Significance
P. tolaasii is the causative agent of brown blotch disease in the common button mushroom Agaricus bisporus and responsible for tremendous losses in mushroom farms every year. The pathogen relies on two different cyclic lipopeptides during infection: the pore forming toxin tolaasin and the motility conferring biosurfactant pseudodesmin. In this study, we show how enzymes produced by helper bacteria of the genus Mycetocola provide protection to the mushroom via lipopeptide cleavage. Using bioactivity guided fractionation of the proteome, we could identify the key enzyme in toxin inactivation. Deciphering such protective interactions paves the way for the development of biocontrol agents for use in both agriculture and medicine.
Keywords: antivirulence, brown blotch disease, cyclic lipopeptides, Mycetocola, tolaasin
Abstract
The bacterial pathogen Pseudomonas tolaasii severely damages white button mushrooms by secretion of the pore-forming toxin tolaasin, the main virulence factor of brown blotch disease. Yet, fungus-associated helper bacteria of the genus Mycetocola (Mycetocola tolaasinivorans and Mycetocola lacteus) may protect their host by an unknown detoxification mechanism. By a combination of metabolic profiling, imaging mass spectrometry, structure elucidation, and bioassays, we found that the helper bacteria inactivate tolaasin by linearizing the lipocyclopeptide. Furthermore, we found that Mycetocola spp. impair the dissemination of the pathogen by cleavage of the lactone ring of pseudodesmin. The role of pseudodesmin as a major swarming factor was corroborated by identification and inactivation of the corresponding biosynthetic gene cluster. Activity-guided fractionation of the Mycetocola proteome, matrix-assisted laser desorption/ionization (MALDI) analyses, and heterologous enzyme production identified the lactonase responsible for toxin cleavage. We revealed an antivirulence strategy in the context of a tripartite interaction that has high ecological and agricultural relevance.
Edible mushrooms are an important food source. To maintain high-yield industrial-scale mushroom farming, it is vital to prevent infections. Numerous bacterial pathogens are known to cause detrimental mushroom diseases, such as cobweb, soft rot, cavity, and brown blotch disease (1). Therefore, many studies have been directed toward understanding the factors promoting virulence, which provides the basis for development of rational measures to prevent mushroom destruction. In general, bacterial pathogens require signal molecules and surfactants to colonize mushrooms (2). To weaken the host and to overcome defense mechanisms, they typically employ virulence factors including chitinolytic enzymes and antifungal agents (3, 4). A set of cyclolipopeptides (CLPs) including tolaasin I (1) has been identified as the major virulence factor of P. tolaasii, the causative agent of brown blotch disease (1, 5–7). The pore-forming toxin (8, 9) causes characteristic browning symptoms in the white button mushroom (A. bisporus), one of the most important cultivated mushrooms worldwide (10, 11). Furthermore, tolaasin-producing bacteria cause diseases of strawberries, cauliflower, and tobacco (12, 13), which calls for unifying strategies to control these pervasive pathogens even beyond mushroom farming.
In contrast to the highly unfavorable use of antibiotics in agriculture, the concept of employing helper bacteria to control pathogens is a potentially less disruptive alternative (Fig. 1). As counterparts to the bacterial pathogen, several antagonistic bacteria have been discovered that either show predatory behavior toward the toxin producer (14) or inactivate its main virulence factor (15–19). For example, the helper bacterium Pseudomonas ‘reactans’ produces a CLP named white-line-inducing principle (WLIP) that precipitates together with tolaasin, thus, forming a characteristic white line when cocultivated with P. tolaasii on solid agar (18). However, WLIP is also toxic against the mushroom host via membrane pore formation and shows hemolytic activities, which precludes its use as a biocontrol agent (20, 21). More promising is the use of helper bacteria that neutralize the effect of tolaasin. Examples of this tolaasin negation activity are provided by members of the genus Mycetocola (M. tolaasinivorans and M. lacteus), which were jointly isolated from the rotting fruiting bodies of mushroom Pleurotus ostreatus (16, 17). The saprophytic gram-positive bacteria, the first members of the novel genus Mycetocola in the family of Microbacteriaceae, are nonpathogenic with respect to the mushroom and were shown to detoxify tolaasin (16). Yet, the mechanism of tolaasin detoxification by these helper bacteria has remained elusive. Here, we report the molecular basis of the antivirulence strategy of two different Mycetocola species. We show that they not only efficiently inactivate the antifungal agent, but also cleave an important bacterial swarming mediator, thus, blocking both pathogenicity and motility of the pathogen.
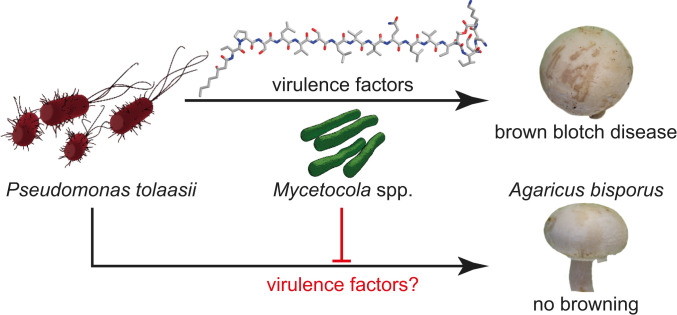
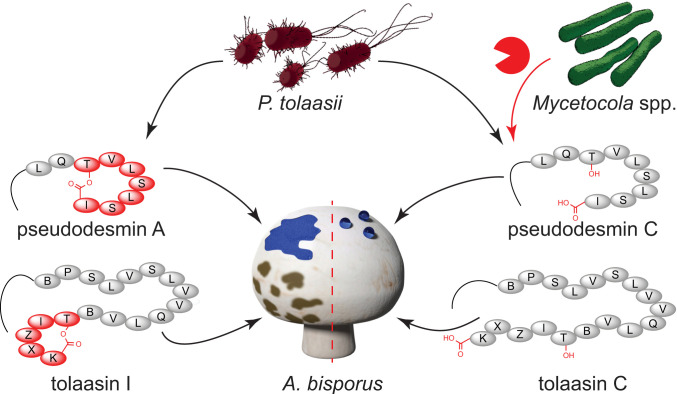
Fig. 1.
Tripartite interaction of mushroom pathogen, helper bacteria, and fungal host. P. tolaasii produces virulence factors that cause bacterial brown blotch in A. bisporus. Mycetocola spp. can prevent decay of the mushroom cap.
Results and Discussion
To gain insight into the pathogen-helper bacteria interaction, we conducted coinoculation experiments of P. tolaasii with M. tolaasinivorans or M. lacteus on mushroom slices (A. bisporus). Whereas mushroom blocks inoculated with P. tolaasii alone showed brown sunken lesions, the presence of M. tolaasinivorans prevented infection symptoms. Nonetheless, A. bisporus was found to be a suboptimal infection model to monitor the inactivation of tolaasin since inoculation with M. lacteus alone led to slight browning of the mushroom tissue, likely owing to its saprophytic lifestyle. Therefore, we turned to potato tubers that reportedly develop black spots in the presence of the toxin (17). Blackening caused by P. tolaasii alone was completely inhibited in coinoculated potato slices (Fig. 2A), indicating effective protection against tolaasin.
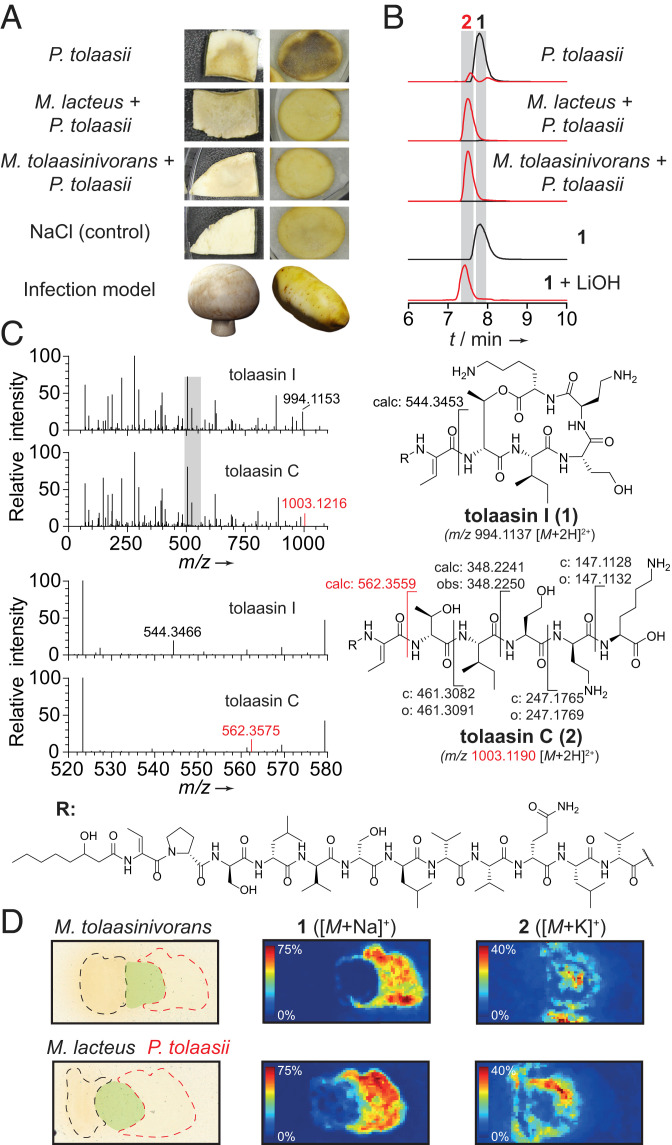
Fig. 2.
Helper bacteria inactivate the toxin produced by the mushroom pathogen. (A) In vivo infection assays with mushroom and potato tuber slices. (B) Extracted-ion chromatogram (EIC) traces of 1 (black, 994.1137 [M + 2H]2+) and 2 (red, 1003.1190 [M + 2H]2+) obtained from high-performance liquid chromatography (HPLC) analysis of culture extracts, isolated 1, and 1 hydrolyzed with 5 eq LiOH, 1 h, RT. (C) Comparison of MS/MS fragmentation patterns of 1 and 2 with highlighted key fragments. Gray bar indicates magnified area. c: calculated; o: observed masses. (D) MALDI imaging of solid cultures of P. tolaasii with M. lacteus, and M. tolaasinivorans, respectively, showing spatial distribution of 1 and 2. Black: colony of M. tolaasinivorans and M. lacteus, respectively; red: colony of P. tolaasii; green: mixed colonies.
To shed light on the chemical fate of the peptide toxin, we monitored the metabolic profiles of liquid cocultures by HPLC-MS. In the axenic P. tolaasii culture, we detected 1 as the main component of a mixture of known tolaasins (22). In contrast, we found that the titers of 1 were drastically reduced in cocultures with each of the helper bacteria (Fig. 2B). The intensity of a double-charged ion with a m/z difference of 9 was dominant. This mass difference of 18 Da in the uncharged molecule could result from the hydration of one of the dehydrobutyrine units or the hydrolysis of one of the amide bonds in the cyclic part of 1. Tandem mass spectrometry (MS/MS) measurements ruled out these possibilities and unequivocally showed that the lactone is cleaved to yield the linearized congener known as tolaasin C (2, Fig. 2C) (22). A similar cleavage potential has been implicated for Microbacterium sp. K3–5 based on MS data (15). We confirmed the identity of 2 by comparison of its retention time with an authentic reference obtained by hydrolysis of 1 with lithium hydroxide (Fig. 2B).
To corroborate these findings on solid media and to dissect microbial interaction processes, we monitored bacterial populations by MALDI imaging. In the colony of the pathogen, 1 is detected as the main product. In addition, small amounts of 2 are colocated with 1, which was not unexpected since 2 occurs naturally in axenic P. tolaasii cultures (22). In the zone where P. tolaasii was mixed with M. tolaasinivorans or M. lacteus, no 1 was detected but high amounts of 2 were (Fig. 2D), clearly indicating that both types of helper bacteria are able to cleave the toxin.
When examining the phenotypes of the cocultures, we noted that the helper bacteria also impair the swarming ability of the pathogen. When grown axenically, P. tolaasii colonies spread uniformly in every direction, forming faint concentric circles that likely indicate different growth states. When coinoculated with M. tolaasinivorans or M. lacteus, however, the swarming diameter of P. tolaasii is reduced to approximately half the size of axenic colonies (Fig. 3A, see also SI Appendix, Fig. S3). Furthermore, marked phenotypic changes, such as zone formation and three-dimensional biofilmlike structures were observed (SI Appendix, Fig. S4).
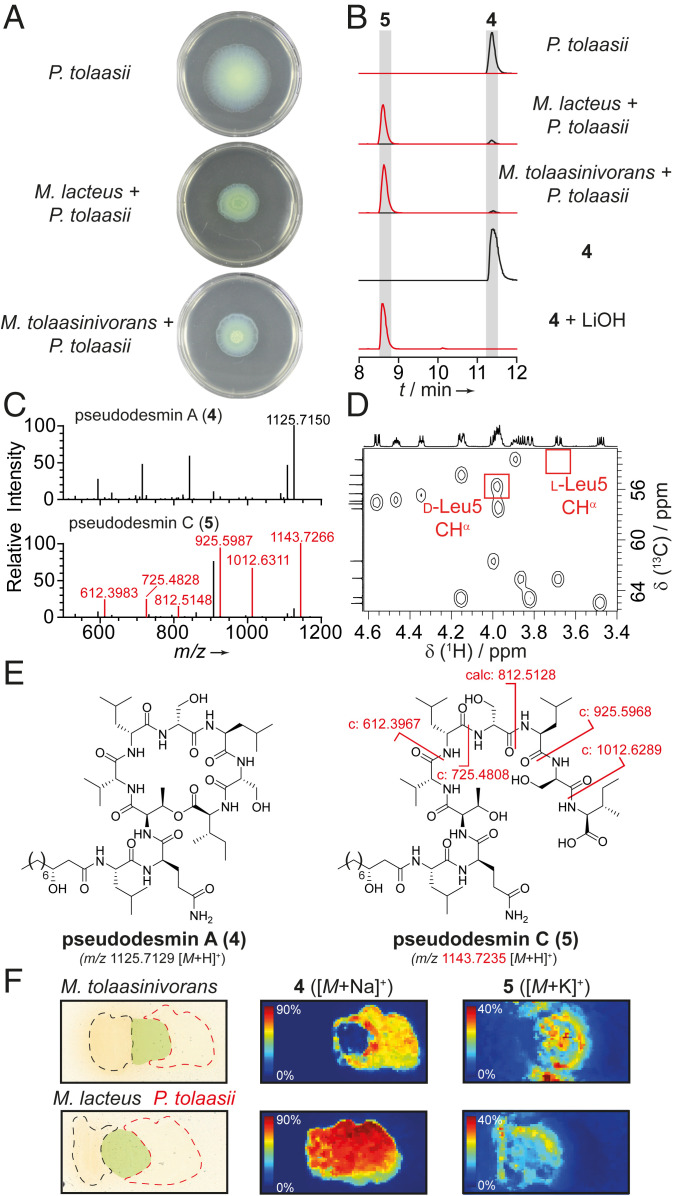
Fig. 3.
Cleavage of 4 impairs dissemination of the pathogen. (A) Swarming assays of P. tolaasii and respective cocultures on soft Kings B agar. (B) EIC traces of 4 (black, 1125.7129 [M + H]+) and 5 (red, 1143.7235 [M + H]+) obtained from HPLC analysis of culture extracts, isolated 4, and 4 hydrolyzed with 5 eq LiOH, 1 h, RT. (C) MS/MS analysis of 4 and 5 with key fragments highlighted in red. (D) Heteronuclear single quantum coherence correlations of the CHα region of 4 with red boxes indicating expected signals for d-Leu CHα and l-Leu CHα in position 5 based on literature data. (E) Structures of 4 and 5 with key fragments highlighted in red. c: calculated masses of fragments. (F) MALDI imaging of cultures of P. tolaasii with M. lacteus or M. tolaasinivorans, showing spatial distribution of 4 and 5. Black: colony of helper bacteria; red: colony of pathogen; green: mixed colonies.
Metabolic profiling and MALDI imaging of the cocultures yielded further insight into the chemical basis of this phenomenon. In liquid cocultures, we observed the emergence of a new compound with m/z 1142 [M – H]– that is not detectable in the requisite axenic cultures (Fig. 3B). MS/MS experiments revealed a fragmentation pattern typical of a peptide and an amino acid sequence that matches with viscosinamide A (3) and pseudodesmin A (4), two related CLPs that only differ in the absolute configuration of the leucine residue at position 5 (SI Appendix, Fig. S5) (23, 24). A mass difference of 18 Da compared to the reported CLPs, and the MS/MS fragmentations (Fig. 3C) implied that the observed species corresponds to the linearized form of one of these molecules. To determine its structure, we isolated the CLP from an up-scaled P. tolaasii culture (2 L). By analysis of the NMR spectra and comparison of 1H and 13C chemical shifts with reference data (24, 25), we found that the isolated CLP is 4 (Fig. 3D, see also SI Appendix), a member of the viscosin family of biosurfactants involved in bacterial swarming and motility (24). We named the linearized congener pseudodesmin C (5) (Fig. 3E) and confirmed its identity further by comparison with an authentic reference generated by hydrolysis of pure 4 with aqueous LiOH (Fig. 3B) as well as by evaluation of NMR data (SI Appendix).
To address whether the cleavage of 4 into 5 in coculture impairs the swarming of P. tolaasii, we scrutinized individual and coinoculated colonies with MALDI imaging (Fig. 3F). In stark contrast to the axenic P. tolaasii colony, intact 4 could not be detected in the zone where P. tolaasii and M. tolaasinivorans interacted. In lieu of the cyclopeptide, high levels of 5 accumulated in this area. In the coculture of P. tolaasii with M. lacteus, we still observed 4 as major congener. However, the linear 5 was also formed in the mixed zone of the cultures. These results show that the presence of helper bacteria leads to the linearization of the surfactant 4 and, therefore, to a reduction of the swarming ability of the pathogen.
To unambiguously demonstrate that intact 4 is essential and sufficient for swarming, we generated a pseudodesmin-deficient P. tolaasii mutant. The putative pseudodesmin biosynthesis gene cluster (BGC) was identified by means of antiSMASH (26) analysis and predicted substrate specificities of the adenylation domains (SI Appendix, Fig. S14). To confirm the cluster identity and to create a nonproducing strain, we deleted a 1.7 kb fragment in the nonribosomal peptide synthetase (NRPS)-encoding gene pseB by homologous double crossover (Fig. 4A). Colonies of the verified null producer (ΔpseB mutant) were unable to swarm (Fig. 4B). To confirm the individual roles of the two CLPs, we also deleted a 1.7 kb segment within the NRPS gene (tolA) of the tolaasin biosynthetic locus that is homologous to the tolaasin BGC in Pseudomonas costantinii (SI Appendix, Fig. S15) (1). The resulting ΔtolA mutant did not cause browning or other visible disease symptoms in any of the tested model systems (Fig. 4B), once again corroborating the key role of 1 in pathogenesis. Whereas the swarming behavior of the tolaasin-deficient mutant was altered, motility was still observed (Fig. 4B). Thus, this surface-active lipopeptide can be considered as a swarming-enhancing factor. In contrast, 4 is essential and sufficient for bacterial motility on soft agar and might be beneficial in colonizing the hydrophobic mushroom caps. The hydrolytic agents produced and released by M. tolaasinivorans and M. lacteus, therefore, not only inactivate the pore-forming toxin secreted by P. tolaasii, but also impair bacterial movement by cleaving its main motility factor.
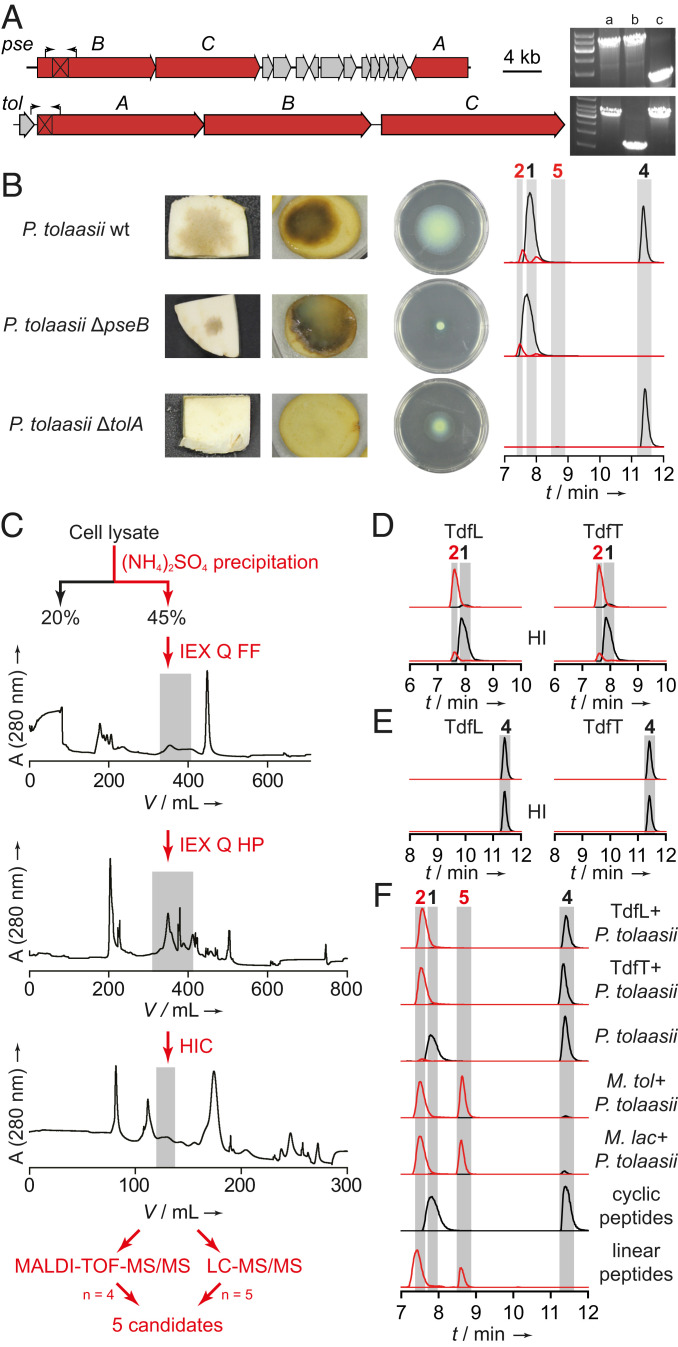
Fig. 4.
Phenotypes of P. tolaasii knockout mutants, and identification of hydrolytic principle. (A) Gene clusters coding for pseudodesmin (pse) and tolaasin (tol) biosynthesis. Black boxes mark deleted areas, arrows indicate primer binding sites used for knockout verification. Agarose gel shows obtained amplicons from (a) P. tolaasii wild type; (b) P. tolaasii ΔtolA; (c) P. tolaasii ΔpseB. (B) Pathogenicity and swarming ability of P. tolaasii strains deficient in the production of tolaasin and pseudodesmin; EIC traces of 1, 2, 4, and 5. (C) Exemplary flow chart of bioactivity-guided fractionation approach. IEX: anion-exchange chromatography; Q: quarternary ammonium anion exchanger; FF: sepharose fast flow; HP: high performance; HIC: hydrophobic interaction chromatography. Gray boxes indicate active fractions that were further fractionated. (D) EIC traces of 1 (black, 994.1137 [M + 2H]2+) and 2 (red, 1003.1190 [M + 2H]2+) obtained from HPLC analyses of in vitro assays of TdfL and TdfT using 1 as a substrate. HI: heat-inactivated. (E) EIC traces of 4 (black, 1125.7129 [M + H]+) and 5 (red, 1143.7235 [M + H]+) obtained from HPLC analysis of in vitro assays of TdfL and TdfT using 4 as a substrate. (F) EIC traces of 1, 2, 4, and 5. In all LC traces: black: cyclic peptides 1 and 4; red: linearized peptides 2 and 5.
In order to identify the hydrolytic factor, we investigated the cell lysate of a mixed culture of M. tolaasinivorans and M. lacteus as these are the native bacterial flora of cultivated mushroom (16). Whereas tolaasins were cleaved by treatment with the crude cell lysate, no conversion was observed using heat-inactivated (HI) aliquots of the same lysate, indicating that an enzyme is responsible for the lactone cleavage. The obtained cytosolic proteome was then fractionated by ammonium sulfate precipitation, and active fractions were further subjected to fast protein liquid chromatography (FPLC)-based separations using different anion exchange columns and hydrophobic interaction chromatography as well as size exclusion chromatography (Fig. 4C).
Active fractions were analyzed using MALDI-time of flight and LC-MS in combination with the in silico proteome generated from genomic sequences of both strains. Then, we filtered the hit lists for basic local alignment search tool (BLAST)-annotated hydrolases/peptidases, and hits occurring in at least four out of nine analyses were chosen for heterologous production in Escherichia coli. The majority of hits derived from M. lacteus and all hits from M. tolaasinivorans were represented by homologs. Therefore, we focused on the production of five different annotated peptidases from M. lacteus (Fig. 4C). All candidates were produced in E. coli using a pET-28-based expression system, yet only one of them cleaves 1 (Fig. 4D, also see SI Appendix, Figs. S16 and S17) and was, thus, named tolaasin-degrading factor of M. lacteus (TdfL). A BLAST search revealed that the closest homologs of TdfL are also encoded in the genomes of Mycetocola tolaasinivorans and Mycetocola saprophilus (pairwise identity of 80.7% and 88.8%, respectively), both identified as tolaasin detoxifiers. TdfL is annotated as a serine protease of the S9 prolyl oligopeptidase family, and we could identify the catalytic triad (Ser, Asp, His) in all three homologs (SI Appendix, Fig. S19).
To prove that these enzymes have the same activity, we heterologously produced the homolog encoded in the M. tolaasinivorans genome, named tolaasin-degrading factor of M. tolaasinivorans (TdfT) and tested the His-tagged protein in the in vitro assay. Like TdfL, TdfT effectively cleaves the lactone bond of 1, whereas the HI aliquot is inactive (negative control) (Fig. 4D). Since neither TdfL nor TdfT showed any hydrolytic activity toward 4 (Fig. 4E), we concluded that these hydrolases are substrate specific detoxifiers and that the helper bacteria harbor at least one other hydrolytic enzyme responsible for the cleavage of the swarming factor 4.
To exclude that 4 is hydrolyzed indirectly by a nonenzymatic reaction, we compared the activity of heat-inactivated cultures of both Mycetocola strains with that of nondenatured cell suspensions toward hydrolysis of 4. For both strains, no formation of 5 could be observed using the heat-treated cultures, indicating that the cleavage of 4 is facilitated by an enzyme (SI Appendix, Fig. S20). We further supplemented growth medium with purified TdfL or TdfT, respectively, prior to inoculation with P. tolaasii to check whether the hydrolytic activity is maintained under cultivation conditions. LC-MS analyses of the culture extracts showed the same activity pattern as in the in vitro assays, i.e., linearization of 1 but not 4 (Fig. 4). These results illustrate that hydrolysis of 4 is performed by an independent enzyme and not indirectly, for example, via change in the pH in the medium (pH of cocultures 7.5–7.7).
Conclusion
In conclusion, we elucidated the role of the mushroom helper bacteria M. tolaasinivorans and M. lacteus that protect A. bisporus from brown blotch caused by P. tolaasii. For both helper strains, we identified homologous enzymes that cleave tolaasins, the major virulence factors of the plant and mushroom pathogen P. tolaasii, yielding the inactive linear forms. M. tolaasinivorans and M. lacteus not only disarm the causative agent of the mushroom disease, but also hamper the colonization of the hydrophobic mushroom cap by linearizing a second cyclic lipopeptide, 4 (Fig. 5). We assigned a biosynthetic gene cluster and a function to 4, which is essential for swarming and is, therefore, crucial for the dissemination of the pathogen. Our findings are an important addition to the currently known enzymatic strategies of bacteria to sabotage microbial virulence. The enzymatic cleavage of a lipopeptide has been reported as a mechanism that confers resistance in competing bacterial species (27), and lactonases are employed by bacteria to interfere with other species' quorum sensing signaling (28). Yet, the observation that helper bacteria halt and disarm pathogens attacking their host is unprecedented. In this ecological context, the enzymatic cleavage of cyclolipopeptides is a remarkable antivirulence strategy, which is in line with rational approaches to block pathogenesis and chemotaxis (29–34). Understanding the mechanism by which helper bacteria protect their host not only sets the basis for the development of biocontrol strains used in agriculture, but also may inspire the design of protective symbionts for use in medicine.
Fig. 5.
P. tolaasii produces pseudodesmins as swarming agents and tolaasins as toxins against A. bisporus. In the presence of protective helper bacteria (Mycetocola spp.) the virulence factors are cleaved by enzymes, such as TdfL or TdfT. One letter code used to represent amino acids in the peptides. B: dehydrobutyrine; X: 2,4-diaminobutanoic acid; Z: homoserine.
Materials and Methods
Detailed materials and experimental methods can be found in the SI Appendix together with additional tables and figures. Genome sequences used in this study can be accessed via NCBI accession numbers NZ_RCUX00000000 (M. tolaasinivorans), NZ_RCUY00000000 (M. lacteus), NZ_PHHD00000000, and NZ_CP020369 (both P. tolaasii).
Supplementary Material
Acknowledgments
We thank A. Perner for LC-HRMS(/MS) measurements, B. Urbansky for support in the generation of expression plasmids, H. Heinecke for NMR measurements, and T. Krüger for tryptic peptide LC-MS/MS. Financial support by the Deutsche Forschungsgemeinschaft (SFB 1127 ChemBioSys, and Leibniz Award to C.H.) and the Alexander von Humboldt Foundation (Postdoctoral Research Fellowship to A.J.K.) is gratefully acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006109117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix. Genome sequences used in this study can be accessed in the National Center for Biotechnology Information (NCBI) GenBank, https://www.ncbi.nlm.nih.gov/genbank/ [accession nos. NZ_RCUX00000000 (M. tolaasinivorans), NZ_RCUY00000000 (M. lacteus), NZ_PHHD00000000, and NZ_CP020369 (both P. tolaasii)].
References
- 1.Scherlach K. et al., Biosynthesis and mass spectrometric imaging of tolaasin, the virulence factor of brown blotch mushroom disease. ChemBioChem 14, 2439–2443 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Thongkongkaew T. et al., Two types of threonine-tagged lipopeptides synergize in host colonization by pathogenic Burkholderia species. ACS Chem. Biol. 13, 1370–1379 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Graupner K. et al., Imaging mass spectrometry and genome mining reveal highly antifungal virulence factor of mushroom soft rot pathogen. Angew. Chem. Int. Ed. Engl. 51, 13173–13177 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Fischer D. et al., Disruption of membrane integrity by the bacterium-derived antifungal jagaricin. Antimicrob. Agents Chemother. 63, e00707–e00719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nutkins J. C. et al., Structure determination of tolaasin, an extracellular lipodepsipeptide produced by the mushroom pathogen, Pseudomonas tolaasii Paine. J. Am. Chem. Soc. 113, 2621–2627 (1991). [Google Scholar]
- 6.Soler-Rivas C., Arpin N., Olivier J. M., Wichers H. J., The effects of tolaasin, the toxin produced by Pseudomonas tolaasii on tyrosinase activities and the induction of browning in Agaricus bisporus fruiting bodies. Physiol. Mol. Plant Pathol. 55, 21–28 (1999). [Google Scholar]
- 7.Soler-Rivas C., Arpin N., Olivier J. M., Wichers H. J., Activation of tyrosinase in Agaricus bisporus strains following infection by Pseudomonas tolaasii or treatment with a tolaasin-containing preparation. Mycol. Res. 101, 375–382 (1997). [Google Scholar]
- 8.Cho K. H., Kim S. T., Kim Y. K., Purification of a pore-forming peptide toxin, tolaasin, produced by Pseudomonas tolaasii 6264. J. Biochem. Mol. Biol. 40, 113–118 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Cho K.-H., Kim Y.-K., Two types of ion channel formation of tolaasin, a Pseudomonas peptide toxin. FEMS Microbiol. Lett. 221, 221–226 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Gea F. J., Navarro M. J., “Mushroom diseases and control” in Edible and Medicinal Mushrooms: Technology and Applications, Diego C. Z., Pardo-Gimenez A., Eds. (John Wiley & Sons Ltd., 2017), pp. 239–259. [Google Scholar]
- 11.Royse D. J., Baars J., Tan Q., “Current overview of mushroom production in the world” in Edible and Medicinal Mushrooms, Diego C. Z., Pardo‐Giménez A., Eds. (John Wiley & Sons Ltd., 2017), pp. 5–13. [Google Scholar]
- 12.Tanprasert P., Reed B. M., Detection and identification of bacterial contaminants from strawberry runner explants. In Vitro Cell. Dev. Biol. Plant 33, 221–226 (1997). [Google Scholar]
- 13.Lo Cantore P., Giorgio A., Iacobellis N. S., Bioactivity of volatile organic compounds produced by Pseudomonas tolaasii. Front. Microbiol. 6, 1082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxon E. B., Jackson R. W., Bhumbra S., Smith T., Sockett R. E., Bdellovibrio bacteriovorus HD100 guards against Pseudomonas tolaasii brown-blotch lesions on the surface of post-harvest Agaricus bisporus supermarket mushrooms. BMC Microbiol. 14, 163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita S. et al., Detoxification process of tolaasins, lipodepsipeptides, by Microbacterium sp. K3-5. Biosci. Biotechnol. Biochem. 82, 1455–1458 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto T., Takeuchi M., Shida O., Murata H., Shirata A., Proposal of Mycetocola gen. nov. in the family Microbacteriaceae and three new species, Mycetocola saprophilus sp. nov., Mycetocola tolaasinivorans sp. nov. and Mycetocola lacteus sp. nov., isolated from cultivated mushroom, Pleurotus ostreatus. Int. J. Syst. Evol. Microbiol. 51, 937–944 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Tsukamoto T., Shirata A., Murata H., Isolation of a gram-positive bacterium effective in suppression of brown blotch disease of cultivated mushrooms, Pleurotus ostreatus and Agaricus bisporus, caused by Pseudomonas tolaasii. Mycoscience 39, 273–278 (1998). [Google Scholar]
- 18.Soler‐Rivas C., Arpin N., Olivier J., Wichers H., WLIP, a lipodepsipeptide of Pseudomonas “reactans,” as inhibitor of the symptoms of the brown blotch disease of Agaricus bisporus. J. Appl. Microbiol. 86, 635–641 (1999). [Google Scholar]
- 19.Lee C.-J. et al., Isolation of the bacterium Pseudomonas sp. HC1 effective in inactivation of tolaasin produced by Pseudomonas tolaasii. Kor. J. Mycol. 41, 248–254 (2013). [Google Scholar]
- 20.Lo Cantore P. et al., Biological characterization of white line-inducing principle (WLIP) produced by Pseudomonas reactans NCPPB1311. Mol. Plant Microbe Interact. 19, 1113–1120 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Coraiola M. et al., WLIP and tolaasin I, lipodepsipeptides from Pseudomonas reactans and Pseudomonas tolaasii, permeabilise model membranes. Biochim. Biophys. Acta 1758, 1713–1722 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Bassarello C. et al., Tolaasins A–E, five new lipodepsipeptides produced by Pseudomonas tolaasii. J. Nat. Prod. 67, 811–816 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Nielsen T. H., Christophersen C., Anthoni U., Sørensen J., Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 87, 80–90 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Sinnaeve D. et al., Structure and X-ray conformation of pseudodesmins A and B, two new cyclic lipodepsipeptides from Pseudomonas bacteria. Tetrahedron 65, 4173–4181 (2009). [Google Scholar]
- 25.Geudens N. et al., Impact of a stereocentre inversion in cyclic lipodepsipeptides from the viscosin group: A comparative study of the viscosinamide and pseudodesmin conformation and self-assembly. ChemBioChem 15, 2736–2746 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Blin K. et al., antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoefler B. C. et al., Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc. Natl. Acad. Sci. U.S.A. 109, 13082–13087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fast W., Tipton P. A., The enzymes of bacterial census and censorship. Trends Biochem. Sci. 37, 7–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escaich S., Antivirulence as a new antibacterial approach for chemotherapy. Curr. Opin. Chem. Biol. 12, 400–408 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Brannon J. R., Hadjifrangiskou M., The arsenal of pathogens and antivirulence therapeutic strategies for disarming them. Drug Des. Devel. Ther. 10, 1795–1806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickey S. W., Cheung G. Y. C., Otto M., Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson B. K., Abramovitch R. B., Small molecules that sabotage bacterial virulence. Trends Pharmacol. Sci. 38, 339–362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakemeyer M., Zhao W., Mandl F. A., Hammann P., Sieber S. A., Thinking outside the box-novel antibacterials to tackle the resistance crisis. Angew. Chem. Int. Ed. Engl. 57, 14440–14475 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Erhardt M., Strategies to block bacterial pathogenesis by interference with motility and chemotaxis. Curr. Top. Microbiol. Immunol. 398, 185–205 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix. Genome sequences used in this study can be accessed in the National Center for Biotechnology Information (NCBI) GenBank, https://www.ncbi.nlm.nih.gov/genbank/ [accession nos. NZ_RCUX00000000 (M. tolaasinivorans), NZ_RCUY00000000 (M. lacteus), NZ_PHHD00000000, and NZ_CP020369 (both P. tolaasii)].