Significance
Immune checkpoint blockade (ICB) has revolutionized treatment of many cancer types, but the majority of treated patients still do not respond to ICB. There is an urgent need to identify predictive biomarkers of response prior to or shortly after therapy initiation, as well as the underlying mechanisms. Here we utilize a model of bilateral tumor implantations followed by resection and immunotherapy-response assessment to study the tumor microenvironment shortly following treatment, identifying biomarkers for response or resistance at early time points. Our biomarker gene signatures derived from CD8+ T cells significantly segregate patients by survival and associate with patient response to ICB. Our findings provide a general approach for studying mechanisms of resistance to ICB and discovering predictive biomarkers of response.
Keywords: immune checkpoint blockade, tumor immune microenvironment, breast cancer, bilateral tumor model, predictive biomarkers
Abstract
Immune checkpoint blockade (ICB) is efficacious in many diverse cancer types, but not all patients respond. It is important to understand the mechanisms driving resistance to these treatments and to identify predictive biomarkers of response to provide best treatment options for all patients. Here we introduce a resection and response-assessment approach for studying the tumor microenvironment before or shortly after treatment initiation to identify predictive biomarkers differentiating responders from nonresponders. Our approach builds on a bilateral tumor implantation technique in a murine metastatic breast cancer model (E0771) coupled with anti-PD-1 therapy. Using our model, we show that tumors from mice responding to ICB therapy had significantly higher CD8+ T cells and fewer Gr1+CD11b+ myeloid-derived suppressor cells (MDSCs) at early time points following therapy initiation. RNA sequencing on the intratumoral CD8+ T cells identified the presence of T cell exhaustion pathways in nonresponding tumors and T cell activation in responding tumors. Strikingly, we showed that our derived response and resistance signatures significantly segregate patients by survival and associate with patient response to ICB. Furthermore, we identified decreased expression of CXCR3 in nonresponding mice and showed that tumors grown in Cxcr3−/− mice had an elevated resistance rate to anti-PD-1 treatment. Our findings suggest that the resection and response tumor model can be used to identify response and resistance biomarkers to ICB therapy and guide the use of combination therapy to further boost the antitumor efficacy of ICB.
Immune checkpoint blockade (ICB) is revolutionizing the treatment of cancer (1). Monoclonal antibodies that block immunosuppressive PD-1/PD-L1 interactions (anti-PD-1: nivolumab, pembrolizumab; anti-PD-L1: atezolizumab, avelumab, and durvalumab) are FDA approved for more than 20 indications, including metastatic melanoma, nonsmall cell lung cancers, kidney cancers, and MSIhi tumors (1–3). However, only a subset of patients responds, and response rates differ substantially in different cancers (4). For example, clinical studies have demonstrated benefits of anti-PD-1/anti-PD-L1 therapies in patients with metastatic triple negative breast cancers (TNBCs) (5–8), as well as advanced ER+/HER− breast cancer (BCs) (9) with an objective response rate ranging from 5 to 24% in patients receiving either anti-PD-1 (5, 9) or anti-PD-L1 treatment (4, 7, 8). This treatment regimen particularly benefitted those with high PD-L1 expression in the tumor microenvironment (TME) (10). However, more than 75% of BC patients who received treatment did not show any objective response. Thus, it is crucial to decipher the resistance mechanisms in these nonresponders to ICB and design more effective treatment strategies to overcome resistance. Additionally, identifying biomarkers that will predict response in BC patients is important for further development of this treatment modality.
There is currently a lack of consensus on what biomarkers are the most useful for predicting response to ICB. Immunohistochemical evaluation of PD-L1 protein expression in the TME (from core biopsies and resected tumors) has been widely used as a predictor of response to anti-PD-1/anti-PD-L1 therapy (1). However, PD-L1 expression is an imperfect biomarker, as some patients with PD-L1 negative tumors respond, while some patients with PD-L1-expressing tumors can be nonresponsive. Similarly, tumor mutational burden has failed to live up to its promise as a biomarker (11). Other potential biomarkers include T cell infiltration and proliferation in a tumor, IFNγ signature, microbiota, and blood vessel normalization (12–18). Thus, there is an urgent need to discover biomarkers for ICB response, as well as identification of markers associated with resistance that could be targeted therapeutically in combination with ICB to improve patient outcomes.
In vivo experimental mouse models that recapitulate human tumors have been valuable tools to elucidate mechanisms of benefit from and resistance to immunotherapy, but identification of predictive biomarkers using mouse models has been challenging. It is difficult to biopsy a mouse tumor due to its small size, and once the entire tumor has been resected, its subsequent response to treatment cannot be assessed. Consequently, higher throughput methods that enable comprehensive analyses of the TME at early time points coupled with evaluation of subsequent response to therapy are sorely needed. Here, we developed a bilateral tumor implantation approach to permit resection as well as evaluation of response in a BC mouse model following treatment at early time points. By implanting two orthotopic BC tumors in the same mouse, we could evaluate and elucidate early determinants of response to ICB using the resected tumor and monitor the response of the nonresected tumor to ICB. A similar bilateral implantation approach was recently used for mesothelioma and kidney cancer models (19). Our approach is distinct from observations of the abscopal effects in which localized radiation treatment of one tumor shrinks a metastatic tumor at a distant site (20, 21). In addition, our approach differs from the instigator and responder model in which two different cell lines are implanted in immunodeficient mice to study dissemination of metastatic cancer cells (22). By using our resection and response model, there is sufficient tumor tissue to perform a number of in depth analyses at early time points (e.g., multiparameter flow cytometry, microscopy, and high throughput sequencing methods such as RNA sequencing [RNA-seq]), while still acquiring full tumor growth kinetics and/or survival data in an unmanipulated tumor.
We used our bilateral tumor implantation model system to identify potential biomarkers of resistance that could differentiate responders from nonresponders to ICB therapy in BC. We found that responder mice had higher levels of CD8+ T cell infiltration in the TME. Transcriptomic analysis of CD8+ tumor infiltrating lymphocytes (TILs) at early time points following ICB treatment revealed distinct gene signatures distinguishing responders from nonresponders. Importantly, these gene signatures for responder and nonresponder tumors significantly correlate with responder and nonresponder gene signatures derived from melanoma patients, as well as with overall survival in a BC patient cohort (23–25). These findings demonstrate the value of this “resection and response” tumor model for identification of novel and clinically relevant biomarkers to predict response to ICB therapy and guide the use of combination therapies for BC and other cancers.
Results
A Model for Studying the Tumor Microenvironment in Early Stages of Immunotherapy.
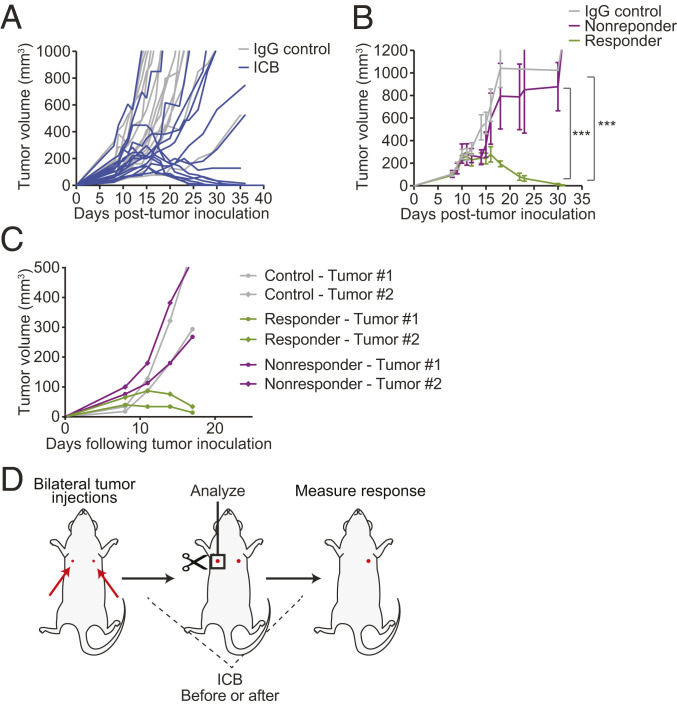
To explore ICB response-associated mechanisms in BC models, we characterized responses to an anti-PD-1 antibody in an orthotopic E0771 breast cancer model in C57BL/6 mice. While the majority of mice responded to therapy and showed tumor remission, we observed a consistent trend in which ∼30 to 40% of the mice did not respond (Fig. 1A). These mice were cohoused from birth in our laboratory’s defined-flora animal facility, eliminating differences in host microbiota as a reason for variability in response to ICB (26, 27). We observed a bimodal response to ICB across the mice, in which mice that responded to therapy showed a sharp reduction in tumor burden which began at ∼7 d following treatment initiation and mice that did not respond showed similar growth rates to IgG-treated (control) mice (Fig. 1 A and B).
Fig. 1.
A model for studying TMEs before or shortly after ICB treatment initiation while maintaining response-to-treatment assessment ability. (A and B) Identical tumor cells implanted in C57BL/6 cohoused littermates give rise to different responses to an anti-PD-1 antibody. Representative results from two independently repeated experiments (n = 15). (A) Tumor growth in ICB-treated (anti-PD-1) and control (IgG) mice bearing orthotopic E0771 breast tumors in C57BL/6 mice. Mice were time and size matched 8 d posttumor inoculation and treated with anti-PD-1 mAb or IgG on days 8, 11, and 14. The treated mice exhibit two distinct trends of growth following ICB therapy, indicative of response or no response. n = 15. Representative results from three independent experiments. (B) Integrated tumor growth behavior of mice bearing E0771 breast tumors. The growth trend of treated mice begins to diverge at day 15, separating them into “responder” or “nonresponder” groups. ***P < 0.0005, by two-way ANOVA with Sidak’s multiple comparisons test. Error bars indicate SEM. (C) The same numbers of E0771 breast tumor cells are implanted orthotopically into the left and right mammary fat pad (MFP) of each mouse. The paired tumors show the same growth trend of either progression or regression on ICB treatment. Representative data of two experiments with n = 10 (treatment) or n = 5 (IgG control) mice. (D) Overview of the bilateral tumor implantation approach to study the TME at early time points in responders versus nonresponders to immunotherapy. The same number of tumor cells are implanted at two distinct locations of each mouse. One tumor is removed and processed for analysis before response can be assessed. The second tumor is monitored to classify the mouse as a responder or nonresponder.
To investigate if the variability in response of tumors to therapy was a tumor-centric or mouse-centric phenomenon, we developed a bilateral tumor system in which two tumors were implanted in genetically identical and cohoused mice, and their response to anti-PD-1 antibody was monitored (Fig. 1C). We observed that the variability in response to ICB in our model was mouse centric, in that two mammary tumors within the same mouse showed the same trend in tumor progression or regression following treatment (Fig. 1C and SI Appendix, Fig. S1 A–C). When responder mice were rechallenged with the same number of E0771 cancer cells, none of the mice developed tumors, suggesting that following treatment a robust host memory response occurred in the responder mice.
Given that response to anti-PD-1 antibody varied in genetically identical and cohoused mice and was mouse centric (both tumors in the same mouse either responded or failed to respond to therapy), our bilateral tumor system seemed ideal for studying the TME before or in early stages of ICB treatment and for monitoring response to ICB (Fig. 1D). We hypothesized that this system could be used to identify early determinants of response, as well as biomarkers that distinguish responders from nonresponders at early stages of ICB treatment. In this system, the same number of tumor cells are implanted into the left and right mammary fat pad of each mouse at the same time. One tumor can be removed for analysis before or shortly after treatment initiation, and the other tumor is monitored to determine whether the mouse is a responder or nonresponder to therapy (Fig. 1D). In establishing this model, we validated that while the total tumor burden decreased upon resection of one tumor as expected (SI Appendix, Fig. S1D), the resection of one of the two tumors did not change the average individual tumor volume of the mouse (SI Appendix, Fig. S1E). Additionally, when we transplanted a responding-tumor into a nonresponder mouse, it remained a responder, pointing to a TME-dominant response. When we transplanted a nonresponding tumor into treatment-naïve mice, it remained a nonresponder. Lastly, if we transplanted a nonresponding tumor into a responder mouse, there was a pseudotumor growth period before all of the tumors regressed, pointing to a host memory response (SI Appendix, Fig. S1F). To test for generalizability of our bilateral tumor approach, we evaluated tumor growth and response to ICB in a B16-F10 melanoma model and observed a similar mouse-centric response, in which both implanted B16-F10 tumors responded to therapy in the same manner (SI Appendix, Fig. S2). Interestingly, B16-F10 tumors were more resistant to treatment and responders eventually relapsed, suggesting that the bilateral tumor approach in the B16-F10 model may be suitable for studying partial responders and tumor relapse. A similar bilateral tumor approach was used in Zemek et al. (19) for mesothelioma and kidney cancer models, further supporting the generalizability of this approach.
Low Tumor Infiltration with CD8+ T Cells before and Shortly after ICB Treatment Is Associated with Poor Response to ICB.
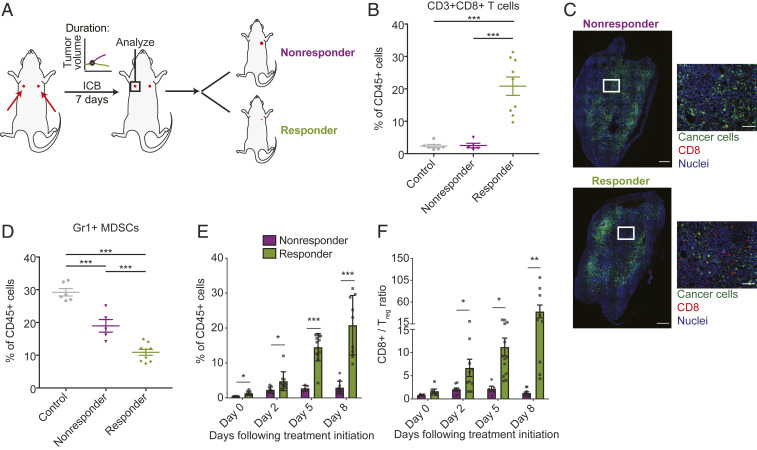
To determine whether response to treatment was associated with differences in the immune TME, we analyzed the immune cell populations in the tumors by flow cytometry. We collected the tumors before the point of growth separation to explore features associated with tumor progression or regression at a time point when outcome cannot yet be assessed by tumor size or growth (Fig. 2A). Responding tumors had significantly higher infiltration of cytotoxic CD8+ T cells (Fig. 2 B and C), consistent with studies reporting that high numbers of preexisting TILs could be indicators of better response to ICB and that increased CD8+ T cell infiltration enhanced antitumor immunity through promoting vessel normalization (18, 28, 29). Immunofluorescence staining of the tumors also showed higher infiltration of CD8+ T cells in the TME of responding tumors (Fig. 2C). In addition, responding tumors had lower numbers of CD11b+Gr1+ myeloid-derived suppressor cells (MDSCs), indicating a less immunosuppressive TME (Fig. 2D) (30, 31).
Fig. 2.
Responder mice have distinct TME characteristics before and shortly following treatment initiation. Bilateral E0771 tumor-bearing mice were treated with anti-PD-1 mAb or IgG (control) on days 8, 11, and 14 after tumor inoculation; n = 6 to 12. Representative data are from three independently repeated experiments. (A) Outline of bilateral tumor implantation experiment to transcriptionally profile CD8+ T cells shortly following anti-PD-1 therapy. The same number of tumor cells were implanted orthotopically into the left and right mammary fat pad (MFP) of each mouse; one tumor was removed at day 7 following treatment initiation for analysis and response was assessed by monitoring growth in the remaining tumor. (B) Relative percentages of CD8+ T cells in total CD45+ populations in tumors resected at day 7 following treatment initiation, as evaluated by flow cytometry. Tumors from responder mice show increased numbers of CD8+ T cells, compared to tumors from nonresponder mice or mice treated with control IgG. ***P < 0.0005, by one-way ANOVA with post hoc Student’s t test. Error bar indicate SEM. (C) Representative immunofluorescence images of tumors from responding and nonresponding mice extracted at day 7 following treatment initiation stained for CD8+ T cells (red) and nuclei (blue). Cancer cells were GFP labeled (green). Tumors from responder mice have higher infiltration and uniformly distributed CD8+ cells in the TME. (Scale bar, 100 μm [for enlarged images] and 500 μm [for whole tumor images].) (D) Relative percentages of CD11b+Gr1+ MDSCs in tumors resected at day 7 following treatment initiation, as evaluated by flow cytometry. Tumors from responder mice show decreased numbers of MDSCs, compared to tumors from nonresponder mice or mice treated with control IgG. *P < 0.05; ***P < 0.0005, by one-way ANOVA with post hoc Student’s t test. Error bar indicate SEM. (E and F) Tumors were collected from the experimental cohort (one tumor collected per mouse) at days 0 (no treatment), 2 (after one dose), 5 (after two doses), and 8 (after three doses) and evaluated for (E) CD8+ T cell infiltration and (F) CD8+/Treg ratio by flow cytometry; n = 4 to 11 from each experiment. T cell infiltration and CD8+/Treg ratio increases in tumors from responders, but not in tumors of nonresponders. *P < 0.05; **P < 0.005, by one-way ANOVA with post hoc Student’s t test. Error bar indicate SEM.
To further investigate whether the increased CD8+ T cells were preexisting in the responding tumors or induced by the treatment, we analyzed the immune cell populations at different time points during the treatment. We isolated and evaluated tumors before and 2 d after each anti-PD-1 dose, at days 0, 2, 5, and 8 following treatment initiation; the tumors had comparable size at the time of analysis. Since pretreatment tumor burden could be a predictor for response to anti-PD-1 antibody (32), we also evaluated the tumor burden prior to treatment initiation. We observed that although there was a trend where responding tumors presented with smaller tumor burden, there were no significant differences in pretreatment tumor size (SI Appendix, Fig. S3A). Notably, the responding tumors had increased numbers of CD8+ T cells prior to therapy (day 0) when compared to the nonresponding tumors (Fig. 2E and SI Appendix, Fig. S3B). The infiltration of CD8+ T cells increased as the treatment progressed for the responding tumors, but not in the nonresponding tumors, as measured by both frequency of CD8+ T cells of total CD45+ cells (Fig. 2E) as well as a CD8/tumor burden ratio (SI Appendix, Fig. S3C). The increase in the CD8/tumor burden ratio (SI Appendix, Fig. S3C) over time demonstrates that the density of CD8+ T cells relative to tumor cells is changing in responders compared to nonresponders, though further work is needed to determine whether there is a certain threshold density associated with response. Although there was an increase in CD8+ T cells in the nonresponding mice after the first dose of treatment, the CD8+ T cell frequency stabilized after this initial increase (Fig. 2E), suggesting that the CD8+ T cells in nonresponders might not be able to mount sufficient immune responses to eradicate the tumors (32, 33). In addition, there was a significant increase in the ratio of CD8+ to CD4+FoxP3+ regulatory T (Treg) cells in the responding mice but not the nonresponding mice (Fig. 2F), further suggesting a more immunostimulatory TME in responders. However, at the time of initial treatment there was no significant difference in the ratio of Treg cells to tumor burden between responders and nonresponders (SI Appendix, Fig. S3D). Additionally, the frequency of Treg cells of the CD45+ population in tumors was similar between responding and nonresponding tumors for the first two doses of anti-PD-1, and then we observed a significant decrease in the frequency of Treg cells in the responding tumor after the third dose of anti-PD-1 therapy (SI Appendix, Fig. S3E). Taken together, our findings suggest that the responder mice have a more immunostimulatory TME with a greater density of CD8+ T cells that increases over time with anti-PD-1, while nonresponding tumors show a lack of effector CD8+ T cell infiltration and activation.
Early-Stage ICB Transcriptional Gene Signatures of Nonresponders Show Association with T Cell Exhaustion.
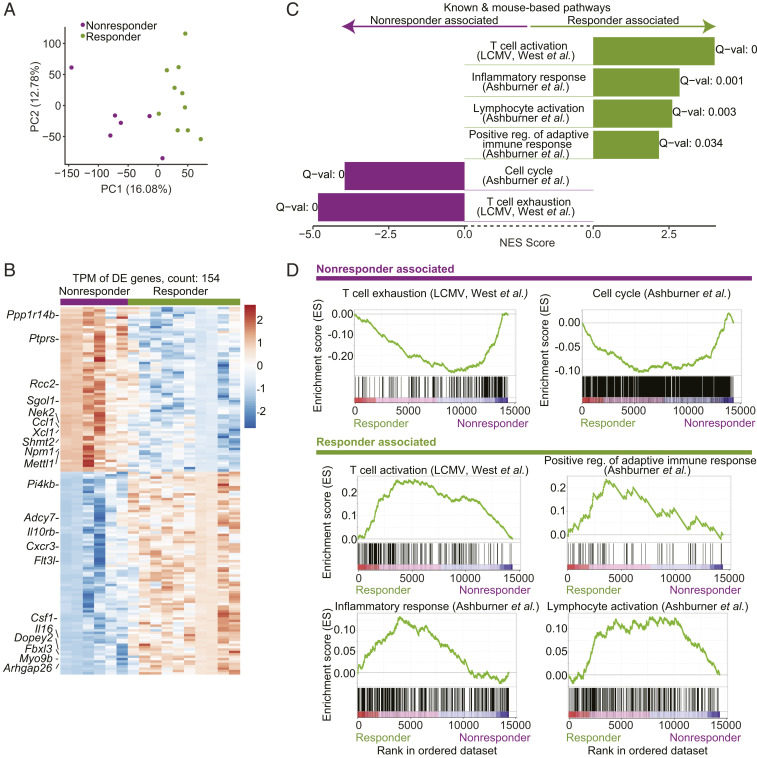
To further characterize the role of T cells in response and resistance to anti-PD-1 therapy, we performed RNA-seq of CD8+ T cells sorted from size-matched tumors isolated before the point of growth separation (Fig. 2A). Mice were treated with ICB for three doses at days 8, 11, and 14 following tumor inoculation, and tumors were isolated at day 15. We observed that the first two principal components, derived from a principal component analysis (PCA) on all genes across all samples, perfectly separated the responder and nonresponder samples, indicating that the strongest factors of variability in the data associate with the responder and nonresponder status (Fig. 3A and Datasets S1 and S2).
Fig. 3.
Transcriptional profiling of CD8+ TILs extracted shortly following treatment initiation yields distinct gene signatures for responders and nonresponders. CD8+ tumor-infiltrating T cells were extracted from tumors surgically removed at day 7 following anti-PD-1 therapy and response was assessed from the remaining paired tumor. (A) PCA of the transcriptional profiles of CD8+ TILs shortly following anti-PD-1 therapy (7 d) shows that the first two principal components separate responders and nonresponders. (B) A heatmap of the 154 differentially expressed genes across responders and nonresponders in CD8+ TILs shortly following anti-PD-1 therapy (7 d). (C) Transcriptional profiles of CD8+ TILs shortly following anti-PD-1 therapy are associated with T cell activation pathways in responders and with T cell exhaustion and cell cycle pathways in nonresponders (GSEA PreRanked analysis). (D) Enrichment plots as output from GSEA for pathways from C.
To establish a gene signature that captures differences in CD8+ T cells across responders and nonresponders that are present shortly following ICB initiation, we conducted a differential-expression analysis and found 85 genes that were up-regulated in responders and 69 genes that were up-regulated in nonresponders and upon inspection showed consistency in expression within each phenotype (Fig. 3B, Dataset S3, and SI Appendix, Methods), constructing early-therapeutic stage responder and nonresponder signatures. Examination of the biological processes that were differentially regulated by the two groups using gene set enrichment analysis (GSEA) (34) showed that the responder signature was highly enriched for pathways related to T cell activation and inflammatory response, consistent with enhancement of immune responses (Fig. 3 C and D and Dataset S4). Additionally, up-regulated genes in responders were enriched for an acute lymphocytic choriomeningitis virus (LCMV) infection CD8+ T cell gene signature as compared to chronic LCMV (24, 35), indicating that within the responding tumors the CD8+ T cells are in a more activated rather than exhausted state (Fig. 3 C and D and Dataset S4).
The nonresponder signature was enriched for a gene signature of CD8+ T cells from a chronic LCMV infection as compared to an acute LCMV infection (24), indicating that within the nonresponding mice the CD8+ TILs are in a more exhausted rather than activated state, even shortly after ICB treatment (Fig. 3 C and D and Dataset S4). In addition, genes in the nonresponder signature were enriched for pathways associated with active cell cycle (Fig. 3 C and D and Dataset S4). While we observed this enrichment for cell cycle pathways at the transcriptional level, we did not observe a difference in the levels of Ki-67 across the responders and nonresponders when measured by flow cytometry (SI Appendix, Fig. S4). Our observations align with studies that have shown that PD-1 could arrest the early cell cycle progression of T cells in the G1 phase (36–38).
High Expression of CXCR3 on Effector T Cells Is a Functional Driver of Initial Response to ICBs.
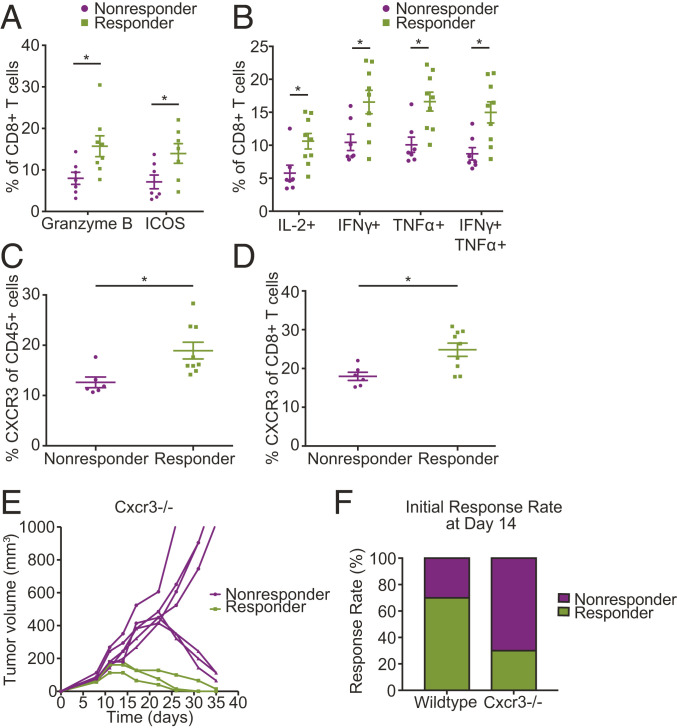
Following our observations that the CD8+ gene signature up-regulated in responder mice is significantly associated with T cell activation (Fig. 3 C and D), we explored whether protein measurements of CD8+ T cells from tumors prior to therapy initiation support this observation. We used our bilateral tumor implantation approach to collect one tumor from mice for pretreatment baseline analysis and to monitor the other tumor after treatment to classify the mice as responders or nonresponders. Using flow cytometry, we found that CD8+ T cells from responder mice expressed higher levels of markers of T cell activation and function, including ICOS, granzyme B (Fig. 4A), and the immunostimulatory cytokines IL2, TNFα, and IFNγ (Fig. 4B), confirming our sequencing analysis. Importantly, the CD8+ T cells from responder mice had higher expression of both TNFα and IFNγ, indicating polyfunctionality (Fig. 4B).
Fig. 4.
Expression of CXCR3 on T cells as a potential biomarker of response to anti-PD-1 therapy. (A and B) Flow-cytometry measurements show CD8+ T cells extracted from tumors prior to treatment initiation express higher levels of (A) markers of T cell activation and function and (B) the indicated cytokines in responders than in nonresponders; n = 6 to 9. P < 0.05 by Student’s t test. Error bars indicate SEM. (C and D) Tumors in mice that respond to anti-PD-1 treatment express higher levels of CXCR3 prior to treatment initiation; n = 6 to 9. Flow-cytometry analysis shows elevated CXCR3 levels in (C) total CD45+ cells (P = 0.012) and in (D) CD8+ T cells (P = 0.010) in tumors extracted from responder mice (by Student’s t test). Error bars indicate SEM. (E) CXCR3 deficiency results in delayed response to ICB. Tumor growth in Cxcr3−/− mice bearing orthotopic E0771 breast tumors treated with anti-PD-1 mAb on days 8, 11, and 14 following tumor inoculation. While up to day 14 posttreatment there is a clear separation between responders and nonresponders, with nonresponders consisting of 70% of the Cxcr3−/− mice, after day 14 posttreatment some of the initial Cxcr3−/− nonresponders show a delayed response to treatment; n = 10 and representative of two independent experiments. (F) Genetic deletion of Cxcr3 leads to a change in the observed response rate at day 14 following anti-PD-1 treatment initiation from 70% in WT to 30% (3/10) in Cxcr3−/− mice. (*P < 0.05) (by Student’s t test). Error bars indicate SEM; n = 10 and representative of two independent experiments.
To evaluate potential causal roles for genes within our transcriptional signatures in driving response to ICB, we evaluated Cxcr3, one of the top 25 differentially expressed genes in our dataset preferentially expressed on CD8+ T cells of responding tumors (SI Appendix, Fig. S5 and Dataset S3). We chose to validate Cxcr3 as a potential biomarker in our model, given its importance in effector T cell activity, as well as response to ICB in other models (39, 40). Effector CD8+ T cells require CXCR3 for trafficking to the tumor vascular interface and functional vasculature is critical for response to immunotherapies (17, 18, 41). Activated T cells often use CXCR3 to migrate to inflammatory sites that secrete high levels of CXCR3 ligands, CXCL9, CXCL10, and/or CXCL11 (42–44). Since we observed significant infiltration of CD8+ T cells in the TME even before treatment (Fig. 2E), we investigated whether CXCR3 expression on CD8+ T cells could be a predictive biomarker for response to anti-PD-1 therapy.
We evaluated CXCR3 protein expression using our bilateral tumor implantation approach, collecting one tumor prior to immunotherapy initiation and monitoring the remaining tumor to assess response to treatment. The frequency of the CXCR3+ CD45+ cell population was higher in the responders compared to nonresponders (Fig. 4C). There was no difference in the CXCR3+ CD45− populations between the groups (SI Appendix, Fig. S6A). Consistent with our RNA-seq analysis, we observed higher expression of CXCR3 on the CD8+ T cells in the responding tumors (Fig. 4D), suggesting CXCR3 may be important for triggering antitumor immunity in the TME. We also evaluated CXCR3 expression in other immune cell populations and observed a trend toward higher CXCR3 expression in CD19+ B cells and CD4+ T cells, but the results were not statistically significant (SI Appendix, Fig. S6B). Thus, the difference in CXCR3 expression between the responders and nonresponders primarily came from the CD8+ T cell population.
To examine the functional significance of CXCR3 expression in response to PD-1 blockade, we examined whether CXCR3 deficiency might lead to lower therapeutic response. We implanted the same numbers of tumor cells orthotopically into Cxcr3−/− or wild-type (WT) mice and treated them with anti-PD-1 antibody. Loss of CXCR3 promoted a nonresponder phenotype early in the response, with only 30% of Cxcr3−/− mice responding to ICB as compared to the typical 60 to 70% seen in WT (Fig. 4 E and F) mice at day 15 posttreatment initiation. However, CXCR3 deficiency was not sufficient to sustain the nonresponder phenotype later in the response. After day 15 posttreatment initiation, 43% of the nonresponsive tumors began to regress. These findings suggest there may be compensatory mechanisms that enable responses to anti-PD-1 in Cxcr3−/− mice. Considering that exhausted CD8+ T cells acquire a fixed differentiation state at the epigenetic level (45, 46), we investigated whether Cxcr3 might be regulated epigenetically. We examined open chromatin regions using assay for transposase-accessible chromatin (ATAC) sequencing data from publicly available datasets (45, 47, 48) in naïve, effector (acute LCMV) day 8 postinfection (p.i.), memory (acute LCMV day 30 p.i.), and exhausted CD8+ T cell subsets (chronic LCMV day 30 p.i., progenitor and terminal exhausted subsets). We found that effector and exhausted CD8+ T cells responding to viral infection had more open chromatin regions in the Cxcr3 locus than naïve or memory (acute LCMV day 30 p.i.) CD8+ T cells (SI Appendix, Fig. S7). Collectively, these data highlight the importance of CXCR3 in promoting antitumor immunity in responder mice, though additional mechanisms can compensate in the absence of CXCR3.
Mouse-Derived Responder and Nonresponder Gene Signatures Are Predictive of Patient Survival and Response to ICB.
Using our CD8+ T cell transcriptional profiles derived from partitioning responder from nonresponder mice (Fig. 2A), we hypothesized that transcriptional gene signatures, incorporating tens to hundreds of genes, may be a powerful tool in predicting responsiveness to immunotherapy. To this end, we tested if our derived responder and nonresponder gene signatures (Fig. 3B) are predictive of response to ICB and general survival in human patients. To test our hypothesis, we conducted three separate lines of analyses based on 1) survival rates of BC patients as they relate to our gene signatures, 2) bulk RNA-seq from BC and melanoma patient samples, and 3) ICB response association based on single-cell RNA-seq of CD8+ T cells from patient samples.
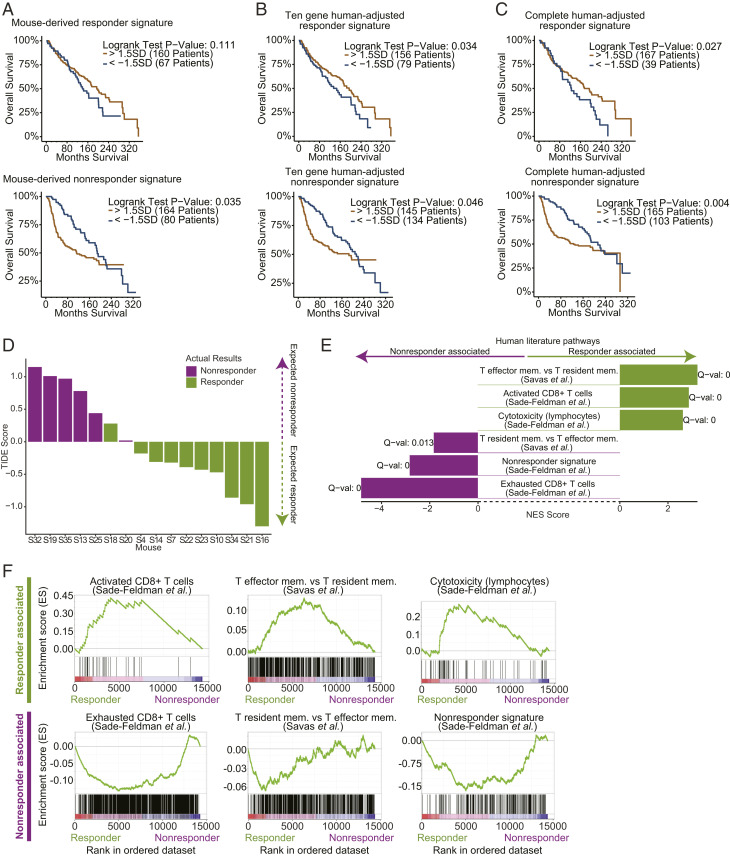
To evaluate our gene signatures as predictive measures within the BC patient landscape we analyzed how our responder and nonresponder signatures align with survival rates in BC patient samples available via the METABRIC database (23). We assessed connections between our gene signatures and general survival in the BC context because comprehensive BC response to ICB datasets is not yet available. To test for associations between our gene signatures and survival in BC patients, we developed a correlation-based method to identify subsets of patient samples that score either highest or lowest for expression of a given signature (e.g., our responder signature) (Materials and Methods). Following the identification of patient subsets from the METABRIC database that were paired with either our responder or nonresponder mouse-derived signatures, we assessed survival rates of BC patients and observed a significant association of the patients scoring highly for the nonresponder signature with worse survival, and a potential trend of the patients scoring highly for the responder signature with better survival (P = 0.035 and 0.1, respectively, Fig. 5A and Dataset S6).
Fig. 5.
Mouse-derived early therapeutic stage responder/nonresponder gene signatures are predictive of patient survival and response to ICB. (A–C) An analysis of association of BC patient survival (METABRIC database) with high or low scores for each of the responder and nonresponder signatures (Materials and Methods). (A) Partitioning BC patients by their high (gold) or low (blue) expression of the mouse-derived responder signature (Top) and nonresponder signature (Bottom). Patients with high values of the responder signature show a trend toward better survival (Top). Patients with high values of the nonresponder signature are significantly associated with worse survival (Bottom). (B and C) Following the adjustment of the mouse-derived early therapeutic stage signatures to the human BC landscape (Materials and Methods), when partitioning BC patients there is a significant association of survival with expression of the responder signature (P = 0.034 for 10-gene signature [B, Top] and P = 0.027 for complete signature [C, Top]) and a significant association of worse survival with expression of the nonresponder signature (P = 0.046 for 10-gene signature [B, Bottom] and P = 0.004 for complete signature [C, Bottom]). (D) An analysis of the mouse-derived transcriptional profiles with TIDE correctly predicts which of the mice in our cohort responded to treatment. (E) Our mouse-derived early therapeutic stage responder and nonresponder signatures shows significant association with T cell signatures annotated from patient data (enrichment analysis performed by GSEA PreRanked tests). (F) Enrichment plots as output from GSEA for select pathways from E.
While the mouse-derived nonresponder signature showed significant associations with BC patient survival (Fig. 5A), we hypothesized that a more accurate human-adjusted signature could be derived by applying a computational strategy to customize the mouse-derived signatures to the human context. Briefly, each mouse signature was used as a “core,” to which the correlation in expression of all human genes was computed. An adjusted human signature was derived by including human genes that had correlation values of three SDs or higher with the mouse signature (Dataset S5). Following this procedure to construct human-adjusted responder and nonresponder signatures, we observed that the responder and nonresponder signatures significantly associated with better and worse survival rates in BC patients, respectively (Fig. 5 B and C and Dataset S6). Last, to ensure that our reported results are consistent within BC types, we restricted our analyses to patient samples with basal-like cancers (since our mouse model was based on such). While these analyses did not result in statistically significant partitions due to the limited patient data available, we identified similar trends of associations between our gene signatures and patient survival in five out of six tests (SI Appendix, Fig. S8). Moreover, when defining the patient subsets by quartiles (rather than SD thresholds) to increase the patient count in each group and hence our statistical power, we observed a significant association of all three responder signatures with survival (P values of 0.046, 0.012, and 0.001 for the mouse-derived, 10-gene human adjusted and complete human adjusted signature, respectively).
To test the relevance of the transcriptional differences we observed in the mouse model in predicting response to ICB in patients, we utilized TIDE, a predictive computational tool based on human melanoma patients, to assess ICB response probability in patients based on their transcriptome (49). First, we ran the TIDE predictor on our mouse CD8+ T cell RNA-seq transcriptional profiles (generated shortly after ICB initiation) and found that the TIDE predictor accurately partitioned the mice into responders and nonresponders (Fig. 5D). The accurate prediction by TIDE of the response within our mouse model to ICB indicates that there are shared transcriptional elements within the predictive landscape of response to ICB in human melanoma patients and the genetically identical mice in our BC model. Next, we investigated whether our derived responder and nonresponder gene signatures associated with response to ICB in patient cohorts independent of The Cancer Genome Atlas (TCGA) dataset. We explored a melanoma patient-derived single-cell RNA-seq dataset (25) and found a significant association between our responder and nonresponder signatures with the melanoma patient-derived CD8+ T cell activation and exhaustion signatures (25), respectively (Fig. 5 E and F and Dataset S4). Interestingly, these patient-derived CD8+ T cell activation and exhaustion signatures were observed to be significantly associated with response and resistance of the melanoma tumors to ICB, respectively (25). Our responder and nonresponder signatures significantly associated with T effector memory and T resident memory signatures derived from treatment-naïve BC tumors (50), respectively (Fig. 5 E and F and Dataset S4).
Taken together, our observations suggest that a strong factor at play in determining and predicting patient responses to ICB is not genetics based, because transcriptional signatures derived from our mouse model significantly overlap with transcriptional signatures associated with patient response. Moreover, our transcriptional signatures were derived from early time points after initiation of ICB treatment, suggesting that at early time points transcriptional information from patients could be used to evaluate prognosis.
Discussion
Although studies have begun to uncover the resistance mechanisms of response to ICBs in solid tumors (51), such studies in BCs are still largely unexplored. TNBCs are of particular significance for study because of the low response rates to ICB alone in patients. A major limitation to interrogating such mechanisms in mice has been the inability to couple comprehensive analyses of tumors early during treatment (e.g., when the tumors are still comparable in size between groups) with the ultimate disease outcome later following treatment. While some methods like intravital microscopy (IVM) have been useful for tracking longitudinal antitumor responses in some settings, IVM has a number of limitations that preclude its ability to be widely used for biomarker studies (42, 52). Here we utilized a model of resection and response evaluation to investigate the responsiveness of breast tumors to anti-PD-1 blockade using a bilateral tumor approach in mice. We observed higher levels of CD8+ TILs in the TME of responder mice. Transcriptomic analysis of CD8+ TILs at early time points following anti-PD-1 treatment identified gene signatures that distinguished responders from nonresponders. Notably, these gene signatures were associated with response or resistance to ICB in patients as well as patient survival rates. These findings demonstrate the potential for this approach to discover mechanisms of response and resistance to ICB therapy and identify clinically relevant biomarkers to predict response to ICB therapy, as well as new therapeutic targets.
We focused on the T cell compartment in this study due to our observation that low levels of CD8+ TILs prior to treatment were associated with nonresponsiveness to PD-1 blockade. Moreover, using our resection and response model, we showed that within CD8+ T cells there were robust gene signatures associated with response and that those aligned with observed signatures in patients. Based on these analyses and our observation that patient survival rates significantly associated with our generated responder and nonresponder signatures, we envision that future studies could construct a classifier that utilizes distinct attributes of these human-adjusted signatures to assist with patient diagnostics and treatment decisions. While in this work we were underpowered to build an explicit classification tool to predict response, our observations indicate that there is a strong potential for building such classifiers based on the significant transcriptional differences in CD8+ T cells shortly following treatment in our mouse model and the associations observed between those transcriptional changes and survival or response to ICB in patients. Although our work highlights the relevance of using CD8+ T cell transcriptional signatures, additional work using histological analysis to determine how “immune hotspots” and the proximity of protective immune components (e.g., activated CD8+ T cells) with tumor cells would also be of interest to determine the density of CD8+ T cells needed for response in this setting. Furthermore, future studies aimed at further dissecting the immune composition of responder versus nonresponder tumors as well as key pathways driving response versus resistance from multiple cell types would be informative for identifying novel targets to improve patient responses to ICB. While our flow cytometry data showed a general decrease in Gr1+ MDSCs and an increase in the CD8/Treg ratio, more comprehensive profiling of both innate and adaptive immune populations would be useful. Consistent with this notion, a recent study by Zemek et al. (19) used a similar technique to identify pretreatment factors that are important for response to ICB treatment in mesothelioma and kidney cancer by profiling a number of both myeloid and lymphoid populations. They found that more activated infiltrating natural killer (NK) cells were present in pretreatment responder tumors, and therapeutic interventions aimed at recruitment of this population into tumors prior to ICB improved response rates. This complementary work highlights the utility of interrogating the entire immune compartment within the TME in a resection and response-type model. Follow up studies in the E0771 BC model used in our study would be useful to elucidate other features within the TME that are predictive and causal of response, including both protective and regulatory immune populations (e.g., CD4+ Foxp3− T cells, Treg cells, MDSCs, etc.) as well as nonimmune populations (e.g., endothelium, cancer-associated fibroblasts [CAFs], cancer cells, etc.).
Our transcriptomic analysis of CD8+ T cells at an early stage of ICB therapy revealed signatures that captured differences in CD8+ T cells across responders and nonresponders that are present shortly following ICB initiation. These mouse-derived responder and nonresponder gene signatures were associated with response or resistance in patients, respectively, as well as with survival rates. However, future studies are needed to validate our transcriptional signatures in breast cancer patients who receive ICB, once the data become available. When comparing our CD8 responder and nonresponder gene signatures to the available ICB-treated cohort databases (in which only bulk RNA-seq of the tumors is available) (19, 43), we did not observe significant associations of our derived gene signatures with response. This observation could be a result of substantial differences across the datasets such as the time of sample collection (pre- vs. shortly post-ICB treatment initiation), the cell population sequenced (bulk tumor vs. the CD8+ component), the difference in the types of tumors modeled, and difference of the ICB therapy administered (anti-PD-1 vs. anti-PD-L1). We further demonstrated that responding tumors had a more immunostimulatory TME by having increased levels of T cell activation markers and cytokines (IL2, TNFα, and IFNγ). Immune checkpoint therapies induce the production of IFNγ, which is critical for enhancing T cell responses (15, 44). Our data suggest that strategies to increase tumor-specific T cell immunity such as vaccine or chemotherapy could lead to greater efficacy. Indeed, combining PD-L1 blockade with nab-paclitaxel achieved an objective response rate of 42% in the patients with triple negative breast cancer, compared to 24% with PD-L1 alone (8). Moreover, new data have shown that the combination of anti-PD-L1 (atezolizumab [Tecentriq]) with chemotherapy including nab-paclitaxel met the primary endpoint of improved pathological complete response as an initial treatment for people with early triple negative breast cancer through the IMpassion031 trial, providing increasing excitement about use of checkpoint blockade in BC. In nonresponding tumors in our study, we observed an enrichment of a signature associated with exhaustion (24), suggesting that the severity of exhaustion could be a mechanism associated with resistance to anti-PD-1 in this BC model. In some models, combinations of antibodies blocking PD-1 with other immune checkpoints such as CTLA-4, LAG-3, or TIGIT showed better response rates than treating with anti-PD-1 alone (53–56). Our previous work with E0771 has shown that combination anti-PD-1 and anti-CTLA-4 resulted in a similar responder versus nonresponder rate (∼30% nonresponders) (57) as anti-PD-1 alone in the current study, suggesting that the addition of CTLA-4 blockade in E0771 would not have been sufficient to overcome exhaustion in this model. However, future studies examining different types of combinations to overcome barriers limiting protective antitumor immunity are warranted.
Focusing on a T cell trafficking signal within our responder signature, we found that CXCR3 expression had functional significance in tumor rejection upon PD-1 blockade, as reported in ref. 40, as most Cxcr3−/− mice did not respond to PD-1 blockade. CXCR3 is a key inflammatory chemokine receptor that plays a major role in immune cell trafficking through binding the IFN-inducible chemokines CXCL9, CXCL10, and CXCL11 (40, 58). Our data showed that a greater frequency of CD8+ T cells expresses CXCR3 in responder mice compared to nonresponder mice. However, it is unclear whether CXCR3 is being regulated at the genomic level, or if it is being regulated at the epigenetic level. Considering that effector and exhausted CD8+ T cells responding to viral infection had more open chromatin regions in the Cxcr3 locus than naïve or memory (acute LCMV day 30 p.i.) CD8+ T cells (SI Appendix, Fig. S7), additional studies examining the role of epigenetic regulation in responders versus nonresponders are warranted. Interestingly, some Cxcr3−/− mice were transiently resistant to anti-PD-1 antibody, suggesting compensatory mechanisms that are highly sensitive to other cytokines may circumvent CXCR3 deficiency (39, 59). One possibility is that other inflammatory trafficking pathways eventually overcome this resistance in Cxcr3−/− mice, such as other chemokine receptors (e.g., CCR5/CCL3,4,5 axis), integrins (e.g., α4β1/VCAM-1 or LFA-1/ICAM-1), or selectins (e.g., PSGL1 binding of P-selectin and/or E-selectin) (60). Another possibility is that without CXCR3, antitumor CD8+ T cells have delayed migration to tumors due to a failure to migrate toward CXCL9 and/or CXCL10 produced by high endothelial venules (HEVs) in the draining lymph node (dLN), which could affect their trafficking to the tumor and/or their differentiation within the LN (61, 62). Future studies in Cxcr3−/− mice, including comparing transcriptional profiles of responders and nonresponders, would be informative for elucidating these compensatory mechanisms. Further investigation of this mechanism and additional pathways governing CD8+ T cell responses within the breast TME is crucial for developing optimal therapies for BCs (63).
In order for chemokine-mediated trafficking to occur, both the ligand (CXCL9 or CXCL10) and receptor (CXCR3) must be present; consequently, regulation at the level of both the ligand and receptor could be contributing to productive antitumor immunity in this setting. Since CXCL11 is not expressed in B6 mice, CXCL9 and CXCL10 are the main drivers of CXCR3-mediated trafficking in this mouse model (58, 64, 65). Upon stimulation with inflammatory cytokines, including IFNγ, CXCL9, and CXCL10, can be produced by a number of different cell types, including monocytes, fibroblasts, endothelial cells, and cancer cells (58, 65). The mechanisms by which the CXCR3/CXCL9-10 trafficking axis contributes to productive antitumor immunity in this setting remain unclear. Considering regulation of the ligand, it is possible that elevated inflammatory cytokines in the TME of responder mice drive higher CXCL9 and CXCL10 production, acting as a stronger chemoattractant for CXCR3+ CD8+ T cells. Alternatively, it is possible that responder and nonresponder tumors have a differential composition of cell types capable of producing CXCL9 and CXCL10. Lastly, it is possible that the nonresponder tumors either specifically lack CXCL9 and CXCL10 (which would be consistent with these tumors being immunologically “cold”) or that these tumors express higher levels of immunosuppressive chemokines that may either repel CD8+ T cells or attract regulatory populations such as MDSCs. Interestingly, Ccl1 and Xcl1 are up-regulated in CD8+ T cells in nonresponder tumors. CCL1 is the ligand for CCR8, a chemokine receptor involved in recruiting monocytes, NK cells, Tregs, DCs, and certain populations of B cells (66). XCL1 is the ligand for XCR1, which is expressed by cross-presenting DCs, and may be involved in helping CD8+ T cells colocalize with antigen presenting cells (67). Additional work is needed to further define how these opposing chemokine pathways are functioning to regulate antitumor immunity in this setting.
Lastly, our data highlight the importance of evaluating the dynamic immune modulation of the TME early during treatment to understand the potential benefit of immunotherapy. Our findings provide a general approach for investigating mechanisms of resistance to ICB and identifying predictive biomarkers of response. For example, this model may be useful in interrogating additional mechanisms by which these cohabiting, inbred mice respond differently to therapeutic interventions in tumors. Studies suggest that even though inbred mouse strains were created to be essentially genetically identical, these mice are made through many generations of brother-sister mating, and minor variations can arise over time that could create differences in the host immune responses (68, 69). Performing whole genome sequencing in this model could be useful for addressing the contributions of minor genetic variations to the responder versus nonresponder phenotype. In addition other “-omics” approaches may be coupled with this method to provide a comprehensive investigation of the dynamic changes within the TME. While our study focused on better understanding CD8+ T cell dynamics in the TME, these studies could be expanded to examine a number of both immune and nonimmune (e.g., CAFs, cancer cells, etc.) compartments within the tumor. In summary, this method presents a way to dissect how different cell types in the TME contribute to response and resistance to immunotherapies before and early during ICB treatment, and has the potential to identify new therapeutic targets and combination therapies.
Materials and Methods
Mouse Tumor Models.
All animal experiments were performed using 6- to 8-wk-old female C57BL/6 mice. For bilateral tumor models, 200,000 cells were implanted at the same time into both third mammary fat pads of a mouse. Tumor sizes were measured with a caliper. All animal procedures were carried out following the Public Health Service Policy on Humane Care of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital.
Histology.
Mice bearing orthotopic E0771 tumors were split into time- and size-matched (∼75 mm3) treatment groups. Tumor sizes were measured with a caliper. The mice were then treated with an anti-PD-1 antibody (200 μg, RMP1-14, BioXcell) or a control IgG antibody (200 μg, 2A3, BioXcell) on days 8, 11, and 14. Tumors were isolated at different time points during the treatment. The tumors were removed, fixed in 4% formaldehyde in phosphate-buffered saline (PBS) (30 min/mm diameter of tissue), incubated in 30% sucrose in PBS overnight at 4 °C, and frozen in optimal cutting temperature compound (OCT) (Tissue-Tek). Frozen sections (20 μm thick) were blocked with 5% normal donkey serum (NDS) and immunostained with primary antibodies. CD8 was stained (BioLegend, 53.6-7, 1:100 dilution) and slides were counterstained with DAPI (Vector Labs).
RNA-Sequencing.
Mice bearing orthotopic E0771 tumors were treated with an anti-PD-1 antibody (200 μg, RMP1-14, BioXcell) or a control IgG (200 μg, 2A3, BioXcell) on days 8, 11, and 14. At day 15, the tumors were then excised and single cell suspensions were made. The suspensions were diluted in PBS buffer with 2% bovine serum albumin (BSA) and 1 mM ethylenediaminetetraacetic acid (EDTA) and the CD45+CD3+CD8+ T cell population was sorted on FACS Aria 2. RNA was extracted using Myone Silane Dynabeads (Thermo Fisher Scientific). The RNA fragments were barcoded using 8-bp barcodes in conjunction with standard Illumina adaptors. Agencourt AMPure XP bead cleanup (Beckham Coulter/Agencourt) and 14 PCR cycles were used to amplify the samples. Sequencing was carried out on a HiSEq. 2000 (Illumina). Using the aligner Salmon (http://salmon.readthedocs.io/en/latest/salmon.html) under default filtering settings, sequencing reads (fastqs) were aligned to, and count estimates calculated for, GenCode-annotated mouse (mm12, vM9) transcripts.
For extended materials and methods please SI Appendix.
Supplementary Material
Acknowledgments
We thank Sylvie Roberge, Julia Kahn, Carolyn Smith, and Tsion Tale for technical assistance. We also thank Vikash Chauhan, Meenal Datta, Dai Fukumura, Peigen Huang, Rosa Ng, and Ethel Pereira for their helpful input. We thank Christina Usher for illustrations. This research was supported by grants from the Ludwig Center at Harvard University to R.K.J. and A.H.S.; grants from the National Foundation for Cancer Research; the Jane’s Trust Foundation; the Advanced Medical Research Foundation; the National Cancer Institute and National Institutes of Health grants P01-CA080124, R35-CA197743, R01-CA208205, U01CA224348 (R.K.J.), R01CA229851 and U54CA224088 (A.H.S.), and P01AI039671 (A.H.S. and M.S.); an Innovation in Cancer Informatics Discovery Grant (M.S.); and by the Department of Defense Breast Cancer Research Program Innovator Award W81XWH-10-1-0016 (R.K.J.). This work was also supported in part by the Parker Institute for Cancer Immunotherapy (M.S.).
Footnotes
Competing interest statement: R.K.J. received an honorarium from Amgen; consultant fees from Merck, Pfizer, SPARC, and SynDevRx; owns equity in Enlight and SynDevRx; and serves on the scientific advisory board of Accurius Therapeutics and boards of trustees of Tekla Healthcare Investors, Tekla Life Sciences Investors, Tekla Healthcare Opportunities Fund, and Tekla World Healthcare Fund. Neither any reagent nor any funding from these organizations was used in this study. A.H.S. has patents on the PD-1 pathway licensed by Roche/Genentech and Novartis, consults for Novartis, is on the scientific advisory boards for Surface Oncology, Sqz Biotech, Elstar Therapeutics, Elpiscience, Selecta, and Monopteros, and has research funding from Merck, Novartis, Roche, and Quark Ventures. R.K.J. and D.A.T. are co-authors on a 2020 Cold Spring Harbor Laboratory meeting report.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002806117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Kalbasi A., Ribas A., Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20, 25–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas A., Wolchok J. D., Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le D. T. et al., Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haslam A., Prasad V., Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open 2, e192535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanda R. et al., Pembrolizumab in patients with advanced triple-negative breast cancer: Phase ib KEYNOTE-012 study. J. Clin. Oncol. 34, 2460–2467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emens L. A. et al., Abstract 2859: Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer (TNBC). Cancer Res. 75 (suppl. 15), 2859 (2015). [Google Scholar]
- 7.Dirix L. Y. et al., Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: A phase 1b JAVELIN Solid Tumor study. Breast Cancer Res. Treat. 167, 671–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid P. et al., Atezolizumab and Nab-Paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Rugo H. S. et al., Safety and antitumor activity of pembrolizumab in patients with estrogen receptor‒positive/human epidermal growth factor receptor 2‒negative advanced breast cancer. Clin. Cancer Res. 24, 2804–2811 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Topalian S. L., Taube J. M., Anders R. A., Pardoll D. M., Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addeo A., Banna G. L., Weiss G. J., Tumor mutation burden-from hopes to doubts. JAMA Oncol. 5, 934–935 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Cristescu R. et al., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P.-L. et al., Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang A. C. et al., A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 25, 454–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minn A. J., Wherry E. J., Combination cancer therapies with immune checkpoint blockade: Convergence on interferon signaling. Cell 165, 272–275 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Matson V. et al., The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian L. et al., Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature 544, 250–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X. et al., Increased vessel perfusion predicts the efficacy of immune checkpoint blockade. J. Clin. Invest. 128, 2104–2115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemek R. M. et al., Sensitization to immune checkpoint blockade through activation of a STAT1/NK axis in the tumor microenvironment. Sci. Transl. Med. 11, eaav7816 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Hu Z. I., McArthur H. L., Ho A. Y., The abscopal effect of radiation therapy: What is it and how can we use it in breast cancer? Curr. Breast Cancer Rep. 9, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngwa W. et al., Using immunotherapy to boost the abscopal effect. Nat. Rev. Cancer 18, 313–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAllister S. S., Weinberg R. A., The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis C. et al.; METABRIC Group , The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West E. E. et al., Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity 35, 285–298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sade-Feldman M. et al., Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivan A. et al., Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vétizou M. et al., Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tumeh P. C. et al., PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savas P. et al., Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 13, 228–241 (2016). [DOI] [PubMed] [Google Scholar]
- 30.O’Donnell J. S., Long G. V., Scolyer R. A., Teng M. W. L., Smyth M. J., Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat. Rev. 52, 71–81 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Highfill S. L. et al., Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 6, 237ra67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang A. C. et al., T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahoney K. M., Rennert P. D., Freeman G. J., Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 14, 561–584 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M. et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latchman Y. et al., PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2, 261–268 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Patsoukis N. et al., Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci. Signal. 5, ra46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patsoukis N., Sari D., Boussiotis V. A., PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle 11, 4305–4309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikucki M. E. et al., Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat. Commun. 6, 7458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow M. T. et al., Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498–1512.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munn L. L., Jain R. K., Vascular regulation of antitumor immunity. Science 365, 544–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cahalan M. D., Parker I., Choreography of Cell Motility and Interaction Dynamics Imaged by Two-Photon Microscopy in Lymphoid Organs. Annu. Rev. Immunol. 26, 585–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariathasan S. et al., TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaretsky J. M. et al., Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen D. R. et al., The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauken K. E. et al., Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller B. C. et al., Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaFleur M. W. et al., PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nat. Immunol. 20, 1335–1347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang P. et al., Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savas P. et al.; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) , Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Restifo N. P., Smyth M. J., Snyder A., Acquired resistance to immunotherapy and future challenges. Nat. Rev. Cancer 16, 121–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okada T., Takahashi S., Ishida A., Ishigame H., In vivo multiphoton imaging of immune cell dynamics. Pflugers Arch. 468, 1793–1801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma P., Allison J. P., Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 161, 205–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakuishi K. et al., Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 207, 2187–2194 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woo S.-R. et al., Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnston R. J. et al., The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 26, 923–937 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Chen I. X. et al., Blocking CXCR4 alleviates desmoplasia, increases T-lymphocyte infiltration, and improves immunotherapy in metastatic breast cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 4558–4566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tokunaga R. et al., CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat. Rev. 63, 40–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett L. D., Fox J. M., Signoret N., Mechanisms regulating chemokine receptor activity. Immunology 134, 246–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slaney C. Y., Kershaw M. H., Darcy P. K., Trafficking of T cells into tumors. Cancer Res. 74, 7168–7174 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Kurachi M. et al., Chemokine receptor CXCR3 facilitates CD8(+) T cell differentiation into short-lived effector cells leading to memory degeneration. J. Exp. Med. 208, 1605–1620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J. K., Kagari T., Clingan J. M., Matloubian M., Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc. Natl. Acad. Sci. U.S.A. 108, E118–E127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammerl D. et al., Clonality, antigen recognition, and suppression of CD8+ T cells differentially affect prognosis of breast cancer subtypes. Clin. Cancer Res. 26, 505–517 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Sierro F. et al., Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc. Natl. Acad. Sci. U.S.A. 104, 14759–14764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Groom J. R. et al., CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity 37, 1091–1103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karin N., Chemokines and cancer: New immune checkpoints for cancer therapy. Curr. Opin. Immunol. 51, 140–145 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Lei Y., Takahama Y., XCL1 and XCR1 in the immune system. Microbes Infect. 14, 262–267 (2012). [DOI] [PubMed] [Google Scholar]
- 68.van der Veeken J. et al., Natural genetic variation reveals key features of epigenetic and transcriptional memory in virus-specific CD8 T cells. Immunity 50, 1202–1217.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chisolm D. A. et al., Defining genetic variation in widely used congenic and backcrossed mouse models reveals varied regulation of genes important for immune responses. Immunity 51, 155–168.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.