Significance
Hematopoiesis involves the control of gene expression that regulates the processes of proliferation and differentiation. We found that the chromatin remodeler CHD7 controls the differentiation process. Knockdown or knockout of CHD7 leads to enhanced hematopoietic differentiation in zebrafish and mice, suggesting that CHD7 acts as a brake on gene expression associated with terminally differentiated blood cells.
Keywords: hematopoiesis, RUNX1, CHD7
Abstract
Hematopoietic stem and progenitor cell (HSPC) formation and lineage differentiation involve gene expression programs orchestrated by transcription factors and epigenetic regulators. Genetic disruption of the chromatin remodeler chromodomain-helicase-DNA-binding protein 7 (CHD7) expanded phenotypic HSPCs, erythroid, and myeloid lineages in zebrafish and mouse embryos. CHD7 acts to suppress hematopoietic differentiation. Binding motifs for RUNX and other hematopoietic transcription factors are enriched at sites occupied by CHD7, and decreased RUNX1 occupancy correlated with loss of CHD7 localization. CHD7 physically interacts with RUNX1 and suppresses RUNX1-induced expansion of HSPCs during development through modulation of RUNX1 activity. Consequently, the RUNX1:CHD7 axis provides proper timing and function of HSPCs as they emerge during hematopoietic development or mature in adults, representing a distinct and evolutionarily conserved control mechanism to ensure accurate hematopoietic lineage differentiation.
Hematopoiesis is established in three waves at different anatomic sites in all vertebrate embryos. The first, primitive wave takes place in the yolk sac and generates primitive erythrocytes, macrophages, and megakaryocytes. In the second, definitive wave, hematopoietic progenitor cells (HPCs) differentiate from hemogenic endothelium in the yolk sac and dorsal aorta (1) and in the placenta, vitelline, and umbilical arteries in mice and humans (2). The third wave, also derived from hemogenic endothelium in the dorsal aorta, vitelline, and umbilical arteries, produces hematopoietic stem cells (HSCs).
A large-scale in vivo reverse genetic screen targeting zebrafish homologs of 425 human chromatin factors with antisense oligonucleotide morpholinos to identify genes controlling embryonic hematopoietic stem and progenitor cell (HSPC) formation uncovered chromodomain-helicase-DNA-binding protein 7 (Chd7) as the only factor that increased the expression of both primitive and definitive hematopoietic genes, including runx1, when inhibited (3). The CHD class of ATP-dependent chromatin-remodeling enzymes alters nucleosome structure and has been implicated in the maintenance of mouse embryonic stem cells, mammalian development, DNA damage response, and transcription regulation (4). Autosomal dominant CHD7 mutations cause the inherited CHARGE and Kallmann syndromes (5). Mutations and copy number variations of CHD7 have been found in hematologic and other cancers (6).
RUNX1 is a master transcription factor absolutely required for hemogenic endothelial specification and the endothelial to hematopoietic cell transition in zebrafish and mice (7–12). Loss of RUNX1 in adult HSCs results in HPC and myeloid lineage expansion and lymphoid lineage depletion (13). Here, we show that CHD7 genetically interacts with RUNX1 during hematopoietic ontogeny and adult hematopoiesis and that disruption of CHD7 leads to enhanced hematopoietic differentiation.
Results
CHD7 Negatively Regulates Hematopoietic Development.
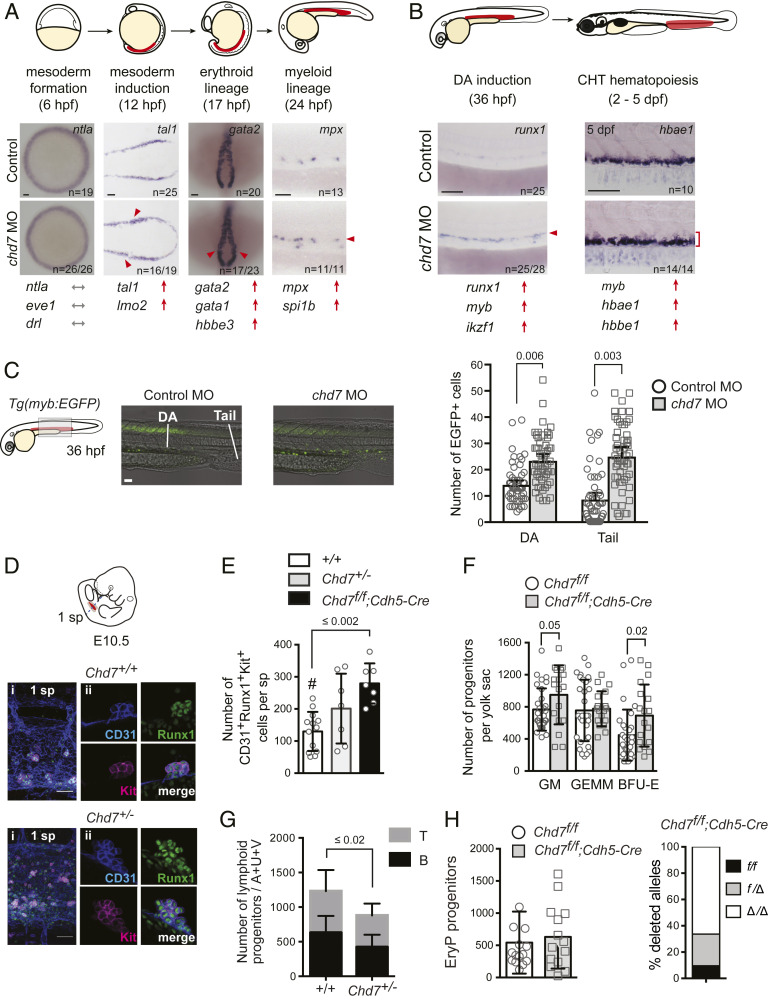
Morpholino (MO) knockdown of chd7 in zebrafish embryos (chd7 morphants) increased the expression of primitive erythroid-specific β-globin e3 (hbbe3) at 17 h post fertilization (hpf), and the definitive hematopoietic markers myb and runx1 in the dorsal aorta (DA) at 36 hpf (SI Appendix, Fig. S1 A–D). Markers of hematopoietic mesoderm at 10– to 12 hpf (tal1 and lmo2) were elevated in chd7 morphants, while earlier expression of the pan-mesodermal markers ntla and eve1 and the early hematopoietic marker drl at 6 hpf were normal (Fig. 1A and SI Appendix, Fig. S2A). Markers of primitive erythroid (gata1, gata2, hbbe3) and myeloid cells (mpx and spi1b) were increased in the posterior lateral mesoderm, and expression of definitive myeloid (myb) and erythroid (hbae1, hbbe1) markers were elevated in the caudal hematopoietic tissue (CHT), a site of HSPC colonization (Fig. 1 A and B and SI Appendix, Fig. S2A). Expression of endothelial kdrl was normal, and analysis of Tg(kdrl:EGFP) chd7 morphants showed no increase in GFP+ endothelial cells (SI Appendix, Fig. S1 E and F). Hence, chd7 regulates both primitive and definitive hematopoietic lineage gene expression in the zebrafish embryo. The increased expression of hematopoietic genes correlated with enhanced hematopoiesis. chd7 Tg(myb:EGFP) morphants had 1.6-fold more myb:EGFP+ cells in the DA and 2.8-fold more in the posterior tail region than control embryos (Fig. 1C). Similar increases were observed in two other hematopoietic transgenic lines, Tg(lmo2:DsRed) and Tg(itga2b:EGFP) (SI Appendix, Fig. S1G) and in an independent study (14). We confirmed a reduction of rag1 expression in the thymus, indicative of a decrease in T lymphocyte progenitors (SI Appendix, Fig. S2A), which was attributed in a previous study to severely impaired thymus organogenesis (14). We conclude that chd7 negatively regulates HSPC formation in the zebrafish embryo.
Fig. 1.
Chd7 negatively regulates embryonic hematopoiesis. (A) Chd7 knockdown increases expression of hematopoietic mesodermal precursor, primitive erythroid and myeloid, but not early mesoderm genes. Representative embryos for whole-mount in situ hybridization are shown, with additional genes shown in SI Appendix, Fig. S2. Regions of blood development are highlighted in red in the embryo schematic. Red arrows and arrowheads indicate an increase. Gray arrows indicate no change. (Scale bars, 50 μm.) Replicates: 2. (B) Chd7 knockdown increases expression of definitive HSPC and definitive myeloid and erythroid genes. Same symbols as in A. (C) Chd7 knockdown in Tg(myb:EGFP) embryos increases EGFP+ cells in the DA and tail region (Left), which is quantified in graph (Right) (n = 53 to 55). Representative embryos shown are from three independent replicates. (D) Chd7 deletion in mice increases Runx1+CD31+Kit+ hematopoietic clusters detected by confocal imaging of E10.5 Chd7+/+, Chd7+/−, and Chd7f/f; Cdh5-Cre AGM regions. Representative clusters shown. (i) One somite pair (sp) area. (ii) Individual cluster. (E) Quantification of data from D (n = 7 to 13). One-way ANOVA, Dunnett’s multiple comparison test; #, comparator. (F) Increased number of burst-forming unit–erythroid (BFU-E) and granulocyte/monocyte progenitors (colony-forming unit for granulocytes and macrophages) in E10.5 Chd7+/− yolk sacs (n = 8 to 14). GEMM, granulocyte/erythrocyte/monocyte/megakaryocyte progenitors. (G) Reduced number of lymphoid progenitors in E10.5 Chd7+/− embryos (n = 10 to 12). A+U+V: AGM, umbilical, and vitelline arteries. (H, Left) The number of erythroid progenitors (EryP) in the yolk sac of Chd7f/f;Cdh5-Cre embryos is not altered (n = 14 to 15). (H, Right) Both Chd7f alleles were deleted in 65% of the EryP colonies, and one allele was deleted in 27% of the colonies; thus Cdh5-Cre was active in the majority of EryPs or their precursors (n = colonies from 6 to 8 yolk sacs). All graphs show mean ± SD, unpaired two-tailed t test unless otherwise specified.
To determine if the function of CHD7 in hematopoiesis is conserved in the mouse, we measured the number of phenotypic HSPCs in the aorta-gonad-mesonephros (AGM) region of Chd7 mutant embryos. HSPCs in mouse embryos briefly accumulate as clusters of Runx1+CD31+Kit+ cells attached to luminal endothelial cells in the major caudal arteries, peaking in number at embryonic day 10.5 (E10.5) (15). Germline deletion of CHD7 caused a developmental delay by E10.5 and lethality by E11.5 (16), preventing accurate assessment of AGM hematopoiesis in null embryos. Therefore, we enumerated Runx1+CD31+Kit+ hematopoietic cluster cells in Chd7+/− embryos, which are viable, and in embryos with Chd7f alleles deleted by Cre driven by vascular endothelial cadherin (Cdh5) regulatory sequences (Cdh5-Cre) (SI Appendix, Fig. S3A). There was a significant increase in Runx1+CD31+Kit+ cells in the AGM region of Chd7f/f;Cdh5-Cre embryos and a trend toward increased numbers in Chd7+/− embryos (Fig. 1 D and E). The numbers of definitive erythroid and myeloid progenitors in the yolk sac of mutant embryos also increased (Fig. 1F), but lymphoid progenitor numbers in the AGM, umbilical, and vitelline arteries (A+U+V) were lower (Fig. 1G). The number of primitive erythroid progenitors in the yolk sac was not altered (Fig. 1H). We conclude that CHD7 restrains the numbers of definitive erythroid and myeloid progenitors differentiating from the yolk sac and phenotypic HSPCs in the AGM region in both zebrafish and mouse embryos.
CHD7 Functions to Regulate Myeloid Output from HSCs.
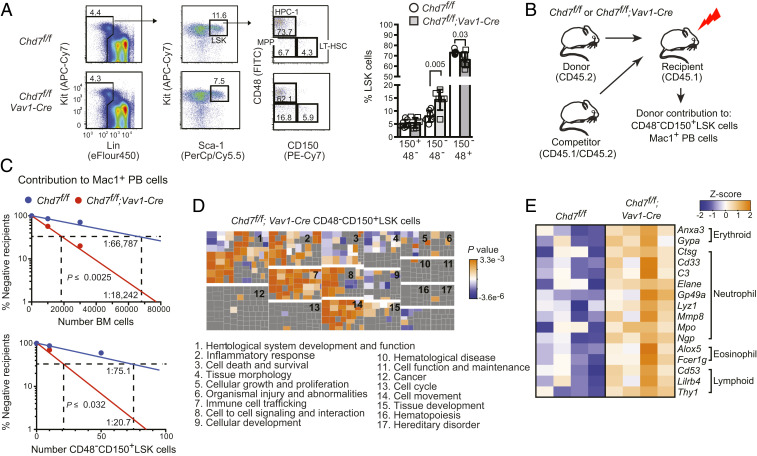
We evaluated the hematopoietic function of CHD7 in the mouse by excising Chd7f alleles with Vav1-Cre (SI Appendix, Fig. S3 A and B). CHD7 is most highly expressed in long-term repopulating HSCs (LT-HSCs) (SI Appendix, Fig. S3C), but deletion of CHD7 did not significantly affect the percentages of phenotypic LT-HSCs (CD48−CD150+ lineage−Sca1+Kit+ [LSK]) or committed hematopoietic progenitors (HPC-1, CD48+CD150−LSK), although there was a significant increase in CD48−CD150−LSK cells, which contain several populations of multipotent progenitors (MPPs) (17) (Fig. 2A). The frequency of functional CHD7-deficient LT-HSCs in both whole bone marrow (BM) and in purified CD48−CD150+LSK cells, determined by limiting dilution transplants, increased more than two-fold when donor contribution to Mac1+ peripheral blood (PB) cells was scored (Fig. 2 B and C). In contrast, no significant increase in LT-HSC frequency was detected when donor contribution to CD48−CD150+LSK cells in the recipient BM was scored (SI Appendix, Fig. S3D), suggesting that CHD7 does not affect the frequency of LT-HSCs, but constrains the myeloid lineage output of LT-HSCs and potentially other downstream progenitors. Serial transplantation of BM cells revealed no differences between control and CHD7-deficient HSCs by the fourth transplant; thus CHD7-deficient LT-HSCs had normal self-renewal capacity (SI Appendix, Fig. S3 E and F). There were no differences in proliferation, quiescence, or apoptosis between CHD7-deficient and control LT-HSCs, MPPs, or HPC-1s in adult mice (SI Appendix, Fig. S3 G–I). The frequencies of phenotypic LT-HSCs and HPC-1s were not differentially affected by stress induced by 5-fluorouracil (FU) injection (SI Appendix, Fig. S3J). We conclude that CHD7 loss does not negatively impact most functional properties of LT-HSCs.
Fig. 2.
CHD7 regulates hematopoiesis cell autonomously. (A) Chd7 deficiency does not affect phenotypic LT-HSCs. Flow cytometry of LT-HSCs (CD48−CD150+), MPPs (CD48−CD150−), and HPC-1s (CD48+CD150−) from LinnegSca1+Kit+ (LSK) bone marrow populations (Left), which is quantified in bar graph (Right) (n = 6 to 7). Mean ± SD, unpaired two-tailed t test. (B) Schematic diagram of mouse limiting dilution transplantation experiments. (C) The frequency of functional CHD7-deficient LT-HSCs increased two-fold in whole BM (Left) and in purified CD48−CD150+LSK cells (Right) when ≥1% donor contribution to Mac1+ PB was scored at 4 mo. LT-HSC frequency was calculated by ELDA (n = 7 to 14 recipients per dose). (D) Loss of CHD7 increases multilineage hematopoiesis. (Top) Functional categories for genes up-regulated in CHD7-deficient mouse LT-HSCs were enriched for hematopoietic-related functions by IPA. Clusters of individual functional gene annotations (squares) belonging to each category are labeled numerically. Replicates: 4. (E) Heatmap of representative lineage-specific genes up-regulated in CHD7-deficient CD48−CD150+LSK cells.
Gene expression profiling, however, did reveal differences between control and CHD7-deficient LT-HSCs. Ingenuity Pathway Analysis (IPA) determined that genes up-regulated in CHD7-deficient LT-HSCs were significantly associated with hematopoietic system development and function, immune cell trafficking, cell-to-cell signaling and interaction, and cellular movement (Fig. 2D and Datasets S1 and S2). Genes representative of each blood lineage, including erythroid (Hbb-b1, glycophorins), myeloid (Mpo, Lyz1, Alox5), and lymphoid (Thy1) were up-regulated in CHD7-deficient LT-HSCs (Fig. 2E and Dataset S1), suggesting that CHD7 deficiency results in LT-HSCs that are more primed for multilineage differentiation. Additional evidence that CHD7 constrains myeloid lineage differentiation includes an elevated frequency of differentiated Gr1+Mac1+ cells in the liver of E14.5 Chd7+/− fetuses (SI Appendix, Fig. S4A) and in adult BM and spleen of Chd7f/f;Vav1-Cre mice (SI Appendix, Fig. S4B) and increased numbers of granulocyte/monocyte progenitors in culture (SI Appendix, Fig. S4C). We conclude that CHD7-deficient LT-HSCs are more primed for differentiation, particularly of myeloid lineage cells.
CHD7 Cooperates with the RUNX1 Transcription Factor.
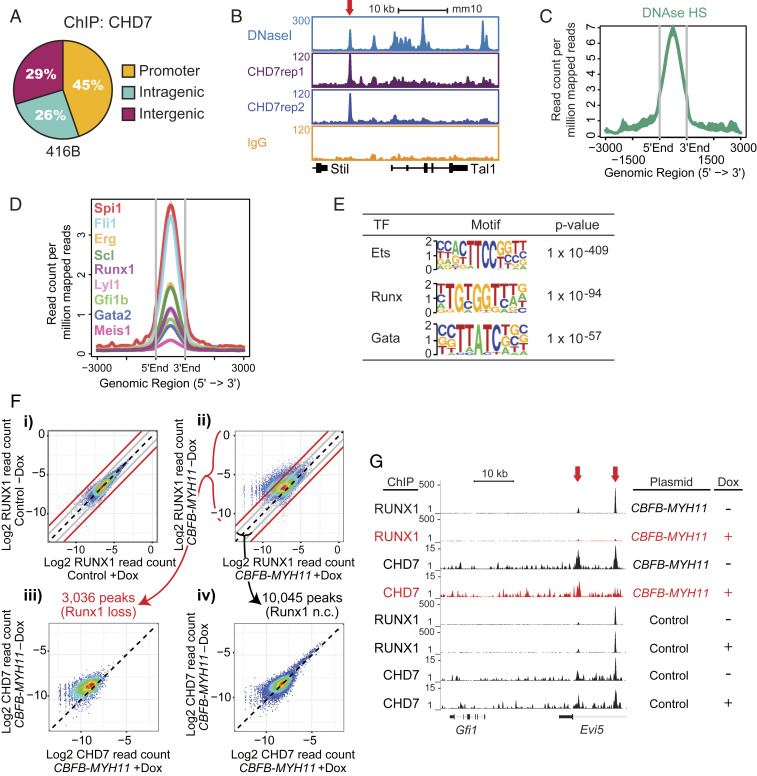
To determine how CHD7 regulates hematopoietic genes, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) to identify CHD7-occupied loci in the murine 416B HPC line (18) (Dataset S3). CHD7-occupied gene regions are DNaseI hypersensitive and colocalize with multiple hematopoietic transcription factors (19) (Fig. 3 A–D). Ets, Runx, and Gata motifs were enriched at CHD7-bound sites, suggesting that CHD7 functions in part through key hematopoietic transcription factors to regulate hematopoiesis (Fig. 3E). Given that RUNX1 was shown to interact with CHD7 (20), we performed ChIP-seq to determine if RUNX1 influenced CHD7 occupancy. We were unable to generate a RUNX1-deficient HPC line, so instead used a Tet-inducible, neomorphic CBFB-MYH11 fusion gene to block RUNX1 activity (SI Appendix, Fig. S5A). CBFβ-SMMHC (encoded by CBFB-MYH11) blocks RUNX1 activity, in part, by sequestering it apart from the DNA (21). Upon induction of CBFB-MYH11 expression (+Dox), we identified peaks with greater than four-fold decreases in RUNX1 binding (Fig. 3F and SI Appendix, Fig. S5B). Of the 3,036 peaks that lost RUNX1 binding, 1,043 (34.4%) showed a greater than two-fold and 270 (8.9%) showed a greater than four-fold loss in CHD7 binding (Fig. 3 F, ii and iii), as illustrated for the Evi5 gene (Fig. 3G). In contrast, of the 10,045 RUNX1 peaks that did not decrease upon CBFB-MYH11 expression, only 781 (7.8%) showed a greater than two-fold and 65 (0.6%) showed a greater than four-fold CHD7-binding loss (Fig. 3 F, ii and iv). These results are reproducible and indicate that CHD7 is selectively lost from regions of the genome where RUNX1 binding is attenuated (Dataset S4). This observation is supported by short-term analysis of CRISPR/Cas9 knockouts for CHD7 and RUNX1 in which initial gene expression changes when either gene is perturbed are positively correlated (SI Appendix, Fig. S5C and Dataset S5) and consistent with previous data showing that ∼30% of genes that are differentially expressed in CBFB-MYH11-expressing cells upon CHD7 loss are direct RUNX1 targets (20). Consequently, CHD7 and RUNX1 function together on target genes important for hematopoiesis in HPCs at the genomic level.
Fig. 3.
CHD7 cooperates with hematopoietic transcription factors to regulate hematopoiesis. (A) CHD7-binding distribution in the murine 416B HPC cell line by ChIP-seq. Replicates: 2. (B) Gene track of CHD7 binding overlaps with DNaseI hypersensitive sites at the Tal1 gene. (C) Overlap of CHD7-binding and DNaseI hypersensitive sites. (D) Overlap of CHD7 binding and hematopoietic transcription factors. (E) CHD7-binding sites are highly enriched for Ets, Runx, and Gata motifs by HOMER motif analysis. (F) CHD7 binding is selectively lost from genomic regions where RUNX1 binding is attenuated by CBFB-MYH11. CBFB-MYH11 expression was induced in myeloid progenitor cells by doxycycline (Dox). RUNX1 occupancy in a (i) control clone and (ii) CBFB-MYH11-expressing clone. Loss of CHD7 occupancy is (iii) higher in regions of greater than four-fold RUNX1 occupancy loss and (iv) minimally changed in regions of less than two-fold RUNX1 occupancy loss. Diagonal black lines indicate no change (n.c.). Gray lines indicate two-fold change. Red lines indicate four-fold change. A replicate experiment is shown in SI Appendix, Fig. S5B. (G) Gene tracks showing loss of RUNX1 and CHD7 binding to Evi5 (red arrows) in Dox-induced CBFB-MYH11-expressing cells.
CHD7 Physically Interacts with RUNX1.
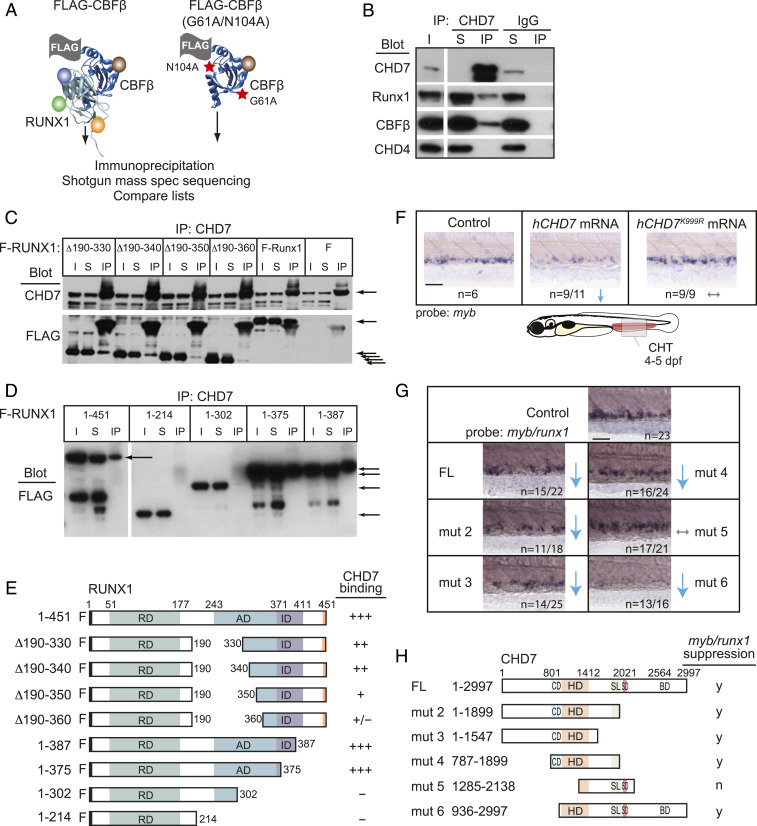
We independently identified CHD7 in an unbiased mass spectrometry screen for proteins that interact with RUNX1 and its non-DNA–binding partner CBFβ (Fig. 4A and SI Appendix, Table S1). We and others confirmed the interaction of endogenous CHD7 with RUNX1 and CBFβ by coimmunoprecipitation in a cell line that expresses high levels of all three proteins (20) (Fig. 4B). CHD7 binds the activation domain of RUNX1, as RUNX1 proteins with deletions impinging on the activation domain do not immunoprecipitate CHD7 (Fig. 4 C–E). Thus, the RUNX1 activation domain, which is essential for RUNX1 function (22), mediates the interaction with CHD7.
Fig. 4.
CHD7 interacts with Runx1 and restrains RUNX1 activity. (A) Scheme for identifying RUNX1-CBFβ–interacting proteins in a murine T-ALL cell line. FLAG-tagged CBFβ containing two amino acid substitutions (red stars) that decrease RUNX1 binding was used as a negative control. (B) CHD7 coimmunoprecipitates RUNX1-CBFβ but not CHD4 in murine T-ALL cells. I, input; S, depleted supernatant following immunoprecipitation; IP, immunoprecipitate. (C) Deletions impinging on the RUNX1 activation domain decrease the interaction between RUNX1 and CHD7. CHD7 was immunoprecipitated, and Western blots were probed with antibodies to CHD7 or FLAG. F-RUNX1, FLAG-RUNX1; ∆ refers to deleted amino acids illustrated schematically in panel E; F, vector expressing FLAG alone. Arrows indicate CHD7 (Top) or full-length and internally deleted RUNX1 proteins (Bottom). (D) C-terminal RUNX1 deletions. (E) Summary of RUNX1 mapping experiments. RD, DNA- and CBFβ-binding Runt domain; AD, transactivation domain; ID, inhibitory domain. (F) Expression of hCHD7 but not the catalytically dead mutant hCHD7K999R in zebrafish embryos reduces myb expression in the CHT by whole-mount in situ hybridization. Representative embryos are shown. Blue arrows indicate a decrease. Gray arrows indicate no change. (Scale bars, 50 μm.) Replicates: 2. (G) Mutation mapping of the hCHD7 domains shows that the ATPase/helicase domain is required to suppress myb and runx1 expression in the CHT. Same symbols as in F. (H) Summary of hCHD7 mapping experiments. FL, full length. CD, chromodomain. HD, ATPase/helicase domain. SL/SD/BD, SLIDE/SANT/BRK domains. y, yes; n, no. Quantification of results from F and G are in SI Appendix, Fig. S2B.
We functionally mapped domains in CHD7 responsible for its ability to constrain definitive hematopoiesis. Overexpression of human CHD7 (hCHD7) messenger RNA (mRNA) (23) suppresses HPCs in the CHT of zebrafish embryos (Fig. 4F). We injected five different hCHD7 truncation mutants (24) into zebrafish embryos to determine which mutant (mut) proteins could suppress myb/runx1 expression. Only hCHD7 mut 5 lacking the N-terminal portion of the ATPase/helicase domain failed to suppress myb/runx1 expression, whereas deletion of the chromodomains (mut 6) or the SLIDE/SANT/BRK domains (mut 2 to 4) had no effect (Fig. 4 G and H and SI Appendix, Fig. S2B). The catalytically dead mutant (23) hCHD7K999R also failed to suppress myb expression in the CHT (Fig. 4F and SI Appendix, Fig. S2B). The ability of CHD7 to suppress hematopoiesis requires its ATPase/helicase activity.
CHD7 Genetically Interacts with RUNX1 to Regulate Hematopoiesis.
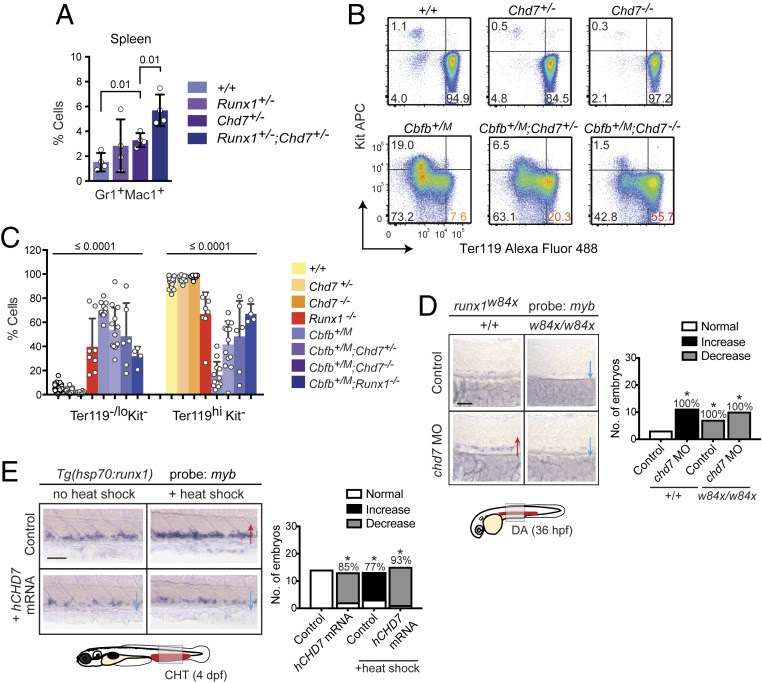
We tested if CHD7 and RUNX1 genetically interact. We observed an increased percentage of Gr1+Mac1+ cells in the spleen of Runx1+/−;Chd7+/− compared to Chd7+/− mice, suggesting that CHD7 represses adult myelopoiesis in collaboration with RUNX1 (Fig. 5A). We also identified a genetic interaction during embryonic hematopoiesis. In the absence of RUNX1, primitive erythropoiesis in the murine yolk sac is delayed, as evidenced by an increase in immature Ter119loKit− cells and a commensurate decrease in mature Ter119hiKit− cells (Fig. 5 B and C). This delay is more pronounced in embryos heterozygous for the neomorphic Cbfb-MYH11 allele (Cbfb+/M) (Fig. 5C). The dominant negative effect of the CbfbM allele requires RUNX1, as it is suppressed by RUNX1 deficiency (Cbfb+/M;Runx1−/−) (25) (Fig. 5B). Deletion of either one or both Chd7 alleles (Chd7+/− or Chd7−/−) did not alter the percentages of Ter119loKit− and Ter119hiKit− cells, but in Cbfb+/M embryos, loss of Chd7 partially restored primitive erythrocyte differentiation (Fig. 5 B and C). Since loss of either Runx1 or Chd7 can suppress the CbfbM allele, they both function in the same genetic pathway and are required for the neomorphic activity of the CBFB-MYH11 fusion gene. This is consistent with a previous study showing that loss of CHD7 delays leukemogenesis caused by CBFB-MYH11 (20).
Fig. 5.
CHD7 interacts genetically with RUNX1 to regulate hematopoiesis. (A) Chd7 and Runx1 interact genetically to repress myelopoiesis in the spleen of adult mice by flow cytometric analysis (n = 4). (B) Restoration of primitive erythrocyte maturation with Chd7 deletion in CbfbM embryos with peripheral blood analysis by flow cytometry (n = 6 to 42). Representative plots are shown. Simplified genotypes are the following: Chd7+/-: Chd7+/f;β-actin-Cre, Chd7-/-: Chd7f/f;β-actin-Cre, Cbfb+/M: Cbfb+/MYH11;β-actin-Cre. (C) Chd7 or Runx1 deletion partially restores normal maturation of primitive erythrocytes in E10.5 embryos expressing the dominant neomorphic Cbfb-MYH11 allele (CbfbM). All values were significantly different as compared to Cbfb+/M. ANOVA and Dunnett’s multiple comparison test. (D) Expansion of myb+ HSPCs in chd7 morphant (MO) embryos is suppressed in runx1w84x mutants expressing truncated Runx1. Whole-mount in situ hybridization of representative embryos is shown, with phenotypic results quantified in bar graph (Right). *P < 0.01 by χ2 test. Red arrows, increase. Blue arrows, decrease. (Scale bar, 50 μm.) Replicates: 2. (E) Overexpression of hCHD7 suppresses the expansion of myb+ HSPCs caused by heat-shock–induced runx1 overexpression. Same descriptions and symbols as in D.
We also examined the functional relationship between chd7 and runx1 during embryonic hematopoiesis in the zebrafish. Definitive hematopoiesis in the DA of zebrafish embryos is impaired by a runx1w84x mutation that truncates the Runx1 protein (26). Knockdown of chd7 in runx1w84x/w84x embryos did not restore myb expression in the DA, as runx1 is necessary for HSPC development, myb expression, and CHD7 activity (Fig. 5D). Therefore, we tested the effect of runx1 overexpression on CHD7 activity. Ectopic expression of hCHD7 mRNA decreases myb expression in the CHT, while overexpression of runx1 from an inducible heat shock promoter increases myb mRNA levels as expected (Fig. 5E). When hCHD7 mRNA was expressed in the context of heat-shock–induced runx1, the ability of runx1 to increase myb expression was suppressed (Fig. 5E). Altogether, the data indicate that chd7 functions to inhibit runx1 activity during embryonic hematopoiesis.
Discussion
We have uncovered a mechanism in hematopoietic differentiation in which the activity of the key transcription factor RUNX1 is regulated by the chromatin-remodeling enzyme CHD7. Expression of CHD7 is not blood specific; nonetheless, it functions to negatively regulate several stages of embryonic and adult hematopoiesis that are well conserved from zebrafish to mouse. Loss of CHD7 enhances HSPC formation and lineage differentiation, particularly of the myeloid lineage. In adult mice, CHD7-deficient LT-HSCs have a gene expression profile suggestive of a primed state that promotes multilineage differentiation, indicating that the enhanced myeloid cell contribution originates from the LT-HSC. Genomic analysis of CHD7 occupancy suggests that it can cooperate with multiple hematopoietic transcription factor genes. Here, we demonstrate that CHD7 genetically and physically interacts with RUNX1 through the RUNX1 transactivation domain and that RUNX1 recruits CHD7 to a subset of its target genes. The outcome of the interaction between RUNX1 and CHD7 depends on the developmental context. RUNX1 and CHD7 have opposing effects during HSPC formation from hemogenic endothelium, with RUNX1 promoting and CHD7 dampening the process. In the adult, however, both RUNX1 and CHD7 function to restrict myeloid lineage output from LT-HSCs (27, 28). This may have more to do with RUNX1’s opposing functions in determining myeloid progenitor numbers in the embryo versus the adult, as we show here that, in multiple contexts, in zebrafish embryos, mouse embryos, and adult mice, CHD7 appears to constrain the differentiation of myeloid lineage cells.
Distinct functions and mechanisms for CHD7 have been described in different cell types and developmental stages. In murine embryonic stem (ES) cells, CHD7 is associated with active chromatin and actively transcribed genes, yet its dominant activity is to suppress gene expression (29). In contrast, CHD7 primarily augments gene expression in preleukemic HSPCs expressing the CBFB-MYH11 protein (20). CHD7’s function in embryonic and adult hematopoiesis is similar to what has been observed in ES cells, i.e., that it primarily dampens the expression of actively transcribed genes.
Mutational analysis showed that the catalytic activity of the ATPase/helicase domain is required for CHD7 to suppress myb expression in zebrafish embryos. Other domains such as the chromodomains and SANT domain, which are required for CHD7’s ATPase/helicase activity in vitro and are mutated in CHARGE syndrome (24), were not required to repress embryonic hematopoiesis. A possible explanation for this paradox is overexpression of CHD7 lacking chromodomains or the SANT domain may bypass the stricter requirement for these domains in biochemical assays using purified proteins, or in vivo where CHD7 is present at physiological levels. ATP binding may be necessary for the ATPase/helicase domain to interact with other proteins. An example of this is the RNA helicase UAP56, which must bind ATP to interact with U2AF in spliceosome assembly (30). Five proteins detected in our CHD7 pulldown are known to be part of, or interact with, the Mi-2/NuRD repressive complex; thus loss of ATP binding by CHD7 could impair the recruitment of repressors at enhancers or gene bodies to regulate gene transcription, leading to increased expression of RUNX1 and other hematopoietic transcription factor targets. A third possibility is that there may be multiple ways by which CHD7 is recruited to chromatin and catalysis is activated; in developmental hematopoiesis, proteins such as RUNX1 may substitute for the chromodomains and SANT domain to recruit CHD7 to chromatin and stimulate its enzymatic activity. Future work elucidating the molecular mechanism of how CHD7 navigates the chromatin to regulate transcription will provide additional insights into how epigenetic regulators function with transcription factors to promote appropriate lineage differentiation.
Materials and Methods
Animal Models.
The zebrafish (Danio rerio) from the Tüebingen strain were bred and maintained according to institutional animal care and use committee guidelines at Boston Children’s Hospital. Morpholino sequences were the following: chd7 exon 3—ACTCGTTTATACTCTACACGTACCT and chd7 exon 4—TTACAAGCAAGTTTACCTGAAC ACC (Gene Tools). Chd7 morpholinos were resuspended in nuclease-free water, and equal amounts of each morpholino were combined (12 to 15 ng) for microinjection at the single-cell stage. Standard control morpholino from Gene Tools was used. Fish lines Tg(myb:EGFP), Tg(lmo2:dsRed), Tg(itga2b:EGFP), Tg(kdrl:RFP), Tg(kdrl:EGFP), Tg(hsp70:runx1;cmlc:dsRed), and runx1w84x/+ were previously described (26, 31–36).
Mouse 129S1/Svlmj and C57BL6/J strains were bred and maintained according to institutional animal care and use committee guidelines at the University of Pennsylvania. Generation of the murine Chd7-targeting vector and electroporation into C57BL/6J ES cells was performed by the InGenious Targeting Laboratory (details available upon request). Chimeric mice were mated to 129S1/SvImj × C57BL6/J F1 mice, and progeny were backcrossed to C57BL/6J mice for six generations. Primers for Chd7 genotyping were the following: JMH61—AAAATGTGGATCTCTCTCCAAACT; JMH65—TTATTTTCTTGAGACAAGGCCTCAC; and JMH66—GGTAACACAC TCCTTTAAACCCAGA. Vav1-Cre mice were provided by Thomas Graf, Centre for Genomic Regulation in Barcelona, Spain (37). Runx1+/− (Runx1tm1Spe), Cbfb+/M (Cbfbtm1lhc), Cdh5-Cre, and β-actin-Cre mice were described previously (28, 38–40). Both male and female animals were used for experiments.
Embryo Staining and Microscopy.
Whole-mount in situ hybridization on zebrafish embryos fixed in 4% paraformaldehyde was performed as described previously (41) with a minimum of two independent replicates performed for each staining. Ratios represent the number of embryos with the indicated phenotype/total number scored. Stained embryos were imaged using a Nikon stereoscope with a Nikon Coolpix 4500 camera or Zeiss camera. Embryos mounted in glycerol were imaged on a Nikon E600 compound microscope. Confocal imaging was performed on a Zeiss spinning disk confocal microscope using Volocity (PerkinElmer) or ZEN (Zeiss) software for image acquisition.
Whole-mount immunostaining of mouse embryos was performed as described (15). Primary antibodies used were rabbit anti-mouse CD117 (Thermo Fisher Scientific catalog #14–1171-82, RRID:AB_467433), rat anti-mouse CD31 (BD Biosciences catalog #550274, RRID:AB_393571), and rabbit anti-human/mouse RUNX1 (Abcam catalog #2593–1, RRID:AB_1580795). Secondary antibodies were purchased from Invitrogen: goat anti-rat Alexa Fluor 647 (Thermo Fisher Scientific catalog #A-21247, RRID:AB_141778), goat-anti rat Alexa Fluor 555 (Thermo Fisher Scientific catalog #A-21434, RRID:AB_2535855), and goat anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific catalog #A-11006, RRID:AB_2534074). Images were collected on a Zeiss LSM 710 confocal microscope equipped with 488-, 543-, and 633-nm wavelengths and a 20× immersion objective (Plan-Apochromat 25×/0.8 numerical aperture). Data were acquired using Zeiss ZEN 2011 and processed using FIJI software (42), LOCI Bio-Formats Importer (https://dev.loci.wisc.edu/fiji/) and the cell counter plugin (version 29, February 2008, Kurt De Vos, http://rsb.info.nih.gov/ij/plugins/cell-counter.html). To count hematopoietic clusters, 2- to 3-μm-thick Z-sections were collected.
Transplantation Assays.
Limiting dilution transplantations were performed using total bone marrow cells and sorted CD48−CD150+LinnegSca1+Kit+ cells from Chd7f/f or Chd7f/f;Vav1-Cre mice (C57BL/6J), along with 2 × 105 total bone marrow competitor cells (129S1/SvlmJ × B6.SJL-PtprcaPepCb/BoYJ F1) into 9-Gy lethally irradiated recipients (B6.SJL-Ptprca Pepcb/BoyJ [B6-LY5.2/Cr]). To purify CD48−CD150+LinnegSca1+Kit+ donor cells, bone marrow cells were lineage depleted using biotinylated antibodies to CD3, CD5, CD19, B220, Gr1, Mac1, and anti-biotin MACS beads on a MACS column (Miltenyi Biotec) and then stained with lineage antibodies (B220, Gr1, Mac1, CD3e, Ter119; eFlour450), Sca1 PerCP/Cy5.5, Kit APC-eFlour780, CD48 APC, and CD150 PE-Cy7 and sorted on a BD FACSAria.
For serial transplantation, 2 × 106 total BM donor cells were transplanted into lethally irradiated B6-LY5.2/Cr recipient mice without competitors as described above. At 4 mo, 2 × 106 BM cells from each primary recipient were transplanted into secondary B6-LY5.2/Cr recipients. The transplant was carried out until tertiary recipients were transplanted into fourth B6-LY5.2/Cr recipients.
Positive engraftment was scored in three different ways: 1) ≥1% of all cell lineages in PB were donor-derived at 4 mo post transplantation; 2) ≥1% Mac1+ cells in PB were donor-derived at 4 mo post transplantation; 3) ≥1% CD48−CD150+LSK cells in BM were donor-derived. ELDA (43) R statistical software was used to assess differences between paired sets of limiting dilution analyses to give the estimate of functional LT-HSC numbers.
To assess response to stress, 150 mg/kg FU (Sigma) was injected intravenously and HSCs were analyzed 7 d later by flow cytometry (27).
Progenitor Assays.
Myeloid methylcellulose assay using bone marrow and fetal cells was described previously (44). Embryonic lymphoid T and B progenitors assays using OP9 and OP9-DL1 stromal cells were performed as described (45, 46). L-Calc (Stem Cell Technologies) was used to calculate the progenitor frequencies.
RNA Isolation, Microarray Processing, and Analysis.
Mouse genome 1.0 arrays were used to perform microarray analysis of Chd7f/f and Chd7f/f;Vav1-Cre CD48−CD150+LSK cells. Following lineage depletion, CD48−CD150+LinnegSca1+Kit+ cells were sorted directly into TRIzol LS (Ambion, 10296028). The complementary DNAs (cDNAs) were generated using the Nugen WT-Ovation Pico system with Exon Module 3 (Nugen, 3300 and 2000–12) and were hybridized to Affymetrix Mouse 430v.2 chip (Affymetrix Mouse Gene 2.0ST). Intensity CEL files were normalized with the Robust Multichip Average algorithm. Expression value fold change cutoff was set at two-fold, and P value set as 0.05 for IPA using default parameters.
MouseGene 2.0 ST arrays were used to perform microarray analysis of LT-HSCs (CD48−CD150+CD34−LSK), ST-HSCs (CD48−CD150+CD34+LSK), and MPP (CD48+CD150−CD34+LSK) populations from 8-wk-old C57BL6J mice for Chd7 expression. Differentially expressed genes were assessed as those with at least a log-fold expression change of 1 and a false discovery rate (FDR) based adjusted P value of <0.1.
CHD7 deletion mutants cloned in pcDNA3.1 vector (24) were used to synthesize mRNA for microinjection in zebrafish embryos. Constructs were linearized with AvrII (NEB) and then purified by ethanol precipitation after phenol:chloroform extraction for mRNA synthesis using mMessage mMachine T7 Ultra kit (Ambion).
Flow Cytometry.
Flow cytometry was performed on BD LSRII or FACSAria, and data were analyzed with Flowjo (Tree Star). For analysis of Tg(kdrl:EGFP) embryos, embryos were manually dissociated in phosphate-buffered saline (PBS) containing Liberase (Sigma-Aldrich) and then washed and filtered in 0.9× PBS/2% fetal bovine serum.
Monoclonal antibodies used for analysis of adult and embryonic blood cells in mice were as described above (27) with the following additions/exceptions: CD19 (BD Biosciences catalog #561738, RRID:AB_10893995) and CD71 (BD Biosciences catalog #553266, RRID:AB_394743). Staining and flow cytometric analysis of the embryonic peripheral blood was performed as described previously (47, 48). For fetal liver HSPC analysis, lineage antibodies included CD3, CD5, CD19, B220, and Gr1, but not Mac1.
Cell-Cycle, Proliferation, and Apoptosis Analysis.
Bromodeoxyuridine assays were performed as described previously (27). Antibodies to Ki-67 (BioLegend catalog #652405, RRID:AB_2561929) and Annexin V (Thermo Fisher Scientific catalog #88–8005-72, RRID:AB_2575162) were used to analyze proliferation and apoptosis.
RT- and qPCR.
Pools of 20 to 50 embryos were homogenized in TRIzol, and RNA extracts were prepared according to the manufacturer’s protocol (Life Technologies). Genomic DNA was removed with TURBO DNA-free kit (Ambion), followed by cDNA synthesis using SuperScript III First Strand synthesis kit according to the manufacturer’s protocol (Life Technologies). Primers used for RT-PCR were the following: chd7 ex2 forward—5′‐GGGCACCTACTCACCAATCA‐3′ and chd7 ex4 reverse—5′- GCCTCTTTCTTGGTGCTGTT‐3′; chd7 ex3 forward—5′-TCCCAAGACACCCAAAGAAC-3′ and chd7 ex5 reverse—5′-GCCTCTTTCTTGGTGCTGTT-3′; and ef1α forward—5′-ATCTACAAATGC GGTGGAAT-3′ and ef1α reverse—5′-ATACCAGCCTCAAACTCACC-3′. qPCR was performed using SsoFast EvaGreen Supermix on a BioRad C1000 CFX-384 real-time PCR machine. qPCR primers used were the following: runx1 forward-1—5′-CCGACAGAAGCCGGATGA-3′ and runx1 reverse-1—5′- TGGCACTTCGCCTCAACTG-3′; runx1 forward-2—5′-CGTCTTCACAAACCCTCCTCAA-3′ and runx1 reverse-2—5′ GCTTTACTGCTTCATCCGGCT-3′. ΔCt values were normalized to ef1α.
Western Blot and Liquid Chromatography with Tandem Mass Spectrometry Protein Identification.
Nuclear extracts from murine T-ALL cell line 720 (49) were incubated with protein A Dynabeads coupled with anti-CHD7 (Abcam catalog #ab31824, RRID:AB_869129), RUNX1 (Millipore catalog #PC284, RRID:AB_2254229), CBFβ [β141.1, (50)], Flag (Sigma-Aldrich catalog #A2220, RRID:AB_10063035), or CHD4 (Abcam catalog #ab72418, RRID:AB_1268107) antibodies overnight at 4 °C or at room temperature for 1 h.
For liquid chromatography with tandem mass spectrometry (LC-MS/MS), FLAG-tagged CBFβ was expressed in and immunoprecipitated from extracts of the 720 T-ALL cell line (49). The CBFβ immunoprecipitates contain RUNX subunits and other RUNX-CBFβ–interacting proteins. As a control, a FLAG-tagged CBFβ containing two amino acid substitutions that decrease RUNX1 binding (51) was immunoprecipitated as a negative control to subtract out proteins that bound to FLAG-CBFβ in the absence of RUNX. Identification of protein–protein interactors was performed by excision of regions of sodium dodecyl sulfate/polyacrylamide gel electrophoresis-separated immunoprecipitates, in-gel digestion, and peptide shotgun sequencing as described (52). Briefly, gel regions were destained to clarity, dehydrated, digested with trypsin, extracted, and analyzed by LC-MS/MS on an LTQ Orbitrap mass spectrometer. The resulting MS/MS spectra were data searched using the SEQUEST algorithm (53) against a mouse proteome database (International Protein Index) and curated to a <1% FDR using the target-decoy strategy (54). Proteins were required to have a minimum of three peptides to be declared as a positive identification.
ChIP-seq.
ChIP-seq and DNase I hypersensitivity mapping in 416B cells were performed as described previously (19, 55). De novo motif analysis was performed using HOMER (56).
RNA-Sequencing.
Transcriptional profiles of 416B cells expressing single guide RNAs (sgRNAs) were analyzed using a protocol based on the Smart-Seq2 system (57) with the following modifications: pools of 75 cells expressing each sgRNA were sorted into 96-well plates containing lysis buffer, and cDNA was amplified with 13 PCR cycles. Each experimental condition was analyzed in quadruplicate. Illumina sequencing was perfomed on a HiSeq 4000 instrument. Reads were aligned against the mouse reference genome GRCm38/mm10 using GSNAP, and those overlapping exons (ENSEMBL m38.81) were counted using HTSeq (58). Differential gene expression was performed using the DESeq2 package (59).
Cell Lines.
To generate CBFB-MYH11–expressing cells, the mouse myeloid progenitor cell line 416B was cotransfected with the following: 1) a plasmid containing the tetracycline transcription silencer, the tetracycline transactivator, and blasticidin resistance under the control of a Ef1α promoter and 2) a plasmid containing the entire Cbfb-Myh11 type A cDNA in frame with an F2A element and mCherry protein under the control of a tetracycline-responsive element. As a control, cells were alternatively transfected with a plasmid lacking the Cbfb-Myh11 cDNA. Transposase PL623 (60), which was kindly donated by Pentao Liu, Sanger Institute, Cambridge, UK, was also transiently expressed to promote simultaneous stable integration of the constructs. Plasmids were transfected into 416B cells by electroporation using a BioRad electroporator (220 V, 900 μF). After 24 h, cells were selected in 1 μg/mL of blasticidin (InvivoGen). After 14 d, mCherry-negative single cells that did not stain with DAPI (Sigma) were sorted into 96-well plates using a BD Influx sorter and cultured for typically 2 wk. Clonal cultures were then tested for induction and expression levels of Cbfb-Myh11 using 1 μg/mL of doxycycline. Induction of Cbfb-Myh11 for ChIP experiments, performed in replicate, was confirmed by flow cytometry for mCherry expression on a BD Fortessa.
The 416B cells expressing Cas9 protein were obtained by transduction with pKLV2-EF1a-Cas9Bsd-W lentivirus (Addgene #68343) and selected with 10 μg/mL of blasticidin (InvivoGen). Three different Chd7 and one Runx1 sgRNAs were cloned into the Perturb-seq Guide Barcodes library backbone (Addgene #85968), and lentivirally transduced BFP+7AAD− cells were sorted after 4 d by flow cytometry for RNA-sequencing or genotyping. Efficiency of CRISPR editing was confirmed by high-throughput sequencing of genomic DNA from 2 × 105 cells. Targeting sgRNAs sequences were the following: Chd7 sgRNA1—AGACGCCAATCCGTTCCCG, Chd7 sgRNA2—TGGTACCTGAACGGCCCGG, Chd7 sgRNA3—GACATGCCCATAAACGAACG, and Runx1 sgRNA1—GCGCACTAGCTCGCCAGGG.
All cell lines tested negative for mycoplasma contamination.
Statistics.
Quantitative data are shown as mean ± SD with P values calculated using unpaired two-tailed Student’s t test or analysis of variance (ANOVA). No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgments
We thank A. D. Yzaguirre for technical assistance with confocal microscopy, C.-Y. Tsai for help with identifying Runx1-interacting proteins, T. Gao and M. Matthews for help with ES cell culture, J. Tobias for help with IPA microarray data analysis, and C. Lobry for help with heatmap visualization of microarray data. We also thank the I. Aifantis, H. Knaut, J. Torres-Vasquez, and R.M. White laboratories in New York for use of laboratory and fish facility space to complete this work. This work was supported by NIH Grants R01 HL089969, R01 HL091724, and U01HL100405 (to N.A.S.); R01 HL04880, PPG-P015PO1HL32262-32, 5P30 DK49216, 5R01 DK53298, 5U01 HL10001-05, and R24 DK092760 (to L.I.Z.); and R01 HG002668 (to R.A.Y.). R.K.H. and P.P.L. are supported by the Intramural Research Program of the National Human Genome Research Institute and NIH Grant 4R00CA148963 (to R.K.H.). N.K.W., V.M., F.J.C.-N., and B.G. were supported by Bloodwise Grant 12029, Leukemia and Lymphoma Society Grant 7001-12, Cancer Research UK Grant C1163/A12765, and core infrastructure support by the Medical Research Council (MRC) of the Wellcome Trust and MRC Cambridge Stem Cell Institute Grant 097922/Z/11/Z. J.H. was supported by NIH Grants 5-T32-HL-007439-34 and K12CA076931. H.-T.H. was supported by NYSTEM Training Grant C026880. L.I.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: L.I.Z. is founder and stockholder of Fate, Scholar Rock, and Camp4 therapeutics and a scientific advisor for Stemgent.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003228117/-/DCSupplemental.
Data Availability.
Expression profiling by array and genome binding/occupancy profiling by high-throughput sequencing data have been deposited in the Gene Expression Omnibus database under accession numbers GSE84136 and GSE83956. All study data are included in the article and supporting information.
Change History
September 16, 2021: The text of this article has been updated; please see accompanying Correction for details. The SI Appendix has been updated to coincide with a formal Correction.
References
- 1.Orkin S. H., Zon L. I., Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dzierzak E., Speck N. A., Of lineage and legacy: The development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang H. T.et al., A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat. Cell Biol. 15, 1516–1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims J. K., Wade P. A., SnapShot: Chromatin remodeling: CHD. Cell 144, 626–626.e621 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Layman W. S., Hurd E. A., Martin D. M., Chromodomain proteins in development: Lessons from CHARGE syndrome. Clin. Genet. 78, 11–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes S. A.et al., COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 39, D945–D950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tober J., Maijenburg M. W., Speck N. A., Taking the leap: Runx1 in the formation of blood from endothelium. Curr. Top. Dev. Biol. 118, 113–162 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bertrand J. Y.et al., Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kissa K., Herbomel P., Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Boisset J. C.et al., In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Eliades A.et al., The hemogenic competence of endothelial progenitors is restricted by Runx1 silencing during embryonic development. Cell Rep. 15, 2185–2199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yzaguirre A. D., Howell E. D., Li Y., Liu Z., Speck N. A., Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis. Development 145, dev158162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Link K. A., Chou F. S., Mulloy J. C., Core binding factor at the crossroads: Determining the fate of the HSC. J. Cell. Physiol. 222, 50–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z. Z.et al., Chd7 is critical for early T-cell development and thymus organogenesis in zebrafish. Am. J. Pathol. 188, 1043–1058 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Yokomizo T.et al., Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 7, 421–431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurd E. A.et al., Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome 18, 94–104 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Oguro H., Ding L., Morrison S. J., SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 13, 102–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dexter T. M., Allen T. D., Scott D., Teich N. M., Isolation and characterisation of a bipotential haematopoietic cell line. Nature 277, 471–474 (1979). [DOI] [PubMed] [Google Scholar]
- 19.Schütte J.et al., An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife 5, e11469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhen T.et al., Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood 130, 2431–2442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shigesada K., van de Sluis B., Liu P. P., Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene 23, 4297–4307 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Okuda T.et al., Biological characteristics of the leukemia-associated transcriptional factor AML1 disclosed by hematopoietic rescue of AML1-deficient embryonic stem cells by using a knock-in strategy. Mol. Cell. Biol. 20, 319–328 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajpai R.et al., CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouazoune K., Kingston R. E., Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. U.S.A. 109, 19238–19243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyde R. K., Zhao L., Alemu L., Liu P. P., Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia 29, 1771–1778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sood R.et al., Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood 115, 2806–2809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai X.et al., Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS One 6, e28430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q.et al., Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 93, 3444–3449 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnetz M. P.et al., CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen H.et al., Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 22, 1796–1803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.North T. E.et al., Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H.et al., Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev. Biol. 281, 256–269 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Traver D.et al., Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Huang H., Zhang B., Hartenstein P. A., Chen J. N., Lin S., NXT2 is required for embryonic heart development in zebrafish. BMC Dev. Biol. 5, 7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi J.et al., FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev. Biol. 304, 735–744 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing L.et al., Adenosine signaling promotes hematopoietic stem and progenitor cell emergence. J. Exp. Med. 212, 649–663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadtfeld M., Graf T., Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development 132, 203–213 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Landrette S. F.et al., Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood 105, 2900–2907 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A., Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewandoski M., Meyers E. N., Martin G. R., Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 62, 159–168 (1997). [PubMed] [Google Scholar]
- 41.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Schindelin J.et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Y., Smyth G. K., ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Tober J., Yzaguirre A. D., Piwarzyk E., Speck N. A., Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765–3776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y.et al., Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597–2612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohtashami M., Zarin P., Zúñiga-Pflücker J. C., Induction of T cell development in vitro by delta-like (Dll)-expressing stromal cells. Methods Mol. Biol. 1323, 159–167 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Hyde R. K.et al., Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood 115, 1433–1443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamikubo Y.et al., The C-terminus of CBFβ-SMMHC is required to induce embryonic hematopoietic defects and leukemogenesis. Blood 121, 638–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Neil J.et al., Activating Notch1 mutations in mouse models of T-ALL. Blood 107, 781–785 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, et al., The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87 (4), 697–708, 10.1016/s0092-8674(00)81389-6 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Tang Y.-Y.et al., Energetic and functional contribution of residues in the core binding factor beta (CBFbeta ) subunit to heterodimerization with CBFalpha. J. Biol. Chem. 275, 39579–39588 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Kettenbach A. N.et al., Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faherty B. K., Gerber S. A., MacroSEQUEST: Efficient candidate-centric searching and high-resolution correlation analysis for large-scale proteomics data sets. Anal. Chem. 82, 6821–6829 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elias J. E., Gygi S. P., Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson N. K.et al., Integrated genome-scale analysis of the transcriptional regulatory landscape in a blood stem/progenitor cell model. Blood 127, e12–e23 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Heinz S.et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picelli S.et al., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Anders S., Pyl P. T., Huber W., HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W.et al., Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. U.S.A. 108, 18283–18288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression profiling by array and genome binding/occupancy profiling by high-throughput sequencing data have been deposited in the Gene Expression Omnibus database under accession numbers GSE84136 and GSE83956. All study data are included in the article and supporting information.