Significance
PCNA is an essential protein in DNA replication and repair, and these functions rely on multiple posttranslational modifications, including small ubiquitin-like modifiers SUMO1 and SUMO2. SUMO2-conjugated PCNA has a distinct function in maintaining genomic stability from SUMO1-conjugated PCNA. Therefore, different SUMO E3 ligases are needed to provide the specificity to covalently attach either SUMO1 or SUMO2 to PCNA in response to different replication stress. However, even though SUMO-conjugated PCNA molecules have been observed in human cells since 2012, to date, the SUMO E3 ligases for PCNA have yet to be identified. This paper reports a SUMO E3 ligase that directly conjugates SUMO2 to PCNA with high specificity to prevent transcription induced DNA breaks in human cells.
Keywords: TRIM28, PCNA, SUMO2, RECQ5, DNA replication
Abstract
In human cells, the DNA replication factor proliferating cell nuclear antigen (PCNA) can be conjugated to either the small ubiquitinlike modifier SUMO1 or SUMO2, but only SUMO2-conjugated PCNA is induced by transcription to facilitate resolution of transcription–replication conflict (TRC). To date, the SUMO E3 ligase that provides substrate specificity for SUMO2-PCNA conjugation in response to TRC remains unknown. Using a proteomic approach, we identified TRIM28 as the E3 ligase that catalyzes SUMO2-PCNA conjugation. In vitro, TRIM28, together with the RNA polymerase II (RNAPII)-interacting protein RECQ5, promotes SUMO2-PCNA conjugation but inhibits SUMO1-PCNA formation. This activity requires a PCNA-interacting protein (PIP) motif located within the bromodomain of TRIM28. In cells, TRIM28 interaction with PCNA on human chromatin is dependent on both transcription and RECQ5, and SUMO2-PCNA level correlates with TRIM28 expression. As a consequence, TRIM28 depletion led to RNAPII accumulation at TRC sites, and expression of a TRIM28 PIP mutant failed to suppress TRC-induced DNA breaks.
PCNA is a DNA replication factor that forms a homotrimeric ring on DNA and interacts with and anchors DNA polymerases at the replication fork to enhance their processivity. In addition to its role in normal DNA replication, PCNA also orchestrates several cellular processes to regulate the cell cycle and maintain genomic stability. These functions depend on distinct posttranslational modifications of PCNA lysine residue 164 (K164). For example, in undamaged cells, PCNA is monoubiquitinated at K164 by the ubiquitin E3 ligase CRL4(CDT2) to antagonize the ubiquitin hydrolase USP1 (1) and facilitate proteasome-dependent degradation of p21 and CDT1 to ensure proper cell cycle progression (2). In cells treated with DNA damaging agents, monoubiquitination of PCNA K164, mediated by the ubiquitin E3 ligase RAD18, recruits error-prone DNA polymerases to the stalled replication fork and bypasses DNA lesions (3). K164 can also be polyubiquitinated to recruit ZRANB3 for DNA damage bypass through template switching (4–6). Posttranslational modification of PCNA K164 is not limited to ubiquitination (7). In both yeast and human cells, SUMOylation of PCNA K164 recruits antirecombination helicases (i.e., Srs2, PARI, FBH1, and RTEL) to the replication fork to suppress homologous recombination (8–14).
Unlike yeast, which only contains one SUMO gene, there are four SUMO paralogs in human cells (i.e., SUMO1–4). Both SUMO1- and SUMO2-conjugated PCNA are found in human cell extracts (12, 15, 16). SUMO1-PCNA recruits the helicase PARI to suppress unwanted homologous recombination at stalled replication forks (12, 15). However, whether SUMO2-PCNA plays a redundant role to SUMO1-PCNA in suppressing homologous recombination for maintaining replication fork stability was not clear until our recent study, which showed that SUMO2-conjugated PCNA but not SUMO1-conjugated PCNA was induced by transcription during DNA replication (17). SUMO2-PCNA conjugation is dependent on RECQ5 (17), a DNA helicase that functions as a tumor suppressor and is a member of the RNAPII complex (18–24). Transcription-induced SUMO2-PCNA is important for resolving TRC (17), which, if not resolved, is a major cause of DNA breaks and instability at common fragile sites (CFSs) (25, 26). SUMO2-PCNA achieves TRC resolution by dissociating active RNAPII (RNAPIIo) via chromatin remodeling by enriching the histone chaperones CAF1 and facilitates chromatin transaction (FACT) at the replication fork (17). CAF1 deposits new histones that contain repressive histone marks at the replication fork, and FACT removes parental histones ahead of the replication fork (27, 28). Through this mechanism, SUMO2-PCNA promotes chromatin remodeling to establish compact chromatin structure and destabilize RNAPIIo from TRC sites to allow replication fork progression (17). Because CAF1A, the catalytic subunit of CAF1, interacts with SUMO2 but not SUMO1 in vitro (29), this preference for SUMO2 binding likely explains why TRC specifically induces SUMO2-PCNA conjugation to facilitate CAF1-mediated chromatin remodeling.
The distinct functions of SUMO1-PCNA and SUMO2-PCNA indicate that these modifications are mediated by different sets of enzymes. In general, SUMOylation is initiated by the conjugation of a SUMO protein to the E1 ligase Sae1/2, which transfers SUMO to the E2 ligase UBC9. A SUMO E3 ligase then simultaneously binds SUMO-UBC9 and the target substrate to promote SUMO conjugation of the substrate (30). Although cells contain only one SUMO E1 and E2, there are several E3 ligases, which provide substrate specificity. Most likely, a distinct SUMO E3 ligase and/or its cofactor(s) is required to provide the specificity needed to conjugate SUMO2 but not SUMO1 to PCNA in a transcription-dependent manner. Even though the presence of SUMO1- and SUMO2-conjugated PCNA in human cell extracts has been reported since 2012 (12, 15, 16), to date, the E3 ligases for these different PCNA SUMO modifications remain unclear.
In the current study, we describe our identification of TRIM28, also known as KRAB-associated protein 1 (KAP1), as the E3 ligase that specifically conjugates SUMO2 but not SUMO1 to PCNA. TRIM28 interacts with PCNA through its PIP motif to promote SUMO2 conjugation, and this interaction depends on both transcription and RECQ5, which may act as a sensor for TRC by interacting with both components of the replication and transcriptional complexes when they are in proximity. Our identification of TRIM28 as the SUMO2-PCNA E3 ligase is further supported by our demonstration that TRIM28 depletion leads to the accumulation of RNAPII and DNA damage at TRC sites, and that DNA breaks formed in the absence of TRIM28 are suppressed by the expression of a SUMO2-PCNA fusion protein to bypass the requirement for TRIM28 in SUMO2-PCNA conjugation.
Results
TRIM28-PCNA Interaction Takes Place during DNA Replication and Is Dependent on Transcription.
The SUMO E3 ligase that promotes SUMO2 conjugation of PCNA is expected to interact with PCNA. Therefore, we searched for the presence of any known SUMO E3 ligase(s) in our mass spectrometric analysis of FLAG-tagged PCNA protein complexes purified from human chromatin prepared from HEK293T cells (17). Our mass spectrometric analysis detected the presence of the E3 ligases RANBP2 (20 peptides) and TRIM28 (16 peptides) in the purified FLAG-PCNA complex (Fig. 1A) as well as limited PIAS1 (1 peptide) and PIAS2 (1 peptide). Western blot analysis of the purified FLAG-PCNA complex confirmed the presence of the two top candidates, RANBP2 and TRIM28 (SI Appendix, Fig. S1A). Each of these SUMO E3 ligases is capable of conjugating SUMO2 to its substrate and is a potential candidate for the SUMO2-PCNA conjugation reaction (31–34).
Fig. 1.
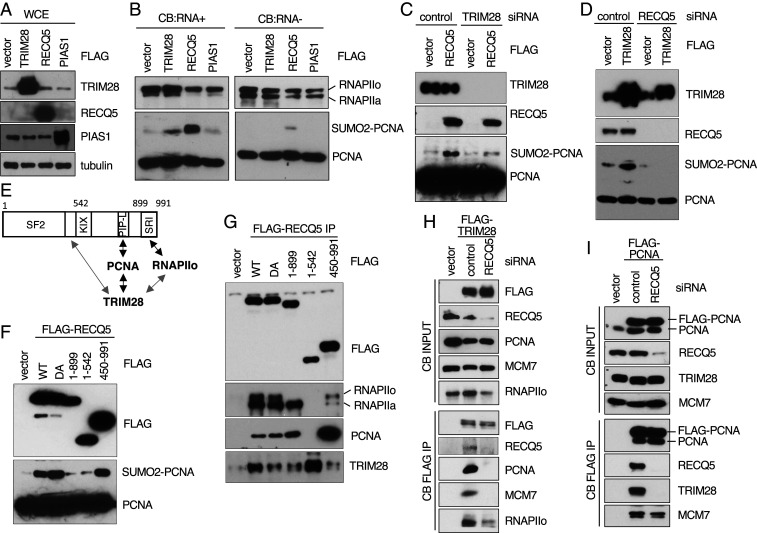
TRIM28-PCNA interaction on human chromatin is dependent on transcription. (A) Molecular weight (MW) and number of peptides detected by mass spectrometry for each of the indicated proteins that copurified with FLAG-tagged PCNA isolated from the chromatin-bound (CB) fraction of HEK293T cells as performed previously (17). (B) MW and number of peptides detected by mass spectrometry for each of the indicated proteins that copurified with FLAG-tagged RECQ5 isolated from the CB fraction of HEK293T cells. (C) Western blot analysis of the indicated proteins in the CB fractions (Left) and the FLAG-RECQ5 complexes immunopurified from CB fractions (Right) prepared from HEK293T cells with or without exogenously expressed FLAG-RECQ5. (D) Western blot analysis of the indicated proteins in the CB fractions (Left) and the endogenous TRIM28 protein complexes immunopurified using an α-TRIM28 antibody from CB fractions (Right) prepared from HEK293T cells. (E and F) Western blot analysis of the indicated proteins in the CB fractions (E) and the FLAG-PCNA complexes immunopurified from CB fractions (F) prepared from HEK293T cells with or without exogenously expressed FLAG-PCNA and with or without DRB treatment. (G) Western blot analysis of the indicated proteins in whole cell extract (WCE) of HEK293T cells containing an inducible TRIM28 shRNA construct with or without DOX treatment. Tubulin and MCM7 were used as loading controls. (H) Western blot analysis of the indicated proteins associated with transcriptionally active open chromatin (CB:RNA+; Left) and proteins bound to transcriptionally low or inactive DNA regions (CB:RNA-; Right) prepared from HEK293T cells shown in G. MCM7 was used as a loading control.
RECQ5 is a critical factor for preventing TRC-induced DNA breaks in multiple human cell lines, including HEK293T cells, and the level of SUMO2-PCNA on human chromatin is dependent on RECQ5 (17). Importantly, SUMO2 conjugation of PCNA requires a direct interaction between RECQ5 and PCNA (17). Therefore, to narrow down the potential SUMO E3 ligase candidates, we determined which of these SUMO E3 ligases was also associated with RECQ5 in an abundant amount on human chromatin. For this, we immunopurified FLAG-tagged RECQ5 from the CB fraction of HEK293T cells and identified RECQ5-associated proteins using mass spectrometry (SI Appendix, Table S1). In addition to known RECQ5-interacting proteins (i.e., RNAPII [represented by RPB1–3 and 9], PCNA, and topoisomerase I) (18, 24, 35, 36), we found that TRIM28 was the most abundant SUMO E3 ligase candidate that copurified with RECQ5 and there was also a limited RANBP2 associated with RECQ5 (Fig. 1B). Western blot analysis of the FLAG-RECQ5 complexes purified from the CB fraction confirmed the association of RECQ5 with TRIM28 (Fig. 1C). In contrast, RANBP2 was present at a much lower level in the purified FLAG-RECQ5 complexes (Fig. 1C). Using an antibody specific to TRIM28, we further validated that the interactions were observed among endogenous TRIM28, PCNA, and RECQ5 proteins (Fig. 1D).
SUMO2-PCNA conjugation takes place during DNA replication and is induced by RNAPII-dependent transcription (17). Indeed, similar to DNA replication factors FEN1 and MCM2–7 (represented by Western blot for MCM7), we found that TRIM28 was enriched in the CB FLAG-PCNA complex during the S phase (SI Appendix, Fig. S1 B and C). We further asked if RECQ5-TRIM28-PCNA interaction is regulated by transcription. Consistent with our previous observation that RECQ5-PCNA interaction primarily takes place in the transcriptionally active chromatin fraction, treatment with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), which inhibits RNAPIIo (Fig. 1E) (37), weakened PCNA interaction with RECQ5 and TRIM28 but not replication factors, such as MCM2–7 helicase (represented by Western blot for MCM2 and MCM7; Fig. 1F and SI Appendix, Fig. S1 D and E). These decreased protein–protein interactions among PCNA, TRIM28, and RECQ5 correlated with a reduced SUMO2-PCNA level on chromatin (Fig. 1F).
The ability of TRIM28 to copurify with both RECQ5 and the PCNA-containing replisome in a transcription-dependent manner suggested that TRIM28 is the E3 ligase for SUMO2-PCNA conjugation. Indeed, when we treated HEK293T cells stably integrated an inducible short hairpin RNA (shRNA) construct specific to TRIM28 with doxycycline (DOX) to induce TRIM28 shRNA and to knockdown (KD) expression of TRIM28 (Fig. 1G), we found that the level of SUMO2-PCNA, which was primarily found in the transcriptionally active chromatin fraction (i.e., CB:RNA+), was reduced in TRIM28 KD cells compared to control cells (Fig. 1H). These analyses indicated that SUMO2-PCNA conjugation is dependent on TRIM28.
TRIM28 Conjugates SUMO2 but Not SUMO1 to PCNA.
Next, we wished to determine if TRIM28 is the E3 ligase that specifically catalyzes the conjugation reaction of SUMO2-PCNA but not SUMO1-PCNA. For this, we performed in vitro SUMOylation assays using purified recombinant Strep-tagged PCNA and either His-SUMO1 or His-SUMO2 in the presence of Sae1/2 E1 ligase and UBC9 E2 ligase. We found that Sae1/2 and UBC9 alone (in the absence of RECQ5 or TRIM28) were capable of conjugating either SUMO1 or SUMO2 to PCNA (Fig. 2 A, Bottom, lanes b and f). Importantly, the level of SUMO2-PCNA was greatly enhanced by the addition of either purified recombinant RECQ5 or TRIM28 with further increase when both RECQ5 and TRIM28 were present in the reaction (Fig. 2A, compare lane f to lanes g–i). In contrast to SUMO2-PCNA conjugation, we found that the presence of TRIM28 or RECQ5 not only did not enhance SUMO1-PCNA conjugation, but also exhibited an inhibitory effect on SUMO1-PCNA conjugation, despite the same experimental conditions (Fig. 2A, compare lane b to lanes c–e). We further confirmed that the SUMO2 conjugation of PCNA took place at the K164 residue of PCNA as wild type (WT) PCNA, but not a mutant in which K164 was mutated to arginine (R) was SUMOylated in RECQ5- and TRIM28-dependent manners (Fig. 2B). In addition, we showed that WT TRIM28 (Fig. 2C, lanes d–f) but not SUMO E3 ligase defective C651F mutant (Fig. 2C, lanes g–i) (38) stimulated SUMO2-PCNA conjugation. Interestingly, at higher concentration, TRIM28 C651F mutant further inhibited the stimulative effect by RECQ5 (Fig. 2C, compare lanes c and i). This result concludes that TRIM28 SUMO E3 ligase activity is required for the conjugation of SUMO2 to PCNA.
Fig. 2.
TRIM28 and RECQ5 catalyze SUMO2-PCNA conjugation but inhibit SUMO1-PCNA formation. (A) Western blot analysis using anti-StrepII antibody to detect recombinant StrepII-PCNA after in vitro SUMOylation assays in the presence or absence of His-tagged SUMO1 or SUMO2, RECQ5, and TRIM28. Long exposure (Bottom) shows the formation of SUMO1-PCNA in the absence of a SUMO E3 ligase; this conjugation decreased in the presence of TRIM28 or RECQ5. All reactions contained Sae1/2 and UBC9. (B) Western blot analysis using anti-StrepII antibody to detect recombinant StrepII-PCNA WT (Left) and K164R mutant (Right) after in vitro SUMOylation assays in the presence or absence of RECQ5 and TRIM28. All reactions contained SUMO2, Sae1/2, and UBC9. (C) Western blot analysis using anti-StrepII antibody to detect recombinant StrepII-PCNA after in vitro SUMOylation assays in the presence of His-tagged SUMO2, RECQ5 and increasing amounts of recombinant TRIM28 WT (d–f) or C651F mutant (g–i). (D) Western blot analysis using anti-StrepII antibody to detect recombinant StrepII-PCNA after in vitro SUMOylation assays in the presence of His-tagged SUMO1 or SUMO2 and increasing amounts of FLAG-TRIM28 protein complex purified from the CB fraction of HEK293T cells.
To validate the specificity of TRIM28 on SUMO2 but not SUMO1 conjugation of PCNA, we immunopurified FLAG-TRIM28 protein complexes from the CB fraction of HEK293T cells stably expressing FLAG-TRIM28 proteins and performed in vitro SUMOylation reactions. We found that, similar to the results of reactions using purified recombinant proteins (Fig. 2A), FLAG-TRIM28 complexes purified from human chromatin also stimulated SUMO2 conjugation of PCNA while inhibiting the formation of SUMO1-PCNA (Fig. 2D). To summarize, we identified TRIM28 as the SUMO E3 ligase that works with RECQ5 to promote SUMO2-PCNA conjugation but inhibits SUMO1-PCNA conjugation.
PIP Motif of TRIM28 Is Required for SUMO2-PCNA Conjugation and Suppresses Transcription-Induced DNA Breaks.
We next determined the residues on TRIM28 that are involved in direct binding to PCNA (i.e., the substrate) and are important for subsequent conjugation to SUMO2. PCNA-interacting proteins are known to bind to PCNA via a PIP motif (39). We identified two PIP motifs within TRIM28: PIP1 among residues 342–348 and PIP2 among residues 786–792 (Fig. 3A), then, performed mutagenesis to make TRIM28 PIP1 and PIP2 mutants. When we expressed TRIM28 PIP1 or PIP2 mutants as FLAG-tagged proteins in HEK293T cells, we found that PCNA copurified with the PIP1 mutant (Fig. 3B). In contrast, PCNA showed reduced interaction with the PIP2 mutant, suggesting that PIP2 is responsible for TRIM28 interaction with PCNA. In addition, MCM2-7 was copurified with TRIM28 (Fig. 1D), but the amount of MCM2–7 associated with the TRIM28 PIP2 mutant was also reduced (Fig. 3B), suggesting that TRIM28 interacts with other components of the replisome through PCNA. PIP2 is located within the bromodomain of TRIM28 (Fig. 3A); together with an upstream PHD finger, this bromodomain has been shown to possess SUMO E3 ligase activity (38, 40). Therefore, to determine if TRIM28 binding to PCNA via its PIP2 motif is required for conjugating SUMO2 onto PCNA, we purified TRIM28 PIP1 and PIP2 mutants as recombinant proteins from Escherichia coli and tested the ability of each to promote SUMO2 conjugation of PCNA in vitro. Indeed, we found that SUMO2-PCNA failed to form in reactions containing the TRIM28 PIP2 mutant (Fig. 3C, lanes f–h). In contrast, the TRIM28 PIP1 mutant induced SUMO2 conjugation of PCNA in a concentration-dependent manner (Fig. 3C, lanes i–k) similar to the WT TRIM28 (Fig. 3C, lanes c–e). Taken together, these results suggest that TRIM28 interacts with PCNA via its PIP2 motif and that this motif, located within the bromodomain of TRIM28, is required for SUMO2 conjugation of PCNA.
Fig. 3.
Identification of the PIP motif of TRIM28 as important for SUMO2-PCNA conjugation. (A) Diagram of human TRIM28. The PHD domain and bromodomain (light gray box) important for its SUMO E3 ligase function, PIP1 and PIP2 residues are shown. (B) Western blot analysis of the indicated proteins in the CB fractions (Left; input) and in the FLAG-TRIM28 complexes immunopurified from CB fractions (Right) prepared from HEK293T cells expressing FLAG-tagged WT, PIP1 mutant, or PIP2 mutant TRIM28 proteins. (C) Western blot analysis using anti-StrepII antibody to detect recombinant StrepII-PCNA (Bottom) after in vitro SUMOylation assays in the presence of RECQ5 and increasing amounts of WT, PIP1, or PIP2 mutant TRIM28 proteins (Top). (D) Western blot analysis of the indicated proteins in the WCE of TRIM28-shRNA-containing HEK293T cells with or without DOX treatment and expressing indicated FLAG vector (V), FLAG-TRIM28, or FLAG-PCNA constructs. Actin was used as a loading control. (E) Average tail moments (horizontal lines) measured using neutral comet assay in the cells shown in D. Each dot represents the tail moment of a single cell. At least, 150 cells were analyzed per experiment. The graph is representative of one experiment; the result was reproduced in two independent comet assays.
SUMO2-PCNA is important for resolving TRC to prevent DNA breaks (17). To demonstrate that TRIM28 as the SUMO E3 ligase for SUMO2-PCNA is required for preventing TRC-induced DNA breaks, we transiently expressed WT, PIP1 mutant, or PIP2 mutant TRIM28 as FLAG-tagged proteins in TRIM28 shRNA KD cells (Fig. 3 D, Top). We then measured DNA break frequencies in these cells using neutral comet assays. After DOX induction to deplete TRIM28, we observed an expected increase in tail moments (indicative of DNA breaks); this increase was suppressed by the expression of either the WT or the PIP1 mutant TRIM28 (Fig. 3E, comparing columns 1–4 from the left). In contrast, the increase in tail moments was sustained after complementation with the PIP2 mutant (Fig. 3E, third column from the right). To determine if TRIM28-mediated SUMO2 conjugation of PCNA is required for suppressing DNA breaks in TRIM28 KD cells, we expressed the PCNA K164R mutant (i.e., KR; Fig. 3D, third lane from the left), and confirmed that the PCNA KR mutant failed to suppress DNA breaks by TRIM28 KD (Fig. 3E, last column from the right). In contrast, in TRIM28 KD cells expressing a SUMO2-PCNA KR fusion protein (i.e., S2-KR; Fig. 3D, fourth lane from the left), which functionally mimics PCNA that has been SUMO2 conjugated at K164 (17), suppressed the increase in tail moments in the TRIM28 KD cells (Fig. 3E, second column from the right), confirming that S2-KR but not KR PCNA could bypass the requirement for TRIM28 to suppress DNA breaks.
TRIM28 Prevents RNAPIIo Accumulation at CFSs.
SUMO2-PCNA resolves TRC by destabilizing RNAPIIo from TRC sites (17). Therefore, to validate the role of TRIM28 in facilitating SUMO2 conjugation of PCNA, we analyzed RNAPII occupancy at FRA7K, a known CFS located within the IMMP2L gene, in TRIM28 shRNA KD cells. To do this, we performed chromatin immunoprecipitation (ChIP) using anti-RNAPIIo antibody (4H8) in HEK293T cells with or without TRIM28 shRNA induction (Fig. 4 A and B). After reversing the crosslinks between proteins and DNA, we measured the amount of RNAPIIo bound to FRA7K by qPCR using primers complementing different regions of the IMMP2L gene. We found that RNAPIIo accumulated within the FRA7K region of the IMMP2L gene, but there was no significant accumulation of RNAPIIo in the IMMP2L coding gene region outside of FRA7K (Fig. 4C). The significant accumulation of RNAPIIo within FRA7K was accompanied by an increased level of γH2AX in the same cells, indicative of DNA damage (Fig. 4D). Importantly, the accumulation of RNAPIIo (Fig. 4E) and the increased γH2AX level (Fig. 4F) were suppressed by overexpressing PCNA S2-KR to bypass the requirement of TRIM28. We further confirmed that the elevated RNAPIIo levels were observed at other CFS loci (i.e., FRA3B, FRA16D, and FRA7I) after TRIM28 KD, and RNAPIIo accumulation was suppressed in all fragile sites by overexpressing PCNA S2-KR (SI Appendix, Fig. S2). On the other hand, overexpressing PCNA KR showed little or reduced efficiency in suppressing RNAPIIo accumulation at CFSs (SI Appendix, Fig. S2). To summarize, these data together support a role for TRIM28 in TRC resolution via SUMO2 conjugation of PCNA.
Fig. 4.
TRIM28 deficiency leads to the accumulation of RNAPIIo and γH2AX at CFS. (A) Western blot analysis of the indicated proteins in formaldehyde-treated TRIM28-shRNA-containing HEK293T cells with or without DOX treatment. Tubulin was used as a loading control. (B) Western blot analysis of RNAPIIo that was immunopurified using an α-RNAPII phosphor-CTD 4H8 antibody or a control immunoglobulin G antibody from cells shown in A. The blot was probed using an α-RNAPII A10 antibody. (C) Schematic (Top) of regions containing DNA breaks associated with FRA7K in the IMMP2L gene. The immunopurified RNAPIIo shown in B was used for ChIP analysis (Bottom) of RNAPIIo occupancy at the indicated regions of the IMMP2L gene using primers derived from the indicated regions of the gene. Each bar represents the average value ±SD calculated from triplicate qPCR reactions per one representative experiment. Indicated P values were calculated using t test for statistically significant differences. (D) ChIP analysis of γH2AX occupancy at the indicated regions of the IMMP2L gene using an α-γH2AX antibody using the same cells shown in A. Each bar represents the average value ±SD calculated from triplicate qPCR reactions per one representative experiment. Indicated P values were calculated using t test for statistically significant differences. (E and F) ChIP analysis of RNAPIIo (E) and γH2AX (F) occupancy at FRA7K (IMMP2L3b) of the IMMP2L gene using TRIM28-shRNA-containing HEK293T cells with or without DOX treatment and with or without expressing PCNA S2-KR fusion protein.
RNAPIIo-Associated RECQ5 Promotes TRIM28-PCNA Interaction for SUMO2-PCNA Conjugation.
In cells, SUMO2-PCNA conjugation requires both TRIM28 and RECQ5 (Fig. 1H) (17). Indeed, overexpressing RECQ5 or TRIM28 (Fig. 5A) enhanced SUMO2-PCNA conjugation in the CB:RNA+ fraction (Fig. 5B) (17). Although PIAS1 has been shown to contribute to SUMOylation of PCNA in human cells (41), overexpressing PIAS1 (Fig. 5A) had little effect on the SUMO2-PCNA level on chromatin (Fig. 5B), confirming the specificity of SUMO2-PCNA by TRIM28. To confirm that RECQ5 and TRIM28 function synergistically on chromatin to promote SUMO2-PCNA conjugation, we transiently KD TRIM28 using siRNA and suppressed the enhancement by RECQ5 overexpression (Fig. 5C). The enhanced SUMO2-PCNA conjugation by TRIM28 overexpression was also abolished after RECQ5 KD using a siRNA specific to RECQ5 (Fig. 5D).
Fig. 5.
TRIM28-PCNA interaction on human chromatin is dependent on RNAPIIo-associated RECQ5. (A) Western blot analysis of the indicated proteins in WCE prepared from HEK293T cells overexpressing the indicated FLAG constructs. Tubulin was used as a loading control. (B) Western blot analysis of the indicated proteins in CB:RNA+ (Left) and CB:RNA fractions (Right) of HEK293T cells overexpressing the indicated FLAG proteins. The ratio of the active form of RNAPII (RNAPIIo) relative to the inactive form of RNAPII (RNAPIIa) was used as the fractionation control for CB:RNA+ and CB:RNA- fractions. (C) Western blot analysis of the indicated proteins in WCE prepared from HEK293T cells with or without overexpression of FLAG-RECQ5 and treated with control or TRIM28 siRNA. (D) Western blot analysis of the indicated proteins in the CB:RNA+ fraction prepared from HEK293T cells with or without overexpression of FLAG-TRIM28 and treated with control or RECQ5 siRNA. (E) Diagram of human RECQ5. The SF2, KIX, PIP-L, and SRI domains are shown. Double arrows indicate protein–protein interactions among TRIM28, PCNA, RNAPIIo, and different domains of RECQ5. (F) Western blot analysis of the indicated proteins in WCE prepared from HEK293T cells with or without overexpression of FLAG-RECQ5 WT and mutants. (G) Western blot analysis of the indicated proteins in the FLAG-RECQ5 WT or mutant complexes immunopurified from CB fractions prepared from HEK293T cells with or without exogenously expressed FLAG-RECQ5 WT or mutant proteins as shown in F. (H) Western blot analysis of the indicated proteins in the CB fractions (Top) and the FLAG-TRIM28 complexes immunopurified from CB fractions (Bottom) prepared from HEK293T cells with or without exogenously expressed FLAG-TRIM28 and treated with control or RECQ5 siRNA. (I) Western blot analysis of the indicated proteins in the CB fractions (Top) and the FLAG-PCNA complexes immunopurified from CB fractions (Bottom) prepared from HEK293T cells with or without exogenously expressed FLAG-PCNA and treated with control or RECQ5 siRNA.
We next wished to determine the role of RECQ5 in promoting TRIM28-mediated SUMO2-PCNA conjugation. Previously, we identified a novel PIP-like (PIP-L; Fig. 5E) motif at the C terminus of RECQ5, and this motif is important for PCNA interaction and SUMO2-PCNA conjugation (17). Indeed, the overexpressed RECQ51–542 fragment lacking the PIP-L motif (Fig. 5 F, Top) failed to enhance SUMO2-PCNA formation (Fig. 5 F, Bottom). Overexpressing the RECQ5450–991 fragment lacking the N-terminal superfamily helicase domain 2 (SF2; Fig. 5E) enhanced SUMO2-PCNA formation (Fig. 5 F, Bottom), indicating that RECQ5 helicase activity is not required for SUMO2-PCNA formation. Indeed, overexpressing the RECQ5 ATPase-defective D157A (DA) mutant efficiently induced SUMO2-PCNA conjugation (Fig. 5 F, Bottom). RECQ5 also interacts with inactive RNAPII (RNAPIIa) through the KIX domain and RNAPIIo via the SET2-RPB1-interction (SRI) domain (Fig. 5E) (18, 20, 42, 43). Interestingly, overexpressing the RECQ51–899 fragment, which interacts with PCNA and RNAPIIa but not RNAPIIo (Fig. 5G), failed to enhance SUMO2-PCNA formation (Fig. 5 F, Bottom). This result indicated that SUMO2-PCNA conjugation requires RECQ5 interactions with both PCNA and RNAPIIo.
TRIM28 copurified with WT RECQ5 and all of the truncation mutants, including RECQ51–542 and RECQ5450–991 fragments (Fig. 5G), suggesting a potential direct interaction between TRIM28 and residues 450–542 of RECQ5. Alternatively, TRIM28 is a component of the transcriptional elongation complex to regulate RNAPIIo release from the pausing site (44), and we found that RNAPIIo copurified with TRIM28 (Fig. 1D). It is possible that TRIM28 interacts either directly with the N terminus of RECQ5 or indirectly through their mutual interactions with RNAPIIo or both (Fig. 5E, gray arrows). Indeed, when we immunopurified the FLAG-TRIM28 complex from the CB fractions of the control and RECQ5 KD cells, we found that RECQ5 depletion only weakened but not abolished TRIM28-RNAPIIo interaction on chromatin (Fig. 5H). Importantly, we found that PCNA and MCM2–7 failed to copurify with TRIM28 in RECQ5-depleted cells (Fig. 5H), and we further confirmed this effect by immunopurifying FLAG-PCNA in the control and RECQ5 KD cells (Fig. 5I). In contrast, the interaction of PCNA with MCM2–7 helicase was not affected after RECQ5 depletion (Fig. 5I). Most likely, RECQ5 promotes the interaction between TRIM28 and its substrate PCNA as part of the replisome. Indeed, when we incubated equal amounts of purified recombinant TRIM28 with Strep-PCNA immobilized on Strep-Tactin beads in the presence of increasing amounts of RECQ5, we found that the amount of TRIM28 pulled down by Strep-PCNA was directly proportional to the amount of RECQ5 present in the reaction (SI Appendix, Fig. S3). Taken together, our results identify multiple interactions among RECQ5, RNAPIIo, PCNA, and TRIM28 on human chromatin (Fig. 5E), and this RECQ5-RNAPIIo-PCNA-TRIM28 complex is crucial for SUMO2-PCNA conjugation.
Discussion
SUMO modification of the DNA replication factor PCNA was first observed in human cells in 2012 (12, 15, 16). However, identifying the SUMO E3 ligase(s) that catalyzes this reaction has been difficult, due to the fact that, in vitro, PCNA can be efficiently SUMOylated in the absence of a SUMO E3 ligase, leaving the requirement for an E3 ligase that facilitates SUMO conjugation of PCNA in human cells ambiguous (45, 46). Using an unbiased proteomic approach, our study identified TRIM28 as the SUMO E3 ligase that promotes SUMO2-specific conjugation of PCNA on human chromatin. Consistent with the role of SUMO2-PCNA conjugation in resolving TRC (17), TRIM28 deficiency led to an accumulation of spontaneous DNA breaks, which could be suppressed by exogenously expressing SUMO2-PCNA fusion protein to bypass the requirement for TRIM28 in SUMOylating PCNA. Importantly, PIP2 mutation, which disrupted the interaction of TRIM28 with PCNA and impaired SUMO2-PCNA conjugation, failed to suppress DNA breaks in TRIM28 KD cells, arguing that TRIM28 plays a significant role as the E3 ligase for SUMO2-PCNA in genome maintenance. In addition to the PIP2 motif, phosphorylation of TRIM28 serine 473 (S473) has been shown to enhance TRIM28 association with the replication complex to reestablish heterochromatin after DNA replication (47). It would be a great interest in the future to explore potential crosstalk between S473 phosphorylation and TRIM28-dependent SUMO2-PCNA conjugation.
SUMO2-PCNA resolves TRC by recruiting CAF1 and FACT to promote histone exchanges to establish repressive chromatin and destabilize RNAPII from the TRC site to facilitate replication fork progression (17). Of particular relevance to our study, TRIM28 has been shown to functionally interact with CAF1 for retroviral gene silencing (48). TRIM28 also contributes to transcriptional gene silencing via the recruitment of a H3K9 deacetylase (CHD3) and a H3K9 methyltransferase (SETDB1) to promote an inactive chromatin structure (49, 50). These functions of TRIM28, coupled with its newly demonstrated role as the SUMO E3 ligase for PCNA, are consistent with the role of SUMO2-PCNA in recruiting CAF1 to the replication complex and increasing H3K9me2/3 repressive marks (17).
In addition to the association with RNAPII, RECQ5 also interacts with PCNA (35, 36), and this interaction is required for SUMO2 conjugation of PCNA (17). In the present study, we further revealed the mechanism by which RECQ5 promotes SUMO2 conjugation of PCNA at transcriptionally active chromatin. We showed that RNAPIIo-associated RECQ5 mediates SUMO2-PCNA conjugation. We suggest that RECQ5 acts as a sensor to promote TRIM28-dependent SUMO2 conjugation of PCNA when RECQ5 simultaneously interacts with components of replisome (i.e., PCNA) and transcriptional elongation complex (i.e., RNAPIIo and TRIM28) when they are in proximity. Alternatively, additional TRIM28 may be recruited to TRC for SUMO2-PCNA conjugation through a direct interaction with the N terminus of RECQ5. Interestingly, our previous mass spectrometric analysis revealed that components of the RNAPII are the most abundant proteins copurified with RECQ5 on human chromatin (18). Our new mass spectrometric analysis of the RECQ5 complexes purified from the CB fraction of HEK293T cells pretreated with NEM, a ubiquitin and SUMO hydrolase inhibitor, revealed that the associations of replication factors including MCM2–7 and FACT histone chaperone with RECQ5 were greatly stabilized by NEM as indicated by the high number of the peptides derived from these proteins (SI Appendix, Table S1). It would be a great interest to determine if the interactions of RECQ5 with these enzymes are dependent on the SUMOylation or ubiquitination of RECQ5 or the partner proteins.
The contribution of TRIM28 to TRC resolution suggests that, similar to RECQ5, TRIM28 may function as a tumor suppressor. Indeed, TRIM28 haploinsufficiency has been linked to Wilms tumor (51). Interestingly, CFS instability within the WWOX gene, which can be suppressed by SUMO2-PCNA (17), has also been shown to be associated with Wilms tumor (52). It would be of great interest to establish animal models to determine if TRIM28 in its role as the E3 ligase for SUMO2-PCNA directly contributes to Wilms tumor pathogenesis in patients with TRIM28 haploinsufficiency.
Materials and Methods
Plasmids.
TRIM28 complementary DNA (cDNA) was PCR-amplified from a HeLa cDNA library, then, cloned into p3xFLAG CMV7.1 (Sigma-Aldrich) between the HindIII and the EcoRI sites to generate an N-terminal FLAG-tagged TRIM28 mammalian expression construct. The TRIM28 cDNA was also cloned into pTXB1 (NEB) between the NdeI and the EcoRI sites to generate a C-terminal chitin-binding domain-tagged TRIM28 bacterial expression construct. The TRIM28 PIP motif mutants were generated by mutagenesis using the WT plasmid as the template and the following primers: PIPM1: 5′-CCAAGATCCAGAAGCACGCGGAGCACGCTCTGCGCGCTGCCTCTTGGGCTCTGG-3′ and PIPM2: 5′-TGCAGTCCATCATCGGCGCGCAGCGCGCCGCCGAGACGCGCATGAACG-3′. The primer for generating the TRIM28 C651F mutant was: 5′-GTTTCCACCTGGACTTTCA CCTGCCGGCCCT-3′. PCNA K164R mutagenesis was performed as previously described (17). Complementary oligonucleotides containing shRNA targeting TRIM28 was dimerized and cloned into the pLKO-Tet-On vector. The target sequence for the TRIM28 shRNA construct was: 5′-CCTGGCTCTGTTCTCTGTCCT-3′. pET11-SUMO1 and pET11-SUMO2 were kindly provided by Dr. Yuan Chen (City of Hope) and used for expression and purification of bacterial His-SUMO1 and His-SUMO2. The StrepII-PCNA, pTXB1-RECQ5, pBiFC-VN173-PCNA WT, VN173-PCNA K164R (PCNA KR), VN173-SUMO2-PCNA K164R (S2-KR), pCMV-FLAG-PIAS1, and pCMV-FLAG-RECQ5 constructs were generated during our previous study (17). All plasmid sequences were confirmed by DNA sequencing.
Antibodies.
Rabbit α-TRIM28 (no. 2,521; 1:5,000) was from ProSci Incorporated. Mouse α-PCNA PC10 (sc-56, 1:5,000), mouse α-tubulin (sc-8,035; 1:3,000), rabbit α-H3 (sc-10,809), goat α-actin (sc-1,616; 1:1,000), mouse α-RanPB2 (sc-74,518; 1:1,000), mouse α-His (sc-8,036; 1:1,000), mouse RECQ5 (sc-515,050), and mouse α-RNAPII A10 (sc-17,798, 1:1,000) were from Santa Cruz Biotechnology. Mouse α-RNAPII phospho-CTD (phospho S5; 4H8; C49,196; 1:5,000) was from Lifespan Biosciences, Inc. Rabbit α-GAPDH (no. 2,118; 1:5,000) and rabbit α-PIAS1 (no. 3,550; 1:1,000) were from Cell Signaling. Rabbit α-MCM7 (ab52,489; 1:5,000) was from Abcam. Mouse α-NWSHPQFEK tag (StrepII tag; A01,732; 1:3,000) was from GeneScript. Rabbit α-FLAG (F7,425; 1:5,000) was from Sigma-Aldrich. Mouse α-γH2AX (NP002,096) for ChIP was from EMD Millipore. Rabbit α-Trim28 for ChIP (15,202-1-AP) was from Proteintech. Rabbit α-PCNA (1:2,000) was kindly provided by Dr. Robert Hickey (City of Hope). Rabbit α-RECQ5 (1:3,000) was generated during our previous study (20).
Cell Culture, Cell Transfection, and Cell Cycle Synchronization.
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and streptomycin/penicillin (1,00 U mL−1). TRIM28 siRNA (sc-38550) was purchased from Santa Cruz. RECQ5 stealth small interfering RNA (siRNA) 5′-UAGACUUGGCAAUAUUCCAAUGGGC-3′ was purchased from Invitrogen. Plasmids and siRNAs were transfected using continuum transfection reagent (GEMINI). When DRB and DOX were used, the concentrations were 50 μM and 25 ng mL−1, respectively. For synchronization, cells were cultured in DMEM with 50 ng/mL nocodazole for 22 h and, then, released by washing 2× with complete DMEM. Fluorescence-activated cell sorter analysis was carried out using a standard propidium iodide method.
Cell Fractionation and Immunoprecipitation.
Cells were lysed (30 min on ice) in three volumes of cytoplasmic buffer (10 mM 2-amino-2-hydroxymethyl-1,3-propanediol-Cl [Tris⋅Cl] pH 7.5, 0.34 M sucrose, 3 mM CaCl2, 2 mM MgCl2, 0.1 mM [ethylenedinitrilo]tetraacetic acid [EDTA], 1 mM dithiothreitol [DTT], and 0.5% Nonidet P-40, 40 mM NEM) containing protease and phosphatase inhibitors. The nuclear pellet was collected by centrifugation (2,400 × g, 5 min). Nuclei were then resuspended in three volumes of nuclear buffer (20 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [Hepes] pH 7.5, 1.5 mM MgCl2, 1 mM EDTA, 150 mM KCl, 0.1% Nonidet P-40, 1 mM DTT, and 10% glycerol) and homogenized with a 21G1/2 needle. The intact chromatin pellet was collected after centrifugation (18,000 × g, 30 min). To obtain the CB fraction, the chromatin pellet was incubated with two volumes of nuclease buffer (20 mM Hepes pH 7.5, 1.5 mM MgCl2, 1 mM EDTA, 150 mM KCl, 10% glycerol, and 0.5 U µL−1 benzonase) overnight at 4 °C, and the supernatant was collected as the CB fraction. Alternatively, to obtain separate CB:RNA+ and CB:RNA fractions, the chromatin pellet was first incubated with RNase A in RNase A buffer (50 mM Tris⋅Cl pH 8.0, 10 mM EDTA, 150 mM NaCl, and RNase A 10 µg mL−1) for, at least, 2 h to overnight at 4 °C. The supernatant was collected as the CB:RNA+ fraction. The remaining pellet was then digested with benzonase for, at least, 2 h to overnight at 4 °C in the nuclease buffer, and the solubilized proteins were collected as the CB:RNA fraction. To immunopurify FLAG-tagged protein complexes, chromatin extracts were incubated overnight with M2-agarose (Sigma-Aldrich) at 4 °C. After binding of the protein complexes, beads were washed extensively with FLAG-A binding buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 0.3 M NaCl, 10 mM KCl, 0.2% Triton X-100, and 10% glycerol). The purified FLAG-tagged protein complexes were eluted by using either sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) loading buffer or FLAG elution A buffer (10 mM Hepes pH 7.9, 0.2 M NaCl, 0.2 mM EDTA, 0.05% Triton X-100, 0.3 mg mL−1 FLAG peptide, and 10% glycerol). All subsequent mass spectrometry analyses were conducted by the Taplin Mass Spectrometry Facility at Harvard University. To purify endogenous TRIM28 protein complexes, CB fractions were incubated overnight with a TRIM28 antibody conjugated to protein A/G agarose beads at 4 °C. After binding of the protein complexes, beads were washed extensively with IP buffer (20 mM Hepes pH 8.0, 1.5 mM MgCl2, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM EDTA, 1 mM DTT, and 10% glycerol) before analysis by Western blotting.
ChIP.
For RNAPIIo and γH2AX ChIP analyses, cells were fixed with 1% formaldehyde for 10 min at room temperature, and the reaction was stopped by the addition of 0.125 M glycine. Cells were resuspended in buffer I (5 mM Pipes pH 8.0, 85 mM KCl, and 0.5% Nonidet P-40), followed by homogenization using a Dounce homogenizer. The chromatin pellet was isolated by centrifugation at 2,350 × g for 5 min at 4 °C. The pellets were resuspended in ChIP lysis buffer (1.0% SDS, 10 mM EDTA, and 50 mM Tris pH 8.0) plus protease inhibitors, and chromatin was sheared by sonication to generate DNA fragments of <1 kb. Chromatin was diluted 10 times in ChIP dilution buffer (16.7 mM Tris pH 8.0, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, and 167 mM NaCl) plus protease inhibitors, then, precleared with protein A/G beads (Thermo Scientific) for 1 h at 4 °C. Precleared samples were incubated overnight at 4 °C with antibodies. For RNAPIIo ChIP, the beads were washed sequentially twice with low salt buffer A (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8.0, and 0.15 M NaCl), high salt buffer A (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8.0, and 0.5 M NaCl), LiCl buffer A (0.25 M LiCl, 1.0% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris pH 8.0), and TE buffer. The RNAPIIo–DNA complexes were eluted using 300 μL elution buffer (0.1 M sodium bicarbonate and 1.0% SDS) for 15 min at room temperature, reverse cross-linked by adding 20 μL 5 M NaCl, and incubated overnight at 65 °C. The DNA was digested using RNase A and proteinase K and purified by phenol-chloroform and ethanol precipitation. For γH2AX ChIP, beads were washed once in dialysis buffer (2 mM EDTA, 50 mM Tris pH 8, and 0.2% sarkosyl) and four times in wash buffer (100 mM Tris pH 8.8, 500 mM LiCl, 1% Nonidet P-40, and 1% sodium deoxycholate). After washing, the beads were resuspended in 200 μL TE buffer, and formaldehyde cross-links were reversed in the presence of 0.5% SDS overnight at 70 °C. DNA was purified using phenol/chloroform and ethanol precipitation. qPCR primers against IMMP2L were previously described (17). qPCR was performed using an ABI 7500 fast real-time PCR system and SYBR Green. Enrichment was calculated using the comparative Ct method. The IMMP2L and WWOX primers used for qPCR are as described previously (17). The qPCR primers for FHIT4 are as follows: 5′-TGGCATATTGGACAGGGGAGGT-3′ and 5′-CTAGTGCGGGACCTGGCACA-3′. The qPCR primers for CNTNAP2 are as follows: 5′-GTGCTGGGGGATGGTCTCCA-3′ and 5′-CCAGTTTCTCCCTGTGTCGCTGT-3′.
In Vitro SUMOylation and Pull-Down Assays.
In vitro SUMOylation was carried out for 4 h at 37 °C using a SUMOylation kit (Enzo Life Sciences, Inc.). For each reaction presented in Fig. 2A, StrepII-PCNA (200 nM) was incubated with the following proteins: SUMO E1 (25 nM), SUMO E2 (50 nM), SUMO1 or SUMO2 (300 nM), TRIM28 (1 nM), and RECQ5 (1 nM). Similar in vitro SUMOylation assays were performed to generate data presented in Figs. 2D and 3C, except that the TRIM28 complex was prepared from the CB fraction of HEK293T cells expressing FLAG-TRIM28. For pull-down assay, StrepII-bound and StrepII-PCNA-bound chitin beads were blocked with 1 mg mL−1 bovine serum albumin in FLAG-binding buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 250 mM NaCl, 0.1% Triton X-100, and 10% glycerol) for 30 min at 4 °C. TRIM28 and RECQ5 were then added to the chitin beads and incubated for 2 h at 4 °C. The bound proteins were washed extensively with FLAG-binding buffer, boiled in SDS sample buffer, separated by SDS/PAGE, and analyzed using Western blots.
Statistics and Reproducibility.
For all experiments, unless stated otherwise, representative analyses from a minimum of three independent experiments are shown. All in vitro analyses using purified recombinant proteins were performed, at least, three times using two different sets of purified proteins. For qPCR, each value represents mean ± SD calculated from triplicate qPCR reactions for one representative experiment. P values were calculated using two-tailed Student’s t tests for statistically significant differences.
Additional methods and materials for protein purification and neutral comet assays are available in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Sarah Wilkinson for her comments and expert editing of this paper. This work was supported by NIH Grant R01 CA225843 to Y.L. Research reported in this publication included work performed in the Analytical Cytometry Core and Drug Discovery & Structural Biology Core supported by the NCI of NIH under Grant P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004122117/-/DCSupplemental.
Data Availability.
All study data are included in the article and the SI Appendix.
References
- 1.Terai K., Abbas T., Jazaeri A. A., Dutta A., CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell 37, 143–149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havens C. G., Walter J. C., Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25, 1568–1582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannouche P. L., Wing J., Lehmann A. R., Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ciccia A. et al., Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell 47, 396–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weston R., Peeters H., Ahel D., ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26, 1558–1572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J., Ghosal G., Chen J., The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47, 410–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung W., Baxley R. M., Moldovan G. L., Bielinsky A. K., Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes 10, E10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papouli E. et al., Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Barber L. J. et al., RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135, 261–271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fugger K. et al., Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186, 655–663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moldovan G. L. et al., Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacquin A. et al., The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 41, 6501–6513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkovics P. et al., The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res. 44, 3176–3189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gali H. et al., Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 40, 6049–6059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendriks I. A., D’Souza R. C., Chang J. G., Mann M., Vertegaal A. C., System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 6, 7289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M. et al., SUMO2 conjugation of PCNA facilitates chromatin remodeling to resolve transcription-replication conflicts. Nat. Commun. 9, 2706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aygün O., Svejstrup J., Liu Y., A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. U.S.A. 105, 8580–8584 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aygün O. et al., Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 284, 23197–23203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., Xu X., Liu Y., The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol. Cell. Biol. 31, 2090–2099 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saponaro M. et al., RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157, 1037–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y. et al., Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 25, 3431–3442 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y. et al., RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M., Pokharel S., Wang J. T., Xu X., Liu Y., RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat. Commun. 6, 6720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmrich A., Ballarino M., Tora L., Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Barlow J. H. et al., Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alabert C., Groth A., Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Prado F., Maya D., Regulation of replication fork advance and stability by nucleosome assembly. Genes 8, E49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uwada J. et al., The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci. Biochem. Biophys. Res. Commun. 391, 407–413 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Gareau J. R., Lima C. D., The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein U. R., Haindl M., Nigg E. A., Muller S., RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell 20, 410–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Q. et al., Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 187, 4754–4763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Rourke J. G. et al., SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 4, 362–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schou J., Kelstrup C. D., Hayward D. G., Olsen J. V., Nilsson J., Comprehensive identification of SUMO2/3 targets and their dynamics during mitosis. PLoS One 9, e100692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P., Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34, 5217–5231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aygün O., Svejstrup J. Q., RECQL5 helicase: Connections to DNA recombination and RNA polymerase II transcription. DNA Repair 9, 345–353 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Yankulov K., Yamashita K., Roy R., Egly J. M., Bentley D. L., The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J. Biol. Chem. 270, 23922–23925 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Ivanov A. V. et al., PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boehm E. M., Washington M. T., R.I.P. to the PIP: PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. BioEssays 38, 1117–1122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng L. et al., Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 15, 626–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohiuddin M. et al., SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines. Proc. Natl. Acad. Sci. U.S.A. 115, 12793–12798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izumikawa K. et al., Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 413, 505–516 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Islam M. N., Fox D. 3rd, Guo R., Enomoto T., Wang W., RecQL5 promotes genome stabilization through two parallel mechanisms–interacting with RNA polymerase II and acting as a helicase. Mol. Cell. Biol. 30, 2460–2472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bunch H., Calderwood S. K., TRIM28 as a novel transcriptional elongation factor. BMC Mol. Biol. 16, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choe K. N., Moldovan G. L., Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell 65, 380–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W. S., Campbell M., Kung H. J., Chang P. C., In vitro SUMOylation assay to study SUMO E3 ligase activity. J. Vis. Exp. 131, 56629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang S. M. et al., KAP1 facilitates reinstatement of heterochromatin after DNA replication. Nucleic Acids Res. 46, 8788–8802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang B. X. et al., Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163, 230–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz D. C., Friedman J. R., Rauscher F. J. 3rd, Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 15, 428–443 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J. 3rd, SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diets I. J. et al., TRIM28 haploinsufficiency predisposes to Wilms tumor. Int. J. Cancer 145, 941–951 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Płuciennik E. et al., Genetic alterations of WWOX in Wilms’ tumor are involved in its carcinogenesis. Oncol. Rep. 28, 1417–1422 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and the SI Appendix.