Significance
Our results show that the experience of being alone has immediate consequences on brain gene activity. Clearly this is important for the human context, where loneliness (i.e., perceived social isolation) is a risk factor for psychological and neurodegenerative disorders. As the response occurs in brain areas involved in higher-order cognition and perception, these changes in gene activity may influence the ongoing processing of experience. Prior studies in humans have measured effects of social isolation in circulating leukocytes, but not in brain, and studies in mammalian models have typically focused on effects of much longer isolation periods (weeks or more). The results also bear on interpretation of animal experiments where short-term isolation is used to establish the baseline reference condition.
Keywords: zebra finch, RNA-seq, DNA methylation, isolation, forebrain
Abstract
Prolonged social isolation has negative effects on brain and behavior in humans and other social organisms, but neural mechanisms leading to these effects are not understood. Here we tested the hypothesis that even brief periods of social isolation can alter gene expression and DNA methylation in higher cognitive centers of the brain, focusing on the auditory/associative forebrain of the highly social zebra finch. Using RNA sequencing, we first identified genes that individually increase or decrease expression after isolation and observed general repression of gene sets annotated for neurotrophin pathways and axonal guidance functions. We then pursued 4 genes of large effect size: EGR1 and BDNF (decreased by isolation) and FKBP5 and UTS2B (increased). By in situ hybridization, each gene responded in different cell subsets, arguing against a single cellular mechanism. To test whether effects were specific to the social component of the isolation experience, we compared gene expression in birds isolated either alone or with a single familiar partner. Partner inclusion ameliorated the effect of solo isolation on EGR1 and BDNF, but not on FKBP5 and UTS2B nor on circulating corticosterone. By bisulfite sequencing analysis of auditory forebrain DNA, isolation caused changes in methylation of a subset of differentially expressed genes, including BDNF. Thus, social isolation has rapid consequences on gene activity in a higher integrative center of the brain, triggering epigenetic mechanisms that may influence processing of ongoing experience.
The experience of prolonged social isolation has well-known effects on multiple indicators of health, well-being, and brain function in humans and many other animals (1–6). Less is known about how the sense of isolation is initially perceived, or how that perception translates over time into lasting biological and behavioral effects. In the zebra finch, a highly social songbird, social interactions are often mediated through vocal signals (7), and exposure to vocal signals can cause large changes in gene expression in portions of the caudomedial forebrain (nidopallium and mesopallium) where neurophysiological responses selective for complex auditory signals are first evident (8). These responses are also sensitive to social context and past history (8–11), so the caudomedial forebrain may be a node in the generation of social awareness.
Overnight isolation of adults in sound attenuation chambers is a common practice in zebra finch experiments as a way to normalize experience and facilitate presentation of auditory stimuli and recording of vocal responses (e.g., refs. 9 and 12–15). Typically, the bird is isolated not only from external sounds, but also from other conspecifics. However, the collateral effects of this acute social isolation have not been directly considered before. With the sequencing of the zebra finch genome (16) and advances in high-throughput RNA and DNA sequencing technologies, it is now feasible to do a comprehensive analysis of gene expression in specific brain areas with replications sufficient to account for natural biological variation. In addition to direct measurement of mRNA levels, it is also possible to test for specific epigenetic changes on the underlying DNA that may contribute to long-term persistence of acute effects on gene expression. Here we set out to determine whether acute social isolation by itself can have direct effects on gene expression in the caudomedial forebrain.
Results
A Neurogenomic Response to Overnight Isolation.
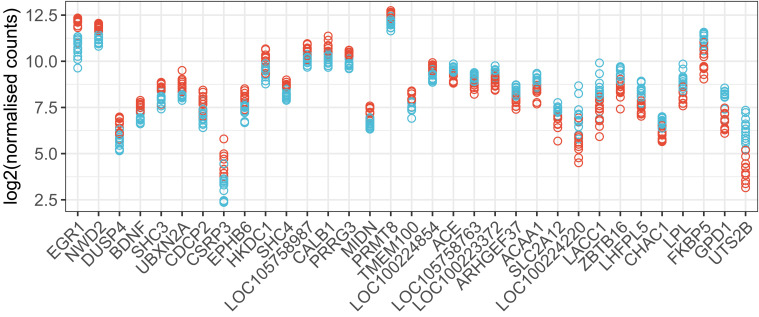
To determine if overnight isolation in a sound attenuation chamber alters gene expression in the caudomedial forebrain the following day, we began with an RNA sequencing (RNA-seq) analysis of 24 adult female zebra finches that had been together in a same-sex aviary (University of Illinois). Twelve were removed and placed alone overnight in a sound isolation chamber, then killed. The other 12 were killed immediately after removal from the aviary. Principal components analysis of sample variance indicated 1 outlier sample (SI Appendix, Fig. S1), which was excluded from subsequent analyses. From 14,687 genes measured by DESeq2 (Dataset S1), we detected significant differential expression [false discovery rate (FDR) < 0.05] for 328 genes (SI Appendix, Fig. S2). Fig. 1 shows the genes with greatest effect sizes.
Fig. 1.
Top genes from differential gene expression analysis (RNA-seq) in female auditory lobule after overnight isolation. Normalized counts for 33 genes with most significant changes in expression between aviary and solo conditions (FDR < 0.001) and the largest fold-changes in expression [abs(log-twofold change) > 0.4]. Data are plotted for 23 individual female zebra finches (11 aviary and 12 solo, red and blue symbols, respectively). Genes are sorted left to right by fold change.
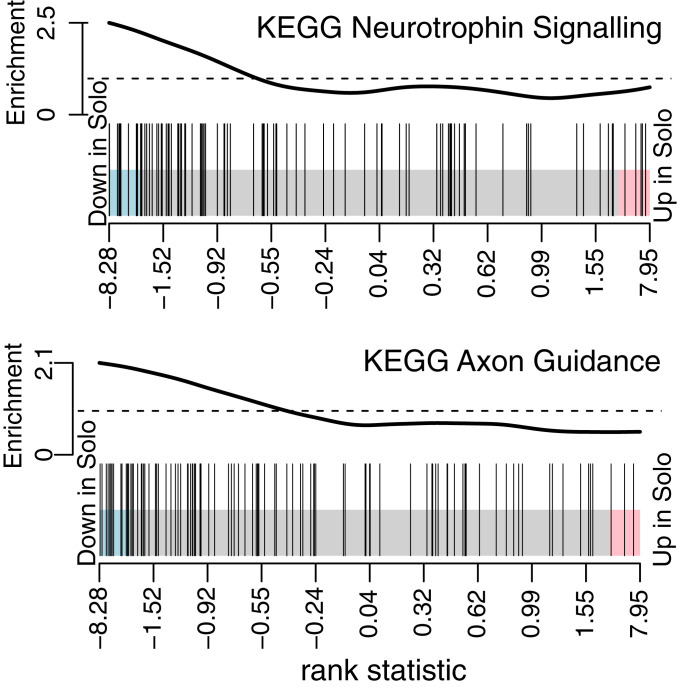
The genes that responded to isolation represent a diverse set of individual functions. To ascertain the biological pathways most influenced by overnight isolation, we employed a competitive gene set enrichment analysis (17) with the RNA-seq data. This analysis tests for significant shifts in the aggregate expression of functional groups of genes; here we mapped our data to 186 human gene sets representing the KEGG Pathway Database (18–20). Two (and only 2) gene sets were found to be significantly more affected by social isolation than other gene sets (Fig. 2): “KEGG Neurotrophin Signaling Pathway” (97 genes, net decrease in solo condition, FDR = 0.015) and “KEGG Axon Guidance” (102 genes, net decrease in solo condition, FDR = 0.045).
Fig. 2.
Competitive gene set enrichment analysis. Gene sets representing 186 KEGG pathways were tested for sensitivity to solo isolation, using cameraPR and the Wald test statistic from DESeq2 to rank the gene sets by relative expression. Two gene sets were down-regulated significantly relative to other gene sets, and the barcode plots show the ranking statistics for the individual genes (vertical bars) in each set. Ranking statistics are displayed with genes down-regulated in the solo condition toward the left and up-regulated toward the right. The wavy line at the top shows enrichment of vertical bars in each part of the plot. Pink and blue shading: P < 0.05 threshold for differential up- or down-regulation of individual genes.
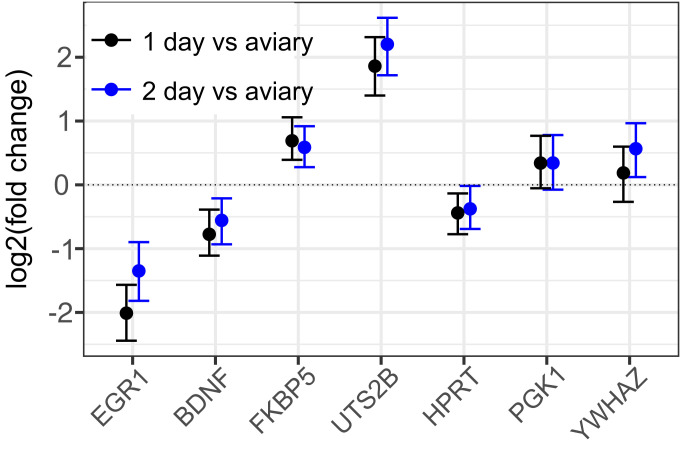
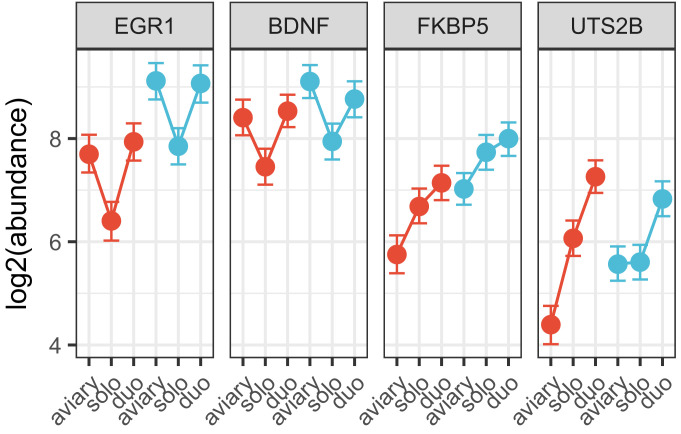
For further detailed studies of the response, we chose 4 genes at the 2 extremes of the distribution. EGR1 showed the greatest magnitude of decrease (Fig. 1) and has been extensively studied in the context of songbird vocal learning and communication (8). BDNF also decreased significantly in the isolated animals (Fig. 1) and is a major neurotrophin implicated in both developmental and adult neural plasticity (21). Among genes that increase in expression upon isolation, we chose FKBP5, a glucocorticoid receptor chaperone that has been associated with stress responses (22), and UTS2B, a neuropeptide gene with diverse functions (23) but implicated especially in vasoregulation (24). We developed primers for qRT-PCR analysis of these and several other targets (SI Appendix, Table S1 and Fig. S3), including 3 genes used as stable reference genes in other songbird qRT-PCR studies (25). To model changes in gene expression, we used the method implemented in the R package MCMC.qpcr (26). With this approach, we determined that a similar change in gene expression had also occurred in male zebra finches (n = 12) that had been collected in the United States at the same time as the females in Fig. 1 (SI Appendix, Fig. S4). Additionally, we replicated the effect in another set of females (n = 10) collected 2.5 y later in the United Kingdom (SI Appendix, Fig. S4). As 1 more test of reproducibility and to establish whether neurogenomic effects of isolation are limited to the first day after capture, we analyzed yet another set of UK females, comparing birds taken directly from the aviary (0 d) or after either 1 or 2 d of solo isolation (Fig. 3). EGR1 and BDNF were both down-regulated after 1 d in solo isolation, and both FKBP5 and UTS2B were up-regulated at this time point, as observed in the original RNA-seq study. Moreover, this pattern of altered expression was sustained after 2 d of isolation, indicating that it was not a transient response to the initial period of capture and chamber introduction.
Fig. 3.
Gene expression changes persist after 2 d in isolation. Changes in expression of EGR1, BDNF, FKBP5, and UTS2B were independently replicated with qRT-PCR for the solo (“1 d” solo isolation, n = 9 females) versus aviary (n = 10 females) comparison, with the addition of a group housed alone for an additional day (“2 d” solo isolation, n = 9 females), and modeled with MCMC.qpcr. Fixed effects of solo isolation (relative to aviary) are plotted for 1 d (black) and 2 d (blue); points represent posterior means with 95% credible intervals. The relative stability of HPRT, PGK1, and YWHAZ is reflected in the model (which is naive to the prediction that these are “control” genes).
Different Anatomical Response Patterns for Different Genes.
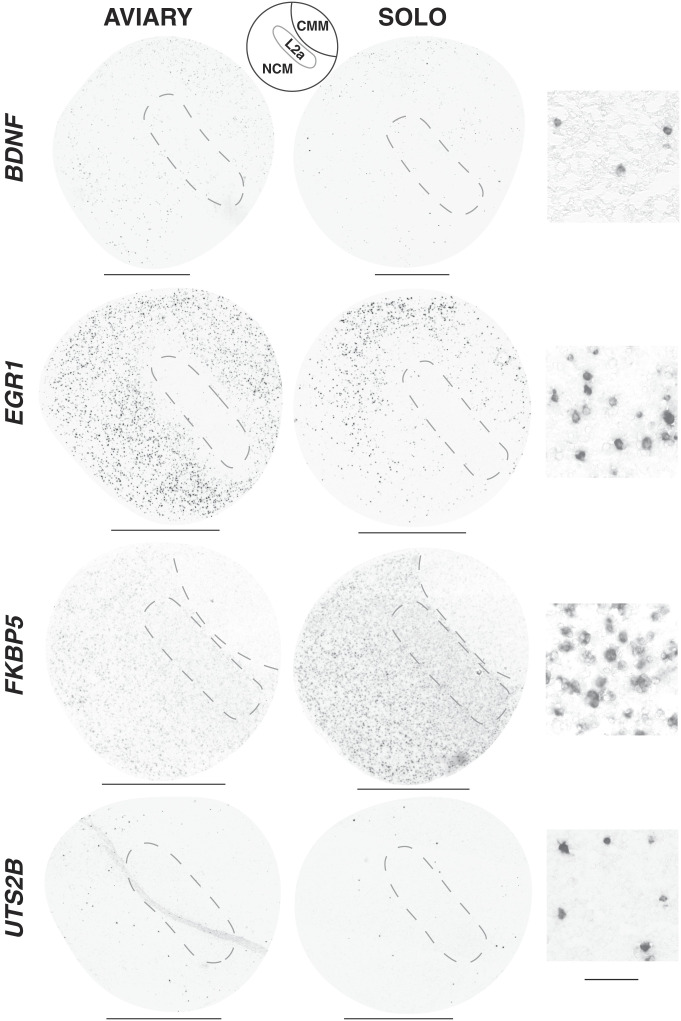
The results above were based on extracts using the auditory lobule (AL) dissection (Materials and Methods), which collects 3 major anatomical subregions of the caudomedial forebrain [caudomedial nidopallium (NCM), caudomedial mesopallium (CMM), and L2a], each of which contains diverse cell types. To determine whether the observed genomic responses occurred in a distinct cell type or were localized to a particular subregion, we used in situ hybridization to map and compare the anatomical distributions of our 4 focal RNAs in sagittal sections near the midline where all 3 subregions are evident (Fig. 4). Each gene of interest showed a different anatomical pattern of expression and response. EGR1 was readily detectable in numerous cells throughout both NCM and CMM in the aviary condition, and the density of labeled cells declined markedly upon solo isolation, especially in the ventral portions of these subregions; no labeling in Field L2a was apparent in either condition. BDNF RNA was also detected in NCM and CMM but at lower levels and in much smaller numbers of cells, and the density of labeled cells also appeared to decrease upon isolation. Conversely, FKBP5 was detected in the aviary condition in numerous cells throughout both NCM and Field L2a, but not in CMM. Upon isolation, the intensity of FKBP5 labeling in each cell appeared to increase. The UTS2B probe produced intense labeling of small cells scattered sparsely within NCM and CMM; an obvious difference in cell density or label intensity in the 2 conditions was not visibly apparent.
Fig. 4.
Anatomical expression patterns of mRNAs within the auditory lobule. Shown are images from in situ hybridizations with riboprobes (4 genes labeled on the Left), from adult females (2015 experiment). The AVIARY and SOLO columns show sections in the sagittal plane (0.2 to 0.5 mm from midline). (Scale bar: 1 mm.) Column 3 shows high-magnification images (40×) illustrating the cellular labeling pattern in NCM for condition of highest expression (BDNF and EGR1: AVIARY; FKBP5 and UTS2B: SOLO). (Scale bar: 50 µm.)
Specific Effects of Social Condition on EGR1 and BDNF.
The results so far establish that placement of a single bird overnight in a sound attenuation chamber alters gene expression in the forebrain, but these changes could be due either to the new chamber environment or to the altered social environment. To distinguish between these factors, we next asked whether the neurogenomic effects of sound chamber isolation could be ameliorated by inclusion of a second bird (of the same sex) in the chamber throughout the procedure. In both males and females, EGR1 and BDNF were significantly down-regulated in the solo group, but not in the animals that were housed with a partner bird (duo, Fig. 5). Thus, for these 2 genes, the presence of a partner prevented the effect of chamber isolation. For FKBP5, in contrast, partner presence did not block the increase upon chamber isolation. Nor did partner inclusion block the increase of UTS2B—indeed, in this experiment we did not detect an increase in the solo condition but still observed it in the duo condition.
Fig. 5.
Including a partner mitigates some, but not all, effects of solo isolation. Female (red) and male (blue) zebra finches were housed overnight in sound attenuation chambers either alone (solo) or with a partner of the same sex (duo). qRT-PCR measured expression of EGR1, BDNF, FKBP5, and UTS2B, and changes were modeled with MCMC.qpcr. Data for HPRT and PGK1 were also incorporated in the model to improve estimation of random effects (Materials and Methods). Points represent inferred transcript abundances, and whiskers represent 95% credible intervals.
To examine to what degree these changes in gene expression might be explained by general stress, we also measured corticosterone (CORT) levels in blood from these same animals, collected at the time of sacrifice. In both males and females, CORT was elevated in both chamber housing conditions (solo and duo) compared with aviary, but no differences were detected between solo and duo conditions in either sex, and no aggregate differences were detected between males and females (SI Appendix, Fig. S5). Given that both FKBP5 and UTS2B were also elevated in both chamber conditions, we next asked whether CORT levels correlated with gene expression of individual animals within treatment groups. We observed such a correlation only for BDNF expression in the solo housed group (Pearson coefficient = −0.45, P = 0.047, SI Appendix, Fig. S6); the negative correlation is consistent with prior evidence that BDNF expression is suppressed by glucocorticoids (27).
Social Isolation Affects DNA Methylation in the Caudomedial Forebrain.
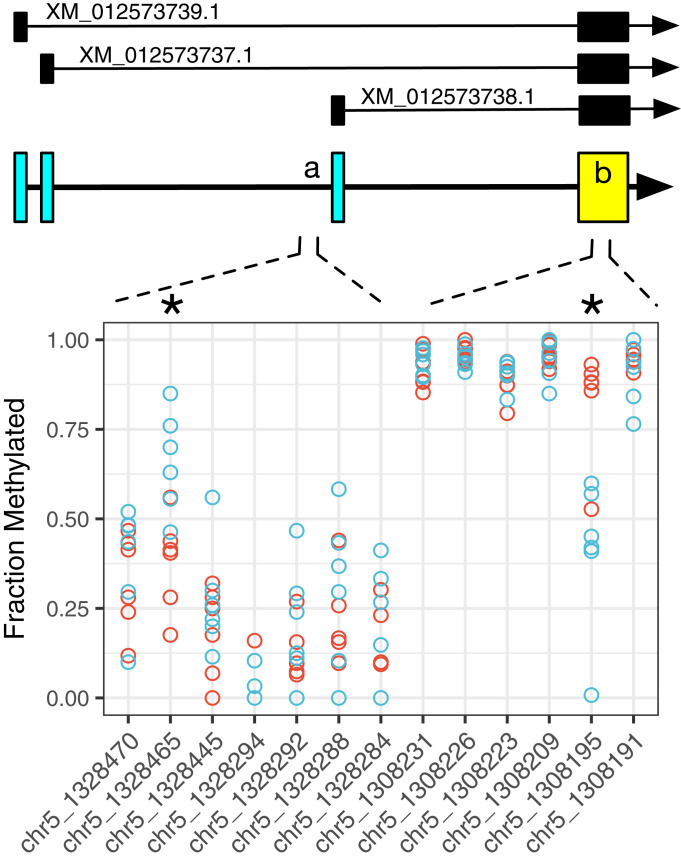
To assess whether DNA methylation might co-occur with observed changes in gene expression after chamber isolation, we conducted a targeted analysis of a dataset collected in Germany which includes Reduced Representation Bisulfite Sequence (RRBS) (28) for the caudomedial forebrain (AL dissections) from 12 male zebra finches (6 housed in a group aviary and 6 placed in solo isolation for 2 nights). The RRBS data were filtered to examine the 328 genes that were differentially expressed in the original RNA–seq experiment (Dataset S1), including gene bodies plus 10 kb upstream of the transcription start site. Of this set, we analyzed the 301 genes that are mapped in the current zebra finch reference genome (Taeniopygia_guttata-3.2.4). In these, we detected a total of 2,197 associated CpG sites, each with a read depth >10. (Note that, by design, RRBS only samples a reduced fraction of the genome; the lack of RRBS coverage does not prove the absence of functional methylation.) After correction for multiple testing, 63 of these sites, associated with 40 genes, showed strong evidence of differential methylation between treatment groups (q < 0.0001, SI Appendix, Fig. S7). As an exemplar, BDNF showed significant differential methylation in the solo condition at 2 sites, both consistent with decreased RNA expression in the solo condition (Fig. 6).
Fig. 6.
Solo isolation leads to altered methylation of the BDNF gene. Schematic gene model shows the 3 currently annotated transcripts (Top). Each circle is the value in 1 animal for the fraction of DNA methylated at that site from RRBS of auditory lobule DNA (solo isolation for 2 d, blue; group aviary, red). Two sites are marked with an asterisk (*): chr5:1328465 (methylation difference +26%, q = 3.03E-05) is localized to a region (“a”) immediately upstream and adjacent to the start site for XM_012573738.1; chr5:1308195 (methylation difference −41%, q = 1.70E-72) is within the BDNF protein coding exon (“b”). Note that both changes are consistent with decreased transcription of the BDNF gene, based on precedents in mammals (54, 55): increased methylation at BDNF exonic start sites (as in A) has been linked to transcriptional suppression (56), whereas increased methylation in coding exons (A) is associated with more active transcription and/or regulation of alternative splicing (57).
Discussion
In humans, social context affects many aspects of cognition, and prolonged social isolation has been shown to cause subtle alterations in gene transcription and DNA methylation measured in peripheral tissues (6). Here we have shown that, in the zebra finch, even a single period of overnight chamber isolation alters the expression of hundreds of genes in higher forebrain centers involved in social communication, suppressing expression of gene sets involved in neurotrophin signaling and axon guidance in particular. This suggests the potential for lasting structural and functional changes in the brain resulting from short periods of isolation. We measured consistent neurogenomic effects of acute isolation in birds sampled on 5 different occasions and in 3 different countries, using 4 different molecular assays (RNA-seq, qRT-PCR, in situ hybridization, bisulfite sequencing). Thus, we believe the phenomenon is likely to be a general one and not restricted to particular populations or rearing environments. For the zebra finch, some behavioral effects of acute solo isolation have been defined, e.g., altered call production (29), but our results provide insight into the immediate neurogenomic consequences.
Our results indicate that chamber isolation evokes a complex multifactorial response where different elements of the experience may be “encoded” in different genomic response components. For EGR1 and BDNF, changes in expression were prevented by inclusion of a second bird in the chamber. This distinction between effects of solo and duo housing suggests that the bird’s perception of “aloneness” may contribute to the regulation of these genes and can be distinguished from the general effects of handling stress and chamber environment which were common in both the solo and duo conditions. In contrast, FKBP5 showed no effect of a partner, suggesting that its expression increase may be a direct response to the physical environment of the chamber or a persisting residual of the initial capture experience. The response of UTS2B was actually potentiated when two birds were in the same chamber, perhaps tracking some parameter related to density (e.g., humidity).
In our experiments, we explicitly focused on a part of the brain that generates higher-order representations of auditory experience and social context. Within this focal brain region we observed different anatomical response patterns for different genes, further suggesting a complex multifactorial response to the isolation experience. Moreover, our in situ mapping experiments produced images that suggest that gene expression changes may not be limited to the caudomedial forebrain (SI Appendix, Figs. S8–S10). In solo housed birds, increased FKBP5 expression was visually evident throughout the nidopallium whereas EGR1 decreased in patches throughout the rostral and lateral telencephalon and in the medial arcopallium, loosely analogous to the mammalian amygdala (30). Although not typically associated with auditory functions, some of these areas also show genomic or neurophysiological responses to vocal signals (31, 32). Analysis of the broader neuroanatomical response patterns of individual genes could provide insight into the various neural processes that may be affected by different social conditions and environmental parameters.
How much of this response might be explained as a simple and direct manifestation of stress, as mediated by the glucocorticoid response? Clearly, being captured and placed in a cage is a stressful experience, as shown in many previous studies documenting a sharp but transient rise in CORT in the first 30 min following capture (33, 34). We did not measure CORT at the point of capture but observed a significant elevation in baseline CORT a day later in all of the chambered birds (but not in ones taken directly from the aviary). Although all of the chambered birds experienced the same capture stress, we saw sustained differences in gene expression that depended on the social conditions maintained in the cage after capture. We also saw only a poor correlation between CORT and gene expression across our individual subjects (SI Appendix, Fig. S6). Thus, we suggest that increased CORT may well support aspects of the chamber response, perhaps as a permissive factor for altered gene expression (35), but alone is insufficient to account for all of the effects on gene expression that we observed.
These experiments raise questions about how isolation is perceived: What sensations or sensory pathways contribute to awareness of whether a bird is alone or not? For songbirds, audition is a primary channel for social communication and perception. Our social manipulation was not explicitly auditory in nature, but of course the auditory environment is different for a bird alone in a cage compared with a bird with a partner in the cage. It will be interesting in future experiments to tease apart the specific sensory channels and experiential factors responsible for the effects of isolation—for example, providing other sounds to the solo bird, simulating a normal aviary auditory environment in the isolation chamber; providing a visual (but silent) surrogate; or leaving a bird alone in a normal aviary flight cage instead of a small cage in a sound isolation chamber. Possible differences in behavior (e.g., gross motor activity, sleep/wake cycle, call production) should also be evaluated as potential contributors to the neurogenomic pattern.
EGR1, which showed the largest decrease after isolation (Fig. 1), has been extensively used as an indicator of neural activity associated with learning, memory, and perception of salience (35). In songbird research, the EGR1 ortholog is commonly referred to as ZENK (12). Many studies have documented increases in EGR1/ZENK in songbird forebrain after experimental presentation of salient conspecific sounds or in vocal motor controls during singing (8). In almost all of these cases, the response is judged against a baseline measured in silence after overnight isolation. Our results here show that this baseline has been suppressed as a result of such isolation. Thus, the apparent induction after exposure to novel song playbacks might be regarded in part as a restoration of the expression level maintained in a more normal social context. Nevertheless, exposure to a novel salient acoustic experience even in a social context can still trigger a further increase in EGR1 expression, as shown in studies of song sparrows in the field (36) and zebra finches group-housed in the laboratory (10).
BDNF, a canonical neurotrophin (21), has been implicated in support of juvenile song learning in male zebra finches (37) and also in adult neurogenesis in the songbird forebrain (38) and rodent hippocampus (39). Prolonged social isolation suppresses survival of new neurons in the adult songbird forebrain (40), and persistent suppression of BDNF in social isolation could underlie this effect. BDNF gene methylation has been proposed as a mechanism underlying life-long consequences of early exposure to adversity in humans and rodents (41, 42). FKBP5, like BDNF, has emerged in studies of the epigenetic response to stress and adversity (35). The FKBP5 protein acts as a cochaperone of the glucocorticoid receptor complex, suppressing effects of glucocorticoids. It also suppresses DNA methyltransferase 1 (DNMT1) activity and may thereby prevent epigenetic suppression of BDNF (43).
The involvement of DNA methylation indicates 1 more level of integration to be considered in the way social experiences and perceptions are built up and represented over the life course. In principle, DNA methylation may support long-lasting changes in gene expression levels, isoform processing, and responsiveness to specific signals. It may ultimately be possible to identify specific methylation signatures or interactions between individual methylation sites and other regulatory elements to gain insight into the molecular logic of different neurogenomic states. How long does the state that we have documented here persist after the animal has been returned to a normal social environment? With even longer periods of isolation, do additional effects on other genes eventually emerge (4)? Ultimately, what is the function of the response? Does it help the animal buffer subsequent stresses, or does it make the animal more vulnerable? Consideration of neurogenomic activities in social animals like the zebra finch may help address fundamental questions about social perception and the links between social circumstance and long-term health and behavior.
Materials and Methods
Animals.
Experiments were conducted with adult zebra finches (>120 d post hatch) in 3 different aviaries over 4 y. Details of each experiment are presented in SI Appendix. Within each experiment, treatment groups were balanced, e.g., for sacrifice order and isolation chamber used, and all samples were collected within the same 1-h midday window. In the United States, procedures were conducted with protocols approved by the University of Illinois Institutional Animal Care and Use Committee. Birds were taken (June 2012) either directly from group aviaries or after a single night of solo isolation in a sound attenuation chamber in a different room and immediately killed by decapitation. Brains were removed and then either immediately dissected (AL dissection; see below) followed by flash freezing in liquid nitrogen or frozen in plastic molds. In the United Kingdom, birds were collected from a colony established at Queen Mary University of London from UK stocks. Animal procedures were conducted under Procedures Project License PPL70-8183. The time course experiment (Fig. 3) ran from November to December 2016, and the partner experiment (Fig. 5) ran from February to March 2016. For the primary qRT-PCR replication (SI Appendix, Fig. S4, performed in January 2015), females were killed by isoflurane overdose and decapitation. Animals for RRBS experiments were collected at the Max Planck Institute for Ornithology in Seewiesen, Germany, in August 2015. Animal housing and welfare were in compliance with the European directives for the protection of animals used for scientific purposes (2010/63/EU), with protocols approved by the Government of Upper Bavaria. Male zebra finches (range 147 to 170 d old; mean age in each treatment group 156 and 157 d, respectively) were collected either directly from a group aviary or placed in sound chambers between 1600 and 1700 hours and killed 2 d later by decapitation.
AL Dissection.
RNA-seq, qRT-PCR, and RRBS analyses were all based on tissues collected using the AL dissection of the caudomedial telencephalon (44). This dissection collects the major loci of the zenk/egr1 response to song playback (the NCM and the CMM), along with nonresponsive Field L2a.
RNA Sequencing and Analysis.
AL samples (US females) were processed by the Barts and the London Genome Centre at Queen Mary University of London, which performed RNA extraction, prepared indexed sequencing libraries (TruSeq, Illumina) and provided sequencing (HiSeq, Illumina) at an average read depth of 31.7 million 100-bp paired-end reads per sample (SI Appendix). Reads were then processed with Trimmomatic (45), mapped to the zebra finch transcriptome with Salmon (46), collapsed to genes with tximport (47), and imported into DESeq2 (48) for analysis of differential gene expression and gene set enrichment analysis (SI Appendix).
qRT-PCR.
Primer sets for qRT-PCR were selected using National Center for Biotechnology Information Primer-BLAST or from a study of reference genes for qRT-PCR (25). qRT-PCR data were analyzed with the R package MCMC.qpcr (26), which models gene expression in a joint GLMM. Primers, reaction conditions, assay validation, and data analysis are described in SI Appendix.
Corticosterone Radioimmunoassay.
Each animal was captured by hand, immediately decapitated, and the body placed into a 50 mL plastic tube containing 10 µL K2EDTA (anticoagulant) to collect the trunk blood (∼1 mL). Blood collection was completed within 1 min of capture, and corticosterone in plasma was quantified by radioimmunoassay (49) with modeling of treatment effects using GLM (SI Appendix).
In Situ Hybridization.
To compare general anatomical patterns of the 4 RNAs of interest, 864 sections of adult female brains were examined from 3 replications of the aviary-solo experiment, each with 6 animals (3 aviary, 3 solo): United States 2012 (coronal sections); United Kingdom 2015 (sagittal sections); United Kingdom 2016 (2 coronal and 1 sagittal for each group). Cryostat sections (10 µm) were postfixed in 3% paraformaldehyde and hybridized to digoxigenin-labeled riboprobes (SI Appendix) using standard methods (50). Digital images were obtained using a Nanozoomer whole slide scanner (Hamamatsu), and images were examined in NDP.view2 (Hamamatsu), comparing to histological reference images for the zebra finch brain (51).
RRBS.
RRBS libraries were prepared as described previously (28) and sequenced by the Biomedical Sequencing Facility at the Research Center for Molecular Medicine in Vienna. Data were preprocessed with Trimgalore for removal of adapters and low-quality sequence (28). Reads were aligned to the zebra finch genome (Taeniopygia_guttata-3.2.4) with BSMAP (52). Data were filtered to extract gene alignments with samtools v1.9, and the R package methylKit (53) was used to filter for coverage and assess differential methylation (details are in SI Appendix).
Supplementary Material
Acknowledgments
We thank the animal care staffs at the University of Illinois, Queen Mary University of London, and the Max Planck Institute for Ornithology (Seewiesen, Germany); Vardhman Rakyan and Rob Lowe for technical advice on RRBS, which was performed by Amelie Kuchler under Christoph Bock (Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna); the Barts and the London Genome Centre; and the Apocrita High-Performance Computing facility supported by Queen Mary University of London Research-IT. Financial support was provided by the NIH (R01NS045264 and 1RC1GM091556); the Leverhulme Trust; the Canadian Institute for Advanced Research; and the UK Biotechnology and Biological Sciences Research Council (BB/L023164/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.B.S. is a guest editor invited by the Editorial Board.
Data deposition: The RNA sequencing data reported in this paper are deposited in the European Nucleotide Archive, accession no. PRJEB28085. Reduced representation bisulfite sequencing are deposited under accession no. PRJEB28656, sample nos. ERS2719622–ERS2719628 and ERS2719634–ERS2719639.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820841116/-/DCSupplemental.
References
- 1.Cacioppo J. T., et al. , Loneliness across phylogeny and a call for comparative studies and animal models. Perspect. Psychol. Sci. 10, 202–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., et al. , Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat. Neurosci. 15, 1621–1623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scaccianoce S., et al. , Social isolation selectively reduces hippocampal brain-derived neurotrophic factor without altering plasma corticosterone. Behav. Brain Res. 168, 323–325 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Zelikowsky M., et al. , The neuropeptide Tac2 controls a distributed brain state induced by chronic social isolation stress. Cell 173, 1265–1279.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grippo A. J., et al. , Social isolation induces behavioral and neuroendocrine disturbances relevant to depression in female and male prairie voles. Psychoneuroendocrinology 32, 966–980 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S. W., Human social genomics. PLoS Genet. 10, e1004601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zann R. A., The Zebra Finch: A Synthesis of Field and Laboratory Studies (Oxford University Press, 1996). [Google Scholar]
- 8.Clayton D. F., The genomics of memory and learning in songbirds. Annu. Rev. Genomics Hum. Genet. 14, 45–65 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Avey M. T., Phillmore L. S., MacDougall-Shackleton S. A., Immediate early gene expression following exposure to acoustic and visual components of courtship in zebra finches. Behav. Brain Res. 165, 247–253 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Vignal C., Andru J., Mathevon N., Social context modulates behavioural and brain immediate early gene responses to sound in male songbird. Eur. J. Neurosci. 22, 949–955 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Woolley S. C., Doupe A. J., Social context-induced song variation affects female behavior and gene expression. PLoS Biol. 6, e62 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mello C. V., Vicario D. S., Clayton D. F., Song presentation induces gene expression in the songbird forebrain. Proc. Natl. Acad. Sci. U.S.A. 89, 6818–6822 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terpstra N. J., Bolhuis J. J., Riebel K., van der Burg J. M. M., den Boer-Visser A. M., Localized brain activation specific to auditory memory in a female songbird. J. Comp. Neurol. 494, 784–791 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Drnevich J., et al. , Impact of experience-dependent and -independent factors on gene expression in songbird brain. Proc. Natl. Acad. Sci. U.S.A. 109 (suppl. 2), 17245–17252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monbureau M., Barker J. M., Leboucher G., Balthazart J., Male song quality modulates c-Fos expression in the auditory forebrain of the female canary. Physiol. Behav. 147, 7–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren W. C., et al. , The genome of a songbird. Nature 464, 757–762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D., Smyth G. K., Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A., et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bramham C. R., Messaoudi E., BDNF function in adult synaptic plasticity: The synaptic consolidation hypothesis. Prog. Neurobiol. 76, 99–125 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Matosin N., Halldorsdottir T., Binder E. B., Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: The FKBP5 model. Biol. Psychiatry 83, 821–830 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Bell Z. W., et al. , Urotensin-related gene transcripts mark developmental emergence of the male forebrain vocal control system in songbirds. Sci. Rep. 9, 816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaudry H., et al. , Urotensin II, from fish to human. Ann. N. Y. Acad. Sci. 1200, 53–66 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Olias P., Adam I., Meyer A., Scharff C., Gruber A. D., Reference genes for quantitative gene expression studies in multiple avian species. PLoS One 9, e99678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matz M. V., Wright R. M., Scott J. G., No control genes required: Bayesian analysis of qRT-PCR data. PLoS One 8, e71448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suri D., Vaidya V. A., Glucocorticoid regulation of brain-derived neurotrophic factor: Relevance to hippocampal structural and functional plasticity. Neuroscience 239, 196–213 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Klughammer J., et al. , Differential DNA methylation analysis without a reference genome. Cell Rep. 13, 2621–2633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma S., Maat A. T., Gahr M., Power-law scaling of calling dynamics in zebra finches. Sci. Rep. 7, 8397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello C. V., Kaser T., Buckner A. A., Wirthlin M., Lovell P. V., Molecular architecture of the zebra finch arcopallium. J. Comp. Neurol. 10.1002/cne.24688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mello C. V., Clayton D. F., Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J. Neurosci. 14, 6652–6666 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii T. G., Ikebuchi M., Okanoya K., Auditory responses to vocal sounds in the songbird nucleus taeniae of the amygdala and the adjacent arcopallium. Brain Behav. Evol. 87, 275–289 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wingfield J. C., Smith J. P., Farner D. S., Endocrine responses of white-crowned sparrows to environmental stress. Condor 84, 399–409 (1982). [Google Scholar]
- 34.Cockrem J. F., Stress, corticosterone responses and avian personalities. J. Ornithol. 148, 169–178 (2007). [Google Scholar]
- 35.Clayton D. F., et al. , The role of the genome in experience-dependent plasticity: Extending the analogy of the genomic action potential. Proc. Natl. Acad. Sci. U.S.A. 117, 23252–23260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarvis E. D., Schwabl H., Ribeiro S., Mello C. V., Brain gene regulation by territorial singing behavior in freely ranging songbirds. Neuroreport 8, 2073–2077 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dittrich F., et al. , Maximized song learning of juvenile male zebra finches following BDNF expression in the HVC. Eur. J. Neurosci. 38, 3338–3344 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rasika S., Alvarez-Buylla A., Nottebohm F., BDNF mediates the effects of testosterone on the survival of new neurons in an adult brain. Neuron 22, 53–62 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Aimone J. B., et al. , Regulation and function of adult neurogenesis: From genes to cognition. Physiol. Rev. 94, 991–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipkind D., Nottebohm F., Rado R., Barnea A., Social change affects the survival of new neurons in the forebrain of adult songbirds. Behav. Brain Res. 133, 31–43 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Roth T. L., Lubin F. D., Funk A. J., Sweatt J. D., Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatry 65, 760–769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundakovic M., et al. , DNA methylation of BDNF as a biomarker of early-life adversity. Proc. Natl. Acad. Sci. U.S.A. 112, 6807–6813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gassen N. C., et al. , Chaperoning epigenetics: FKBP51 decreases the activity of DNMT1 and mediates epigenetic effects of the antidepressant paroxetine. Sci. Signal. 8, ra119 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Cheng H.-Y., Clayton D. F., Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J. Neurosci. 24, 7503–7513 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wingfield J. C., Farner D. S., The determination of five steroids in avian plasma by radioimmunoassay and competitive protein-binding. Steroids 26, 311–321 (1975). [DOI] [PubMed] [Google Scholar]
- 50.Carleton J. B., et al. , An optimized protocol for high-throughput in situ hybridization of zebra finch brain. Cold Spring Harb. Protoc. 2014, 1249–1258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karten H. J., et al. , Digital atlas of the zebra finch (Taeniopygia guttata) brain: A high-resolution photo atlas. J. Comp. Neurol. 521, 3702–3715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinf. 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akalin A., et al. , methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aid T., Kazantseva A., Piirsoo M., Palm K., Timmusk T., Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 85, 525–535 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruunsild P., Kazantseva A., Aid T., Palm K., Timmusk T., Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics 90, 397–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lubin F. D., Roth T. L., Sweatt J. D., Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 28, 10576–10586 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.