Abstract
Biological embedding occurs when life experience alters biological processes to affect later life health and well-being. Although extensive correlative data exist supporting the notion that epigenetic mechanisms such as DNA methylation underlie biological embedding, causal data are lacking. We describe specific epigenetic mechanisms and their potential roles in the biological embedding of experience. We also consider the nuanced relationships between the genome, the epigenome, and gene expression. Our ability to connect biological embedding to the epigenetic landscape in its complexity is challenging and complicated by the influence of multiple factors. These include cell type, age, the timing of experience, sex, and DNA sequence. Recent advances in molecular profiling and epigenome editing, combined with the use of comparative animal and human longitudinal studies, should enable this field to transition from correlative to causal analyses.
Keywords: epigenetic mechanisms, gene–environment interplay, biological embedding of experience, epigenome, translational research
Early experiences impact children’s future outcomes through biological embedding: the process whereby experiences produce lasting changes in the function of a biological system with consequences for development, behavior, and health (1–3). Early life social experiences (e.g., early caregiving, trauma, maternal mental health) are known to contribute to individual differences in susceptibility and resilience for a range of physical and mental health outcomes (4–7). In this primer, we explore epigenetic systems as candidate mechanisms for the biological embedding of experience. For clarity, we adopt an inclusive definition of epigenetics proposed by the NIH Epigenomics Roadmap Project initiative, which states, “Epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable” (ref. 8, although see refs. 9 and 10).
Many animal and human studies show correlations between experience and epigenetic modifications. However, the identification of causal links is challenging for a number of reasons (11): 1) Many of the experiences that we are interested in understanding (e.g., exposure to trauma and adversity) cannot ethically be randomly assigned in humans; 2) it is difficult to accurately capture and measure the complex environments that humans inhabit as well as to distinguish specific and often co-occurring adversities or protective factors (12); and 3) epigenetic processes are complex and influenced by genetic variants (13), chronological age, sex, and cell type (11), such that epigenetic states in accessible tissues are not necessarily informative of brain-based phenotypes.
We discuss DNA modifications and considerations for animal and human studies. We then consider biological factors that play a role in the association of DNA methylation and experience as well as less well-studied epigenetic modifications (broadly defined) that warrant future attention. We end with a discussion of advances in molecular profiling and future directions. We focus on the nervous system and brain-based phenotypes but acknowledge that additional systems (e.g., immune, microbiome) are likely also involved in biological embedding (e.g., refs. 14 and 15). This primer is not a comprehensive review; we use selected examples from the literature to illustrate key points of interest.
DNA Modifications
DNA Methylation and the Biological Embedding of Experience.
DNA methylation typically refers to the addition of methyl groups to cytosine residues on the DNA, although methylation can occur on other DNA residues (16, 17) (see Additional DNA Modifications and Fig. 1A). In humans, cytosine methylation (mC) is frequent, occurring most often at CpG sites (cytosine followed by a guanine base in the DNA sequence) (16, 18). DNA methylation is also found at cytosines followed by a nonguanine base, i.e., adenine, cytosine, or thymine, such non-CpG methylation is an abundant modification in neural tissues and increases during development (19).
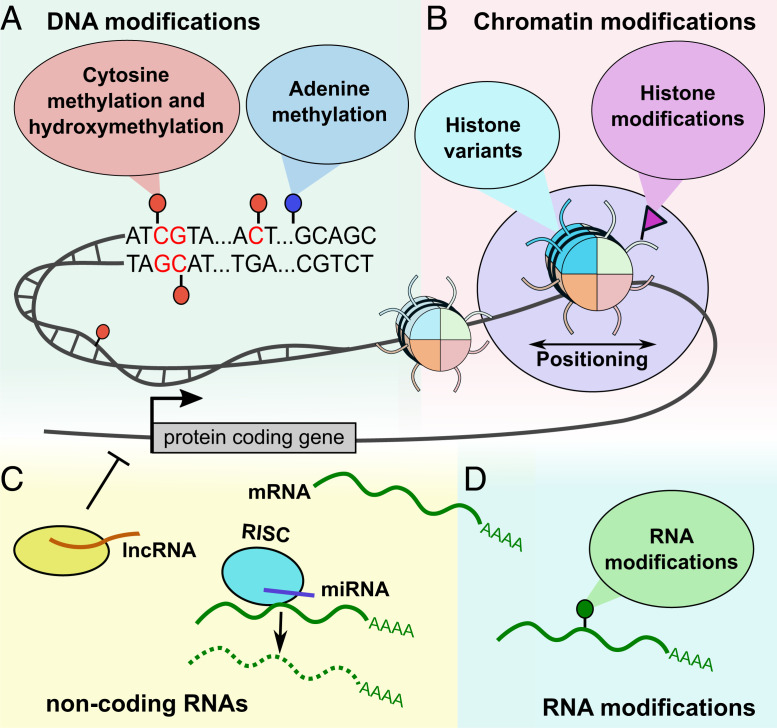
Fig. 1.
Complex landscape of epigenetic mechanisms. (A) DNA can be modified at cytosine and adenine residues by the addition of chemical groups. Cytosines can be modified by methylation, hydroxymethylation (hmC), formylation (fC), and carboxylation (caC), while adenines are modified by methylation. (B) Nucleosomes, composed of DNA wrapped around histone proteins, can change position to increase or decrease DNA accessibility. In addition, nucleosomes can be modified by the incorporation of histone variants and the addition of posttranslational modifications. (C) Noncoding RNAs play an important role in transcription regulation and are sometimes considered an epigenetic mechanism. Within RNA-induced silencing complexes (RISCs), miRNAs mediate the recognition and binding of RNAs that become targeted for degradation. lncRNAs are associated with other complexes and can activate or repress transcription. (D) All RNA nucleotides can be modified by the addition of chemical groups. The list of RNA modifications includes N6-methyladenosine (m6A) and over 160 other chemical modifications.
Animal and human research indicates that experience is associated with differences in mC (20–23). In an early example, high maternal care in rats (high maternal licking and grooming of pups) was associated with reduced mC at the promoter of the glucocorticoid receptor (GR) gene (Nr3c1) involved in the regulation of the stress response (20, 21). Experimental manipulation of maternal care through cross-fostering of pups from low- to high-care mothers produced a significant decrease in mC at Nr3c1, suggestive of a causal association. This maternal care-induced DNA demethylation involves increased binding of nerve growth factor-inducible protein A (NGFI-A), a transcription factor that acts at the Nr3c1 gene (20). The notion that experience shapes epigenetic variation via altered activity/binding of transcription factors has been supported by independent studies in rodents (24–26) and humans (27). Indeed, transcription factors play an essential role in shaping epigenomic variation across multiple levels, including DNA accessibility (28) and mC (reviewed in ref. 29).
Human studies also correlate the social environment with variation in mC, either at specific sites or broadly across the epigenome (16, 30, 31–35). Reminiscent of findings in rodents, McGowan et al. (35) found that child maltreatment was associated with increased mC within the NR3C1 promoter in hippocampal tissue of adults who committed suicide. Attempts to extend such findings using samples from accessible peripheral tissues have shown mixed results (30). Indeed, one of the largest human studies to date (involving blood samples from ∼1,600 twin pairs) failed to detect associations between childhood and adolescent chronic social stress (CSS; including physical maltreatment) and mC using an unbiased genome-wide or candidate gene approach (36). This limited reproducibility likely reflects the tissue specificity of epigenetic states and the contribution of other factors (e.g., timing of exposure) that influence the impact of an exposure on mC (11). Furthermore, inconsistencies in measurement of seemingly equivalent exposures also contribute to problems with reproducibility [e.g., maltreatment measures based on subject interviews (36) vs. mother reports (37)].
Timing is emerging as an important factor in the study of biological embedding. Early social experiences are more often correlated with variation in mC than experiences later in life, even when the latter are more proximal to the time of biological sample collection (14, 27, 31, 32, 37). In an early example by Lam et al. (14), early life, but not current socioeconomic status (SES), correlated with mC in peripheral blood samples. More recently, Dunn et al. (37) found that exposure to various adversities was significantly associated with mC at 38 sites from a genome-wide scan of blood samples from a UK-based child cohort (n ∼ 700). Adversity-associated changes in mC were generally modest (i.e., ≤5%). Interestingly, almost all sites (35 of 38) tended to be associated with adversity occurring at ≤3 y of age compared with more recent events (37). These studies suggest that the timing of an early experience plays a role in biological embedding, although we note that the biological embedding of experience can occur across the lifespan (38).
Considerations for Human Studies.
Accurate measurement of the multifaceted environments that humans inhabit is difficult. This complicates attempts to isolate the influences of specific aspects of early experience on the epigenome. Randomization of participants across exposures is considered the gold standard but is rarely achieved in large-scale human studies. Research designs that compare across siblings help control for a wide range of exposures (e.g., family environment, neighborhood characteristics) and factors (e.g., genetic variants) that may confound associations of interest. Using a large cohort, Marzi et al. (36) identified genome-wide significant associations between adolescent self-reported measures of maltreatment and mC at 2 sites in the genome (cg03960390, annotated to RER1, and cg07146173, annotated to ALKBH5). Leveraging the twin design strategy unexpectedly revealed that mC at these genomic sites did not differ between twin pairs discordant for maltreatment levels. Thus, cohort design strategies can help account for unmeasured confounding factors (genetic and environmental) to identify more robust findings. Longitudinal studies that follow individuals over time can take advantage of unanticipated events that are exogenous to cohort members. For example, the recession of 2008 greatly increased the likelihood of job loss, eviction, and wealth depreciation in the United States, facilitating a series of analyses on the impact of SES on cardiovascular risk factors (39). Regression discontinuity, a strategy that leverages a cutoff or threshold (e.g., date of implementation of a new policy) to estimate the effect of an intervention in the absence of randomization also offers opportunities to facilitate causal inference in studies where repeated assessments of epigenetic marks were collected.
Disasters like the Dutch Hunger Winter or the Holocaust provided evidence that epigenetic mechanisms are associated with the biological embedding of experience and individual differences in susceptibility to depression and posttraumatic stress disorder (PTSD). The Dutch Hunger Winter, a period of famine at the end of World War II, gave rise to a range of negative metabolic and mental health outcomes in later life. Heijmans et al. (40) found reduced mC within a gene implicated in growth and metabolism (insulin growth factor 2 [IGF2]) in blood samples from adults born to women exposed to famine in the periconceptual period. Similarly, parental PTSD following exposure to the Holocaust is associated with increased psychiatric risk and reduced cortisol production in the next generation, effects that are most pronounced in children conceived closer in time to the Holocaust (41). In a small study, Yehuda et al. (42) reported reduced mC within a region of the FKBP5 gene in blood samples from adult offspring of Holocaust survivors, which correlated with reduced cortisol levels at waking. Such studies provide proof of principle that severe exogenous stressors can be used to describe the biological embedding of experience at a molecular level. However, because biological samples are not available from before the onset of the stressor in many of these studies, it is difficult to determine if the changes occurred in response to the experience. Even for studies with samples collected before and after a stressor, a comparable unexposed control group is needed to distinguish the effects of the exposure from biological processes that could already be in motion, unrelated to the exposure of interest (e.g., development, disease progression, industrialization, environmental pollution). Thus, isolating the influence of a single exposure or the combinatorial influence of correlated exposures on epigenomic variation during periods of hardship is challenging but likely to provide rich insights into the biological embedding of experience.
Human cohorts are becoming comprehensive data resources with large sample sizes and phenotyping across multiple levels (i.e., environmental, behavioral, genomic) over time (43). New technologies and tools for capturing the physical and social environment in which individuals live are rapidly accumulating (44). This wealth of data provides exciting opportunities for the development of innovative computational and analytic approaches to identify specific, or cumulative, effects of different life experiences and their effects on epigenetic variation and health outcomes. For example, Wang et al. (45) report significantly increased power (∼6-fold increased prediction of mental health phenotypes) of integrative models that combine multiple measures of the genome and its function versus models based on genetic variation alone. Finally, given the limitations of human cohort designs for testing causality, significant insights can be gained by pairing human studies with experimental animal studies, which present unique opportunities for discovery.
Biological Factors That Play a Role in the Association of DNA Methylation and Experience.
Tissue/cell type, age, and sex.
Each tissue and cell type has unique DNA methylation profiles (methylome), making it challenging to extrapolate findings obtained in one tissue to another (ref. 11; comparisons of methylomes across tissues are provided in refs. 46–48). This tissue specificity underscores the importance of considering cellular heterogeneity of biological samples when investigating mC across individuals (49). mC can also differ according to the age of the organism (17, 50, 51), indicating that mC measures at one age might differ from those obtained earlier or later in an individual’s life. The relationship between age and the epigenome may, in part, explain why the timing of an experience is a strong predictor of its biological embedding (14, 33, 37). Sex differences in mC, beyond those found on the sex chromosomes, are also common (52). These biological factors (tissue/cell type, age, and sex) contribute to variation in most epigenetic mechanisms, and thus should be considered in all epigenetic analyses (53–55).
Genome/genotype.
DNA sequence (genotype) is a key modulator of the methylome (18, 48, 56–58). The presence or absence of cytosines in the DNA sequence can influence mC by introducing or removing possible modifiable sites (e.g., refs. 59 and 60). Genotypic differences can also affect the binding of transcription factors, some of which influence epigenetic modifications (57, 61, 62). For instance, DNA sequence variation that results in increased transcription factor binding and transcriptional activity can decrease mC at proximal sites (29). Not surprisingly, genetic variation and ancestry show strong associations with mC profiles (11). The relationship between the genome and epigenome is complex and bidirectional. For instance, methylated cytosines are prone to mutation, changing the genome with potential consequences for individuals during their lives and species over evolutionary time (63, 64).
The interdependency between the genome and the methylome also has potential consequences for behavior and health. For example, mating behavior in prairie voles is regulated by the vasopressin 1a receptor (Avpr1a), a gene known to play a role in vertebrate social behavior. Increased neuronal gene expression of Avpr1a is associated with greater sexual fidelity, with differences in Avpr1a expression arising from an allele-specific effect on mC; voles that carry genetic variants that give rise to a greater number of CpG sites show higher levels of mC and lower overall Avpr1a gene expression compared with voles that carry fewer CpG sites (59). In humans, Klengel et al. (27) found allele-specific demethylation of the GR coregulator FKBP5 in adults exposed to childhood trauma. This effect was likely mediated by increased binding of the GR to a binding site within FKBP5. Genetic variants in FKBP5 influence local chromatin structure and mC levels, which correlate with measures of brain structure (hippocampal volume) and PTSD symptoms in individuals exposed to childhood trauma (27). Similarly, Teh et al. (65) showed that the interaction of local genetic variants and aspects of the prenatal environment best explained variation in mC at 75% of genomic regions in human umbilical cord samples, a finding that is consistent with recent findings in neonatal blood (66). Such allele-specific epigenetic effects may provide a biological mechanism to explain gene-by-environment interactions (27). Thus, it is imperative to consider measures of DNA sequence variation in analyses of epigenetic variation when investigating the biological embedding of experience.
The Meaning of DNA Methylation Marks.
Early studies reported a negative relationship between mC levels at a gene’s promoter and measures of gene expression from that gene (64). Paired with evidence that mC was involved in gene- or chromosome-silencing events (e.g., imprinting, X-chromosome inactivation), this gave rise to the idea that mC was a repressive epigenetic modification. However, technological advances that enabled the measurement of epigenome-wide mC and genome-wide gene expression levels (67) now reveal a more nuanced relationship between mC and gene expression: in some cases, they are associated, whereas in others they are not (14, 61). Additionally, when mC is associated with gene expression, the direction of the relationship (positive or negative) depends on where in the genome mC is deposited (e.g., within a protein-coding or regulatory region). Furthermore, the presence of mC can influence the binding of transcription factors (61, 68), which, in turn, affects gene expression. Thus, the role of mC in gene expression needs further study.
Forecasting Future Transcriptional Responses.
Stressful experiences in early life can have lasting effects that are only detectable during future stressful conditions (69). In this issue, Provençal et al. (70) explore the biological embedding of glucocorticoid exposure within human hippocampal neuronal progenitor cells. The authors report widespread changes in mC (increases and decreases) that depend, in part, on the timing of exposure to glucocorticoids (i.e., before or after the progenitors differentiate into neuronal cells). Glucocorticoid-induced changes in mC did not correlate with baseline gene expression, but were strongly associated with a heightened transcriptional response to a second glucocorticoid challenge (70). This genomic “priming” is in line with previous observations in hippocampal tissues from rodents treated with glucocorticoids and subsequently exposed to stress (71). Similar findings are observed in humans, where mC at specific sites predicts the future transcriptional response to immune stimulation in peripheral blood monocytes (14). Thus, epigenetic signals may function to prime sites for a future transcriptional response to experience. The increased appreciation that transcription can be regulated by long-range effects acting over long periods of time is vital to the concept of biological embedding (72). Such effects, which arise within an individual over time, are distinct from those that may occur between individuals across generations. The notion of intergenerational epigenetic inheritance posits that epigenetic states established in one generation can be transmitted to the next. Despite enthusiasm (and skepticism) for the idea of intergenerational transmission of epigenetic states, it should be noted that there is no clear evidence for such effects in humans (reviewed in refs. 10, 73, and 74).
Less Well-Characterized Epigenetic Modifications
Next, we highlight less well-studied epigenetic modifications that warrant future attention.
Additional DNA Modifications.
In addition to DNA methylation, DNA hydroxymethylation (hmC), formylation (fC), and carboxylation (caC) have been observed (Fig. 1A). hmC is prevalent in the nervous system and stem cells (75, 76), and it is sensitive to early life experience in animals. hmC differs in the prefrontal cortex of mother- versus peer-reared (without a mother) adult rhesus macaques at a subset of candidate genes; peer rearing is known to be stressful (77). Similarly, global decreases in hmC are observed in the amygdala of male rats exposed to early life maltreatment (78). However, no effect of early life maltreatment is observed in female rats, highlighting the potential role of sex in modulating hmC. Whether fC and caC contribute to the biological embedding of experience is unknown. Adenine residues on DNA can also be modified by methylation, a modification that is rare compared with mC (79). The prefrontal cortex of mice exposed to 2 h of daily physical restraint for 14 consecutive days shows altered adenine methylation (6mA) across the genome (80).
Chromatin Modifications.
Chromatin, the main DNA packing structure, is composed of nucleosomes, DNA wrapped around a core of histone proteins (81) (Fig. 1B). Chromatin structure is not fixed or rigid and can be modified by protein complexes that specify the location, composition, and modification state of nucleosomes, ultimately regulating access to the underlying DNA sequence.
Chromatin Accessibility.
A broad measure of chromatin structure is how accessible the underlying DNA sequence is to proteins that bind it. Chromatin accessibility is the product of many mechanisms, including those that deposit, modify, and reposition histones along the DNA (Fig. 1B). In humans, differences in chromatin accessibility are found in neurons at different developmental stages (82) as well as in the prefrontal cortex of schizophrenia patients compared with controls (83). Furthermore, genetic loci previously implicated in schizophrenia are enriched within regions of accessible chromatin (83).
In mice, CSS (exposure to an aggressor for 10 min each day for 10 days) alters chromatin accessibility in the nucleus accumbens, a brain region involved in motivated behavior (84). Moreover, mice that developed depression-like behavior following exposure to CSS had increased messenger RNA (mRNA) and protein levels of BAZ1A, a subunit of the adenosine 5′-triphosphate (ATP)–utilizing chromatin assembly and remodeling complex, compared with unexposed control mice. Genome-wide mapping of the histone protein H3 within the nucleus accumbens identified over 70,000 genomic regions that differ in nucleosome density and position between CSS-exposed and control mice, suggesting widespread chromatin remodeling following exposure to CSS. Translating these rodent findings to humans, the authors observed increased BAZ1A gene expression in the nucleus accumbens of depressed patients compared with controls. This study highlights how pairing mouse models with human research can provide molecular insights on human mental health.
Histone Modifications.
Posttranslational modification of histone proteins also alters chromatin function and DNA accessibility. A large number of known chemical modifications, including acylation, methylation, phosphorylation, and ubiquitination, occur along the tails and globular domains of histone proteins (85) (Fig. 1B). Histones and nucleosomes can be decorated by multiple posttranslational modifications, highlighting the complexity of this epigenetic landscape (86). Collectively, these chemical modifications regulate how tightly DNA adheres to the nucleosome, influencing the accessibility of the underlying DNA to transcription factors or enzymes that regulate epigenetic states (e.g., mC, hmC). Histone modifications, are also affected by underlying genetic variation (13, 87), as well as contribute to DNA sequence maintenance (88) (e.g., histone H2Ax phosphorylation and histone 4 lysine 20 methylation), highlighting a bidirectional relationship between genetic variation and histone modifications.
In model organisms, genome-wide differences in histone H3 lysine 9 acetylation and histone 3 lysine 27 acetylation (H3K9ac and H3K27ac, respectively) are found in relation to experience (89–93). In rats, increased levels of H3K9ac are observed in the hippocampus of individuals exposed to acute restraint stress compared to controls, a change that maps to mobile and repetitive elements within DNA and whose mRNA levels decreased upon stress exposure (89). In bees, sticklebacks, and mice, differences in genomic locations marked by H3K27ac are observed when comparing neural tissue from individuals exposed to an intruder compared to unexposed controls, with some of the altered locations mapping to genes whose mRNA levels changed in response to an intruder (90–92).
Histone modification states may also regulate behavioral phenotypes. In ants, inhibition of the Rpd3 histone deacetylase (HDAC) is sufficient to induce foraging and increase the number of ants displaying scouting behavior (94). Interestingly, the effects of HDAC inhibition in ants are caste- and age-specific, highlighting the ability of HDACs to shape behavioral differences between individuals and across their lifespan (94). Similarly, experimental inhibition of hippocampal HDAC4 activity in mice exposed to chronic stress (repeated forced swim tests) increases acetylcholinesterase gene expression and reduces stress-induced anxiety-like behavior (95). In Drosophila, G9a, a histone methyltransferase, interacts with the genetic background to regulate specific transcripts of the foraging gene and modulate individual differences in foraging behavior (87). Anreiter et al. (87) show that behavioral differences can be abolished by transgenically manipulating the foraging transcripts regulated by G9a. Thus, carefully controlled animal studies can build a strong case for a causal role of histone-modifying enzymes in the regulation of behavioral states. This study in Drosophila also highlights the importance of genome–epigenome interactions, as histone methylation at the foraging promoter is allele-specific (87).
Histone Variants.
Most nucleosomes are composed of 2 copies of the core canonical histones H2A, H2B, H3, and H4, whose expression is tightly linked to DNA replication. However, nucleosomes can be modified by the replacement of canonical histones with histone variants (e.g., H3.3, H2A.X, H2A.Z) (Fig. 1B), which differ in structure and function and can affect the activity of the genomic region around which they are located.
Histone variants are associated with an organism’s experience and behavior. For instance, elevated mRNA levels of histone H3.3 are observed in the nucleus accumbens of nonmedicated individuals diagnosed with depression compared with medicated patients and controls (96). Increased H3.3 mRNA levels were also found in mice with depression-like symptoms that were exposed to CSS compared to controls (96).
Noncoding RNAs.
Initially believed to be a transcriptional error, noncoding RNAs are now known to play central roles in transcriptional regulation (97).
MicroRNAs.
MicroRNAs (miRNAs) are short (∼22 nucleotides in length), single-stranded, noncoding RNAs (98) that interact with proteins to form RNA-induced silencing complexes, which regulate mRNAs via degradation or inhibition of their translation into proteins (99) (Fig. 1C). Interestingly, the mRNA–miRNA relationship is bidirectional. For example, the binding of the mRNA NREP to miR-29 leads to the destruction of the miRNA, a mechanism involved in motor- and contextual fear-learning in mice (100).
miRNAs are linked to behavioral phenotypes and the response to adverse social experiences in humans and animals. The sperm of men exposed to a larger number of adverse childhood experiences (ACEs; e.g., child maltreatment, exposure to violence, parental loss) show decreased levels of a subset of miRNAs, including miR-449a and miRNA-34c-5p, compared with sperm from men with fewer ACEs (101). In agreement with this human research, decreased levels of miRNA-449a and miRNA-34c-5p are found in the sperm of adult mice exposed to chronic social instability during adolescence (changing littermates twice a week for 7 wk) compared with controls (101). Similarly, exposure to CSS in mice alters the expression of several miRNAs and a key miRNA processing factor called Dicer in the nucleus accumbens (102). In mice, a reduction in Dicer results in social avoidance upon exposure to a mild form of CSS, an effect not observed in control mice. Also in mice, genetically manipulating the levels of mir137, a miRNA implicated in neuropsychiatric disorders, is sufficient to induce synaptic overgrowth, memory deficits, repetitive behaviors, and altered social interactions (103). The expression of miR-125b-1-3p is also reduced in hippocampal progenitor cells exposed to the stress hormone cortisol and in the hippocampus of prenatally stressed rats (104). Likewise, reduced miR-125b-1-3p is observed in blood samples from patients with schizophrenia who experienced child maltreatment compared with controls (104). Furthermore, several reported risk loci for mental health outcomes map within miRNAs or regions of the genome that code for miRNAs target sequences (105). For instance, a single-nucleotide polymorphism (SNP; rs17228616) in the miRNA target region of the acetylcholinesterase gene modulates the binding of a miRNA (miRNA-608). This SNP also correlates with the neural response to military, medical, or neutral images in soldiers reporting chronic military stress during a functional magnetic resonance imaging task (106). In summary, there is convergent evidence from animal and human studies that implicates miRNAs in susceptibility to stress-related phenotypes and their biological embedding.
Long noncoding RNAs.
Long noncoding RNAs (lncRNAs) are a large family (∼50,000 in the human genome) of RNAs that are >200 base pairs in length (107) (Fig. 1C). lncRNAs are abundant in neural tissues but detected at very low levels elsewhere. lncRNAs reduce transcription, regulate the processing of mRNAs to modulate the abundance of subtly different transcripts from the same gene (alternative splicing), and influence the activity of miRNAs (108). Although evidence implicating lncRNAs in the biological embedding of experience is lacking, they are linked to mental health conditions in humans (109, 110).
RNA Modifications.
Like DNA, RNA nucleotides can be modified by the addition of chemical groups (111). To date, over 160 different RNA modifications are known, including methylation and hmC (Fig. 1D), and these can modulate many aspects of RNA biology, including RNA structure, stability, and processing. N6-methyladenosine (m6A) is the best-studied and most abundant mRNA modification (112), with evidence linking it to behavioral phenotypes and possibly biological embedding. For instance, increased m6A is detected in the mRNAs of genes related to key neuronal functions (dendritic and postsynaptic regulation) in the prefrontal cortex of mice exposed to fear conditioning compared with naive controls (113). Similarly, differences in m6A and N6,2′-O-dimethyladenosine (m6Am) are observed at 20 different genes in cortical samples from mice exposed to restraint when compared with unrestrained mice (114). Interestingly, the effect of restraint on m6A is brain region-specific. The prefrontal cortex shows decreased m6A levels, while the amygdala shows increased levels following exposure to stress. In the same study, the authors report a dynamic decrease in m6A in blood samples from healthy volunteers following exposure to glucocorticoids, a response that is absent in patients with current depression (114). Thus, the role of RNA modifications in biological embedding of experience is a new and likely important direction for future research.
Advances in Molecular Profiling
New molecular methods provide the ability to survey the epigenome in greater depth and breadth than previously possible. Such advances permit the integrated analyses of different epigenetic states in a single sample and, increasingly, within a single cell. For example, nucleosome occupancy methylome sequencing describes DNA methylation and nucleosome position across the genome in a single analysis (115). Similarly, the availability of new methods, based on mass spectrometry, now allow parallel analysis of histone variants and modifications to histone proteins (116). Collectively such advances move the field beyond the analysis of a single epigenetic state to provide a better understanding of the multiple facets of the epigenome, and its contribution to gene regulation and brain function. Likewise, the emergence of single-cell techniques [e.g., methylome profiling (117), chromatin accessibility (118)] can begin to answer if epigenetic profiles observed in bulk tissues are conserved at the individual cell level. Such single-cell analyses directly address issues of cellular heterogeneity, which can confound studies of the biological embedding of experience, and will help identify specific cell types that are most affected by early experiences.
Epigenome editing makes use of guide proteins or RNAs that can be targeted to specific sites in the genome, coupled to effector enzymes that can alter epigenetic modifications. Tools exist for increasing or decreasing DNA methylation, histone methylation, or histone acetylation (a comprehensive review is provided in ref. 119). Such tools allow a direct analysis of the role of the epigenome in experience-dependent changes in gene expression and its consequences for behavior. For example, Sase et al. (120) used epigenome editing (targeted histone acetylation) to examine how regulation of Cdk5 gene expression functioned in sex-specific fear memory recall following exposure to a foot shock paradigm in mice. The authors show that targeted histone acetylation increases Cdk5 expression in females to levels comparable to males, and abolishes the sex-specific differences observed in fear memory observed between untreated female and male mice (120). In an elegant example, Liu et al. (121) used epigenetic editing, targeting DNA demethylation of the FMR gene in human neurons derived from fragile X syndrome patients; this reactivated the silenced FMR gene, normalizing neuronal activity within these neurons to levels comparable to those from healthy subjects.
These findings suggest that site-specific manipulation of epigenetic states can directly influence gene expression and cellular function. However, there are limitations to epigenome editing technologies. First, there is potential for such molecular modifiers to influence the epigenetic states of unintended genes (off-target effects). In the study by Liu et al. (121), epigenome editing of the FMR gene produced decreases in mC ≥ 10% at 28 off-target sites, although this led to minimal effects on the expression of those genes. Second, altering a single epigenetic state may alter other features of the chromatin landscape. In the study by Liu et al. (121), decreasing mC within the FMR gene increased histone modifications (H3K4me3 and H3K27ac) associated with “active” or expressed genes. Finally, existing epigenome editing methods seek to target a single epigenetic modification at a single site; refinements of such approaches may help us understand phenotypes that arise from epigenetic variation across the genome.
Future Directions
The question of how epigenetic mechanisms might contribute to the biological embedding of experience presents us with both challenges and opportunities. We provide a number of limitations in measurement and study design for consideration as the field moves forward.
Epigenetic mechanisms function in concert within a cell and may interact with each other (122, 123). Thus, a systems level approach is needed to understand the biological embedding of experience. Mapping efforts like ENCODE and PsychENCODE are uncovering the interplay across epigenetic marks through deep characterization and the merging of different levels of analysis (45, 124). Sophisticated new technologies now make it possible to measure multiple epigenetic domains (mC and chromatin accessibility) and transcription in tandem within the same cell population (125). These approaches promise to improve our understanding of interdependencies in the epigenetic landscape and its links to genome function (126). Technological advances are also likely to identify novel epigenetic modifications and transcriptional regulators (e.g., families of miRNAs). Importantly, epigenetic studies need to factor in genetic variation and developmental timing. Future studies should also consider interactions between biological systems, such as the brain and gut microbiota, which are sensitive to environmental stress (127) and can contribute to epigenetic variation (128).
Without a fundamental understanding of epigenetic change in relevant biological samples and their relationship to more accessible tissues, any attempts to establish causal relationships in humans are likely to be in vain. One approach is to use human postmortem brain samples; however, details about the individual’s experiences are usually lacking. Another approach is to use human-derived neurons produced from somatic tissues such as skin, urine, or blood. However, such neurons, while derived from adult biosamples, are phenotypically more similar to neurons in the early fetal brain (129). The pairing of human cohort studies with animal models, where mechanistic investigations within the brain and other tissues are feasible, provides an additional meaningful way forward. In these model systems, emerging state-of-the-art epigenetic editing technologies hold great promise to determine causal relationships between epigenetic states and altered activity of neural processes and developmental trajectories (e.g., refs. 87 and 130–132).
Overcoming some of the current limitations in human research may be possible through a paradigm shift to within-person comparisons over time, which, by design, holds constant a number of fixed genetic and environmental factors that are likely to confound environmental and epigenetic associations. By providing data at multiple and different time points before and after events, longitudinal studies are likely to be more informative of epigenetic changes induced by circadian rhythms (133), developmental transitions (e.g., puberty), and naturally occurring stressors or exogenous “shocks” (e.g., a recession). Ongoing ecological momentary assessment and other intensive longitudinal study designs should consider integrating repeated (daily/monthly) blood or buccal cell collections alongside experimental manipulations of mild and ethically feasible stressors, for example, the Trier social stress task, social exclusion experimental paradigms, or the identification of other naturally occurring and time-linked shocks [e.g., reports of violent crime or gunshots in the neighborhood temporally linked to sampling (e.g., ref. 134)]. The current paucity of longitudinal epigenetic datasets across childhood means that there is currently no description of “typical” versus “atypical” epigenetic maturation or variation. Thus, adopting a person-specific paradigm shift (135) also places considerations of timescales in human development at the forefront of building conceptual models and study designs. We acknowledge that repeated sampling and analysis of biological samples in human cohorts is challenging and costly, presenting significant barriers to this strategy.
Nevertheless, creative and longitudinal designs are needed in order to help answer key questions in the field: What are the most relevant timescales for capturing epigenetic changes? How do epigenetic patterns change within individuals across development and adulthood? Are experiential and epigenetic correlations more pronounced during known sensitive periods of development? The answers to these questions will move the field forward by delineating the molecular processes that underlie the biological embedding of experience. The ability to identify, and quantify, how experience gets “under the skin” has important implications for our understanding of how experience shapes individual differences in human health and development.
Acknowledgments
We thank all members of CIFAR’s Child and Brain Development (CBD) Program for helpful discussions of this manuscript. A.G., C.L.O., E.B.B., M.S.K., M.B.S., S.M., and T.W.M. are fellows in the CBD Program. M.J.A. is supported by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship. K.J.O. is a CIFAR Azrieli Global Scholar in the CBD Program. M.B.S. is a CIFAR Weston Fellow.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.T.B. is a guest editor invited by the Editorial Board.
References
- 1.Shenhar-Tsarfaty S., et al. , Fear and C-reactive protein cosynergize annual pulse increases in healthy adults. Proc. Natl. Acad. Sci. U.S.A. 112, E467–E471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen E., Miller G. E., Kobor M. S., Cole S. W., Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol. Psychiatry 16, 729–737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hertzman C., Boyce T., How experience gets under the skin to create gradients in developmental health. Annu. Rev. Public Health 31, 329–347, 3 p following 347 (2010). [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell K. J., Glover V., Barker E. D., O’Connor T. G., The persisting effect of maternal mood in pregnancy on childhood psychopathology. Dev. Psychopathol. 26, 393–403 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Edwards V. J., Holden G. W., Felitti V. J., Anda R. F., Relationship between multiple forms of childhood maltreatment and adult mental health in community respondents: Results from the adverse childhood experiences study. Am. J. Psychiatry 160, 1453–1460 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Gur R. E., et al. , Burden of environmental adversity associated with psychopathology, maturation, and brain behavior parameters in youths. JAMA Psychiatry 76, 966–975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phua D. Y., et al. ; Growing Up In Singapore Towards Healthy Outcomes Study Group , Positive maternal mental health during pregnancy associated with specific forms of adaptive development in early childhood: Evidence from a longitudinal study. Dev. Psychopathol. 29, 1573–1587 (2017). [DOI] [PubMed] [Google Scholar]
- 8.NIH Roadmap Epigenomics Mapping Consortium , Overview of the Roadmap Epigenomics Project. http://www.roadmapepigenomics.org/overview. Accessed 14 June 2019. [DOI] [PMC free article] [PubMed]
- 9.Greally J. M., A user’s guide to the ambiguous word ‘epigenetics’. Nat. Rev. Mol. Cell Biol. 19, 207–208 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Cavalli G., Heard E., Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Jones M. J., Moore S. R., Kobor M. S., Principles and challenges of applying epigenetic epidemiology to psychology. Annu. Rev. Psychol. 69, 459–485 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Manrai A. K., et al. , Informatics and data analytics to support exposome-based discovery for public health. Annu. Rev. Public Health 38, 279–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng B., et al. , An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam L. L., et al. , Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. U.S.A. 109 (suppl. 2), 17253–17260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDade T. W., et al. , Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. Proc. Natl. Acad. Sci. U.S.A. 114, 7611–7616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lister R., et al. , Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister R., et al. , Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziller M. J., et al. , Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz M. D., et al. , Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver I. C., et al. , Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004). [DOI] [PubMed] [Google Scholar]
- 21.McGowan P. O., et al. , Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One 6, e14739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedrosian T. A., Quayle C., Novaresi N., Gage F. H., Early life experience drives structural variation of neural genomes in mice. Science 359, 1395–1399 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Araki R., et al. , Epigenetic regulation of dorsal raphe GABA(B1a) associated with isolation-induced abnormal responses to social stimulation in mice. Neuropharmacology 101, 1–12 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Singh-Taylor A., et al. , NRSF-dependent epigenetic mechanisms contribute to programming of stress-sensitive neurons by neonatal experience, promoting resilience. Mol. Psychiatry 23, 648–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curley J. P., Champagne F. A., Influence of maternal care on the developing brain: Mechanisms, temporal dynamics and sensitive periods. Front. Neuroendocrinol. 40, 52–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan P., Fleming A. S., Lawson D., Jenkins J. M., McGowan P. O., Within- and between-litter maternal care alter behavior and gene regulation in female offspring. Behav. Neurosci. 128, 736–748 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Klengel T., et al. , Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 16, 33–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swinstead E. E., et al. , Steroid receptors reprogram foxa1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schübeler D., Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Turecki G., Meaney M. J., Effects of the social environment and stress on glucocorticoid receptor gene methylation: A systematic review. Biol. Psychiatry 79, 87–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyce W. T., Kobor M. S., Development and the epigenome: The ‘synapse’ of gene-environment interplay. Dev. Sci. 18, 1–23 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Gassen N. C., Chrousos G. P., Binder E. B., Zannas A. S., Life stress, glucocorticoid signaling, and the aging epigenome: Implications for aging-related diseases. Neurosci. Biobehav. Rev. 74, 356–365 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Essex M. J., et al. , Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Dev. 84, 58–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labonté B., et al. , Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry 69, 722–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGowan P. O., et al. , Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 12, 342–348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzi S. J., et al. , Analysis of dna methylation in young people: Limited evidence for an association between victimization stress and epigenetic variation in blood. Am. J. Psychiatry 175, 517–529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn E. C., et al. , Sensitive periods for the effect of childhood adversity on dna methylation: Results from a prospective, longitudinal study. Biol. Psychiatry 85, 838–849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz J., et al. , Social history and exposure to pathogen signals modulate social status effects on gene regulation in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 117, 23317–23322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boen C., Yang Y. C., The physiological impacts of wealth shocks in late life: Evidence from the Great Recession. Soc Sci Med 150, 221–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heijmans B. T., et al. , Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U.S.A. 105, 17046–17049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yehuda R., et al. , Parental posttraumatic stress disorder as a vulnerability factor for low cortisol trait in offspring of holocaust survivors. Arch. Gen. Psychiatry 64, 1040–1048 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Yehuda R., et al. , Holocaust exposure induced intergenerational effects on fkbp5 methylation. Biol. Psychiatry 80, 372–380 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Allen N. E., Sudlow C., Peakman T., Collins R., Biobank U. K., UK biobank data: Come and get it. Sci. Transl. Med. 6, 224ed4 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Wild C. P., Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Wang D., et al. ; PsychENCODE Consortium, Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R. D., Jones M. J., Meaney M. J., Turecki G., Kobor M. S., BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry 7, e1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hannon E., Lunnon K., Schalkwyk L., Mill J., Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 10, 1024–1032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Islam S. A., et al. , Integration of DNA methylation patterns and genetic variation in human pediatric tissues help inform EWAS design and interpretation. Epigenetics Chromatin 12, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., et al. , Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner M., et al. , Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew. Chem. Int. Ed. Engl. 54, 12511–12514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zannas A. S., et al. , Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. U.S.A. 116, 11370–11379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomon O., et al. , Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics 13, 655–664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ameling S., et al. , Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med. Genomics 8, 61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Somel M., et al. , MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 9, e1001214 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stefanelli G., et al. , Learning and age-related changes in genome-wide h2a.z binding in the mouse hippocampus. Cell Rep. 22, 1124–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stadler M. B., et al. , DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Husquin L. T., et al. , Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 19, 222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lienert F., et al. , Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 43, 1091–1097 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Okhovat M., Berrio A., Wallace G., Ophir A. G., Phelps S. M., Sexual fidelity trade-offs promote regulatory variation in the prairie vole brain. Science 350, 1371–1374 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Tay N., et al. , Allele-specific methylation of SPDEF: A novel moderator of psychosocial stress and substance abuse. Am. J. Psychiatry 176, 146–155 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Lea A. J., et al. , Genome-wide quantification of the effects of DNA methylation on human gene regulation. eLife 7, e37513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldmann A., et al. , Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 9, e1003994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernando-Herraez I., Garcia-Perez R., Sharp A. J., Marques-Bonet T., DNA methylation: Insights into human evolution. PLoS Genet. 11, e1005661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deaton A. M., Bird A., CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teh A. L., et al. , The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 24, 1064–1074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Czamara D., et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium , Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nat. Commun. 10, 2548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kundaje A., et al. ; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin Y., et al. , Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q., Shelton R. C., Dwivedi Y., Interaction between early-life stress and FKBP5 gene variants in major depressive disorder and post-traumatic stress disorder: A systematic review and meta-analysis. J. Affect. Disord. 225, 422–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Provençal N., et al. , Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 117, 23280–23285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gray J. D., Rubin T. G., Hunter R. G., McEwen B. S., Hippocampal gene expression changes underlying stress sensitization and recovery. Mol. Psychiatry 19, 1171–1178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dekker J., Mirny L., The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim J. P., Brunet A., Bridging the transgenerational gap with epigenetic memory. Trends Genet. 29, 176–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez M. F., Lehner B., Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Kriaucionis S., Heintz N., The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Penn N. W., Suwalski R., O’Riley C., Bojanowski K., Yura R., The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 126, 781–790 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Massart R., et al. , Hydroxymethylation and DNA methylation profiles in the prefrontal cortex of the non-human primate rhesus macaque and the impact of maternal deprivation on hydroxymethylation. Neuroscience 268, 139–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doherty T. S., Forster A., Roth T. L., Global and gene-specific DNA methylation alterations in the adolescent amygdala and hippocampus in an animal model of caregiver maltreatment. Behav. Brain Res. 298, 55–61 (2016). Correction in: Behav. Brain Res 312, 431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu T. P., et al. , DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao B., et al. , DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 8, 1122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGinty R. K., Tan S., Nucleosome structure and function. Chem. Rev. 115, 2255–2273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de la Torre-Ubieta L., et al. , The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172, 289–304.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bryois J., et al. , Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat. Commun. 9, 3121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun H., et al. , ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat. Med. 21, 1146–1153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang H., Sabari B. R., Garcia B. A., Allis C. D., Zhao Y., SnapShot: Histone modifications. Cell 159, 458–458.e1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sadeh R., Launer-Wachs R., Wandel H., Rahat A., Friedman N., Elucidating combinatorial chromatin states at single-nucleosome resolution. Mol. Cell 63, 1080–1088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anreiter I., Kramer J. M., Sokolowski M. B., Epigenetic mechanisms modulate differences in Drosophila foraging behavior. Proc. Natl. Acad. Sci. U.S.A. 114, 12518–12523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paquin K. L., Howlett N. G., Understanding the histone DNA repair code: H4K20me2 makes its mark. Mol. Cancer Res. 16, 1335–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hunter R. G., et al. , Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc. Natl. Acad. Sci. U.S.A. 109, 17657–17662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bukhari S. A., et al. , Temporal dynamics of neurogenomic plasticity in response to social interactions in male threespined sticklebacks. PLoS Genet. 13, e1006840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saul M. C., et al. , Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 27, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shpigler H. Y., et al. , Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav. 16, 579–591 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Fitzsimons H. L., Scott M. J., Genetic modulation of Rpd3 expression impairs long-term courtship memory in Drosophila. PLoS One 6, e29171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simola D. F., et al. , Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 351, aac6633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sailaja B. S., Cohen-Carmon D., Zimmerman G., Soreq H., Meshorer E., Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc. Natl. Acad. Sci. U.S.A. 109, E3687–E3695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lepack A. E., et al. , Aberrant H3.3 dynamics in NAc promote vulnerability to depressive-like behavior. Proc. Natl. Acad. Sci. U.S.A. 113, 12562–12567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001). [DOI] [PubMed] [Google Scholar]
- 98.Kim Y. K., Kim B., Kim V. N., Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 113, E1881–E1889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beveridge N. J., et al. , Maturation of the human dorsolateral prefrontal cortex coincides with a dynamic shift in microRNA expression. Schizophr. Bull. 40, 399–409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bitetti A., et al. , MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 25, 244–251 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Dickson D. A., et al. , Reduced levels of miRNAs 449 and 34 in sperm of mice and men exposed to early life stress. Transl. Psychiatry 8, 101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dias C., et al. , β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng Y., et al. , Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci. 21, 1689–1703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cattane N., et al. , Identification of a miRNAs signature associated with exposure to stress early in life and enhanced vulnerability for schizophrenia: New insights for the key role of miR-125b-1-3p in neurodevelopmental processes. Schizophr Res. 205, 63–75 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Devanna P., et al. , Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol. Psychiatry 23, 1375–1384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin T., et al. , Intensified vmPFC surveillance over PTSS under perturbed microRNA-608/AChE interaction. Transl. Psychiatry 6, e801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zuo L., et al. , Long noncoding RNAs in psychiatric disorders. Psychiatr. Genet. 26, 109–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li L., Zhuang Y., Zhao X., Li X., Long non-coding RNA in neuronal development and neurological disorders. Front. Genet. 9, 744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ziats M. N., Rennert O. M., Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 49, 589–593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Y., et al. , Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl. Psychiatry 8, 44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu Y., Dominissini D., Rechavi G., He C., Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Widagdo J., et al. , Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Engel M., et al. , The role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99, 389–403.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kelly T. K., et al. , Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bastle R. M., Maze I., Chromatin regulation in complex brain disorders. Curr. Opin. Behav. Sci. 25, 57–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Luo C., et al. , Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen X., Miragaia R. J., Natarajan K. N., Teichmann S. A., A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 9, 5345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kungulovski G., Jeltsch A., Epigenome editing: State of the art, concepts, and perspectives. Trends Genet. 32, 101–113 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Sase A. S., et al. , Sex-specific regulation of fear memory by targeted epigenetic editing of Cdk5. Biol. Psychiatry 85, 623–634 (2019). [DOI] [PubMed] [Google Scholar]
- 121.Liu X. S., et al. , Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172, 979–992.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lövkvist C., Sneppen K., Haerter J. O., Exploring the link between nucleosome occupancy and dna methylation. Front. Genet. 8, 232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kobor M. S., Lorincz M. C., H2A.Z and DNA methylation: Irreconcilable differences. Trends Biochem. Sci. 34, 158–161 (2009). [DOI] [PubMed] [Google Scholar]
- 124.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Clark S. J., et al. , scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rizzardi L. F., et al. , Neuronal brain-region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nat. Neurosci. 22, 307–316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zijlmans M. A. C., Korpela K., Riksen-Walraven J. M., de Vos W. M., de Weerth C., Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology 53, 233–245 (2015). [DOI] [PubMed] [Google Scholar]
- 128.Krautkramer K. A., et al. , Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Amiri A., et al. ; PsychENCODE Consortium , Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362, eaat6720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gervain J., et al. , Valproate reopens critical-period learning of absolute pitch. Front. Syst. Neurosci. 7, 102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sokolowski H. M., et al. , The Drosophila foraging gene human orthologue PRKG1 predicts individual differences in the effects of early adversity on maternal sensitivity. Cogn. Dev. 42, 62–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sokolowski M. B., Social interactions in “simple” model systems. Neuron 65, 780–794 (2010). [DOI] [PubMed] [Google Scholar]
- 133.Oh G., et al. , Circadian oscillations of cytosine modification in humans contribute to epigenetic variability, aging, and complex disease. Genome Biol. 20, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sharkey P., The acute effect of local homicides on children’s cognitive performance. Proc. Natl. Acad. Sci. U.S.A. 107, 11733–11738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Molenaar P. C. M., Campbell C. G., The new person-specific paradigm in psychology. Curr. Dir. Psychol. Sci. 18, 112–117 (2009). [Google Scholar]