Abstract
Background
The role of the microbiome in liver transplantation (LT) outcome has received a growing interest in the past decades. In contrast to bacteria, the role of endogenous viral communities, known as the virome, is poorly described. Here, we applied a viral metagenomic approach to study the dynamic evolution of circulating viruses in the plasma of LT recipients and its effect on the clinical course of patients.
Methods
Patients chronically infected with hepatitis B virus (HBV) that received a LT due to endstage liver disease were included in this study. Longitudinal plasma samples were collected pre- and post-LT. Intact viral particles were isolated and sequenced on an Illumina HiSeq 2500 platform. Short read libraries were analysed with an in-house bioinformatics pipeline. Key endpoints were the dynamics of viral families and post-LT complications.
Findings
The initiation of immunosuppression induced a bloom of the Anelloviridae that dominated the post-LT plasma virome. A variety of post-LT complication were observed. Nephrotoxicity was reported in 38% of the patients and was associated with a high abundance of anelloviruses. Besides nephrotoxicity, 16 (67%) patients experienced flares of viral or bacterial infections in post-transplant follow-up. These flares were recognized by an increased burden of anelloviruses (p < 0.05). Interestingly, no mortality was observed in patients infected with human pegivirus.
Interpretation
These findings suggest a diagnostic potential for the Anelloviridae family in post-LT complications. Furthermore, the impact of human pegivirus infection on post-transplant survival should be further investigated.
Funding
This trial was supported by Gilead Sciences grant number BE-2017-000133.
Keywords: Virome, Liver transplantation, Plasma, Transfusion, Metagenomic, Anellovirus
Research in context.
Evidence before this study
Immunosuppressive therapies have significantly improved the survival of organ transplant patients. However, these therapies also increase the chance of post-transplant complications including secondary infections and de novo malignancies. Mounting evidence suggest an important role of the microbiome in organ transplant survival and patient longevity. The viral part of the microbiome, called the virome, has been associated with the functioning of the immune system. The crosstalk between the virome and immune system could have important implications in organ transplantation medicine. However, limited information is available concerning the peculiar role of these ‘commensal’ viruses.
Added value of this study
Patients that received a liver transplantation due chronic HBV infection were included in the study.
Viral metagenomic analyses of longitudinal plasma samples collected before and after liver transplantation revealed the presence of a divergent virus community. The initiation of immunosuppressive therapy induced a bloom of anelloviruses in both absolute abundance and species diversity. A variety of post-transplant complications were observed, including renal failure and infections. Remarkably, patients diagnosed with nephrotoxicity and post-transplant infections had a higher abundance of the Anelloviridae family in the blood samples. Both multi- and univariate analyses implied that the anellovirus abundance could be a proxy for immunosuppression and the possible risk of post-transplant complications. Besides the Anelloviridae, also human pegivirus (Flaviviridae) was observed. Previous studies have subscribed immunomodulatory effects to this virus in different clinical backgrounds. In our study, no mortality was observed in patients that were found positive for pegivirus. These findings clearly demonstrate the potential clinical relevance of the virome in post-liver transplantation complications.
Implications of all the available evidence
The virome remains an understudied component of the human microbiome. This study continued to further explore the implications of viruses in human health. Based on our findings, specific members of the human blood virome could be used as a diagnostic marker for post-transplant complications. Little is known about the ‘commensal’ viruses that inhabit the blood stream and their interactions with the immune system. However, based on previous findings and the data presented in this study, these viruses could be attractive for developing innovative biomarkers and support clinicians in developing personalized treatment strategies.
Alt-text: Unlabelled box
1. Introduction
In patients receiving liver transplantation (LT), organ rejection, post-transplant infections and complications related to long-term immunosuppression such as nephrotoxicity and de novo malignancies are the main substantial threats [1]. All these complications are potentially related to viral infections or viral reactivations. Consequently, viral infections impose an adverse impact on the survival rate after LT [2]. For instance, cytomegalovirus (CMV) is the most common infectious pathogen that leads to graft failure in liver transplant recipients [3]. Therefore, liver transplanted patients are regularly monitored for CMV viremia and subjected to preventive antiviral therapy in case of high-risk serological constellations [4,5]. Notably, patients diagnosed with viral hepatitis-related hepatocellular carcinoma (HCC) have a higher risk of adverse post-transplant outcomes compared to the non-viral HCC patients because of possible recurrent infections [6].
Over the past decades, there has been a growing interest in the role of endogenous microbes for the outcome of organ transplantation. Studies of the gut microbiota revealed that dysbiosis following liver transplantation is associated with impaired graft survival [7]. On a functional level, increased intestinal permeability, decreased abundance of beneficial commensal bacteria and an increase in pathogenic species may contribute to post-transplant infections and acute rejection [8]. However, there is still a large gap of information concerning the role of the virome in liver transplantation.
The virome is the most abundant and genetically diverse fraction of the human microbiome [9]. The virome interacts with host genetic factors and other microbiome components to shape the immunophenotype [9,10]. This phenotype can affect the clinical outcome of infections with pathogens, inflammatory illnesses and interventions like organ transplantation [10]. Viral metagenomic studies in organ transplant settings demonstrated that some plasma virome components take advantage of the reduced immunocompetence imposed by immunosuppressive agents [4]. These findings suggest that the viral abundance of particular viruses could be a marker of immunocompetence in immunosuppressed patients. Besides the potential role as a prognostic biomarker of adverse events, virome components are also related to positive outcomes in specific clinical manifestations. For example, the co-infection of human pegivirus (HPgV) with HIV has a beneficial impact on the survival of HIV infected individuals [11].
We hypothesized that the plasma virome and its dynamic evolution after transplantation might be associated with the clinical course of liver transplanted patients. To fill the current gap, we evaluated the impact of LT on the virome composition and dynamics before and after transplantation in a cohort of hepatitis B virus (HBV) infected individuals undergoing liver transplantation. HBV-infected patients provide a unique opportunity to study such virome-host interactions. They typically cure their HBV infection due to the liver transplantation that is accompanied by antiviral therapy and HBV hyperimmunoglobulin (HBIG) administration. However, these patients have a higher risk of hepatic malignancies, while the risk for liver transplant-related events (rejection, cirrhosis, re-transplantation) and death after transplantation are similar to non-HBV exposed recipients [12]. By analysing longitudinal plasma samples via next-generation sequencing and corresponding clinical data, we provide a comprehensive analysis of the dynamic evolution of the circulating virome and its associations with the clinical course in LT patients.
2. Methods
2.1. Study population and ethics statement
HBV infected patients diagnosed with end-stage liver diseases who received a liver transplantation at UZ Leuven (Belgium) between 2007 and 2014 were consecutively enrolled in this study. These patients are part of the chronic HBV cohort that is followed in UZ Leuven. Indications for liver transplantation other than HBV infection and a history of previous organ transplantations were used as exclusion criteria. This study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics committee research UZ/KU Leuven, Belgium. All participants gave informed consent and were able to withdraw from the study at any moment.
Serial plasma samples were collected from the HBV biobank. In total, six longitudinal samples were collected per patient, two plasma samples before (samples A and B) and four (samples C, D, E and F) after liver transplantation. The two samples (A and B) before transplant were collected at 41 ± 45 (A) and 2 ± 1 (B) months pre-transplant, while four samples (C, D, E and F) after transplant were taken at 0.5 ± 0.2 (C), 5 ± 1 (D), 13 ± 6 (E), and 23 ± 11 (F) months post-transplant. Biochemical, serological, and virologic markers were assayed by the hospital laboratories. Moreover, demographic data including age, sex, country of origin, and medication were obtained from the patient clinical record.
2.2. Post-liver transplant treatment protocol
All patients received immunosuppressive therapy that consisted of tacrolimus (e.g. 2 × 1 mg/d), mycophenolate mofetil (e.g. 2 × 250 mg/d) and methylprednisolone. Dosages of immunosuppressive therapies were adapted according to the therapeutic window and clinical signs of side-effects. Initially, methylprednisolone was prescribed in high dose of 2 × 20 mg/d and tapered in the following months until discontinuation after three months, depending on clinical conditions of the patients. Infection prophylaxis included a regimen of antibiotic and antiviral compounds. Prophylactic therapy with Valganciclovir (450 mg/d) was prescribed in case of donor and recipient CMV mismatch. Since all patients received a liver transplantation due HBV related end-stage liver disease, HBV prophylaxis was prescribed post-liver transplantation. Anti-HBV therapy consisted of intravenous injection of HBV antibodies and HBV nucleoside analogues.
2.3. Plasma sample processing and viral nucleic acid extraction
Plasma samples were treated with an optimized viral enrichment protocol. First, samples were sequentially filtered with 5.0 µm polypropylene (Millipore) and 0.8 µm polyethersulphone (Sartorius) filters. To reduce free-floating nucleic acids, filtrated plasma samples were treated with a cocktail of micrococcal nuclease (New England Biolabs) and benzonase (Millipore) and subsequently extracted with the QIAmp Viral RNA Mini Kit (Qiagen) without carrier RNA. To increase the yield of nucleic acids content, a random amplification step was performed using the Whole Transcriptome Amplification Kit 2 (WTA2, Sigma Aldrich) with an initial denaturation step at 95 °C instead of 70 °C. The extracted nucleic acids were subjected to 20 amplification cycles and purified using the MSB SPIN PCRAPACE kit (Stratec).
2.4. Sequencing library preparation and sequencing
NGS library preparation was performed using the Nextera XT DNA Library Preparation kit (Illumina) with a decreased tagmentation time to 4 min (input DNA of 1.2 ng/ µl) and halved reagent quantities to increase average DNA fragment sizes. The average fragment sizes of DNA libraries were evaluated using the High Sensitivity DNA Kit (Agilent) on a Bioanalyzer 2100 (Agilent). Libraries were quantified with the KAPA Library Quantification kit (Kapa Biosystems). Finally, sequencing was performed on a HiSeq 2500 platform (Illumina) for 2 × 150 cycles, with a total of 10 million paired end reads per sample.
2.5. Bioinformatic analysis
NGS reads were analysed with an in-house bioinformatics pipeline. Raw reads were trimmed with Trimmomatic [13]. To remove non-viral sequences, reads were mapped against the human genome and a non-redundant database of contaminating sequences with Bowtie2 [14]. The remaining reads were de novo assembled with metaSPAdes and the assembled contigs were annotated by DIAMOND with the sensitive option using the Genbank's non-redundant database [15,16]. Moreover, an additional annotation was performed using BLASTn against the nucleotide reference database (NCBI database of January 2018). Bacteriophages were annotated using both MetaPhinder2 and VirSorter in ‘virome decontamination’ mode [17,18]. The assembled contigs were clustered to generate genome bins with 95% nucleotide identity and a minimal coverage of 80% using nucmer from the MUMmer package [19]. Finally, reads were mapped back against the clustered genomes with BBMap and genome bins that mapped less than 100 reads were discarded from further analysis [20].
2.6. Anellovirus quantification
qPCR was carried out on 1:10 diluted extracted DNA/RNA samples. Universal forward (ACWKMCGAATGGCTGAGTTT) and reverse (CCCKWGCCCGARTTGCCCCT) primers were used targeting the UTR of the Anelloviridae. The qPCR-assay was developed for detecting and quantifying anelloviruses with a SYBR-green approach. Oligonucleotides (Eurogentec, Belgium) with known concentration were used for the standard curve to determine the sample concentrations. Per sample, the master mix consisted of 10 µl qPCRBIO SyGreen Blue Mix Lo-ROX (Sopachem, Belgium), 2.5 µl of both primers (10 µM), 4 µl dNTPs, and 1 µl H2O. Samples were analysed on a 7500 Fast real-time PCR system (Applied Biosystems). PCR cycle parameters were as follows: 95°C for 90 seconds, forty cycles at 94°C for 15 seconds, and 68°C for 1 minute.
2.7. Phylogenetic analysis
The Anelloviridae family constitutes a highly heterogenous group of viruses that are classified based on the genetic similarity of the open reading frame 1 (ORF1). The current demarcation criterion for Anelloviridae species is set at 35% nucleotide sequence identity of the ORF1 by ICTV taxonomy. We extracted all ORF1 sequences with ORF Finder from the dataset and clustered these at 35% cut-off to generate a non-redundant anellovirus database [21]. Amino acid alignment was performed with MUSCLE in MEGA7.0 [22]. A maximum-likelihood (ML) phylogenetic tree was generated from the extracted ORF1 amino acid sequences in MEGA7.0 with LG + F model. Support for ML trees was assessed by 1000 bootstrap replications.
2.8. Statistics
All statistical analyses were performed using R-software [23]. The non-parametric Wilcoxon rank-sum test was used to test statistically significant differences of Anelloviridae abundance and sum of species in patients with or without nephrotoxic side-effects. A similar approach was applied for determining ‘statistical significance between virome data and post-LT complications. Spearman's rank correlation coefficient was calculated to determine the correlation between the qPCR and metagenomic sequencing results. Fisher's exact test was used to determine significant differences in viral presence and absence count data and categorical variables. The Friedman test was used for repeated measures with a post-hoc Dunn's-test analysis with Bonferroni correction for multiple testing. Kaplan-Meier curves were constructed using the survival package and FactoMineR was utilized to perform factor analysis of mixed data [24,25]. The lme4 package was used for logistic regression analysis [26]. Statistical significance was assessed at an alpha level of < 0.05. The ggplot2 package was used for designing figures [27].
2.9. Role of the funding source
The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report.
3. Results
3.1. Participants
Twenty-four patients that received a liver transplantation due to HBV related end-stage liver diseases were included in this study and we collected a total number of 142 longitudinal samples. These individuals were part of the chronic HBV cohort that are followed in UZ Leuven, Belgium. Patients were chronically infected with HBV and three were co-infected with hepatitis D virus (HDV) before LT. Both recipient and donor characteristics are summarized in Table 1. The diagnosis for LT in these individuals were either liver cirrhosis (N = 10) or hepatocellular carcinoma (N = 14) due to a chronic HBV infection. The study population consisted of 19 males and 5 females with a median age of 58 (range, 39–70) at the time of liver transplantation (Table 1). The median lab MELD score was 15 and the median donor age 52. Furthermore, we retrieved blood transfusion data from 23/24 patients. In these 23 patients, a variety of blood product were given during the course of surgery, including erythrocyte concentrates, fresh-frozen plasma and platelet concentrates. No differences were observed in demographic data and patients diagnosed with liver cirrhosis or HCC.
Table 1.
Demographic overview of the study population. The numbers are expressed as absolute values (percentage) or median (range).
| Recipients characteristics | |||
|---|---|---|---|
| Gender | Male | 19 | (79%) |
| Female | 5 | (21%) | |
| Age | 58 | (39–70) | |
| Disease | Cirrhosis | 10 | (42%) |
| HCC | 14 | (58%) | |
| LabMELD | 15 | (7–34) | |
| Antiviral therapy | HBV-nucleoside analogues | 24 | (100%) |
| Lamivudine | 13 | (54%) | |
| Tenofovir | 7 | (29%) | |
| Adefovir + Lamivudine | 3 | (13%) | |
| Entecavir | 1 | (4%) | |
| HBV Immunoglobulins (post-LT) | 24 | (100%) | |
| CMV-nucleoside analogues | 5 | (21%) | |
| Valganciclovir | 5 | (100%) | |
| HDV coinfection | 3 | (13%) | |
| Blood transfusion | 23 | (96%) | |
| Pre-LT | AST | 62 | (13–157) |
| ALT | 52 | (14–115) | |
| Hospital stay (days) | ICU | 5 | (1–23) |
| Off ICU | 16 | (7–43) | |
| Ischemic perfusion time (hours) | Cold | 7 | (4–11) |
| Warm | 0.7 | (0.5–1.3) | |
| Donor characteristics | |||
| Gender | Male | 18 | (75%) |
| Female | 6 | (25%) | |
| Age | 52 | (16–57) | |
The clinical data revealed that all patients received nucleoside analogues before LT, including lamivudine, tenofovir, entecavir, or adefovir and lamivudine dual treatment. Anti-HBV treatment was complemented with the administration of HBIG for all patients post-LT, which was part of the standard of care protocol for HBV related LT. Furthermore, five patients received valganciclovir as anti-CMV treatment. Patients followed a strict therapeutic protocol including immunosuppressive drugs and anti-inflammatory treatment post-LT. This protocol included a standard initiation therapy with tacrolimus, mycophenolate mofetil and methylprednisolone. In three patients that suffered from liver cirrhosis, tacrolimus was combined with everolimus.
3.2. Sequencing data
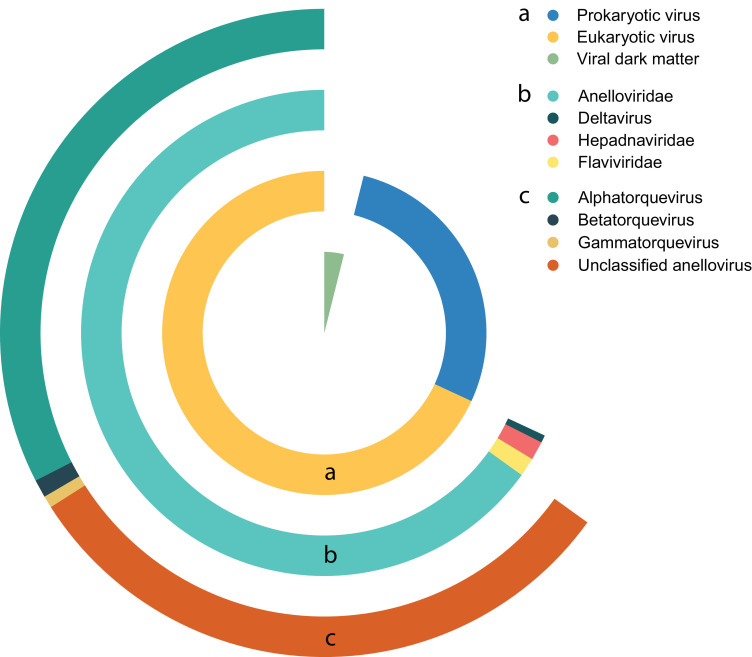
Illumina sequencing generated ∼1.4 billion reads, which was reduced by removing low quality and human genome reads to ∼0.5 billion reads. In total, 65 million reads were annotated as viral and corresponded to 4.7% of all the generated reads. The majority of viral reads were attributed to eukaryotic viruses (70.9%), followed by prokaryotic viruses (28.9%) and unclassified viral reads (0.2%) (Fig. 1). Three known bloodborne viral families that infect humans were detected, including Anelloviridae, Hepadnaviridae, and Flaviviridae, together with HDV (prototype virus of the deltavirus genus). Different genera of the Anelloviridae consisting of alphatorque, betatorque, and gammatorquevirus, as well as a group of unclassified anelloviruses were the most frequently identified viruses and widely present in the blood stream of the study population.
Fig. 1.
Distribution of the annotated viral reads over different taxonomical ranks.
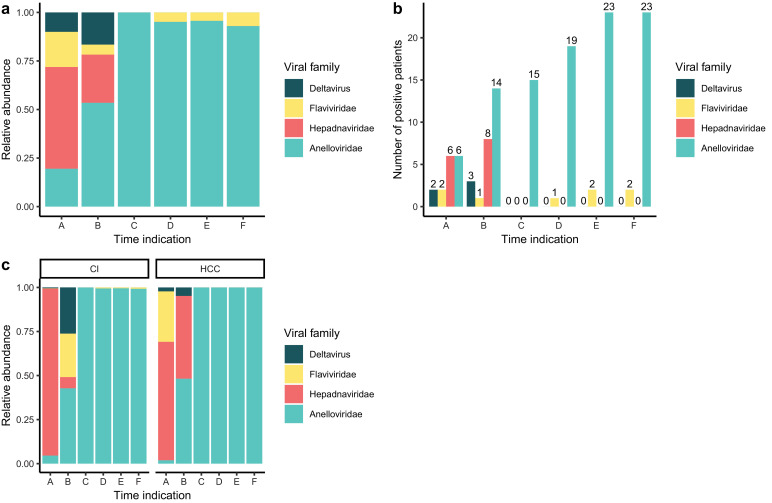
The virome composition was determined in plasma samples at different time points before and after LT. The relative abundance of eukaryotic viral families before (A and B) and after (C, D, E, and F) LT (Fig. 2(a)) were strikingly different, indicating an impact by the initiation of immunosuppressive therapy post-LT. Before transplant (samples A and B), the eukaryotic viral component is majorly composed of the Hepadnaviridae family (52%). Sample A corresponds to the first sample that was collected in UZ Leuven after which all patients received anti-HBV therapy. The impact of therapy is illustrated by the decline of Hepadnaviridae abundance (24%) seen in sample B, shortly before LT. In total, six and eight patients out of 24 were positive for HBV reads and were detected before transplant in samples A and B respectively. Also, members of the deltavirus genus (HDV in two and three patients in samples A and B respectively) were detected in three patients. Furthermore, infections with Flaviviridae were observed in two patients at timepoint A and one patient in timepoint B, making up 18% and 5% of the relative abundance of all eukaryotic viruses respectively (Fig. 2(b) and (a)).
Fig. 2.
(a) Relative abundance of the detected viral families in different timepoint before (A and B) and after (C, D, E and F) liver transplant. (b) Number of patients that are positive for the different viral families before (A and B) and after (C, D, E and F) liver transplant. (c) Relative abundance of the detected viral families in CI and HCC patients. CI: cirrhosis, HCC: hepatocellular carcinoma.
Towards transplantation, the relative abundance of the Anelloviridae increases. While in sample A, the Anelloviridae family constituted only 19% of the eukaryotic viral reads, in sample B the abundance increased up to 53%. After transplant (samples C, D, E, and F) the virome is dominated by the Anelloviridae (Fig. 2(a) and (b)).
Besides anelloviruses, also Flaviviridae were detected with 4% of the annotated reads in both samples D and E, and 7% in sample F. The Anelloviridae make up the remaining eukaryotic viral reads. After transplant, the number of patients that were found positive for Anelloviridae increased up to 23 out of 24 patients in samples E and F (Fig. 2(b)). Differentiating the study population based on the clinical indication for LT, shows a sustained HBV abundance in HCC patients before transplant while in cirrhotic patients the abundance decreases. However, this trend did not reach statistical significance (Wilcoxon rank-sum: p > 0.05).
To further explore the viral presence in liver cirrhosis and HCC patients, we generated cross-tables (Table 2) for the different viral families and patient diagnosis. For the Anelloviridae, the patients were categorized based on the tertiles of the average anellovirus abundance in high, intermediate and low burden groups. Although, a Fisher's exact test did not supportdifferences of Flaviviridae presence between cirrhosis and HCC patients (Fisher's exact test: p > 0.05), an increasing trend was observed in cirrhosis patients. Finally, patients diagnosed with HCC had a higher prevalence of the Hepadnaviridae (Fisher's exact test:p < 0.05 ).
Table 2.
Cross tables for the presence of viral families in patients diagnosed with liver cirrhosis or hepatocellular carcinoma. Numbers are expressed in frequencies and the percentage of patients positive for the virus within the disease group.
|
Anelloviridae |
Hepadnaviridae* |
Flaviviridae |
Deltavirus |
||||||
|---|---|---|---|---|---|---|---|---|---|
| N (%) | High | Medium | Low | Present | Absent | Present | Absent | Present | Absent |
| CI | 4 (40) | 3 (30) | 3 (30) | 1 (10) | 9 (90) | 3 (30) | 7 (70) | 1 (10) | 9 (90) |
| HCC | 4 (28) | 5 (36) | 5 (36) | 8 (57) | 6 (43) | 1 (7) | 13 (93) | 2 (14) | 12 (86) |
Fisher's exact test p < 0.05.
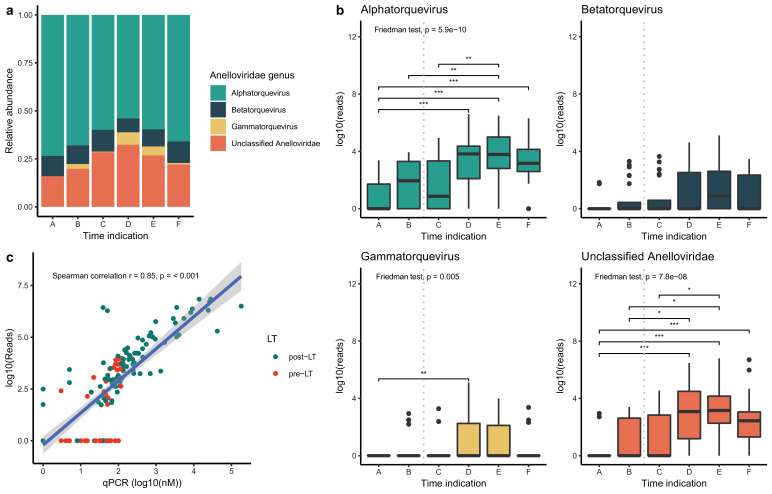
3.3. Anelloviridae
Due to the large abundance of Anelloviridae in the transplant patients, we further characterized their genera. The anelloviruses identified in this study were annotated in three genera and an additional group of unclassified anelloviruses. No significant changes were detected in the relative abundance of the three genera (Fig. 3(a)). However, the absolute number of reads of alphatorque, gammatorque and unclassified anelloviruses increased after transplant (Fig. 3(b)). To confirm the NGS results for the anelloviruses, an in-house designed qPCR assay was used on the original samples. A linear correlation was found between the number of reads and the qPCR results (Fig. 3(c), Spearman's rank correlation r= 0.85, p < 0.001).
Fig. 3.
(a) Relative abundance of the Anelloviridae genera before (A and B) and after (C, D, E and F) liver transplantation. (b) Logarithmic value of the absolute number of reads for the different Anelloviridae genera before and after liver transplantation. Grey dotted line: liver transplantation. (c) Linear model describing the correlation between NGS and qPCR data for Anelloviridae abundance. Red: pre-liver transplantation, blue: post-liver transplantation. Vertical grey dotted line: liver transplantation. Dunn's test: * p < 0.05 ** p < 0.01 *** p < 0.001.
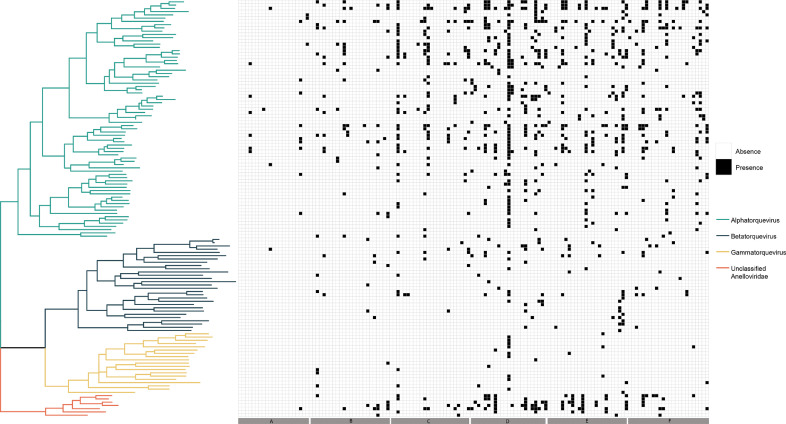
Anellovirus ORF1 was extracted from the non-redundant contig dataset to perform phylogenetic analysis. A ML phylogenetic tree was inferred from the aligned amino acid data and shows the clustering of the ORF1 sequences in the three main genera and unclassified Anelloviridae sequences (Fig. 4). To determine the presence or absence of the different ORFs in the samples, a threshold of 100 reads was set. An increasing number of anellovirus species were detected in the samples after transplant, with the majority of species derived from the alphatorque genus.
Fig. 4.
ML phylogenetic tree of the extracted amino acid ORF1 sequences with a corresponding presence and absence heatmap ranked according to the different timepoints.
3.4. The virome and post-transplant complications
The clinical data revealed incidences of post-LT side-effects in the study population. Primarily, cases of mortality, post-LT infection, nephrotoxicity and new-onset type II diabetes (NO-TIID) were reported (Table 3). The frequency of post-LT complications was compared between cirrhosis and HCC patients. The highest discrepancy was observed in mortality, with six patients in the HCC group compared to one in the cohort of patients diagnosed with liver cirrhosis (Fig. S1). However, no statistically significant differences were observed across the different post-LT complications (Fisher's exact test: p > 0.05).
Table 3.
Overview of the post-liver transplant complications.
| Variable | N | Percentage | |
|---|---|---|---|
| Mortality | 7 | (29) | |
| Recurrent HCC | 2 | (29) | |
| HCC metastasis | 4 | (57) | |
| Pneumonia | 1 | (14) | |
| Infection | 16 | (67) | |
| Viral | CMV | 2 | (11) |
| EBV | 2 | (11) | |
| Polyomavirus | 1 | (5) | |
| Bacterial | Urinary tract | 7 | (37) |
| Positive blood culture | 4 | (21) | |
| Gastroenteritis | 2 | (11) | |
| Nephrotoxicity | 9 | (38) | |
| NO-TIID | 7 | (29) | |
3.5. Nephrotoxicity
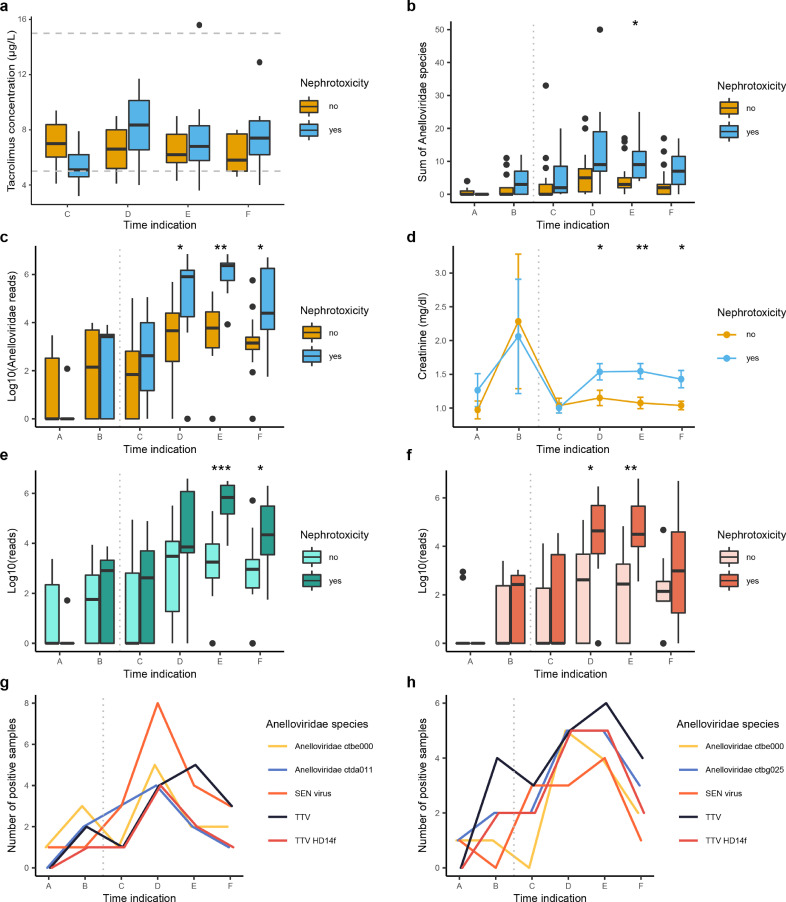
Nine patients (38%) were diagnosed with symptoms of nephrotoxic side-effects post-LT. We compared the number of Anelloviridae reads in patients that developed kidney failure to patients that did not report impaired kidney function. The tacrolimus level remained between the reference values (Fig. 5(a)) (5–15 µg/L) and did not differ between both patient groups. However, both the absolute number of Anelloviridae species and the viral abundance indicate a significant increase of this family in patients suffering from nephrotoxic side-effects post-LT (Fig. 5(b) and (c), Dunn's-test). Interestingly, the increase in anellovirus preceded and paralleled increases in serum creatinine concentrations, supporting the association between anellovirus and renal insufficiency (Fig. 5(d)). Besides an increase of the Anelloviridae read abundance on family level, both reads attributed to the alphatorque genus and unclassified anelloviruses were higher for patients with renal complications in post-LT samples (E/F and D/E respecively, Dunn's-test). To evaluate the individual Anelloviridae species in patients with different clinical courses, we compared the OTU presence in both patient groups (Fig. 5(g) and (h)). This analysis indicated that 8/9 (89%) of patients with nephrotoxic symptoms carried the SEN-virus in timepoint D (Fig. 5(e)), compared to 3/15 (20%) non-nephrotoxic patients (Fig. 5(f)) (Fisher's exact test: p < 0.05).
Fig. 5.
(a) Distribution of the tacrolimus concentration in patients diagnosed with or without nephrotoxic side-effects. (b) Sum of all Anelloviridae species divided in patients with and without nephrotoxic side effects. (c) Differences in the number of Anelloviridae reads (logarithmic) between patients with and without nephrotoxic side effects. (d) Differences in blood creatinine concentrations between patients with and without nephrotoxic side effects. (e) The number of reads attributed (logarithmic) to the alphatorque genus. (f) The number of unclassified Anelloviridae reads (logarithmic). (g) The most abundant viral species in patients with nephrotoxic side-effects. (h) The most abundant viral species in patients without nephrotoxic side-effects. Blue: with nephrotoxic side effects, yellow: without nephrotoxic side effects. Vertical grey dotted line: liver transplantation. Horizontal grey dotted line: reference values. Wilcoxon rank-sum test: * p < 0.05 ** p < 0.01 *** p < 0.001.
3.6. Infections
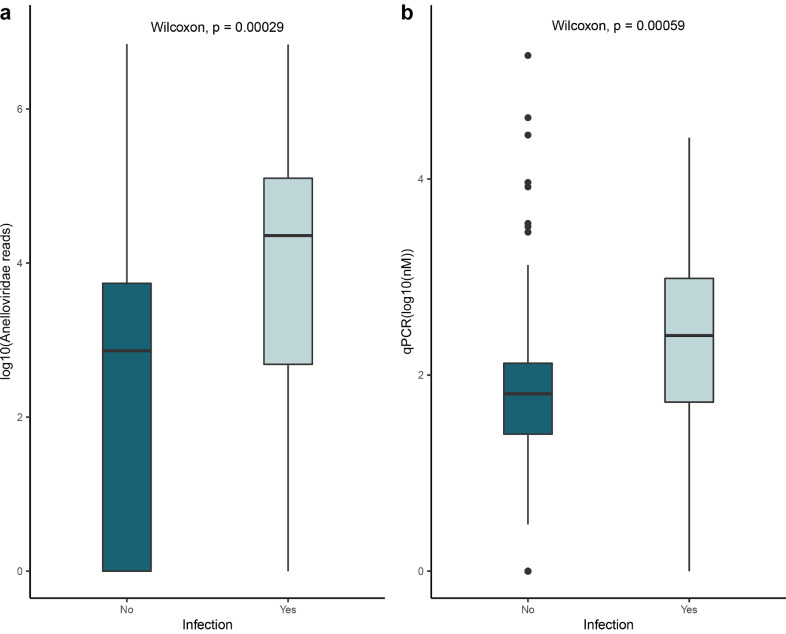
Analysis of routine laboratory results and patients’ clinical data revealed a substantial number of infections occuring after LT. A total of 16 (67%) patients experienced flares of infection in post-transplant samples, either bacterial or viral infections (Fig. 6 and Table 3). Infections were documented during regular and emergency consultations in post-transplant care. We analyzed the anellovirus load in samples close to the diagnosis of infection. The data indicates that in both the number of reads as well as the qPCR data, the Anelloviridae abundance is increased during infection compared to non-infection timepoint (Wilcoxon rank-sum: p < 0.05).
Fig. 6.
(a) The Anelloviridae reads abundance in samples diagnosed with or without infections. (b) qPCR results of the Anelloviridae family in samples diagnosed with or without infections. Only significant p-values are shown (Wilcoxon rank-sum test).
3.7. Survival analysis
A 5-year survival analysis was performed to determine the impact of initial diagnosis on post-LT survival (Fig. S2). No significant difference was found in post-LT survival after five years (log rank test: p > 0.05). In total, three patients died in the cirrhosis group, with pneumonia being the cause of death in two patients and cancer in one patient. In the group of individuals that received an LT because of HBV induced HCC, six patients died of recurrent HCC or HCC metastasis and one individual of intestinal ischemia. Remarkably, post-LT survival for individuals that died during follow-up was lower for patients with HCC (median survival 24.5 months) compared to cirrhosis patients (median survival 98 months).
A more detailed analysis was performed to study the relationship between viral families and 5-year post-LT survival. All patients had post-LT samples that were positive for anelloviruses through the course of follow-up. Therefore, to study the impact of anellovirus burden on patient survival, we classified the population according to the average post-LT presence of anelloviruses (Fig. S2(b)). Between the different groups (high, medium and low Anelloviridae abundance) no difference was found in 5-year post-LT survival (log rank test: p > 0.05). In patients that were tested positive for HBV in pre-LT samples, 56% of the patients were still alive after five years compared to 80% in HBV negative patients (Fig. S2(c)). Furthermore, HDV co-infected patients (N=3) were still alive after five years. Patients that were found positive for HPgV in follow-up samples did not report mortality cases (N=4) post-LT (Fig. S2(d)).
3.8. New-onset type II diabetes
Besides nephrotoxicity, seven patients (29%) were diagnosed with new-onset tye II diabetes (NO-TIID). Tacrolimus concentration in patients were not different in NO-TIID patients (Fig. S3(a), Wilcoxon rank-sum: p > 0.05). However, there was an increasing trend in the Anelloviridae abundance shortly after LT (Fig. S3(b)). Furthermore, comparing the most abundant OTUs of patients with and without NO-TIID (Figs. S3(c) and (d), repectively) did not show significant changes (Wilcoxon rank-sum: p > 0.05).
3.9. Clinical relevance of the Anelloviridae
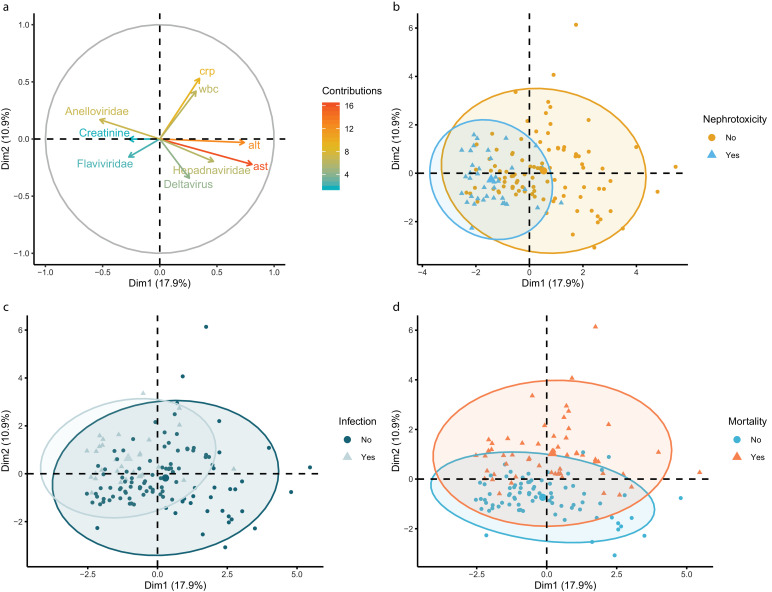
To study the relationships between individuals and different quantitative and qualitative variables, a factor analysis of mixed data was applied (Fig. 7). In multiple panels, a clustering of patients diagnosed with post-transplant complications were observed. The two dimensions in the analysis account for 28.8 % of the variance of the dataset. Scattering across the second dimension is primarily driven by clinical markers of hepatitis and the logarithm of the Anelloviridae reads in the opposite direction (Fig. 7(a)). Serum AST an ALT (R2: 0.79 and 0.71 respectively, p < 0.05) are positively correlated and the Anelloviridae abundance is negatively correlated (R2: −0.52, p < 0.05) with dimension one. The vertical distribution is mainly related to metastasis and mortality (Fig. 7(d)). Clustering of samples diagnosed with infection and nephrotoxicity along the horizontal axis partially overlap the distribution of the Anelloviridae abundance (Fig. 7(a)–(c)). These findings indicate that the Anelloviridae abundance might have discriminative power to identify nephrotoxicity and infection in clinical samples.
Fig. 7.
(a) Contribution plot of the quantitative variables across the first two dimensions. (b) Clustering of individual samples of patients diagnosed with nephrotoxicity. (c) Clustering of individual samples of patients diagnosed with post-liver transplantation infections. (d) Clustering of individual samples of patients that died during follow-up.
To further elucidate the diagnostic potential of the Anelloviridae abundance, a univariate logistic regression analysis was applied. The clinical outcome was used as the outcome variable and the logarithm of the Anelloviridae reads as the independent variable with patient ID as random effect. We observed a significant odds ratio (OR) for infection (OR 1.58, 95% CI 1.24–2.15, logistic regression: p < 0.001) and renal failure (OR 1.59, 95% CI 1.21–2.20, logistic regression: p < 0.01) (Table 4). However, no significant OR was observed for mortality (OR 1.04, 95% CI 0.11–11.70, logistic regression: p > 0.05) and NO-TIID (OR 1.63, 95% CI 0.93–3.47, logistic regression: p > 0.05).
Table 4.
Relationship between Anelloviridae abundance and post-liver transplant complications.
| Odds ratio | 95% CI | ||
|---|---|---|---|
| Anelloviridae | Infection | 1.58⁎⁎ | 1.24–2.15 |
| Nephrotoxicity | 1.59* | 1.21–2.20 | |
| Mortality | 1.04 | 0.11–11.70 | |
| NO-TIID | 1.63 | 0.93–3.47 | |
Logistic regression analysis
p < 0.01
p < 0.001
4. Discussion
Metagenomic studies have provided a strong link between the clinical status of patients and fluctuation of microorganism populations living and circulating in different biological niches of the human body [4,28]. This research added fundamental knowledge for finding disease aetiology, therapeutic and prophylactic measures. For the first time, we studied the fluctuation of the blood virome in LT recipients before and after surgery in a set of longitudinal samples.
4.1. Viral persistence (HBV)
The implemented antiviral strategy to prevent resurgence of HBV infection seemed to be effective considering that no HBV was detected post-LT in the study population. The majority of patients positive for HBV were diagnosed with HCC. Among these individuals, the primary cause of mortality was related to recurrent HCC or HCC metastasis. This supports previous data describing HBV related HCC as a risk factor for post-LT mortality [29,30]. The increased presence of HBV in HCC patients could be explained by the decreased availability of hepatocytes for viral replication in liver cirrhosis patients [31]. Also, highly active tumour cells in HCC conditions could function as a viral replication site and act as a potential reservoir [32]. However, providing HBV prophylaxis reduces the risk of recurrent HBV infection as demonstrated in this study.
4.2. Post-LT complications
An increase in the abundance and number of Anelloviridae species was observed post-LT. Furthermore, multivariate and univariate analyses further confirmed the potential clinical relevance of the Anelloviridae abundance in diagnosing post-LT complications. These observations corresponded with the initiation of immunosuppressive therapy. The interaction between immunosuppression and blooming of the anellovirus population in LT recipients approves previous studies in other organ transplantation settings [4,33].
The Anelloviridae family constitutes a highly heterogeneous group of viruses that are widely distributed in healthy individuals [34]. Anellovirus infection is acquired shortly after birth from which it establishes a possibly lifelong infection. Currently, no pathogenic effects have been subscribed to these viruses, albeit an association with autoimmune diseases and cancers has been implied in the past [35].
Previous studies demonstrated an association between the anellovirus load and solid organ rejection [4]. Of note, a lower anellovirus abundance corresponded to an increased risk of organ rejections, while a high Anelloviridae load translated in an elevated susceptibility of opportunistic infections and secondary cancers. Although, no cases of liver transplant rejection were observed, 23/24 patients suffered from post-LT complications.
4.3. Post-liver transplantation infections
In 16 patients we observed bacterial or viral post-transplant infections. The risk of infection is the result of two factors, the pathogenic exposure and the patient immune status [5]. Bacterial infections that occurred shortly after transplantation were mainly related to surgical procedures and hospital environment exposure, i.e. nosocomial exposure. The reduced immunocompetence after samples C (± 3-6 months post-LT) accommodated the occurrence of opportunistic viral infections, such as CMV and EBV reactivation. Importantly, the samples that were collected during the course of infections, contained a higher Anelloviridae load, both in terms of read number and qPCR value (Wilcoxon rank-sum: p < 0.05). This data suggests that anellovirus kinetics might be indicative for post-transplant infections, which is consistent with previous reports in different types of organ transplantations [4,36,37]. In contrast to previous findings, this study focussed at intact viral particles in an untargeted manner, that allows a broader exploration of the viral landscape in LT patients. Therefore, a more detailed assessment of the wide variety of anelloviruses in immunosuppressed patients was achieved covering multiple genera of the Anelloviridae family. In current clinical practice, the reduced signs of inflammation and possible drug interactions complicate the development of optimized diagnostic and treatment strategies in immunocompromised patients respectively [5]. Our findings support the potential use of anelloviruses as a surrogate marker of the net immune status in immunocompromised patients [4,38]. However, these findings should be interpreted with caution, as not all infectious episodes were accompanied by high Anelloviridae loads and vice versa. A careful longitudinal follow-up of organ transplant patients is required to validate the diagnostic relevance of anelloviruses in post-transplant care.
4.4. Post-liver transplantation nephrotoxicity
Besides post-LT infections, nine patients suffered from nephrotoxic side-effects after liver transplantation. An impaired renal function was related to a higher anellovirus load after LT compared to non-renal failure patients. Multiple scenarios might explain this clinical observation. The administration of immunosuppressive calcineurin inhibitors (CNIs), like tacrolimus, have significantly improved transplantation success and patient survival. However, nephrotoxicity is a common side-effect of long-term exposure to these compounds [39]. Therefore, the reported increase of Anelloviridae in patients with an impaired kidney function can be a direct consequence from the elevated tacrolimus concentration, which also negatively affects renal function. However, no significant differences were observed in the plasma tacrolimus concentrations in patients with or without renal toxicity. In contrast, the Anelloviridae load corresponded to an increase in the plasma creatinine concentration. Creatinine clearance is a biomarker that indicates kidney functioning [40]. Although, it is not considered a highly sensitive biomarker, our data demonstrates that serum creatinine is elevated in patient with renal failure. Furthermore, the data suggests that the Anelloviridae could be used as a proxy for nephrotoxic side-effects, based on the increased abundance in patients suffering from renal failure. A more detailed analysis of the individual anellovirus species revealed that some species occurred more often in patients suffering from renal failure than others. As such, SEN-virus was detected in 8/9 timepoint D samples for patients with reduced renal function. SEN-virus infection has been related to non-A-E hepatitis, albeit the data is highly inconsistent [41]. Considering the epidemiological pattern of this virus in different populations, it is unlikely that it exerts detrimental consequences for the host. Studies that demonstrated the presence of SEN-virus in healthy blood donors imply that this virus can be transmitted through blood transfusions [42,43]. However, the viral abundance did not correspond to the frequency or type of blood transfusions. Another explanation for the increase of SEN-virus post-LT, could be related to the increased Anelloviridae abundance in immunosuppressed conditions. Therefore, the incremental replication causes the viral load to pass the detection threshold of NGS post-LT. Future studies should focus at characterizing the dynamics and elucidating the role of SEN-virus in post-transplant complications and blood transfusion practice.
4.5. Post-liver transplantation new onset type II diabetes
Besides the emergence of nephrotoxic side-effects, a substantial number of patients reported post-LT NO-TIID (7/24). Multiple factors are known to contribute to the development of NO-TIID like pre-existing insulin resistance. Furthermore, drugs administered in post-LT standard care regimen negatively affect glucose control of the patient and instigates the progression to NO-TIID [44]. For instance, corticosteroids are widely used in high doses shortly after transplant and are known for causing hyperglycaemia. However, tapering of the drug improves glucose tolerance and restores the normal homeostasis. Besides corticosteroids, CNIs (e.g. tacrolimus) have been associated with the development of NO-TIID. In this study, we analysed Anelloviridae kinetics in patients with or without NO-TIID. We detected an increasing trend in the Anelloviridae abundance in NO-TIID patients, albeit no statistically significant result was found. Also, NO-TIID did not seem to correlate with other post-LT outcomes, in contrast to previous findings [45].
The plasma concentration of CNIs is an important indicator of immunosuppression and possible risk of side-effects [46]. However, we demonstrated that the level of CNIs did not discriminate between individuals diagnosed with or without post-LT complications. Although, CNIs are considered as the main contributor to post-transplant toxicities, a possible synergistic scenario between immunosuppressive drugs and viral families have not been evaluated. For instance, it has been shown that viruses such as the BK polyomavirus are involved in renal failure of non-renal solid organ transplant recipients who received immunosuppressive agents [47]. Although, we did not report the presence of BK polyomavirus in our samples, the clinical impact of other viral families found in this study should be further explored.
4.6. Anelloviridae
The majority of clinical studies have focused on the presence the alphatorquevirus genus of the Anelloviridae family. The alphatorquevirus genus is often targeted by qPCR in different clinical manifestation and is considered a potential marker for immunocompetence in organ transplant patients [48,49]. Although, this approach seems promising, a well-validated anellovirus biomarker is still lacking. Our findings indicate that a single patient can be coinfected with multiple species that belong to different genera of the Anelloviridae family. This complicates the development of a unique biomarker for clinical interpretation. Future studies should regard the high heterogeneity of the Anelloviridae family to validate the clinical importance of these viruses as biomarkers for immunocompetence.
4.7. Human pegivirus
Besides anelloviruses, we detected human HPgV in samples of four patients (three patients with cirrhosis and one patient with HCC). All patients that were found positive for HPgV were still alive 5-years post-LT, which might indicate a protective role of HPgV in post-LT survival. HPgV infection has received a growing attention by the research community because of its immune modulating potential and positive outcomes in other viral infections. Patients coinfected with HIV and HPgV demonstrated a prolonged survival with decreased HIV viral load and higher CD4+ T-cell counts [50], [51], [52]. Furthermore, HIV infected patients that acquired HPgV through blood transfusion demonstrated an improved survival compared to non-HPgV infected patients [53]. These characteristics have inspired researchers to explore the use of HPgV as a bio vaccine to complement anti-HIV therapy [11,54]. Besides the positive impact on HIV-outcome, HPgV co-infection demonstrated a potential protective role in Ebola virus infected patients [55]. Despite a single study that reported a decreasing trend in HBV DNA in HBV and HIV coinfected individuals with HPgV, the role of HPgV in HBV infected individuals is still unexplored [56]. Furthermore, the impact of HPgV and post-LT mortality should be further investigated in future studies. In contrast to the positive impact on HIV and Ebola patients’ survival, the immune modulating effect of the virus may contribute to the development and progression of non-Hodgkin's lymphoma [57,58]. Therefore, additional research is warranted to study the long-term impact of HPgV infection in healthy, HBV infected and organ transplanted patients in a larger population.
In this study, the samples were collected in a retrospective manner from the HBV cohort in UZ Leuven with a limited population size. The relatively small number of patients could have underpowered parts of the analysis. Therefore, future research should confirm the observed clinical relevance of the Anelloviridae abundance in a larger population. Furthermore, exploring the virome in different clinical settings is recommended. For instance, studying the virome in other organ transplant or blood transfusion settings will provide a more detailed insight in virome-modulating factors that could attenuate the relationship between these viruses and clinical outcomes.
In conclusion, in the assessment of longitudinal plasma samples, multiple viral families have been detected in the blood virome of liver transplant recipients. The metagenomic approach provided an unprecedented insight in the viral communities. These findings support that the clinical status and pharmacological perturbation of the immune system can have a profound impact on the presence and abundance of viral populations. The notion of an increased Anelloviridae abundance and diversity in patients suffering from post-LT side-effects calls for a further exploration of the relationship between these viruses and immunocompromised individuals. The identification of HPgV in a subgroup of our study population, highlights the necessity of studying the impact of this virus on the clinical outcome of HBV infected patients.
Data sharing statement
Raw reads were deposited in NCBI's Sequence Read Archive (SRA) database under accession no. PRJNA660895.
Declaration Competing Interest
We declare no competing interests.
Author contributions
MRP and MVR designed the study. MT, MRP, WL and FN recruited the patients, collected the samples and clinical data. MT and CKY did the NGS sequencing. MT, LB and WD analysed the NGS data. MT did the statistical analysis over the clinical data and NGS findings. MT and FT provided the first draft of manuscript and developed it. LB, WD, CKY, FN, MRP and MVR edited the manuscript and improved it. All authors read and approved the final version of the manuscript.
Funding
MT is a strategic based PhD fellow at the Research Foundation Flanders (FWO, Belgium). MRP is supported by a postdoctoral grant from the FWO. LB was funded by FWO. CKY was funded by the Interfaculty Council for the Development Cooperation (IRO) from the KU Leuven. The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103009.
Appendix. Supplementary materials
Supplementary figure 1. Post-liver transplant complications in patients diagnosed with liver cirrhosis (yellow) and hepatocellular carcinoma (blue). CI: cirrhosis, HCC: hepatocellular carcinoma, NOD: new-onset type II diabetes.
Supplementary figure 2. Kaplan-Meier plots of 5-year post-liver transplant survival.
Supplementary figure 3. (a) Distribution of the tacrolimus concentration in patients diagnosed with or without NO-TIID. (b) Differences in the number of Anelloviridae reads (logarithmic) between patients with and without NO-TIID. (c) The most abundant viral species in patients diagnosed with NO-TIID. (d) The most abundant viral species in patients without NO-TIID. Vertical grey dotted line: liver transplantation. Horizontal grey dotted line: reference values. Blue: with NO-TIID, grey: without NO-TIID. Only significant p-values are shown.
References
- 1.Burra P, Burroughs A, Graziadei I, Pirenne J, Valdecasas JC, Muiesan P. EASL clinical practice guidelines: liver transplantation. J Hepatol. 2016;64(2):433–485. doi: 10.1016/j.jhep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Tsai HI, Yu HP. A review of nationwide population study of organ transplantation in Taiwan. Acta Anaesthesiol Taiwan. 2016;54(2):70–74. doi: 10.1016/j.aat.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Liu PY, Cheng SB, Lin CC, Lin CH, Chang SN, Cheng CY. Cytomegalovirus disease after liver transplantation: a nationwide population-based study. Transpl Proc. 2014;46(3):832–834. doi: 10.1016/j.transproceed.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 4.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155(5):1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishman J. Infection in organ transplantation. A J Transpl. 2017;17(4):856–879. doi: 10.1111/ajt.14208. [DOI] [PubMed] [Google Scholar]
- 6.Younossi ZM, Stepanova M, Saab S, Ahmed A, Lam B, Srishord M. The impact of viral hepatitis-related hepatocellular carcinoma to post-transplant outcomes. J Viral Hepatitis. 2016;23(1):53–61. doi: 10.1111/jvh.12449. [DOI] [PubMed] [Google Scholar]
- 7.Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59(1):328–339. doi: 10.1002/hep.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doycheva I, Leise MD, Watt KD. The intestinal microbiome and the liver transplant recipient: what we know and what we need to know. Transplantation. 2016;100(1):61–68. doi: 10.1097/TP.0000000000001008. [DOI] [PubMed] [Google Scholar]
- 9.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157(1):142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14(7):654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenhalgh S, Schmidt R, Day T. Fighting the public health burden of AIDS with the human pegivirus. Am J Epidemiol. 2019 doi: 10.1093/aje/kwz139. [DOI] [PubMed] [Google Scholar]
- 12.Chen PH, Limketkai BN, Trilianos P, Pirtini‐Cetingul M, Woreta TA, Kim B. Effect of prior hepatitis B virus exposure on long-term risk of liver-related events after liver transplantation. Clin Transpl. 2016;30(5):579–588. doi: 10.1111/ctr.12723. [DOI] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat methods. 2012;9(4):357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 17.Jurtz VI, Villarroel J, Lund O, Larsen MV, Nielsen M. MetaPhinder—identifying bacteriophage sequences in metagenomic data sets. PloS One. 2016;11(9) doi: 10.1371/journal.pone.0163111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolduc B RS.Clustering viral genomes in iVirus. 2017 [cited 2018 25 May]. Available from: https://www.protocols.io/view/clustering-viral-genomes-in-ivirus-gwebxbe.
- 20.Bushnell B. BBTools software package. URL http://sourceforgenet/projects/bbmap. 2014.
- 21.Stothard P.The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. 2000. [DOI] [PubMed]
- 22.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Team RC. R: a language and environment for statistical computing. 2013.
- 24.Therneau T.A package for survival analysis in R. 2020;R package version 3.1–11.
- 25.Husson F, Josse J, Le S, Mazet J. FactoMineR: multivariate exploratory data analysis and data mining with R. R Package Version. 2013;1(1.29) [Google Scholar]
- 26.Bates D, Sarkar D, Bates MD, Matrix L. The lme4 package. R Package Version. 2007;2(1):74. [Google Scholar]
- 27.Wickham H. Springer; 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 28.Lecuit M, Eloit M. The human virome: new tools and concepts. Trends Microbiol. 2013;21(10):510–515. doi: 10.1016/j.tim.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baganate F, Beal EW, Tumin D, Azoulay D, Mumtaz K, Black SM. Early mortality after liver transplantation: Defining the course and the cause. Surgery. 2018;164(4):694–704. doi: 10.1016/j.surg.2018.04.039. [DOI] [PubMed] [Google Scholar]
- 30.Burra P, Germani G, Adam R, Karam V, Marzano A, Lampertico P. Liver transplantation for HBV-related cirrhosis in Europe: an ELTR study on evolution and outcomes. J Hepatol. 2013;58(2):287–296. doi: 10.1016/j.jhep.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Liu C, Wang L, Xie H, Zhang L, Wang B, Luo C. The relationship between serum hepatitis B virus DNA level and liver histology in patients with chronic HBV infection. PloS one. 2018;13(11) doi: 10.1371/journal.pone.0206060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roche B, Roque-Afonso AM, Nevens F, Samuel D. Rational basis for optimizing short and long-term hepatitis B virus prophylaxis post liver transplantation: role of hepatitis B immune globulin. Transplantation. 2015;99(7):1321. doi: 10.1097/TP.0000000000000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blatter JA, Sweet SC, Conrad C, Danziger-Isakov LA, Faro A, Goldfarb SB. Anellovirus loads are associated with outcomes in pediatric lung transplantation. Pediatr Transpl. 2018;22(1):e13069. doi: 10.1111/petr.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Villiers E-M, Zur Hausen H. Springer Science & Business Media; 2008. TT viruses: the still elusive human pathogens. [PubMed] [Google Scholar]
- 35.Freer G, Maggi F, Pifferi M, Di Cicco ME, Peroni DG, Pistello M. The virome and its major component, anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front Microbiol. 2018;9:686. doi: 10.3389/fmicb.2018.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frye BC, Bierbaum S, Falcone V, Köhler TC, Gasplmayr M, Hettich I. Kinetics of torque teno virus-DNA plasma load predict rejection in lung transplant recipients. Transplantation. 2019;103(4):815–822. doi: 10.1097/TP.0000000000002436. [DOI] [PubMed] [Google Scholar]
- 37.Maggi F, Focosi D, Statzu M, Bianco G, Costa C, Macera L. Early post-transplant torquetenovirus viremia predicts cytomegalovirus reactivations in solid organ transplant recipients. Sci Rep. 2018;8(1):1–8. doi: 10.1038/s41598-018-33909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz P, Martínez-Picola M, Santana M, Muñoz J, Pérez-del-Pulgar S, Koutsoudakis G. Torque teno virus is associated with the state of immune suppression early after liver transplantation. Liver Transpl. 2019;25(2):302–310. doi: 10.1002/lt.25374. [DOI] [PubMed] [Google Scholar]
- 39.Naesens M, Kuypers DR, Sarwal M. Calcineurin inhibitor nephrotoxicity. Clin J Am Soc Nephrol. 2009;4(2):481–508. doi: 10.2215/CJN.04800908. [DOI] [PubMed] [Google Scholar]
- 40.Edelstein CL. Biomarkers of Kidney Disease. Elsevier; 2017. Biomarkers in acute kidney injury; pp. 241–315. [Google Scholar]
- 41.Mrzljak A, Tabain I, Premac H, Bogdanic M, Barbic L, Savic V. The role of emerging and neglected viruses in the etiology of hepatitis. Curr Infect Dis Rep. 2019;21(12):51. doi: 10.1007/s11908-019-0709-2. [DOI] [PubMed] [Google Scholar]
- 42.Akiba J, Umemura T, Alter HJ, Kojiro M, Tabor E. SEN virus: epidemiology and characteristics of a transfusion-transmitted virus. Transfusion. 2005;45(7):1084–1088. doi: 10.1111/j.1537-2995.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 43.Fred HL, Thangam M, Aisenberg GM. Baylor University Medical Center Proceedings. Taylor & Francis; 2018. Pathogens transmitted in red blood cell transfusions: an up-to-date table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shivaswamy V, Boerner B, Larsen J. Post-transplant diabetes mellitus: causes, treatment, and impact on outcomes. Endocrine Rev. 2016;37(1):37–61. doi: 10.1210/er.2015-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv C, Zhang Y, Chen X, Huang X, Xue M, Sun Q. New-onset diabetes after liver transplantation and its impact on complications and patient survival. J Diab. 2015;7(6):881–890. doi: 10.1111/1753-0407.12275. [DOI] [PubMed] [Google Scholar]
- 46.Jia J-J, Lin B-Y, He J-J, Geng L, Kadel D, Wang L. ''Minimizing tacrolimus''strategy and long-term survival after liver transplantation. World J Gastroenterol: WJG. 2014;20(32):11363. doi: 10.3748/wjg.v20.i32.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuppachi S, Kaur D, Holanda DG, Thomas CP. BK polyoma virus infection and renal disease in non-renal solid organ transplantation. Clin Kidney J. 2015;9(2):310–318. doi: 10.1093/ckj/sfv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaksch P, Kundi M, Görzer I, Muraközy G, Lambers C, Benazzo A. Torque teno virus as a novel biomarker targeting the efficacy of immunosuppression after lung transplantation. J Infect Dis. 2018;218(12):1922–1928. doi: 10.1093/infdis/jiy452. [DOI] [PubMed] [Google Scholar]
- 49.Focosi D, Maggi F. Torque teno virus monitoring in transplantation: the quest for standardization. Am J Transpl. 2019;19(5):1599–1601. doi: 10.1111/ajt.15194. [DOI] [PubMed] [Google Scholar]
- 50.N'Guessan KF, Anderson M, Phinius B, Moyo S, Malick A, Mbangiwa T, editors. The impact of human pegivirus on CD4 cell count in HIV-positive persons in Botswana. Open forum infectious diseases. Oxford University Press; US: 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vahidnia F, Petersen M, Stapleton JT, Rutherford GW, Busch M, Custer B. Acquisition of GB virus type C and lower mortality in patients with advanced HIV disease. Clin Infect Dis. 2012;55(7):1012–1019. doi: 10.1093/cid/cis589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC. Infection with GB virus C and reduced mortality among HIV-infected patients. N England J Med. 2001;345(10):715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- 53.Lanteri MC, Vahidnia F, Tan S, Stapleton JT, Norris PJ, Heitman J. Downregulation of cytokines and chemokines by GB virus C after transmission via blood transfusion in HIV-positive blood recipients. J Infect Dis. 2015;211(10):1585–1596. doi: 10.1093/infdis/jiu660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gretch D. Oxford University Press; 2012. Editorial commentary: advocating the concept of GB virus c biotherapy against AIDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lauck M, Bailey AL, Andersen KG, Goldberg TL, Sabeti PC, O'Connor DH. GB virus C coinfections in West African Ebola patients. J Virol. 2015;89(4):2425–2429. doi: 10.1128/JVI.02752-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.N'Guessan KF, Boyce C, Kwara A, Archampong TN, Lartey M, Sagoe KW. Human pegivirus (HPgV) infection in Ghanaians co-infected with human immunodeficiency virus (HIV) and hepatitis B virus (HBV) Virus Genes. 2018;54(3):361–367. doi: 10.1007/s11262-018-1555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang CM, Stapleton JT, Klinzman D, McLinden JH, Purdue MP, Katki HA. GBV-C infection and risk of NHL among US adults. Cancer Res. 2014;74(19):5553–5560. doi: 10.1158/0008-5472.CAN-14-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krajden M, Yu A, Braybrook H, Lai AS, Mak A, Chow R. GBV-C/hepatitis G virus infection and non-Hodgkin lymphoma: a case control study. Int J Cancer. 2010;126(12):2885–2892. doi: 10.1002/ijc.25035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Post-liver transplant complications in patients diagnosed with liver cirrhosis (yellow) and hepatocellular carcinoma (blue). CI: cirrhosis, HCC: hepatocellular carcinoma, NOD: new-onset type II diabetes.
Supplementary figure 2. Kaplan-Meier plots of 5-year post-liver transplant survival.
Supplementary figure 3. (a) Distribution of the tacrolimus concentration in patients diagnosed with or without NO-TIID. (b) Differences in the number of Anelloviridae reads (logarithmic) between patients with and without NO-TIID. (c) The most abundant viral species in patients diagnosed with NO-TIID. (d) The most abundant viral species in patients without NO-TIID. Vertical grey dotted line: liver transplantation. Horizontal grey dotted line: reference values. Blue: with NO-TIID, grey: without NO-TIID. Only significant p-values are shown.