Significance
Rhizobia are soil-dwelling bacteria that form symbioses with legumes and provide biologically useable nitrogen as ammonium for the host plant. High-throughput DNA sequencing has led to a rapid expansion in publication of complete genomes for numerous rhizobia, but analysis of gene function increasingly lags gene discovery. Mariner-based transposon insertion sequencing has allowed us to characterize the fitness contribution of bacterial genes and determine those functionally important in a Rhizobium–legume symbiosis at multiple stages of development.
Keywords: Rhizobium, legume, N2 fixation, nodulation, rhizosphere
Abstract
By analyzing successive lifestyle stages of a model Rhizobium–legume symbiosis using mariner-based transposon insertion sequencing (INSeq), we have defined the genes required for rhizosphere growth, root colonization, bacterial infection, N2-fixing bacteroids, and release from legume (pea) nodules. While only 27 genes are annotated as nif and fix in Rhizobium leguminosarum, we show 603 genetic regions (593 genes, 5 transfer RNAs, and 5 RNA features) are required for the competitive ability to nodulate pea and fix N2. Of these, 146 are common to rhizosphere growth through to bacteroids. This large number of genes, defined as rhizosphere-progressive, highlights how critical successful competition in the rhizosphere is to subsequent infection and nodulation. As expected, there is also a large group (211) specific for nodule bacteria and bacteroid function. Nodule infection and bacteroid formation require genes for motility, cell envelope restructuring, nodulation signaling, N2 fixation, and metabolic adaptation. Metabolic adaptation includes urea, erythritol and aldehyde metabolism, glycogen synthesis, dicarboxylate metabolism, and glutamine synthesis (GlnII). There are 17 separate lifestyle adaptations specific to rhizosphere growth and 23 to root colonization, distinct from infection and nodule formation. These results dramatically highlight the importance of competition at multiple stages of a Rhizobium–legume symbiosis.
Biological N2 fixation carried out by bacteria provides ∼65% of the biosphere’s available nitrogen (1), with the single greatest contribution coming from the symbioses between soil-dwelling bacteria of the Rhizobiaceae family and plants of the legume family (1–3). Rhizobia provide the plant with ammonium (NH4+) and receive carbon sources as an energy supply in return (4). This effectively functions as a biofertilizer for the legume host, increasing plant growth without the need for exogenously applied nitrogen (3, 5). During symbiosis, rhizobia must adapt to several different lifestyles. These range from free-living growth in the rhizosphere, through root attachment and colonization, to passage along infection threads, differentiation into bacteroids that fix N2, and, finally, bacterial release from nodules at senescence.
Several decades of intense research have gone into understanding the mechanistic details of signaling between rhizobia and legumes leading to nodulation (6–8). Rhizobia respond to plant flavonoids by synthesizing lipochitooligosaccharides (LCOs) that are detected by plant LysM receptors to initiate nodule morphogenesis and concomitant rhizobial infection (8–11). Rhizobia typically, but not always, attach to the tips of developing root hairs, triggering root hair curling and entrapment of bacteria in an infection pocket (7). The plant cell plasma membrane invaginates to form an infection thread, down which bacteria grow toward the base of the root hair cell through continuous division at the leading edge. Infection is usually a clonal event (12), selecting one Rhizobium and allowing it to multiply to high numbers within the ramifying threads. Infection threads deliver rhizobia to the root cortex of the developing nodule. In symbiosis between Rhizobium leguminosarum bv. viciae and pea, bacteria are internalized by nodule cells and undergo terminal differentiation into N2-fixing bacteroids (10). Nodules provide a protective microaerobic environment to maintain oxygen-labile nitrogenase (6). In exchange for NH4+ and alanine, the legume host provides carbon sources to fuel this process, primarily as dicarboxylic acids (13, 14).
However, nodule infection is only one stage of the lifestyle of rhizobia, and they spend much of their time surviving in the rhizosphere, the zone of soil immediately surrounding roots (15). Plants can release up to a fifth of their photosynthate via their roots (16), making the rhizosphere a nutritionally rich zone, but one where there is strong selective pressure and competition for resources. Attachment and subsequent colonization of roots by rhizobia is almost certainly important for their long-term survival, but it is unclear how important these steps are to infection of legumes (17). Research over several decades has enabled identification of around 8 nif and 27 fix genes required directly for N2 fixation, as well as genes essential for recognition by the plant, differentiation, and energization of N2 fixation. However, it has long been speculated these probably represent a fraction of genes needed by rhizobia for successful survival and growth in competition with other bacteria.
Rhizobiaceae have large genomes that probably reflect their adaptation to the heterogenous soil environment and dramatic lifestyle and developmental changes undergone in symbiosis (18, 19). The 7.75-megabase genome of R. leguminosarum bv. viciae Rlv3841 consists of a circular chromosome (4,788 genes) and 6 plasmids: pRL12 (790 genes), pRL11 (635 genes), the symbiosis plasmid pRL10 (461 genes), pRL9 (313 genes), pRL8 (141 genes), and pRL7 (189 genes). Here, we perform a multistage mariner transposon-insertion sequencing (INSeq) genetic screen of Rlv3841 engaging in symbiosis with pea (Pisum sativum) at four different symbiotic lifestyle stages: 1) growth in the rhizosphere, 2) root colonization, 3) undifferentiated nodule bacteria, and 4) terminally differentiated bacteroids. INSeq is a powerful and robust technique that can be used to functionally annotate bacterial genomes (20–24). Libraries of mariner transposon-insertion mutants are analyzed by high-throughput sequencing to assess the effect of mutation of a gene on growth and survival of the bacterium. This has allowed us to identify the genetic requirements for competitiveness at multiple developmental stages during symbiosis.
Results and Discussion
Mariner Library Construction, Sequencing, and Analysis.
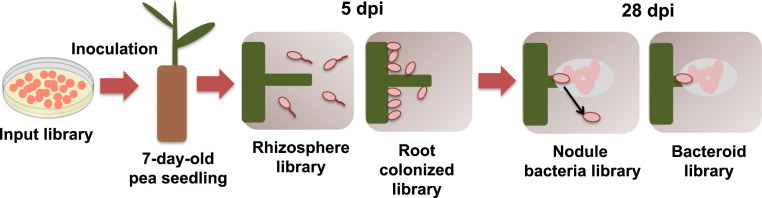
Rlv3841 mariner-insertion mutants (input1) were inoculated onto 7-d-old pea seedlings and subsequently recovered at 5 d postinoculation (dpi) to construct two libraries for INSeq analysis: 1) rhizosphere and 2) roots (Fig. 1). A second mutant library (input2) was inoculated onto 7-d-old pea seedlings and subsequently recovered at 28 dpi from nodules. A total of 150,000 nodules were picked from ∼1,500 plants, and a further two libraries for INSeq analysis were constructed: 3) undifferentiated nodule bacteria, generated by growing bacteria from crushed nodules (which will represent nodule bacteria that are not terminally differentiated along with those recently released from infection threads) for 12 h in rich medium before extracting DNA; and 4) bacteroids, generated from DNA extracted directly from nodules (Fig. 1). A total of six INSeq libraries (input 1, input 2, rhizosphere, roots, nodule bacteria, and bacteroids) were sequenced and analyzed by a Hidden Markov Model (HMM) (25). The HMM assigns genes to one of four classification states based on the fitness of mutants inferred from their population frequency in the sequenced library (25). Each of the 7,357 Rlv3841 genes that contain TA insertion sites (99.9% of the genome) was classified as essential (ES) (tolerates no or very few insertions), defective (DE) (insertion impairs fitness), neutral (NE) (insertion has a neutral impact on fitness), or advantaged (AD) (insertion enhances fitness) (Dataset S1) (25). Genes classified as NE in the respective input library (6,429 for input1 and 6,656 for input2) were considered for each test condition, with genes classified as either ES or DE (ES/DE) regarded as required at that stage of symbiosis and described thus throughout this paper.
Fig. 1.
Collection steps following inoculation of pea plants with the INSeq input libraries. Bacterial DNA was purified from 1) the rhizosphere (5 dpi) and 2) colonized roots (5 dpi) following inoculation with input1; and from 3) nodule bacteria (28 dpi) and 4) bacteroids (28 dpi) following inoculation with input2. These four samples, together with DNA purified from input1 and input2, were used to make libraries for INSeq analysis.
Bacteria with a mutation that results in out-competition at any stage of early nodule formation will occupy fewer nodules on peas and lead to an ES/DE classification. However, many mutants unable to fix N2, such as those defective in nifH (encoding Fe protein of nitrogenase), do not alter the competitiveness for nodule infection. Nonetheless, plant sanctioning results in the formation of smaller nodules (26). We therefore predicted at the outset of this work, and subsequently confirm, that with mutants impaired specifically in N2 fixation but not nodule formation, the result is an ES/DE classification for the gene in bacteroids.
Classification of Symbiotic Fitness Determinants.
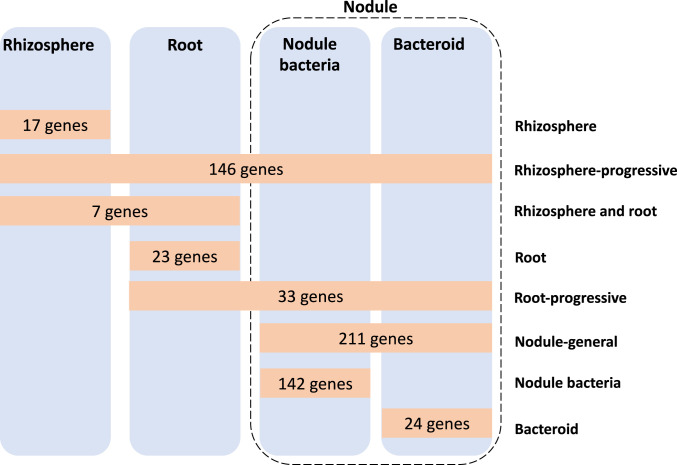
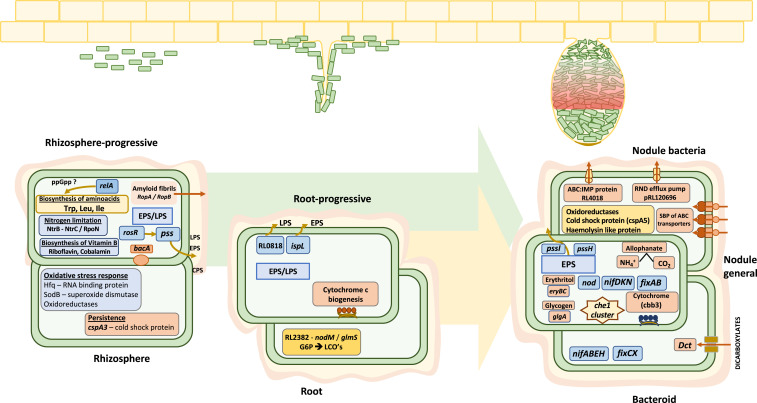
Across all stages of symbiosis, INSeq reveals a total of 603 genetic regions (593 genes, 5 transfer RNAs [tRNAs], and 5 RNA features) are required to competitively nodulate peas (Fig. 2). A total of 17 are required only in the rhizosphere (SI Appendix, Table S1), while 146 are classified as rhizosphere-progressive (SI Appendix, Table S2), required not only in the rhizosphere but also at every subsequent stage. Seven genes are required for both rhizosphere and root colonization (rhizosphere and root) (SI Appendix, Table S3), with 23 genes specifically required for root colonization (root) (SI Appendix, Table S4). Genes classified as rhizosphere- or root-required are important for fitness and persistence in those environments but not linked to nodule infection, per se. There are 33 genes required from root colonization to nodule (root-progressive) (SI Appendix, Table S5), while 211 genes are required by both nodule bacteria and bacteroids (nodule-general) (SI Appendix, Table S6). There are an additional 142 specific for nodule bacteria, i.e., required only for regrowth of bacteria from crushed nodules (SI Appendix, Table S7), with 24 specific for bacteroids (direct isolation from nodules without regrowth) (SI Appendix, Table S8 and Fig. 2).
Fig. 2.
Rlv3841 genes required during the stages of symbiosis with pea. Orange boxes show the number at each symbiotic stage: rhizosphere (17 genes) (SI Appendix, Table S1), rhizosphere-progressive (146 genes) (SI Appendix, Table S2), rhizosphere and root (7 genes) (SI Appendix, Table S3), root (23 genes) (SI Appendix, Table S4), root-progressive (33 genes) (SI Appendix, Table S5), nodule-general (211 genes) (SI Appendix, Table S6), nodule bacteria (142 genes) (SI Appendix, Table S7), and bacteroid (24 genes) (SI Appendix, Table S8). All genes shown were classified as NE in the respective input library.
Rhizosphere Genes.
The rhizosphere, the zone immediately surrounding plant roots, is an oxidative environment formed through the release of reactive oxygen species (27, 28). Mutation of the 17 genetic regions required specifically in the rhizosphere (SI Appendix, Table S1) did not impair subsequent stages of root colonization or nodulation (Fig. 2). Rhizosphere-specific genes involved in the oxidative-stress response, important for survival in this environment, encode a putative oxidoreductase (pRL110539), a superoxide dismutase (RL1340 [sodB]), and an RNA-binding protein Hfq (RL2284) involved in control of posttranscriptional gene regulation. Persistence in the rhizosphere also required RL2964 (cspA3), encoding a cold shock protein. This rhizosphere-specific group also includes a single tRNA (tRNAGln) and five genes predicted to encode metabolic or biosynthetic functions for pRL80090, a fucose operon protein; pRL90136, a glycosyl transferase; pRL90204, an amidase; pRL100145, an acyl-CoA dehydrogenase; and RL3335, a lysophospholipase.
Rhizosphere-Progressive Genes.
The 146 rhizosphere-progressive genes (Fig. 2 and SI Appendix, Table S2), required in the rhizosphere but also at subsequent stages, include genes for the biosynthesis of tryptophan (RL0021 [trpB], RL0022 [trpA], RL2493 [trpD], RL2494 [trpC], and RL3521 [trpE]), riboflavin (RL1621 [ribG] and RL1622 [ribC]), leucine (RL3513 [leuA1], RL4707 [leuB], and RL4732 [leuS]), isoleucine (RL1803 [ilvD1], RL3205 [ilvC], and RL3245 [ilvI]), and cobalamin (pRL110632 [cobF], RL2781 [cobU], RL2781A [cobV], RL2829 [cobO], RL4348 [cobT], and RL4349 [cobS]). There may be a specific requirement for these biosynthetic genes in the rhizosphere, with mutants subsequently uncompetitive at later stages of symbiosis, or, alternatively, they may be required throughout all symbiotic stages. Riboflavin availability influences rhizobial survival in the rhizosphere and root colonization. An R. leguminosarum mutant in ribN (encoding putative riboflavin transporter protein RibN) showed lower nodule occupancy in pea plants (29).
The outer surface of rhizobia, composed of a variety of polysaccharides, including lipopolysaccharide (LPS) and exopolysaccharide (EPS) (17), is critical for both attachment to root hairs and host-specific recognition. Rhizosphere-progressive genes include those encoding putative proteins: O-antigen ligase (pRL90053); EPS production protein (pRL110389), with high homology (97% identity [id]) to PssF of R. leguminosarum TA1 involved in polysaccharide export (30); and O-antigen transporter (RL0821). In addition, both RL3651 (pssG) and RL3664 (pssN) have well-defined roles in EPS biosynthesis (30), and their requirement, alongside pRL110389 (pssF) and RL1600 (ppx), highlights the importance of correct EPS processing not only in nodule infection but in all stages of interaction with the plant. The transcriptional regulator RL1379 (rosR), classified as rhizosphere-progressive, has a role in nodulation competitiveness (31). RosR regulates pss genes encoding EPS, and R. leguminosarum rosR mutants produce only one-third of the EPS of wild type (32).
Other requirements include RL1589 (ropB) and RL2775 (ropA), which are suggested to encode outer membrane amyloid fibrils in R. leguminosarum (33). These are conserved across numerous rhizobia (34, 35) and are possibly required for either root attachment or transport. RL2256 (ntrB) and RL2257 (ntrC), encoding the NtrB/NtrC two component regulatory system, control expression of genes in response to nitrogen limitation. While in this work, pea seedlings were grown in nitrogen-free rooting solution to enable nodulation, this pathway is likely to be important in a natural soil environment where nitrogen availability is low. RL0422 (rpoN), the gene encoding RNA polymerase sigma factor 54, is classified as rhizosphere-progressive. Interestingly, this indicates that while essential for nif gene expression and dicarboxylate transport (36) (processes that occur in nodules), it is required for processes in the rhizosphere. Bacterial uptake of solutes is important, shown by the requirement for genes encoding ATP-binding cassette (ABC) transporters of nitrogen-containing ions (RL4400 to RL4402) and ferric cations (RL4583 [fbpB]). Another ABC system is required (RL1004 to RL1003), but the solute transported is unknown. RL3557 (bacA) encoding the BacA transporter protein is well established to be required for bacteroid development (37). BacA protects rhizobia against the antimicrobial activity of nodule-specific cysteine-rich (NCR) peptides produced by the host pea (38, 39), but clearly it is also important at earlier stages of interaction with the plant. The gene encoding the stringent response protein RelA (RL1506 [relA]), which synthesizes ppGpp, was similarly required across all symbiotic stages, indicating its central importance in response to stress.
Root Colonization Genes.
The root library, formed by collecting bacteria from the entire pea root, consists of colonizing bacteria attached over the whole epidermal surface. The genes specific for rhizosphere and root (7 genes; SI Appendix, Table S3) are required for both bacterial rhizosphere growth and root colonization, while 23 genes are required only for root colonization (SI Appendix, Table S4; designated as “Root” in Fig. 2). This latter group is important for attachment to roots (8, 17, 40) but is not involved in nodule formation, indicating a pathway of root colonization, distinct from that of nodulation.
Root-specific genes include those encoding putative cell envelope-related functions for pRL90052, polysaccharide biosynthesis O-antigen–related protein; and RL3663 (pssO), a polysaccharide transport outer membrane protein. These cell envelope-related functions may actively facilitate adherence to the root surface or may be important for general cell fitness during colonization. RL2512, encoding a putative transmembrane protein, was specifically required for root colonization. RL2512 encodes a putative SecG, which forms part of the general protein secretion (Sec) system in bacteria. The protein encoded by RL2512 shows homology to SecG in Escherichia coli (39% id over the amino-terminal 71 amino acids) and Agrobacterium tumefaciens (73% id over 175 amino acids). Potential orthologs of the Sec system have been identified in Rlv3841 by homology with E. coli proteins (41). However, these chromosomal genes were not located in operons or even close to one another. SecG forms a protein-conducting channel in the SecYEG complex (41). Both the putative Rlv3841 genes encoding SecE (RL1759) and SecY (RL1794) were essential in the input libraries as well as across all stages of symbiosis. The different phenotype observed for RL2512 suggests there may be a second gene encoding a SecG-like function in Rlv3841 (although no gene was found with >26% id).
Root-Progressive Genes.
Thirty-three root-progressive genes affect all stages from root colonization onward (Fig. 2 and SI Appendix, Table S5). They include RL2382 (nodM/glmS), encoding a fructose-6-phosphate aminotransferase that forms glucosamine-6-phosphate. There are two homologous nodM genes in R. leguminosarum, RL2382 and pRL100180, encoded on the chromosome and symbiosis plasmid, respectively (42). Biochemical assays demonstrated either protein provides sufficient glucosamine-6-phosphate (G6P) for LCO Nod factor synthesis (42). A double nodM mutant in R. leguminosarum no longer nodulated pea or vetch (42). The two genes fall into different categories in this study: pRL100180 (nodM) is required only by nodule bacteria, while RL2382 (nodM/glmS) is root-progressive. Their different INSeq phenotypes suggest differential expression during symbiosis, consistent with plant-flavonoid induction of nodM.
This root-progressive group also includes two genes encoding putative cell surface constituents for RL0818, a LPS biosynthesis protein; and RL3677 (ispL), a UDP-glucuronate 5′-epimerase required for glycoconjugation in EPS biosynthesis (43). This root-progressive designation suggests these two cell surface–related genes are required to produce the correct bacterial cell surface needed for root colonization, as well as in subsequent steps, potentially root hair attachment, infection thread ramification, or bacteroid formation in nodules. An ABC transporter (RL4538 to RL4539 [ccmB, cycZ]) showing homology with a system exporting heme-binding proteins to the periplasm in Rhizobium etli (44) is required, together with RL4540 (cycY), a cytochrome c biogenesis protein responsible for interchange of thiol:disulfide bonds. This suggests novel cytochrome biosynthesis and export is important during initial root colonization.
Nodule-General Genes.
A total of 211 genes are classified nodule-general (Fig. 2 and SI Appendix, Table S6). These are genes whose mutation impaired recovery from both nodule bacteria (i.e., regrown bacteria) and bacteroids (i.e., isolated from total nodule rhizobacterial DNA without regrowth). This group includes nine nod genes, encoding components for the biosynthesis and signaling pathways of LCO nodulation (Nod) factors (45). LCOs are diffusible molecules, and nod mutants are still able to initiate symbiosis if coinoculated 1:1 with wild type. However, in mass competition, such as this INSeq experiment, nod mutants may either have delayed establishment or reduced cell numbers within nodules, one or both of which could result in a nodule-general classification. The genes are organized in three operons on pRL10: pRL100180 to pRL100178 (nodMNT), pRL100181 to pRL100183 (nodLEF), and pRL100184 to pRL100189 (nodD-nodABCIJ), and in the latter two clusters, every gene (nine in total) is classified as nodule-general (SI Appendix, Table S6). These genes include biosynthesis of Nod factor as well as export by the ABC transporter encoded by pRL100188 (nodI) and pRL100189 (nodJ).
The nitrogenase complex is encoded by genes pRL100162 to pRL100158 (nifHDKEN) and pRL100196 to pRL100195 (nifAB). These genes are classified as nodule-general (nifDK and nifN) (SI Appendix, Table S6) and bacteroid (nifH, nifE, and nifAB) (SI Appendix, Table S8). Although the classification of these latter four as bacteroid specific suggests that their mutation reduces N2-fixing bacteroids but not the level of undifferentiated bacteria within nodules, as these classifications are closely related, the division of functionally related clusters into separate groups should not be overinterpreted.
Electron-transfer flavoproteins (ETFs) transfer electrons to nitrogenase by bifurcation (46). Genes fixABCX (pRL100200 to pRL100197) encoding ETF FixABCX were required in the nodule, although, as above, they fall into two categories: nodule-general (fixAB) (SI Appendix, Table S6) and bacteroid (fixCX) (SI Appendix, Table S8). Again, we might tentatively conclude that while all four genes are needed for symbiotic N2 fixation, mutation of fixC and fixX does not alter the number of viable nodule bacteria. Mutation of ETF proteins abolishes N2 fixation in several rhizobia–legume symbioses (47).
The microaerobic environment required for the efficient functioning of the nitrogenase complex is achieved by the combined activities of plant leghemoglobin and the bacteroid’s high-affinity CBB3 cytochrome complex. Rlv3841 has two clusters of fix genes that encode the components of CBB3, one on the symbiosis plasmid fixN1O1Q1P1G1H1I1S1 (pRL100205 to pRL100211) and the other on pRL9 (pRL90018 to pRL90012 [fixN2O2Q2P2G2H2I2S2]). Only fixN1 and fixN2 were required (SI Appendix, Tables S2 and S6), suggesting the other duplicated components of the CBB3 complex can compensate for each other and, possibly, that the level of FixN limits the activity of the complex.
Differentiation of rhizobia into bacteroids requires significant reprogramming of the LPS and outer membrane (48). Genes with a predicted role in cell envelope functions formed an important group of nodule-general genes. These include RL3649 (pssI) and RL3650 (pssH), encoding putative glycosyltransferases, and RL3820, encoding a putative EPS-production protein. These proteins may restructure the cell surface during infection and bacteroid differentiation. O-antigen biosynthesis also appears to play a role in these structural changes, with pRL110056, encoding a putative UDP-N-acetyl-d-glucosamine 6-dehydrogenase involved in O-antigen biosynthesis classified as nodule-general. As with growth in the rhizosphere and root colonization, stress- and defense-response genes are also required in nodules. These include RL0824, encoding a putative oxidoreductase; RL0962, encoding a putative dioxygenase; and RL0994 and RL1545, encoding putative reductases.
In terms of metabolism, nodule-general genes include pRL100434 to pRL100433, which are predicted to encode allophanate hydrolase, catalyzing the hydrolysis of allophanate yielding ammonium and CO2. Allophanate can be either derived from cyanuric acid degradation or be an alternative to urease in degradation of urea (49), in which urea is first carboxylated to allophanate. Indeed, pRL100436 and pRL100437 (nodule bacteria-specific; SI Appendix, Table S6) are annotated to encode a biotin-dependent carboxylase, which may be a urea carboxylase. Urea carboxylase belongs to the biotin-dependent carboxylase family of enzymes, which has 40% id to urea carboxylase of Saccharomyces cerevisiae and Oleomonas sagaranensis (50). Overall, this suggests urea catabolism is important in nodule bacteria and bacteroids.
Genes pRL100246 and pRL100247, encoding a putative aldehyde dehydrogenase and adjacent conserved hypothetical protein, were nodule-general. This suggests aldehyde metabolism is important for optimal fitness in both nodule bacteria and bacteroids. Soybean nodules contain aldehydes (31 to 53 nM), which are converted to acetate by aldehyde dehydrogenase (51, 52). Erythritol utilization is also required in nodules: pRL120205 (eryB), encoding erythritol phosphate dehydrogenase; and pRL120206 (eryC), encoding erythrulose 4-phosphate dehydrogenase are nodule-general. This agrees with the known reduction in nodule competitiveness of ery mutants (53). RL4117 (glgA) encoding glycogen synthase is nodule-general. Glycogen is an important carbon store in rhizobia, and its disruption impairs growth on glucose-containing molecules (54).
Various transport systems are required in nodules. Genes required include those encoding pRL110047 (permease of ABC exporter), pRL90262 (ABC of a hydrophobic amino acid transporter [HAAT] ABC uptake system), RL0186 to RL0187 (permease components of peptide transporter [PepT] ABC uptake system), RL0226 and RL0228 (permease and solute binding protein [SBP] of a second PepT family), and RL3333 (ABC of an unclassified transporter). The importance of NCR peptides in development of mature bacteroids within N2-fixing nodules is well known (55). While we speculate that the two PepT uptake systems are involved in delivery of plant-derived peptides crucial for plant-controlled bacteroid development, uptake by systems transporting unknown solutes and export of unwanted compounds is clearly also necessary.
Nodule Bacteria Genes.
Although most genes with known or obvious roles in nodule infection are classified as nodule-general or bacteroid, there are an additional 142 genes specific for nodule bacteria (Fig. 2 and SI Appendix, Table S7). This INSeq library is made up of undifferentiated bacteria, likely to come from inside, or recently released from, infection threads, and capable of growth in tryptone–yeast (TY) (fully differentiated bacteroids are unable to regrow in this way). Classification of genes as specific for nodule bacteria may be for one of two reasons: firstly, that they are needed for optimum bacterial growth within infection threads; or, secondly, that they are required for regrowth in TY medium, with implications for importance in bacterial release as nodules senesce.
This list of genes includes several flagella-related genes that are considered below. A few defense- and stress-related genes, including RL0392 (encoding a putative hemolysin-like protein), RL2102 (cspA5) (encoding a putative cold shock protein), and pRL110509 and RL2526 (encoding putative oxidoreductases), are also specific for nodule bacteria. The gene encoding a resistance–nodule–division (RND) family efflux transporter (pRL120696) is required, and its role is likely to be in defense, exporting toxic/unwanted chemicals from bacterial cells. The gene encoding an ABC export system RL4018 (ABC-permease fusion) may also be involved in a similar task. In fact, genes encoding components of several ABC uptake transporters are classified as nodule bacteria-specific, including pRL100062 (permease of polyamine/opine/phosphonate transporter [POPT] family), pRL100072 (SBP of polar amino acid transporter [PAAT] family), pRL120071 (SBP of PAAT, which may transport galactosamine, glucosamine given its high homology to SMb21135) (56), pRL120560 (ABC carbohydrate uptake transporter 1 [CUT1] family), RL2659 (SBP, class unclassified), and RL3353 (ABC, ferric iron transporter [FeT]). The large number of transporters involved in uptake of amino acids, sucrose, iron, and other unknown solutes may be required for optimum recovery from crushed nodules and subsequent growth in TY and their mutation consequently results in loss from the nodule bacteria library.
Bacteroid Genes.
There are 24 genes specifically required by bacteroids (Fig. 2 and SI Appendix, Table S8). Mutants in these genes have reduced fitness specifically in formation or maintenance of bacteroids but are not affected in bacterial progression through infection threads or regrowth from nodules. They include several nif (pRL100196 and pRL100195 [nifAB], pRL100159 [nifE], and pRL100162 [nifH]) and fix (pRL100198 and pRL100197 [fixCX]) genes that are needed for nitrogenase assembly and microaerobic respiration (57, 58). This confirms our prediction that mutants impaired in N2 fixation would result in an ES/DE classification in the libraries recovered from nodules.
The dicarboxylate transport system (Dct) genes (RL3425 [dctB] and RL3426 [dctD]), enabling dicarboxylic uptake by bacteroids to fuel N2 fixation (6, 59, 60), are bacteroid-specific. Prior to bacteroid differentiation, rhizobia utilize a wide range of carbon sources and dct mutants form well-developed nodules and bacteroids (61). We assume that wild-type and dct mutant strains maintain similar populations of viable bacteria in nodules (reflected in the nodule bacteria results), while dct mutants have severely reduced N2 fixation and numbers of bacteroids. RL3424 (dctA), encoding the Dct system transporter, gave an NE classification in HMM analysis; however, inspection of the insertion sequencing data showed that out of 17 TA sites in dctA, 11 sites had no or very few reads (less than 10). This suggests that 65% of TA sites in dctA do not tolerate insertion, emphasizing the significant role of this transporter. When TA insertions are near the gene termini, it may be that mariner insertion does not fully disrupt gene function. Our previous INSeq screen in Rlv3841 similarly reported the termini of RL3424 (dctA) to tolerate a large number of insertions (62). RL0037 (pckA), encoding phosphoenolpyruvate carboxykinase, which is essential for growth on dicarboxylates (63), was also specifically required in nodule bacteroids (SI Appendix, Table S8). This fits with evidence that dicarboxylates drive bacteroid metabolism but not bacterial growth during nodule infection, where multiple carbon sources are probably available.
The cytochrome bc1 complex is encoded by RL3486 to RL3484 (petABC), with RL3486 (petA) encoding the ubiquinol–cytochrome c reductase iron-sulfur subunit required across all symbiosis growth conditions (rhizosphere-progressive), whereas petB is classified as nodule-general. In contrast, RL3484 (petC), encoding the cytochrome c1 precursor, is specifically required in bacteroids. A mutant of RL3484 (petC) has been demonstrated to be Fix− in R. leguminosarum (64). In addition, RL3577 (guaD), encoding guanine deaminase, an enzyme that catalyzes the hydrolytic deamination of guanine producing xanthine and ammonia, is also bacteroid-specific (SI Appendix, Table S8).
Mutants with Improved Bacterial Colonization or Increased Numbers in Nodules.
When mutation in a gene leads to a bacterial strain with improved fitness for a given condition or environment, its INSeq classification is AD. (AD genes at each stage from rhizosphere to nodule [NE or DE in input library] are summarized in SI Appendix, Fig. S1 and Table S9.) No genes are classified as AD across all four conditions (rhizosphere-progressive) and none are root-progressive. However, there are considerable groups where mutation leads to better recovery from the rhizosphere (84 genes) and/or increased colonization of roots (172 and 33 genes, respectively), as well as leading to an increased population recovered from crushed nodules: nodule-general (11 genes), nodule bacteria-specific (following regrowth; 207 genes), or bacteroid-specific (22 genes) (SI Appendix, Fig. S1 and Table S9). A large proportion (approximately 66%) of AD genes are contiguous and clustered, with many in operons. In contrast to the genes required at a given stage, the reason for an AD classification is far more nuanced. Mutation of genes involved in uptake, sensing, or metabolism of specific compounds can be advantageous to bacteria in a given environment, increasing their fitness, as energy is not expended on pathways or gene expression that is not needed. For example, in contrast to rapid growth in rich broth in vitro, when bacteria are growing slowly, their metabolism will be more restricted, and there will be a greater penalty for synthesizing proteins not absolutely required. AD genes are likely to reflect this. In the rhizosphere, AD-classified genes (84) include ABC homoserine transporter pRL80088, Major Facilitator Superfamily (MSF) nitrate transporter NarK (RL1992), ABC nitrate transporter (NitT) family SBP RL1993, and nitrate reductase transcriptional regulator NasT (RL1994), as well as a large region of 26 genes (RL1870 to RL1895), including genes encoding sensor regulators, heat shock proteins, among others.
Rhizosphere and root AD genes (172; SI Appendix, Fig. S1 and Table S9) include ABC transporters involved in uptake of quaternary amines (QAT) pRL100078 to pRL100083 (qatV6W6X6); peptides, PepT-family pRL110054 to pRL110053 (optDA), pRL110281, and pRL120330; amino acids, pRL120404 (braC2) involved in uptake of branched chain amino acids (65) (HAAT), general l-amino uptake system RL2204 to RL2203 (aapJQ, polar amino acid [PAAT] family) and pRL80060 (PAAT SBP with homology to a mimosine transporter of Rhizobium sp. TAL 1145) (66); carbohydrates RL2895 to RL2896 (CUT1); and phosphate uptake transporter (PhoT) RL1683 and polyamine/opine/phosphonate uptake (POPT) family RL2009 to RL2014. Under these specific experimental conditions, in the rhizosphere and on roots, bacteria can clearly survive without the wide range of compounds that these transporters import. Export systems, RND-family efflux proteins pRL100287 to pRL100286, and multidrug efflux transporter RL3414 (norM) are also classified AD under these conditions.
There is a small group of nodule-general AD transport genes, including CUT1 uptake system RL0749 (aglK) transporting alpha-glucosides and exporter RL3632. This latter is part of a cluster of five genes (RL3628 to RL3632), which includes a sugar decarboxylase (RL3628) and three glycosyl transferases (RL3629 to RL3631).
When we consider nodule bacteria, even slight changes in the terminal differentiation to bacteroids will affect the proportion of bacteria recovered from nodules. Mutation of a gene that leads to delay of differentiation might lead to an AD gene classification. However, as preparation of the nodule bacteria sample includes recovery in TY broth, any conclusions should be tentative as the influence may be to increase fitness for regrowth in a nutrient-rich medium. Nodule bacteria AD genes (207) include numerous ABC and other transporter systems; CUT1 family systems pRL100440 to pRL100443, contiguous with an inositol-degradation protein (pRL100439); pRL120006 to pRL120007; pRL120557 to pRL120558, contiguous with a glycosyl hydrolase (pRL120559); and RL4378 to RL4375; CUT2 family systems eryEFG (pRL120202 to pRL120200), together with eryH (pRL120203), a periplasmic lipoprotein involved in erythritol uptake and/or metabolism; pRL90246 to pRL90247; HAAT family components pRL120095 and RL3745 (braC) (involved in uptake of branched chain amino acids; ref. 65); and PepT complete systems pRL110517 to pRL110513 and pRL90249 to pRL90253, which could be involved in uptake of NCR peptides as without their uptake, differentiation into bacteroids might be reduced leading to increased bacterial recovery. Tripartite ATP-independent transporter (TRAP) import system RL4359 to RL4361 is likely to be involved in the uptake of methyl pyruvate or pyruvate by homology with Sinorhizobium meliloti (56). For uptake of metals, the following genes are classified AD in nodule bacteria; Iron chelate uptake transporter (FeCT) components RL2713 to RL2715; FeT family RL3350 to RL3352; Manganese/Zinc/Iron chelate transporter (MZT) family components RL3885 to RL3886 (sitAB), likely to import manganese (from homology to S. meliloti transporter; ref. 56), and MgtE-like family cationic transporter RL2550. In addition, AD genes include clusters encoding three extracytoplasmic function (ECF) sigma factors and their associated proteins involved in signal transduction from the external environment: RL1471 to RL1477, including ecfDdgpfD1D2D3; RL3233 to RL3235, including ecfElppE; RL3508 to RL3512, including lppFecfFasfF. These sigma factors control, often large, networks of genes, with their mutation meaning energy is not consumed by unrequired gene expression, thus providing an advantage in the test environment. Genes involved in motility and chemotaxis classified as AD are discussed in detail in Importance of Motility and Chemotaxis during Symbiosis.
Bacteroid AD genes (22) include another ECF sigma factor, EcfO (pRL110418), together with nearby gene pRL110416 (rhaI) encoding a rhamnose isomerase involved in competition for nodulation in R. leguminosarum bv. trifolii (67). Contiguous gene pRL110415 (rhaD) encodes a dehydrogenase required for nodulation (SI Appendix, Table S6), although the contiguous rhamnose transporter (pRL110413 to pRL110410), which might have been expected to be required for nodulation based on the results with R. leguminosarum bv. trifolii, is not required at any stage examined (Dataset S1). This is presumably because there is redundancy in Rlv3841 where another uptake system (most likely a CUT2) imports rhamnose, but the metabolism carried out by pRL110415 is needed for successful nodule formation.
Importance of Motility and Chemotaxis during Symbiosis.
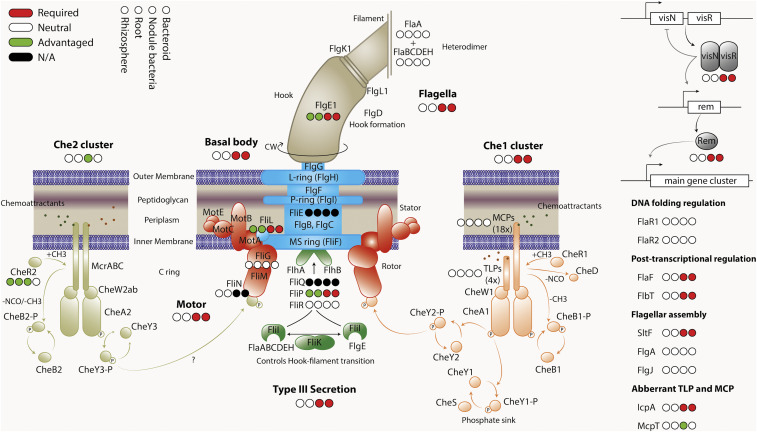
The genome of Rlv3841 contains up to 93 genes with putative roles in chemotaxis and motility, arranged in 7 clusters of 2 genes or more, and 28 single genes (SI Appendix, Table S10). Genes encoding putative proteins for chemotaxis (che [16 genes]), methyl-accepting chemotaxis receptors (mcp [24 genes] and mcr [3 genes]), and flagellar biosynthesis and motor function (fla [10 genes], flg [16 genes], flh [2 genes], fli [12 genes] and mot [5 genes]) have a variety of classifications throughout symbiosis, although the majority seem to fall into 3 clear categories: either not required for symbiosis (39 genes), required for nodule formation and function (nodule-general [39 genes]; SI Appendix, Table S6), or their mutation is advantageous in some way for growth in the rhizosphere and root colonization (5 genes) (SI Appendix, Table S10).While 10 motility-related genes are encoded on plasmids in Rlv3841 (7 on pRL12, and one each on pRL8, RL10, and pRL11), only chromosomally encoded motility-related genes are required for symbiosis.
Each of the individual genes in che1 cluster (RL0686 to RL0693 [icpAcheX1Y1A1W1R1B1Y1D1]) is classified as nodule-general (SI Appendix, Tables S6 and S10), which means that they are all needed for nodule function and also, perhaps counter intuitively, that the cluster is not required for optimal colonization of the rhizosphere or roots (Fig. 3). This confirms the results of Miller et al., who found the che1 cluster promotes competitive nodulation of peas (68). However, the INSeq results also suggest that Che1 is either required for chemotaxis specifically to root hairs or early progression into infection threads but is not essential for colonization of the root surface. In contrast, all genes in the che2 cluster RL4036 to RL4028 (cheY3A2W2mcrCBAcheW3R2B2) are classified AD in nodule bacteria (Fig. 3 and SI Appendix, Tables S9 and S10). While the reason is not known, we speculate that the che2 gene cluster is involved in the later stages of bacterial movement through infection threads. A mutation that delays progression from infection threads to differentiated bacteroids would result in their overrepresentation in the nodule bacteria library, leading to the genes’ classification as AD.
Fig. 3.
Classification of genes encoding motility and chemotaxis proteins in rhizosphere, root, nodule bacteria, and bacteroid INSeq libraries. Genes required at a given stage are shown in red; genes where mutation has no effect (NE) are shown in white; genes that, when mutated, are advantageous are in green; and those unclassified due to too few TA sites are in black. (INSeq results are shown in SI Appendix, Table S10.) The proteins are colored by functional group, but the color is arbitrary with respect to INSeq phenotype.
Most flagellar assembly and basal body apparatus proteins are important in nodules, both in bacteroids and nodule bacteria (Fig. 3 and SI Appendix, Table S10). The gene encoding critical motor protein FliG (RL0700 [fliG]) is neutral across all stages of symbiosis; however, in contrast, the gene encoding an interacting integral motor protein MotA (RL0703) is required in nodule bacteria and bacteroids (Fig. 3). In addition, the genes encoding master flagella and chemotaxis regulator pair VisNR (RL0696 and RL0697) are required in nodule bacteria and bacteroids (Fig. 3), although the carboxy-terminal GerE-encoding domain of visN displays a dramatic increase in transposon insertions, suggesting its deletion does not impair VisN function and may even enhance it. Several flagellar biosynthesis-encoding genes required in nodule bacteria are not needed, or their mutation is even advantageous, in the rhizosphere and for root colonization. Nonmotile bacteria may become stranded in the rhizosphere or on the root surface. Overall, it suggests that under the experimental conditions, motility and chemotaxis are not essential for colonization of the root surface but have a particularly important role in the nodule infection pathway, for movement toward root hairs or, possibly, progression down infection threads (Fig. 3). However, bacterial chemotaxis has previously been shown to provide a competitive advantage in colonization of plant root surfaces (69, 70). To minimize preselection the INSeq library was inoculated from a frozen culture at stationary phase (likely nonmotile) onto sterile vermiculite-grown plants. However, growth in soil, in competition with a motile, complex microbiome, is likely to impose different motility and chemotaxis selection upon the bacteria, with loss of chemotaxis and motility likely to be deleterious for long-term survival in soil and root colonization (71).
Plant Rescue of Auxotrophic Rhizobia.
While it is generally accepted that terminally differentiated bacteroids are mainly supplied with dicarboxylic acids, the metabolic status of rhizobia during infection remains largely enigmatic. Root exudates contain a large variety of amino acids, sugars, organic acids, purines, and pyrimidines (72, 73); however, it remains unknown what compounds are available in substantial amounts and used by rhizobia. Therefore, we compared INSeq results from Rlv3841 grown in minimal medium (62) with symbiosis to identify auxotrophic mutants rescued by plant-derived compounds (SI Appendix, Table S11). We identified the main biosynthetic pathways for 18 of 20 amino acids and key components for purine, pyrimidine, S-adenosyl-methionine, heme, vitamin B12, CoA, riboflavin and flavin mononucleotide/flavin adenine dinucleotide, pyridoxal, and tetrahydrofolate biosynthesis (SI Appendix, Table S11). These pathways are required throughout all stages tested (i.e., rhizosphere-progressive), with the exception of purine biosynthesis, where initial steps to 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranosyl 5′-monophosphate (AICAR) are only required in nodules and not in the rhizosphere or on roots (SI Appendix, Table S11). As the genes encoding the enzymes of these pathways are rhizosphere-progressive, root exudates presumably contain insufficient amounts of these compounds to sustain competitive growth of these mutants. Only AICAR (or an equivalent metabolite) is released at sufficient levels into the rhizosphere and onto root surfaces to enable bacterial growth, however not in sufficient amounts for growth in infection threads or during bacteroid formation. Interestingly, several genes, NE in minimal or rich (TY) medium are required during stages of symbiosis. Rlv3841 has at least two glutamine synthases, and, of these, RL3549 encoding GlnII, is NE in minimal medium and in colonization of roots or rhizosphere but is ES/DE in nodules and nodule-recovered bacteria (i.e., nodule-general) (SI Appendix, Table S6). This is intriguing and suggests that increased glutamine synthesis may be required during infection. Similarly, there are two redundant copies of argG (RL2987 and RL4515). ArgG catalyzes the condensation of citrulline and aspartate to argininosuccinate in the urea cycle as part of arginine biosynthesis. As RL4515 is nodule-general (SI Appendix, Table S6) but is NE in minimal medium or during colonization, this suggests an increased role for arginine synthesis or degradation during bacteroid formation or function. Intriguingly, urea is also derived from arginine (see above) and urea catabolism is classified nodule-general. This highlights a potential role for arginine and urea catabolism via allophanate (urea-1-carboxylate) in nodule bacteria and/or bacteroids.
Histidine biosynthesis requires formation of imidazole-glycerole-3-phosphate from phosphoribulosyl-formimino-AICAR-phosphate, catalyzed by heterodimeric HisFH. Two copies are encoded in the genome, one of which (RL0042/RL0046) is required for growth in the input conditions. Of the second copy, RL0819 is nodule-general (SI Appendix, Table S6), and RL0820 is rhizosphere-progressive. This indicates an increased need for these reaction steps or possibly an incompatibility with other enzymes under N2-fixing conditions. Glutathione-biosynthesis genes (RL0855 [gshA], RL0338 [gshB], and RL1571 [pepA]; SI Appendix, Table S11) are rhizosphere-progressive (SI Appendix, Table S2) but are dispensable in minimal medium, indicating that when interacting with plants, rhizobia must cope with increased oxidative stress (74, 75).
Validation of INSeq Predictions.
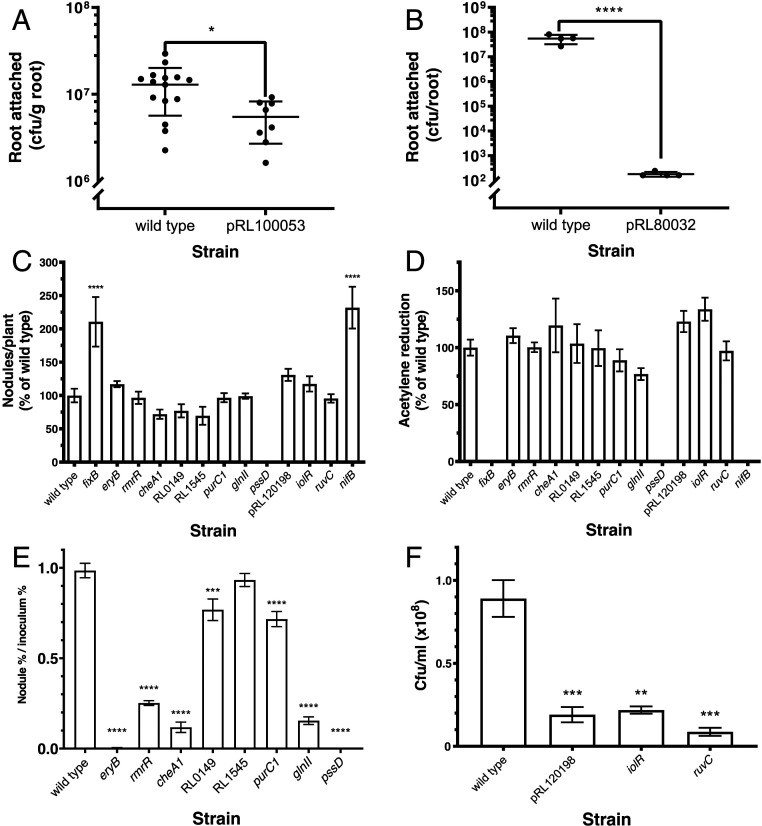
Fifteen genes identified by INSeq as required in different stages of colonization and symbiosis were mutated by pK19 insertion, and their mutant phenotypes were determined experimentally (Table 1). Mutants in genes classified as root-progressive (pRL100053 and pRL80032) showed reduced colonization of roots compared with wild type (Fig. 4 A and B) but no significant reduction in survival in the rhizosphere (wild type, 2.88 × 107 cfu/mL [SEM: 3.84 × 106; n = 15; cfu, colony-forming units], and mutant in pRL100053, 1.81 × 107 cfu/mL [SEM: 2.57 × 106; n = 8]; wild type, 2.13 × 108 cfu/mL [SEM: 3.20 × 107; n = 3], and mutant in pRL80032, 2.22 × 108 cfu/mL [SEM: 3.10 × 107; n = 3]). Mutants in genes classified as nodule-general, together with those specific for nodule bacteria or bacteroids (Table 1), were tested experimentally for the ability to nodulate and to fix nitrogen in single-strain inoculations. Of these 13 mutants, 10 nodulated peas at levels similar to wild type (Fig. 4C). The exceptions were mutants in pssD, a nodule-general mutant that formed no nodules, and nifB (nodule bacteroid-specific) and fixB (nodule-general), which induced approximately twice as many, but small white, nodules as wild type (Fig. 4C and SI Appendix, Fig. S2). Assessing nitrogenase activity, the nifB, fixB, and pssD mutants had no detectable acetylene reduction, while all other mutants showed activity similar (approximately 80 to 130%) to that of wild type (Table 1 and Fig. 4D). These three mutant phenotypes show clearly that pssD, nifB, and fixB are required for nodulation and N2 fixation. It is also apparent why mutants in these genes have been lost from nodule bacteria and bacteroid libraries (Fig. 4C), as less DNA would be present in the nodules collected from strains that form smaller nodules (nifB, fixB) or are unable to form nodules at all (pssD). The reason for the classification of the other seven genes as nodule-general (Table 1) was unclear until experiments were performed to test their mutant’s ability to compete against wild type to form nodules. Mutant strains (marked with gusA) were competed against wild-type celB-marked bacteria by inoculation in a 1:1 ratio (SI Appendix, Fig. S3). Six of the seven nodule-general mutants were significantly impaired in competitive nodulation compared with wild type (Fig. 4E). A competitive disadvantage was evident despite the fact that a 1:1 inoculum provides significantly less competition than the INSeq environment, where up to 105 strains, with mutations in any 1 of over 6,000 genes (6,429/6,656 classified as NE in input1/input2), compete to form nodules. For these six strains, we have shown that the gene mutated in each case is required for effective symbiosis, and these INSeq experiments have identified the stage at which it is needed. Only a single predicted nodule-general strain (RL1545 mutant) showed no reduction in nodule occupancy in inoculation at this competition ratio, and we can speculate that its deficiency (it encodes a dehydrogenase/reductase) may only become apparent under stiffer competition conditions.
Table 1.
Phenotype of mutants affected at different stages of symbiosis
| Gene mutated | Description | Experiment | |||||
| Rhizosphere* | Root† | Nodules‡ | C2H2 reduction§ | Nodule occupancy¶ | Regrowth from nodules# | ||
| Root-progressive | |||||||
| pRL100053 | Transmembrane protein | ++ | + | ++ | ND | ND | ND |
| pRL80032 | LysR family transcriptional regulator | ++ | + | — | ND | ND | ND |
| Nodule-general | |||||||
| pRL100199 (fixB) | Electron-transfer protein FixB | ND | ND | +++ | — | ND | ND |
| pRL120205 (eryB) | Erythritol phosphate dehydrogenase | ND | ND | ++ | ++ | — | ND |
| pRL90058 (rmrR) | TetR family transcriptional regulator of RND family efflux transporter | ND | ND | ++ | ++ | + | ND |
| RL0688 (cheA1) | Chemotaxis protein CheA | ND | ND | ++ | ++ | + | ND |
| RL0149 | XRE family transcriptional regulator | ND | ND | ++ | ++ | + | ND |
| RL1545 | Short-chain dehydrogenase/reductase | ND | ND | ++ | ++ | ++ | ND |
| RL2606 (purC1) | Saicar synthetase | ND | ND | ++ | ++ | + | ND |
| RL3549 (glnII) | Glutamine synthetase II | ND | ND | ++ | ++ | + | ND |
| RL3654 (pssD) | Polysaccharide biosynthesis protein | ND | ND | — | — | — | ND |
| Nodule bacteria | |||||||
| pRL120198 | l-xylulose kinase | ND | ND | ++ | ++ | ND | + |
| RL1496 (iolR) | SIS (sugar isomerase)-RpiR family transcriptional regulator | ND | ND | ++ | ++ | ND | + |
| RL3986 (ruvC) | Holliday junction endoDNase RuvC | ND | ND | ++ | ++ | ND | + |
| Bacteroid | |||||||
| pRL100195 (nifB) | FeMo cofactor biosynthesis protein NifB | ND | ND | +++ | — | ND | ND |
Rhizosphere colonization by a single strain.
Root colonization by a single strain.
Nodule formation by a single strain.
C2H2 (acetylene) reduction when peas inoculated with a single strain.
Nodule occupancy when in competition with wild type bacteria.
Regrowth of bacteria from crushed nodules. ++, wild-type levels; +, reduced levels; +++, increased levels; —, no measurable activity; ND, not determined.
Fig. 4.
Experimental characterization of mutant phenotypes predicted from INSeq data. (A) Root colonization by bacteria (cfu/g root) from a single-strain inoculation (105 cfu) of a 7-d-old pea plant comparing wild type and a mutant in pRL100053 (classified as root-progressive) at 5 dpi (n ≥ 8). An unpaired t test was used to assess statistical significance. (B) Root colonization by bacteria (cfu/root) from a single-strain inoculation (105 cfu) of a 7-d-old pea plant comparing wild type and a mutant in pRL80032 (root-progressive) at 5 dpi (n ≥ 4). An unpaired t test was used to assess statistical significance. (C) Nodule formation on peas (nodules/plant as a percentage of wild type [Rlv3841GusA, 180 ±19.2 nodules per plant]) from a single-strain inoculation (104 cfu) for mutants in nodule-general genes (9), nodule bacteria genes (3), and a bacteroid gene (Table 1) at 28 dpi (n ≥ 3). A Dunnett’s test comparing each mutant to wild type was used to assess statistical significance. (D) Acetylene reduction as a percentage of wild type (Rlv3841GusA, 5.18 ± 0.380 μmol of ethylene per plant per hour) of mutants in nodule-general genes (9), nodule bacteria genes (3), and a bacteroid gene (Table 1) at 28 dpi (n ≥ 3). A Dunnett’s test comparing each mutant to wild type was used to assess statistical significance. (E) Competition for nodule occupancy of mutants with wild type (nodule percentage/inoculum percentage) from 1:1 (mutant:wild type) coinoculation (total, 104 cfu) of pea plants with eight nodule-general mutants harvested at 21 dpi (n ≥ 5). Statistical significance was assessed by a mixed-effects model comparing the proportion of each strain in inoculum and nodules. (F) Bacterial recovery (cfu/mL [×108]) from crushed nodules (10 per plant) and 36 h growth in TY broth at 28 °C for wild type and three nodule bacteria mutants (n ≥ 3). To assess statistical significance, a Dunnett’s test comparing each mutant strain to wild type was used. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars show ±SEM.
For genes classified as nodule bacteria-specific, we tested regrowth in TY medium since the initial preparation of this library involved a recovery step of growth in TY medium after nodule crushing. Wild type and mutants in three nodule bacteria-specific genes (pRL120198, RL1496 [iolR], and RL3986 [ruvC]) were grown in TY after extraction from nodules and compared. All mutants had a significantly lower rate of regrowth from crushed nodules than wild type (Fig. 4F). While this explains the low recovery of each of these mutants in this library, we did not identify any genes that increased the ratio of bacteria to bacteroids in nodules in such a small sample set. Future studies using flow cytometry to measure this ratio could be a powerful approach in such analyses.
Conclusion
Here, we present an in-depth investigation of N2-fixing bacterial interactions with a host legume during symbiosis by examining bacterial gene requirements at different stages of rhizosphere growth, root colonization, and nodulation. Key observations are summarized (Fig. 5), illustrating the progressive nature of many mutations impairing effective symbiosis. It emphasizes that rhizobia experience many different soil and plant environments and is consistent with the Rhizobiales being a large family with many members being symbiotic or commensal bacteria (76). The power of INSeq is that it enables identification of those genes required when in competition with other bacteria. This is important when investigating environmental competition, since a mutation may not be deleterious for subsequent nodule formation or effective symbiosis in the artificial environment of inoculation with a single strain. It is sobering to appreciate that each of the 603 genetic regions required for competitive nodulation is individually just as important for a successful symbiosis as the structural components of nitrogenase. This INSeq dataset provides a powerful resource for better understanding the genetic basis of the crucially important N2-fixing rhizobia–legume symbioses.
Fig. 5.
Rlv3841 genes required at different stages of symbiosis with pea from INSeq analysis. Three environments (rhizosphere, root, and nodule) are shown. Progressive genes alter competition at subsequent stages. Gene requirements at each stage are discussed in Results, and full gene lists can be found in SI Appendix, Tables S1–S8.
Materials and Methods
Bacterial and Plant Growth.
Bacterial strains and plasmids shown in SI Appendix, Table S12 were grown as described previously (64). Mutant strains in R. leguminosarum were constructed as described previously (64), except ligation was performed using a BD In‐Fusion cloning kit (Clontech), according to the manufacturer’s instructions (primers listed in SI Appendix, Table S12). Pea plants (Pisum sativum cv. Avola) were grown as described (77).
INSeq Library Construction and Sequencing.
Construction of the mariner library for INSeq experiments was as described previously (62), producing input libraries from two independent conjugation pools, input1 and input2. Seven-day-old peas grown in boiling tubes were inoculated with 105-cfu mariner-insertion mutants (input1). Rhizosphere and root-colonization samples were recovered from these at 5 dpi. Pea seedlings were grown in boiling tubes whose diameter allowed the rooting system to fully expand throughout the space; the vermiculite surrounding the roots has been defined as the rhizosphere in this study. To recover the rhizosphere population, the vermiculite surrounding the roots was separated, and 10 mL of phosphate-buffered saline (PBS) was added, vortexed (1 min), and filtered through four layers of muslin. To recover the root-colonized population, the root, snipped from the cotyledons, was vortexed (1 min) with 20 mL of PBS, ground by pestle and mortar, and filtered as above. Both were grown on TY (12 h) and spun to pellet cells before DNA extraction and library preparation. Seven-day-old peas grown in 2-L pots were inoculated with 106 cfu mariner-insertion mutants (input2). Nodule bacteria and bacteroid samples were recovered from these at 28 dpi. A total of ∼150,000 nodules were harvested from 1,500 plants and libraries prepared from 1) extract regrown in TY (12 h) before DNA preparation (nodule bacteria) and 2) DNA extracted from nodules (bacteroids). A total of six INSeq libraries (input1, input 2, the rhizosphere, roots, nodule bacteria, and bacteroids) were prepared and sequenced as described previously on an Ion Torrent Proton (62).
Transposon Insertion Analysis with a Four-State HMM.
For sequencing data analysis, sequencing reads were uploaded onto a Linux server, and read-quality filtering and mapping (78, 79) and conversion of mapped data to wig files (for each replicon) were as described previously (62). Wig files were analyzed by TRANSIT software (80) using an HMM analysis method as described in ref. 25. In short, the HMM algorithm uses read-mapping statistics for all of the genes in a replicon and assigns one of four state classifications for each gene (ES, DE, NE, AD) (25, 80). In simplified form, ES genes should tolerate zero or very few insertions, and NE genes will sit within a mean parameter of insertions that do not excel past the boundaries set for DE or AD. For the DE state, the boundary is set at less than 1/100th of the read counts for a statistically significant consecutive stretch of insertion sites (25, 80). For the AD state, the boundary is set at over five times the mean read count for a statistically significant consecutive stretch of insertion sites (25, 80). There were a small number of TA sites where insertions gave very high AD read counts that misled the HMM to predict all subsequent TA sites as ES, as these very high read counts skewed the local mean read count. The values for these TA sites were reduced to next local-highest AD TA read count, and it was these wig files that were used for analysis. In TRANSIT, two analysis modes were used: 1) whole gene and 2) core region (considering 10 to 90% of gene, ignoring 10% at 5′ and 3′ ends), with the following parameters: method, HMM (to assign gene status) (25); and normalization, trimmed total reads and corrected for genome positional bias (Dataset S1). Gene status assignment differed by <0.85% between the two analyses. The 25 gene features that lacked TA sites in their core region were excluded from core-region analysis, and whole-gene analysis was used instead. For disagreements between the two analyses, if a gene was designated ES/DE/AD by whole-gene analysis but NE by core region analysis, then the final designation was based on the whole-gene analysis (i.e., ES/DE/AD). However, when assignment by core-region analysis was not NE, then read counts mapped to genes were checked in Integrated Genome Viewer (81), and designation was manually assigned. A fitness value (FV) for each gene was then calculated from the HMM analysis mapping statistics (62). FV was calculated as follows: FV = (number of insertion sites [TA] in a gene) × (mean insertion density of a gene) × (mean read count of a gene). The FV per gene was then standardized per million library reads (Dataset S1). FV gives a representation of the total gene mutants in a population and a numerical quantification for how well a gene may tolerate insertion (62); however, it should be appreciated that FV does not possess the same statistical rigor for analyzing transposon-insertion sequencing data as the HMM method. The HMM outputs for each gene status classification are provided in Dataset S2.
Characterization of Bacterial Mutants.
Root colonization was assessed at 5 dpi from inoculation of 7-d-old peas with 105 cfu of bacteria. The root, snipped from the seed, was vortexed (1 min) with PBS (25 mL) before being ground (as above) and resuspended in PBS (10 mL). The rhizosphere population was assessed by adding PBS (30 mL) and vortexing thoroughly. Serial dilution and plating on appropriate antibiotics were used to calculate cfu. Acetylene-reduction assays (14) of seed-inoculated peas were performed at 28 dpi, and the same roots, subsequently stained for GusA activity using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylammonium salt (82), were photographed to count blue-stained nodules. Competition between two strains to form nodules was assessed at 21 dpi on seed-inoculated peas (total inoculum, 104 cfu) by double-sequential staining for GusA activity using Magenta-GlcA and CelB activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (82), to count magenta- and blue-stained nodules, respectively. Recovery of bacteria from nodules was assessed by picking the 10 largest nodules from a root, which were surface-sterilized (5 min in 2% bleach, followed by 6× washes in sterile distilled water) and crushed in 100 μL of PSM buffer (64) in a sterile 1.5-mL Eppendorf with a plastic pestle, before increasing the total volume to 1 mL of PSM and vortexing thoroughly; 25 μL was used to inoculate 50 mL of TY broth and an optical density at 600 nm (OD600) followed for 36 h of growth, with shaking (140 rpm) at 28 °C (OD600 of 1 = 8 × 108 cfu).
Supplementary Material
Acknowledgments
This work was supported by Natural Environment Research Council Grant NE/L501530/1, Biotechnology and Biological Sciences Research Council Grants BB/M011224/1 and BB/N013387/1, and Swiss National Science Foundation Postdoc.Mobility Grant 183901 (to R.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009094117/-/DCSupplemental.
Data Availability.
The data supporting the findings of the study are available in this article and its SI Appendix.
References
- 1.Fowler D. et al., The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields S., Global nitrogen: Cycling out of control. Environ. Health Perspect. 112, A556–A563 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galloway J. N. et al., Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 320, 889–892 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Poole P., Allaway D., Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43, 117–163 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Hardy R. W. F., Havelka U. D., Nitrogen fixation research: A key to world food? Science 188, 633–643 (1975). [DOI] [PubMed] [Google Scholar]
- 6.Udvardi M., Poole P. S., Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Oldroyd G. E., Murray J. D., Poole P. S., Downie J. A., The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Downie J. A., The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Poole P., Ramachandran V., Terpolilli J., Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Radutoiu S. et al., LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26, 3923–3935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Via V. D., Zanetti M. E., Blanco F., How legumes recognize rhizobia. Plant Signal. Behav. 11, e1120396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gage D. J., Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White J., Prell J., James E. K., Poole P., Nutrient sharing between symbionts. Plant Physiol. 144, 604–614 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allaway D. et al., Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36, 508–515 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Walker T. S., Bais H. P., Grotewold E., Vivanco J. M., Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estabrook E. M., Yoder J. I., Plant-plant communications: Rhizosphere signaling between parasitic angiosperms and their hosts. Plant Physiol. 116, 1–7 (1998). [Google Scholar]
- 17.Wheatley R. M., Poole P. S., Mechanisms of bacterial attachment to roots. FEMS Microbiol. Rev. 42, 448–461 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Alves L. M. C., de Souza J. A. M., de Mello Varani A., de Macedo Lemos E. G., The Family Rhizobiaceae, (Springer, 2014), pp. 419–437. [Google Scholar]
- 19.Young J. P. W. et al., The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7, R34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barquist L., Boinett C. J., Cain A. K., Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 10, 1161–1169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cain A. K. et al., A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 21, 526–540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao M. C., Abel S., Davis B. M., Waldor M. K., The design and analysis of transposon insertion sequencing experiments. Nat. Rev. Microbiol. 14, 119–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman A. L., Wu M., Gordon J. I., Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 6, 1969–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry B. J., Yost C. K., Construction of a mariner-based transposon vector for use in insertion sequence mutagenesis in selected members of the Rhizobiaceae. BMC Microbiol. 14, 298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeJesus M. A., Ioerger T. R., A Hidden Markov Model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinf. 14, 303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westhoek A. et al., Policing the legume-Rhizobium symbiosis: A critical test of partner choice. Sci. Rep. 7, 1419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya C. et al., Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211. Front. Microbiol. 8, 411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doornbos R. F., van Loon L. C., Bakker P. A., Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32, 227–243 (2012). [Google Scholar]
- 29.García Angulo V. A. et al., Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J. Bacteriol. 195, 4611–4619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janczarek M., Rachwał K., Kopcińska J., Genetic characterization of the Pss region and the role of PssS in exopolysaccharide production and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Plant Soil 396, 257–275 (2015). [Google Scholar]
- 31.Janczarek M., Jaroszuk-Sciseł J., Skorupska A., Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie van Leeuwenhoek 96, 471–486 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Janczarek M., Kutkowska J., Piersiak T., Skorupska A., Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 10, 284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosolapova A. O. et al., Two novel amyloid proteins, RopA and RopB, from the root nodule bacterium Rhizobium leguminosarum. Biomolecules 9, 694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roest H. P., Bloemendaal C. J. P., Wijffelman C. A., Lugtenberg B. J. J., Isolation and characterization of ropA homologous genes from Rhizobium leguminosarum biovars viciae and trifolii. J. Bacteriol. 177, 4985–4991 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki S. et al., Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl. Environ. Microbiol. 73, 6650–6659 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronson C. W., Nixon B. T., Albright L. M., Ausubel F. M., Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J. Bacteriol. 169, 2424–2431 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karunakaran R. et al., BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 192, 2920–2928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold M. F. F. et al., Genome-wide sensitivity analysis of the microsymbiont Sinorhizobium meliloti to symbiotically important, defensin-like host peptides. MBio 8, e01060-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haag A. F. et al., Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol. Rev. 37, 364–383 (2013). [DOI] [PubMed] [Google Scholar]
- 40.De Troch P., Vanderleyden J., Surface properties and motility of Rhizobium and Azospirillum in relation to plant root attachment. Microb. Ecol. 32, 149–169 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Krehenbrink M., Downie J. A., Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genom. 9, 55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marie C., Barny M. A., Downie J. A., Rhizobium leguminosarum has two glucosamine synthases, GlmS and NodM, required for nodulation and development of nitrogen-fixing nodules. Mol. Microbiol. 6, 843–851 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Quelas J. I. et al., Lack of galactose or galacturonic acid in Bradyrhizobium japonicum USDA 110 exopolysaccharide leads to different symbiotic responses in soybean. Mol. Plant Microbe Interact. 23, 1592–1604 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Aguilar G. R., Soberón M., Cloning and sequence analysis of the Rhizobium etli ccmA and ccmB genes involved in c-type cytochrome biogenesis. Gene 182, 129–135 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Geurts R., Bisseling T., Rhizobium nod factor perception and signalling. Plant Cell 14, S239–S249 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seefeldt L. C., Hoffman B. M., Dean D. R., Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78, 701–722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weidenhaupt M., Rossi P., Beck C., Fischer H. M., Hennecke H., Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch. Microbiol. 165, 169–178 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Carlson R. W., Forsberg L. S., Kannenberg E. L., Lipopolysaccharides in Rhizobium-Legume Symbioses, (Springer, 2010), pp. 339–386. [DOI] [PubMed] [Google Scholar]
- 49.Cheng G., Shapir N., Sadowsky M. J., Wackett L. P., Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl. Environ. Microbiol. 71, 4437–4445 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanamori T., Kanou N., Atomi H., Imanaka T., Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186, 2532–2539 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tajima S., Larue T. A., Enzymes for acetaldehyde and ethanol formation in legume nodules. Plant Physiol. 70, 388–392 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson J. B., LaRue T. A., Soluble aldehyde dehydrogenase and metabolism of aldehydes by soybean bacteroids. J. Bacteriol. 151, 1473–1484 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yost C. K., Rath A. M., Noel T. C., Hynes M. F., Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152, 2061–2074 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Lodwig E. M. et al., Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422, 722–726 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Mergaert P. et al., Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauchline T. H. et al., Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U.S.A. 103, 17933–17938 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer H.-M., Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58, 352–386 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Bruijn F. J., . “Biological nitrogen fixation” in Principles of Plant-Microbe Interactions, (Springer, 2015), pp. 215–224. [Google Scholar]
- 59.Reid C. J., Poole P. S., Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 180, 2660–2669 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yurgel S. N., Kahn M. L., Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28, 489–501 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Ronson C. W., Astwood P. M., Downie J. A., Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J. Bacteriol. 160, 903–909 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheatley R. M. et al., Role of O2 in the growth of Rhizobium leguminosarum bv. viciae 3841 on glucose and succinate. J. Bacteriol. 199, e00572-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulley G. et al., Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation. J. Bacteriol. 192, 4944–4953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karunakaran R. et al., Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J. Bacteriol. 191, 4002–4014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosie A. H. F., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S., Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 184, 4071–4080 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borthakur D., Soedarjo M., Fox P. M., Webb D. T., The mid genes of Rhizobium sp strain TAL1145 are required for degradation of mimosine into 3-hydroxy-4-pyridone and are inducible by mimosine. Microbiology 149, 537–546 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Richardson J. S., Hynes M. F., Oresnik I. J., A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 186, 8433–8442 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller L. D., Yost C. K., Hynes M. F., Alexandre G., The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63, 348–362 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Kawasaki A. et al., Microbiome and exudates of the root and rhizosphere of brachypodium distachyon, a model for wheat. PLoS One 11, e0164533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scharf B. E., Hynes M. F., Alexandre G. M., Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 90, 549–559 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Turnbull G. A., Morgan J. A. W., Whipps J. M., Saunders J. R., The role of motility in the in vitro attachment of Pseudomonas putida PaW8 to wheat roots. FEMS Microbiol. Ecol. 35, 57–65 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Gaworzewska E. T., Carlile M. J., Positive chemotaxis of Rhizobium leguminosarum and other bacteria towards root exudates from legumes and other plants. J. Gen. Microbiol. 128, 1179–1188 (1982). [Google Scholar]
- 73.Haichar F. Z., Santaella C., Heulin T., Achouak W., Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80 (2014). [Google Scholar]
- 74.Santos R., Hérouart D., Sigaud S., Touati D., Puppo A., Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant Microbe Interact. 14, 86–89 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Chang C., Damiani I., Puppo A., Frendo P., Redox changes during the legume-rhizobium symbiosis. Mol. Plant 2, 370–377 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Garrido-Oter R. et al.; AgBiome Team , Modular traits of the Rhizobiales Root microbiota and their evolutionary relationship with symbiotic Rhizobia. Cell Host Microbe 24, 155.e5-167.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karunakaran R. et al., Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 188, 6661–6668 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B., Wilks C., Antonescu V., Charles R., Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 35, 421–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeJesus M. A., Ambadipudi C., Baker R., Sassetti C., Ioerger T. R., TRANSIT–A software tool for Himar1 TnSeq analysis. PLoS Comput. Biol. 11, e1004401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson J. T. et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sánchez-Cañizares C., Palacios J., Construction of a marker system for the evaluation of competitiveness for legume nodulation in Rhizobium strains. J. Microbiol. Methods 92, 246–249 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of the study are available in this article and its SI Appendix.