility in maize. As ZmMs7 is a key regulator of anther and pollen development, and the regulatory pathways by ZmMs7 orthologs are conserved among different plant species, we successfully developed an applicable DMS system by using the p5126-ZmMs7 transgenic element in maize, rice, and Arabidopsis.
ZmMs7 orthologs have been identified in several plant species, including AtMs1 (19–22), OsMs1/OsPTC1 (23, 24, 31), and HvMs1 (25). Nevertheless, the precise molecular mechanisms and direct target genes of these TFs remain largely unknown. Here, Y2H, Co-IP, and BiFC assays revealed that ZmMs7 can interact with ZmNF-Y subunits to form three ZmMs7-NF-YA6-YB2-YC9/12/15 complexes, suggesting that ZmMs7 achieves its transcriptional regulatory function through interaction with other TFs. The NF-Y heterotrimeric TFs are found in all eukaryotes. In plants, NF-Y subunits interact with various types of transcriptional regulators to form multiple kinds of complexes, thereby regulating individual biological processes (35–38). Here we identified a PHD-type TF as a NF-Y interactor though the domain of ZmMs7 required for association with the NF-Ys need to be further defined. PHD finger proteins are versatile epigenome readers that activate or repress gene expression through recruitments of multiprotein complexes of TFs and chromatin regulators (39). Many PHD finger proteins bind to histone H3 tails with trimethylated lysine 4 (H3K4me3), which is a transcription activation mark (40). We speculate that the fundamental function of ZmMs7 may be recognizing epigenetic marks (e.g., H3K4me3) on its target genes. ZmMs7 may then recruit NF-Y subunits to form multiprotein complexes that activate target gene expression. Notably, we cannot rule out the possibility of ZmMs7 interacting with TFs other than the NF-Y subunits. Logically, ZmMs7 may recruit different interactors when regulating different types of genes. Anyway, elucidating the epigenetic reader role and identifying more interaction partners will help to fully understand the regulatory mechanisms of ZmMs7 and its homologous genes in plants.
Premature expression of ZmMs7 in maize driven by p5126 led to dominant male sterility with completely aborted pollen formation. Unlike most of male-sterility mutants, almost no exine structure was detected on the microspore surface in the p5126-ZmMs7M-01 line, and instead only irregular deposition of lipid droplets was observed. The severely defective pollen wall is a main feature of the dominant male sterility in the p5126-ZmMs7M-01 line. A wide range of genes potentially involved in tapetum development and pollen exine formation displayed altered expression patterns in the p5126-ZmMs7M-01 line. Anther development is a complicated and tightly controlled biological process. Obviously, the disturbance on gene networks responsible for tapetum and pollen development by the p5126-ZmMs7 element is the molecular basis underlying the dominant male sterility. Given that the regulatory systems by ZmMs7 and its orthologs are conserved in plants (13, 20, 24, 25), the dominant male sterility of p5126-ZmMs7 transgenic rice and Arabidopsis plants may be caused by similar molecular mechanisms.
Crop heterosis utilization requires preventing self-pollination of the female inbred parents. It is well known that male sterility is the most effective way to ensure cross-pollination and produce pure hybrid seeds (2, 41, 42). The ZmMs7 gene and its mutant ms7-6007 have been reported to develop the multi-control sterility system in our laboratory, and the system can be used to maintain and propagate maize recessive male-sterility lines and has several advantages as described (13), e.g., no transgenic element in male-sterility lines and hybrid seeds. Here, we tested genetic stability of the ms7-6007 male-sterility lines and found that among 403 F2 populations, 6 (1.5%) type I (0 ≤ P values < 0.05 and 1.2 < ratios ≤ 1.6) and 21 (5.2%) type III (0 ≤ P values < 0.05 and 7.0 < ratios ≤ 42.0) populations showed segregation deviated from the expected 3:1 ratio. For the 21 type III populations with the fertile/sterile ratios far more than 3:1, the reason may be that exogenetic fertile pollen unexpectedly participates in the self-pollination of F1 plants. The only 1.5% type I F2 populations showed the fertile/sterile ratios significantly less than 3:1, possibly due to the smaller sample amount of the investigated F2 individuals (35 to 49 plants) or environmental conditions. Nevertheless, the genotypes of the two types of F2 individuals completely matched with their corresponding phenotypes (SI Appendix, Fig. S9 and Table S3). Therefore, the ms7-6007 male sterility is fairly stable under different maize genetic backgrounds. In addition, we investigated the effects of ms7-6007 mutation on grain yield and other agronomic traits in hybrid maize production, and found that ms7-6007 mutation has no obvious negative effects on maize heterosis and field production. Taken together, based on the developed multi-control sterility system (13), the ZmMs7 gene and its mutant ms7-6007 are applicable for hybrid maize breeding and seed production. On the other hand, we developed the p5126-ZmMs7M-based DMS system to create dominant male-sterility lines in maize and rice and found that genetic stability of the p5126-ZmMs7M male-sterility lines is relatively high under different maize genetic backgrounds (SI Appendix, Fig. S15 and Table S7). Notably, the two different male-sterility systems (multi-control sterility and DMS) are developed by using the same gene ZmMs7 with different maize promoters and molecular mechanisms underlying male sterility. For the multi-control sterility system, loss of function of ZmMs7 that encodes a transcriptional activator inactivates its regulated downstream genes related to cutin biosynthesis and tapetal cell PCD, thus causing the delayed tapetal degeneration, abnormal Ubisch body development, and pollen wall formation, and ultimately leads to complete male sterility of the ms7-6007 mutant. Only plants with the homozygous mutation genotype (ms7-6007/ms7-6007) display complete male-sterility phenotypes, and thus are named as recessive male-sterility lines. However, for the DMS system, premature expression of ZmMs7 driven by p5126 induces the altered expression patterns (i.e., early and high expressions) of a wide range of genes potentially involved in tapetum development and pollen exine formation, and thus results in complete male sterility of the p5126-ZmMs7M lines. Plants with the hemizygous genotype (p5126-ZmMs7M/-//ZmMs7/ZmMs7) show complete male-sterility phenotypes but female fertility and normal vegetative growth, and thus are named as dominant male-sterility lines.
The DMS system produces two types of F1 hybrid seeds, i.e., the red fluorescent transgenic dominant male-sterility seeds (DMS hybrid seeds) and normal color nontransgenic male-fertility seeds (SI Appendix, Fig. S13C). For cross-pollinated plants such as maize, sunflower, and Brassica campestris L., the two types of F1 hybrid seeds can be flexibly used for crop field production in different countries. For example, in some countries where planting transgenic crops is prohibited, the nontransgenic male-fertility hybrid seeds can be sorted out and planted in the field, while in other countries where transgenic crops are allowed to grow, both of the two types of F1 hybrid seeds can be mixed up and planted. Although 50% DMS transgenic dominant male-sterility F1 plants are male sterile, the other 50% nontransgenic male-fertility F1 sibling plants can provide enough pollen grains to pollinate the male sterile F1 plants and ensure no impact on the field yield as reported previously (17). On the other hand, for self-pollinated plants such as rice, sorghum, and millet, the nearly 50% nontransgenic male-fertility hybrid seeds can be sorted out and grown in the field. Compared with the CMS and other biotechnology-based male-sterility systems, the DMS system has several potential advantages. First, relative to the CMS system, the DMS lines are created by using premature expression of a single gene ZmMs7 and show the high stability of male sterility under different genetic backgrounds. Second, compared with the seed production technology and multi-control sterility systems (12–14), the DMS system is not limited by the lack of GMS mutants and fertility restorer genes in many crops. Third, compared with the Barnase/Barstar system (2, 43, 44), the DMS system utilizes a plant endogenous gene (ZmMs7) and promoter (p5126) to generate male sterile lines, which has no ethical problems. Together, the DMS system is a simple, cost-effective, and multiple-crop applicable biotechnology.
Materials and Methods
The ms7-6007 mutant (No. 712AA) was sourced from the Maize Genetics Cooperation Stock Center (maizecoop.cropsci.illinois.edu). Methodological details of plant growth, male-sterility stability analysis, cytological observation, RNA-seq, lipidomic analysis, gene expression analysis, dual-luciferase assay, subcellular localization, protein-protein interaction, and protein-DNA interaction assays are described in SI Appendix, Supplemental Materials and Methods. The primers used in this study are listed in SI Appendix, Table S8.
Supplementary Material
Acknowledgments
The National Key Research and Development Program of China (2018YFD0100806, 2017YFD0100304, 2017YFD0102001, and 2017YFD0101201), the National Natural Science Foundation of China (31900610, 31971958, 31771875, and 31871702), the Fundamental Research Funds for the Central Universities of China (06500136), and the Beijing Science and Technology Plan Program (Z191100004019005) supported this work.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010255117/-/DCSupplemental.
Data Availability.
A complete set of RNA-seq raw data has been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (accession no. PRJNA637676). RNA-seq data in ms7-6007 and WT anther transcriptomes as well as p5126-ZmMs7M-01 and WT anther transcriptomes are listed in SI Appendix, Tables S1, S5, and S6. Genotypic data of 3,072 SNP markers in 403 and 392 maize inbred lines used for male-sterility stability analysis of ms7-6007 and p5126-ZmMs7M-01, respectively, are listed in SI Appendix, Tables S3 and S7. Lipidomic data of WT and ms7-6007 anthers are listed in SI Appendix, Table S2. Phenotypic data of the investigated 17 agronomic traits for testing the effects of the ms7-6007 mutation on maize heterosis and field production are listed in SI Appendix, Table S4.
References
- 1.Tester M., Langridge P., Breeding technologies to increase crop production in a changing world. Science 327, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Wan X. et al., Maize genic male-sterility genes and their applications in hybrid breeding: Progress and perspectives. Mol. Plant 12, 321–342 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Chen L., Liu Y.-G., Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kim Y.-J., Zhang D., Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Cheng S. H., Zhuang J. Y., Fan Y. Y., Du J. H., Cao L. Y., Progress in research and development on hybrid rice: A super-domesticate in China. Ann. Bot. 100, 959–966 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J.-Z., E Z. G., Zhang H. L., Shu Q. Y., Workable male sterility systems for hybrid rice: Genetics, biochemistry, molecular biology, and utilization. Rice (N. Y.) 7, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams M.-E., Genetic engineering for pollination control. Trends Biotechnol. 13, 344–349 (1995). [Google Scholar]
- 8.Yuan L., Purification and production of foundation seed of rice PGMS and TGMS lines. Hybrid Rice 6, 1–3 (1994). [Google Scholar]
- 9.Wilson Z.-A., Zhang D. B., From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 60, 1479–1492 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Shi J., Cui M., Yang L., Kim Y. J., Zhang D., Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Wan X., Wu S., Li Z., An X., Tian Y., Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 13, 955–983 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Wu Y. et al., Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14, 1046–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang D. et al., Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 16, 459–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An X. et al., ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 12, 343–359 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Zhu T. et al., Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 132, 2137–2154 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Chang Z. et al., Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. U.S.A. 113, 14145–14150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox T. et al., A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 15, 942–952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Deng X. W., Development of the “third-generation” hybrid rice in China. Genomics Proteomics Bioinformatics 16, 393–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson Z.-A., Morroll S. M., Dawson J., Swarup R., Tighe P. J., The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Ito T. et al., Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19, 3549–3562 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T., Shinozaki K., The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Yang C., Vizcay-Barrena G., Conner K., Wilson Z. A., MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z. et al., OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99, 175–191 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Li H. et al., PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernández Gómez J., Wilson Z. A., A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Vizcay-Barrena G., Wilson Z. A., Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57, 2709–2717 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M., Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Cigan A.-M. et al., Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 14, 135–142 (2001). [Google Scholar]
- 29.Evans M.-M., The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19, 46–62 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nan G.-L. et al., MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 144, 163–172 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Yi J. et al., Defective tapetum cell death 1 (DTC1) regulates ROS Levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 170, 1611–1623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu N. et al., EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4, 1445 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Myers Z.-A., Holt B.-F. 3rd, NUCLEAR FACTOR-Y: Still complex after all these years? Curr. Opin. Plant Biol. 45, 96–102 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z., Li X., Zhang C., Zou H., Wu Z., Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 478, 752–758 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Zhao H. et al., The Arabidopsis thaliana nuclear factor Y transcription factors. Front Plant Sci 7, 2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S., Parida S. K., Agarwal P., Tyagi A. K., Transcription factor OsNF-YB9 regulates reproductive growth and development in rice. Planta 250, 1849–1865 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Hwang K., Susila H., Nasim Z., Jung J. Y., Ahn J. H., Arabidopsis ABF3 and ABF4 transcription factors Act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Bello B. K. et al., NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 17, 1222–1235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez R., Zhou M.-M., The PHD finger: A versatile epigenome reader. Trends Biochem. Sci. 36, 364–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gatchalian J., “PHD fingers as histone readers” in Histone Recognition, Zhou Z.-Z., Ed. (Springer, 2015), pp. 27–47. [Google Scholar]
- 41.Perez-Prat E., van Lookeren Campagne M. M., Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci. 7, 199–203 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Kempe K., Gils M., Pollination control technologies for hybrid breeding. Mol. Breed. 27, 417–437 (2011). [Google Scholar]
- 43.Mariani C. et al., Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347, 737–741 (1990). [Google Scholar]
- 44.Mariani C. et al., A chimaeric ribonuclease-inhibitor gene restores fertility to male sterile plants. Nature 357, 384–387 (1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A complete set of RNA-seq raw data has been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (accession no. PRJNA637676). RNA-seq data in ms7-6007 and WT anther transcriptomes as well as p5126-ZmMs7M-01 and WT anther transcriptomes are listed in SI Appendix, Tables S1, S5, and S6. Genotypic data of 3,072 SNP markers in 403 and 392 maize inbred lines used for male-sterility stability analysis of ms7-6007 and p5126-ZmMs7M-01, respectively, are listed in SI Appendix, Tables S3 and S7. Lipidomic data of WT and ms7-6007 anthers are listed in SI Appendix, Table S2. Phenotypic data of the investigated 17 agronomic traits for testing the effects of the ms7-6007 mutation on maize heterosis and field production are listed in SI Appendix, Table S4.