Significance
Detecting and avoiding noxious stimuli is critical for survival. Nociceptive sensory neurons detect these stimuli and activate neural circuits that elicit stereotyped escape responses. However, individual animals may have genetic predispositions to respond differently to the same stimuli. Furthermore, it is not well understood how experiences during development affect these escape responses. Here, we describe a role for the foraging gene in nociception and show that differences in nociceptive sensitivity may arise from genetic variation of this gene. We also demonstrate that altering synaptic activity in a nociceptive circuit during development changes the sensitivity of the escape response elicited. Our findings suggest that both genetic variation and experiences in development can shape escape responses.
Keywords: nociception, optogenetics, Drosophila, plasticity, genetic variation
Abstract
Painful or threatening experiences trigger escape responses that are guided by nociceptive neuronal circuitry. Although some components of this circuitry are known and conserved across animals, how this circuitry is regulated at the genetic and developmental levels is mostly unknown. To escape noxious stimuli, such as parasitoid wasp attacks, Drosophila melanogaster larvae generate a curling and rolling response. Rover and sitter allelic variants of the Drosophila foraging (for) gene differ in parasitoid wasp susceptibility, suggesting a link between for and nociception. By optogenetically activating cells associated with each of for’s promoters (pr1–pr4), we show that pr1 cells regulate larval escape behavior. In accordance with rover and sitter differences in parasitoid wasp susceptibility, we found that rovers have higher pr1 expression and increased sensitivity to nociception relative to sitters. The for null mutants display impaired responses to thermal nociception, which are rescued by restoring for expression in pr1 cells. Conversely, knockdown of for in pr1 cells phenocopies the for null mutant. To gain insight into the circuitry underlying this response, we used an intersectional approach and activity-dependent GFP reconstitution across synaptic partners (GRASP) to show that pr1 cells in the ventral nerve cord (VNC) are required for the nociceptive response, and that multidendritic sensory nociceptive neurons synapse onto pr1 neurons in the VNC. Finally, we show that activation of the pr1 circuit during development suppresses the escape response. Our data demonstrate a role of for in larval nociceptive behavior. This function is specific to for pr1 neurons in the VNC, guiding a developmentally plastic escape response circuit.
Efficient and rapid escape behavior in reaction to threatening sensory stimuli is vital for defense and, ultimately, survival (1–5). Despite their importance, little is known about how noxious stimuli bring about stereotyped escape behaviors, as well as what factors contribute to variation in the latency to perform these escape responses.
Drosophila melanogaster larvae must forage for food while avoiding noxious stimuli and predators, such as parasitoid wasps (6, 7). Drosophila larvae use class IV (cIV) multidendritic sensory neurons on their cuticle to detect noxious stimuli or an attack by a parasitoid wasp and then escape by eliciting a sequential body bending and corkscrew-like rolling response (8, 9). This escape behavior is also seen in response to mechanical or thermal nociception (10–16). Silencing cIV multidendritic sensory neurons with tetanus toxin eliminates this nociceptive-like escape response (8), whereas optogenetic activation of these neurons induces the curling and rolling escape response (8, 17, 18). These sensory neurons also display experience-dependent plasticity, as prolonged optogenetic activation during development sensitizes these neurons, resulting in larvae that are less likely to roll in response to subsequent optogenetic stimulation (19).
The Drosophila foraging (for) gene encodes a guanosine 3′,5′-cyclic monophosphate–dependent protein kinase (PKG) (20) and affects numerous behaviors (20–25), several types of plasticity (26–29), and synaptic function (30–32). Allelic variants of for (rover and sitter) differ in for messenger RNA (mRNA) levels, foraging behavior (20–22), and their susceptibility to predation by parasitoid wasps (33–35). However, it is not known if for is involved in the nociceptive-like escape response. PKG has been shown to regulate nociception in several organisms (36–38). Here, we use an optogenetic screen to show that activation of a subset of for neurons induces a nociceptive-like escape response. In addition, we show that FOR is required for this nociceptive-like escape response and that this response is influenced by both genetic variation of for and activation of these for neurons during development.
Results
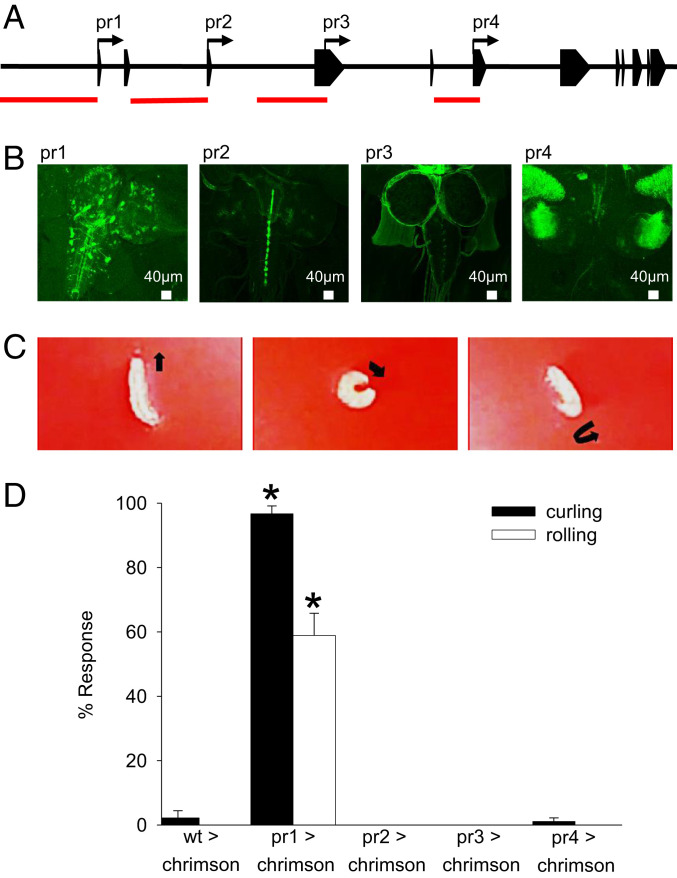
Activation of for pr1 Cells Induces a Curling and Rolling Response.
The for gene has a complex structure that contains four promoters (pr1–pr4) (22). GAL4 lines were previously generated for each of these promoters, and these pr-GAL4s drive expression of GFP in distinct patterns in the larval central nervous system (CNS) (25). We confirmed that pr1-GAL4 and pr4-GAL4 express primarily in neurons in the larval CNS, whereas pr2-GAL4 and pr3-GAL4 express in glia (Fig. 1 A and B). To better understand the functional significance of this expression pattern, we took an optogenetic approach and used the GAL4/UAS system (39) to drive the expression of a red-light–activated channelrhodopsin, chrimson (40), in subsets of cells that are associated with each of the four for promoters (pr1–pr4). Larvae were stimulated for 5 s with red light (617 nm). Activation of pr1 cells induced a curling and rolling behavior (Movie S2) that was not seen in control larvae (Movie S1) or when pr2, pr3, or pr4 cells were activated (Fig. 1 C and D). Thus, activation of pr1 cells results in a curling and rolling behavior.
Fig. 1.
Optogenetic activation of for pr1 neurons induces a curling and rolling response. (A) Schematic of the for gene. The four transcription start sites (pr1, pr2, pr3, and pr4) are indicated with arrows, exons are indicated with boxes, and introns are shown as black lines. Regions used to make pr-GAL4s are indicated in red. (B) Representative images (n = 6 per genotype) of fixed pr1-GAL4 > UAS-mCD8GFP, pr2-GAL4 > UAS-mCD8GFP, pr3-GAL4 > UAS-mCD8GFP, and pr4-GAL4 > UAS-mCD8GFP larval CNSs stained with anti-GFP. (C) pr1-GAL4 > UAS-chrimson larvae curl and roll in a corkscrew-like motion in response to red light. (D) pr1-GAL4 > UAS-chrimson larvae exhibited significantly more curling and rolling than UAS-chrimson/+ larvae when stimulated with red light [one-way ANOVA: curling F(4,145) = 754.7, P < 0.0001; rolling F(4,145) = 72.67, P < 0.0001; n = 30 per genotype]. No significant effect was seen when pr2-GAL4, pr3-GAL4, or pr4-GAL4 was used to drive UAS-chrimson P > 0.05; n = 30 per genotype]. Error bars indicate SEM. wt, wild type.
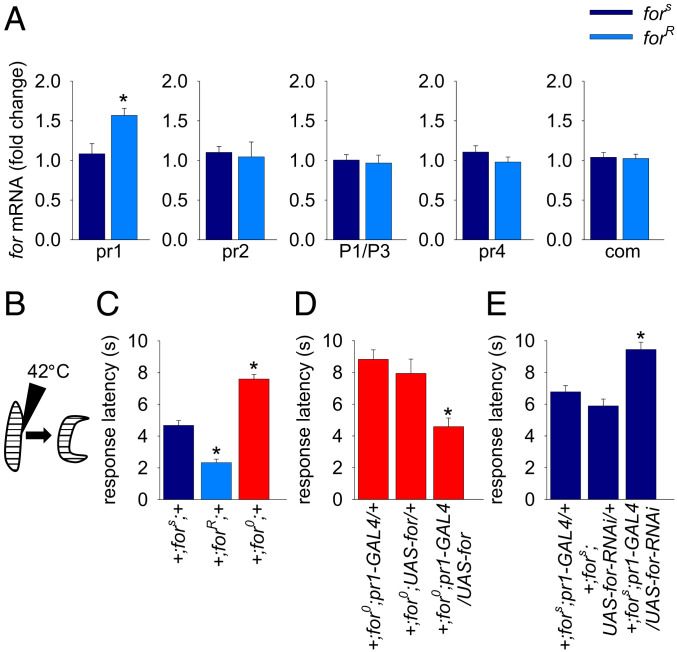
for in pr1 Cells Regulates Thermal Nociception.
The curling and rolling behavior is reminiscent of an escape response that larvae display in response to both a parasitoid wasp attack (8) and thermal nociception (10). To determine if for functions in thermal nociception (Fig. 2B), we examined two allelic variants of for (rover and sitter) that differ in their susceptibility to predation by parasitoid wasps (33–35). We found that the response latency (the time in seconds to the onset of curling and rolling) was significantly shorter for rovers (forR) than for sitters (fors; Fig. 2C). The pr1-GAL4 primarily expresses in the CNS of larvae (25). Consequently, we hypothesized that for pr1 mRNA levels of rover and sitter larvae would differ in dissected larval CNSs. We tested this by performing quantitative RT-PCR (qRT-PCR) to characterize the levels of for mRNA associated with each of for’s four promoters in dissected larval CNSs. We found rovers had significantly higher levels of for pr1 mRNA in comparison to sitters (Fig. 2A). Correspondingly, we detected no differences in the other for pr transcripts between rovers and sitters in the larval CNS (Fig. 2A). We concluded that rovers are more sensitive (shorter response latencies) to thermal nociception and have higher levels of for pr1 mRNA in their larval CNSs than sitters do.
Fig. 2.
for in pr1 neurons regulates thermal nociception. (A) Rovers (+;forR;+) have significantly higher pr1 transcript levels than sitters (+;fors;+) in dissected larval CNSs [unpaired t test: t(10) = 3.12, P = 0.011; n = 6]. No significant differences were found in the levels of common coding, pr2, P1/3, or pr4 transcripts between rovers and sitters (P > 0.05; n = 6). (B) Schematic of thermal nociception assay. Larvae curl and roll in a corkscrew-like motion when touched with a probe heated to 42 °C. (C) Rovers (+;forR;+) displayed a significantly shorter response latency (P < 0.001) in comparison to sitters (+;fors;+). The response latency of for null mutants (+;for0;+) was significantly increased (P < 0.001) in comparison to rovers and sitters [n = 126 for sitters, n = 58 for rovers, and n = 123 for for null mutants; Kruskal–Wallis one-way ANOVA: H(2) = 112.46, P < 0.001]. (D) Increased response latency seen in for null mutants was rescued by expressing for in pr1 neurons (+;for0;pr1-GAL4/UAS-for-flag-4c) [n = 34 for +;for0;pr1-GAL4/+, n = 13 for +;for0; UAS-for-flag-4c/+, n = 18 for +;for0;pr1-GAL4/UAS-for-flag-4c; one-way ANOVA F(2,62) = 10.76, P < 0.001]. (E) Knocking down for in pr1 neurons (+;UAS-Dcr, fors/fors;pr1-GAL4/UAS-for-RNAi) significantly increases response latency in comparison to controls (+;fors;pr1-GAL4/+ and +;UAS-Dcr, fors/fors;UAS-for-RNAi/+) [n = 78 for +;fors;pr1-GAL4/+, n = 73 for +;UAS-Dcr, fors/fors;/UAS-for-RNAi/+, and n = 114 for +;UAS-Dcr, fors/fors;pr1-GAL4/UAS-for-RNAi; Kruskal–Wallis one-way ANOVA: H(2) = 28.74, P < 0.001]. Error bars indicate SEM.
We further studied the role of for in thermal nociception, using genetic and transgenic manipulations of for. We found that for0 null mutants (22) had significantly longer response latencies than both rovers and sitters (Fig. 2C). To determine if the absence of for in pr1 cells was responsible for the longer latency response seen in for0 null mutants, we used the GAL4/UAS system (39) to drive expression of for in pr1 cells in a for0 null mutant background. We found that expression of for in pr1 cells rescued the longer latency response seen in for0 null mutants (Fig. 2D). Next, we used a for RNA interference (RNAi) line to knock down for transcripts (described and validated in SI Appendix, Fig. S1). Knocking down for with RNAi in pr1 cells phenocopied the longer response latencies seen in for0 null mutants (Fig. 2E). Overall, these data demonstrate that for in pr1 cells is required for the curling and rolling response induced by thermal nociception.
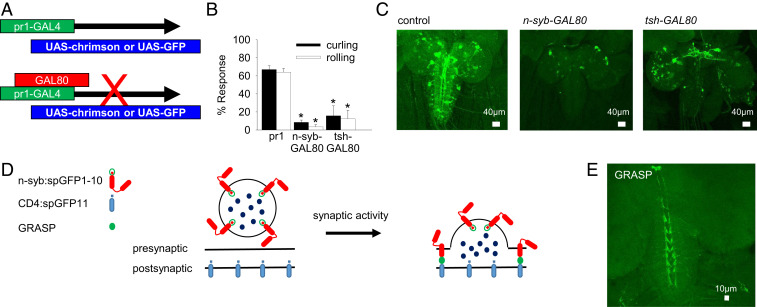
pr1 Neurons in the Ventral Nerve Cord Are Required for the Curling and Rolling Response.
The cIV multidendritic neurons are required for generating the nociceptive-like escape response (8). Therefore, we next determined if pr1-GAL4 expresses in cIV multidendritic neurons by comparing its expression with that of ppk-eGFP (41), a protein known to be expressed in cIV multidendritic neurons. We found that pr1-GAL4 does not express in cIV multidendritic neurons, but appears to express in tracheal multidendritic neurons on the larval body wall (SI Appendix, Fig. S2).
pr1-GAL4 also expresses outside the nervous system in a few enteroendocrine cells in the anterior of the midgut (25). To further refine the expression patterns of GAL4s, we used the GAL4 repressor, GAL80 (42, 43) (Fig. 3A). To determine if pr1 neurons were required for the curling and rolling response, we combined the pan-neuronal GAL4 inhibitor nsyb-GAL80 (44) with pr1-GAL4 and tested whether activation of nonneuronal pr1 cells, such as enteroendocrine cells, could induce the response. Loss of most of the neuronal expression (Fig. 3C) blocked the curling and rolling response (Fig. 3B). Thus, pr1 neurons specifically are required for the curling and rolling response.
Fig. 3.
pr1 neurons in the VNC are required for the curling and rolling response. (A) Schematic of GAL80 screen. (B) pr1-GAL4 > UAS-chrimson with n-syb–GAL80 (n = 68) and tsh-GAL80 larvae (n = 70) showed significantly less curling and rolling behavior than control pr1-GAL4 > UAS-chrimson larvae (n = 124) when stimulated with red light (*P < 0.0001) [one-way ANOVA tests: curling F(2,313) = 100.4, P < 0.0001; rolling F(2,313) = 22.05, P < 0.0001]. (C) Representative images of fixed pr1-GAL4 > UAS-mCD8GFP with n-syb–GAL80 or tsh-GAL80 larval CNSs stained with anti-GFP. (D) Schematic of activity-dependent GRASP. (E) Representative image of fixed +;lexAop-nsyb-spGFP1–10, UAS-CD4-spGFP11/+;/ppk-lexA/pr1-GAL4 larval CNS stained with anti-GFP. Preparations were stimulated with high K+ for 20 min before fixation. Error bars indicate SEM.
The cIV multidendritic neurons are known to synapse onto the ventral region of the ventral nerve cord (VNC) (19, 45–47). We next tested whether pr1 VNC neurons were required for the curling and rolling response by genetically separating pr1-GAL4 brain lobe expression from VNC expression by combining the VNC-specific GAL4 inhibitor tsh-GAL80 (48) with pr1-GAL4. Loss of expression in the VNC (Fig. 3C) blocked the curling and rolling response (Fig. 3B). This demonstrates that pr1 neurons in the VNC are required for the curling and rolling response.
To determine if cIV multidendritic neuron synapse onto pr1 neurons in the VNC, we used activity-dependent GFP reconstitution across synaptic partners (GRASP) (49). This involves the reconstitution of two fragments of a split GFP (spGFP1–10 and spGFP11) across active synapses. The spGFP1–10 is fused to the C terminus of n-synaptobrevin (n-syb) and only binds to its GRASP partner (CD4::spGFP11) when it is exposed to the synaptic cleft following synaptic vesicle (SV) fusion (49). We used ppk-lexA to drive the expression of lexAop-nsyb-spGFP1–10 in cIV multidendritic neurons and pr1-GAL4 to drive the expression of UAS-CD4-spGFP11 in pr1 neurons (Fig. 3D). We used high K+ saline to induce SV fusion and observed a specific GFP signal along the axon tracks of cIV multidendritic neurons (Fig. 3E), demonstrating that cIV multidendritic neurons do, in fact, synapse onto pr1 neurons in the VNC. Collectively, these experiments allow us to conclude that cIV multidendritic neurons, involved in the curling and rolling response, synapse onto pr1 neurons in the larval VNC.
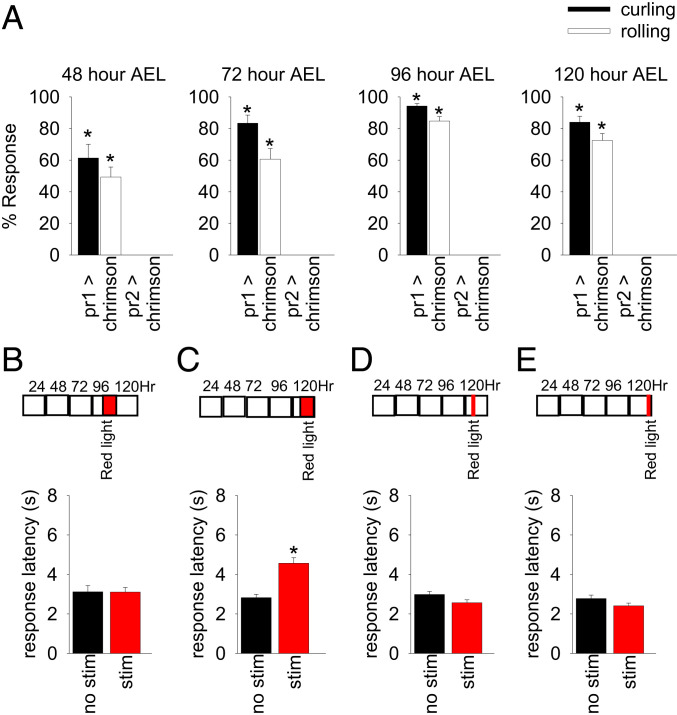
pr1 Neurons Are Part of a Developmentally Plastic Sensory Circuit.
Next, we tested if optogenetic activation of pr1 cells at different larval stages of development could induce a curling and rolling response. We found that optogenetic activation of pr1 neurons, but not pr2 cells, elicited a curling and rolling response in first instar [48 h after egg laying (AEL)], second instar (72 h AEL), early third instar (96 h AEL) and mid-third instar (120 h AEL) larvae (Fig. 4A). This demonstrates that the sensory circuit for the curling and rolling response is functional at all three larval stages of development.
Fig. 4.
Optogenetic activation of pr1 neurons during development reduces the curling and rolling response of larvae in response to thermal nociception. (A) pr1-GAL4 > UAS-chrimson larvae curl and roll in response to red light at different stages of development (n = 50 for 48 h AEL, n = 22 for 72 h AEL, n = 98 for 96 h AEL, n = 75 for 120 h AEL) significantly more than control pr2-GAL4 > UAS-chrimson larvae at 48 h after AEL [Mann–Whitney rank sum test: U = 217, P < 0.001 for curling and U = 294.5, P < 0.001 for rolling; n = 31, 50], 72 h AEL [Mann–Whitney rank sum test: U = 0, P < 0.001 for curling and U = 23, P < 0.001 for rolling; n = 22, 23], 96 h AEL [Mann–Whitney rank sum test: U = 0, P < 0.001 for curling and U = 70, P < 0.001 for rolling; n = 28, 98], and 120 h AEL [Mann–Whitney rank sum test: U = 115.5, P < 0.001 for curling and U = 198, P < 0.001 for rolling; n = 33, 75]. (B–E) pr1-GAL4 > UAS-chrimson larvae were exposed to red light at different times during development and then tested for thermal nociception at 120 h AEL. Control larvae were reared in the same conditions but were not exposed to red light. (B) Activation of pr1 neurons (from 79 to 95 h AEL) did not significantly affect response latency in comparison to controls [unpaired t test: t(223) = 0.0325, P = 0.974; n = 84–141]. (C) Activation of pr1 neurons (103–119 h AEL) significantly increased response latency in comparison to controls [unpaired t test: t(300) = 4.961, P < 0.0001; n = 134–168]. Activation of pr1 for 1 h [D; at 103 h AEL, unpaired t test: t(502) = 1.960 P = 0.051; n = 242–262] or [E; at 118 h AEL, unpaired t test: t(472) = 1.647, P = 0.100; n = 229–245] did not significantly affect response latency in comparison to controls. Error bars indicate SEM. stim, stimulation.
Optogenetic stimulation of cIV multidendritic neurons during development sensitizes cIV multidendritic neurons and makes larvae less likely to curl and roll in response to subsequent optogenetic stimulation (19). Given that pr1 neurons are downstream of cIV multidendritic neurons, we hypothesized that activation of pr1 neurons during development would also suppress the nociceptive-like escape response. We expressed chrimson in pr1 cells and activated it in developing larvae with brief pulses of red light (5 s of illumination followed by 5 min of no illumination). We then used the thermal nociception assay to determine if the nociceptive-like escape response was affected by activation of pr1 cells during third instar larval development. Activation of pr1 cells at 103–119 h AEL (5 s of illumination every 5 min) increased the response latency of larvae (Fig. 4C), whereas activation of pr1 cells at 79–95 h AEL (5 s of illumination every 5 min) had no effect on the response latency of larvae (Fig. 4B). More acute stimulation for 1 h (5 s of illumination every 5 min) at 103 h AEL or 118 h AEL had no effect on response latency (Fig. 4 D and E). Taken together, these results show that activation of pr1 cells during a specific time during larval development (103–119 h AEL) suppressed the nociceptive-like escape response of Drosophila larvae.
Discussion
We establish that FOR is required for a nociceptive-like escape response by Drosophila larvae and that this response is influenced by genetic variation of for. Furthermore, we show that pr1 neurons in the VNC are required for this curling and rolling response and that these pr1 neurons are part of a plastic sensory circuit.
Our data demonstrate that activation of for pr1 cells induces a curling and rolling response and that reduction of FOR in these cells reduces the likelihood that larvae will curl and roll in response to thermal nociception. Together, these data suggest that FOR is required for activation of pr1 cells to induce the nociceptive-like escape response. Presynaptic Drosophila FOR and mammalian PKG maintain sustained neurotransmitter release by facilitating SV endocytosis (32, 50, 51). The inability of for0 null mutants to maintain sustained neurotransmitter release in pr1 neurons, which are downstream of cIV multidendritic sensory neurons, may reduce the likelihood of larvae curling and rolling in response to thermal nociception. Mammalian PKG has also been shown to have a presynaptic function in nociception (37) and is required for activity-dependent nociceptive hypersensitivity in mice (37) and Aplysia (36).
Rovers have a shorter latency to perform the nociceptive-like escape response and have higher levels of for pr1 transcript in the larval CNS than sitters. Several single-nucleotide polymorphisms (SNPs) between rovers and sitters are found in the DNA sequence of pr1, and several putative transcription factor binding sites have been identified in this region (21, 52). A SNP in pr4 was previously found to correlate with differences in foraging behaviors of rover and sitter adult flies (21). We speculate that the SNPs in the DNA sequence of pr1 are responsible for the higher levels of for pr1 transcripts in rovers, which is likely the cause of the shorter latency time seen in rovers in response to thermal nociception in comparison to sitters.
Interestingly, for expression in pr1 neurons was recently shown to be required for larval foraging behavior (25). It seems likely that the higher levels of for pr1 transcript that we find in rovers may also be the cause of the increased foraging trail lengths seen in rovers in comparison to sitters. This increased foraging behavior makes rovers more susceptible to parasitization by parasitoid wasps, such as Ganaspis xanthopoda and Asobara tabida (33–35), that search for larvae by sensing their vibrations. We found that rovers have a shorter latency to perform the nociceptive-like escape response than sitters. A shorter latency may benefit larvae that are more easily detected by parasitoid wasps because they move more and cover a larger area while foraging (33). It may be advantageous for the same neurons or neurons in the same circuit to regulate the nociceptive-like escape response and foraging behavior; however, it is not known if this is the case. Alternatively, different subsets of pr1 neurons in different circuits may regulate these behaviors.
Reconstructed electron microscopy data have identified 13 second-order neurons in the VNC that are targets of cIV multidendritic neurons in Drosophila larvae (18). Of these, several have been shown to trigger the nociceptive-like escape response (18, 46, 47, 53) and one activates a pair of command-like high-order neurons (18). It is not known whether all 13 second-order neurons can trigger the nociceptive-like escape response. Using activity-dependent GRASP, we show that cIV multidendritic neurons synapse onto pr1 neurons in the VNC, which can also trigger the nociceptive-like escape response. The dilp7 second-order neurons have previously been identified to be part of the nociceptive-like escape response circuit (53), and they do not colocalize with pr1 neurons (SI Appendix, Fig. S3). Future studies will address whether the pr1 neurons identified in the present study correspond to the second-order neurons known to trigger the nociceptive-like escape response.
Optogenetic activation of cIV multidendritic neurons during development sensitizes these sensory neurons, resulting in larvae being less likely to curl and roll in response to subsequent optogenetic stimulation (19). We show that cIV multidendritic neurons synapse onto pr1 neurons and that activation of pr1 neurons reduces the likelihood of larvae curling and rolling in response to a thermal probe. This may allow larvae to adapt to continuous noxious stimuli and suppress the nociceptive-like escape response to perform other important behaviors, such as foraging for food.
There are multiple levels of biological organization underlying animal behavior. Drosophila nociception has primarily been studied from a neuronal circuitry perspective (18, 46, 47, 53), but less is known about the genes and factors that influence the functioning of this circuitry. Here, we describe a role for the for gene in modulating the function of a nociceptive circuit. Furthermore, our data suggest that differences in nociceptive sensitivity might arise from genetic variation at the for locus. We show that for regulates nociception and that for pr1 neurons are part of a nociceptive circuit that can be influenced by synaptic activity during development.
Methods
Fly Stocks.
Fly stocks were grown in uncrowded conditions at 25 °C on molasses-based fly medium (54). This fly medium was supplemented with all-trans retinal for optogenetic experiments. Mid-third instar larvae were used for all experiments unless otherwise indicated. Rover (forR) and sitter (fors) strains were previously described (22). The GAL4/UAS system (39) and the lexA/lexAop system (55) were used for tissue-specific expression of transgenes. pr1-GAL4, pr2-GAL4, pr3-GAL4, and pr4-GAL4 (25) were used to drive the expression of UAS-mCD8-GFP or other transgenes in subsets of for cells. The for0 null mutant contains a 35-kb deletion that removes the entire for locus (22). UAS-chrimson was used to express a red-light–activated channelrhodopsin, chrimson (40). UAS-for-flag-4c was used to express FOR with a flag-4c tag on the C terminus in selected tissues or cells (32). FOR-flag-4c was previously shown to be a functional FOR protein that recapitulates FOR function (32). UAS-Dcr, UAS-forRNAi was used to knock down for in selected cells. Knockdown of for was performed in a fors background to get as strong a knockdown as possible, since fors has lower levels of for than forR (Fig. 2A). da-GAL4 was used as a ubiquitous driver (56). ppk-eGFP was used to visualize cIV multidendritic neurons (41). The n-syb–GAL80 was used to repress GAL4 activity in the nervous system (44). tsh-GAL80 was used to inhibit GAL4 activity in the VNC (48). ppk-lexa was used as a cIV multidendritic neuron driver (57). lexAop-nsyb-spGFP1–10 and UAS-CD4-spGFP11 were used for GRASP experiments (49).
Cloning.
A RNAi line targeting all for transcripts was designed for the exon 7 and 8 exon junction. A 376-bp region spanning exons 7 and 8 was amplified from complementary DNA (cDNA) with the following primers: RNAi-forexon7:8-forward (F): AATAGCTAGCATGGACGCGTGGAGGTTTCCC and RNAi-forexon7:8-reverse (R): AATAGCTAGCATCGTCACTCGCACTTTTCC. The primers included a NheI restriction site (underscored in primer sequences), which was used to clone the 5′-3′ fragment into the NheI site of the pWIZ RNAi cloning vector (58) and the 3′-5′ fragment into the AvrII site of pWIZ. P element injections into w1118, performed by BestGene, Inc., resulted in insertion of the transgene on the third chromosome.
qRT-PCR.
Total RNA was extracted using the Arcturus PicoPure RNA Isolation Kit (catalog no. KIT0204; Applied Biosystems) and the RNase-free DNase set (catalog no. 79254; Qiagen), with the following modifications from the manufacturer’s protocol: CNSs of 74 ± 2-h-old larvae (posthatch) were dissected in Schneider’s Insect Medium (catalog no. S0146; Sigma–Aldrich) and immediately placed in 100 μL of RNA extraction buffer and incubated at 42 °C for 30 min. The manufacturer’s protocol for CapSure Macro LCM Caps was then followed starting at step 2, including the optional DNase treatment step. RNA was extracted from three to six biological replicates with n = 4 brains per replicate.
Following extraction, RNA was quantified using a Nanodrop 2000c (Thermo Scientific) and RNA integrity was accessed by gel electrophoresis. cDNA was synthesized with the iScrip Advanced cDNA Synthesis Kit for qRT-PCR (catalog no. 1725037; Bio-Rad), using 250 μg of transfer RNA per sample. qRT-PCR was performed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad), using SsoAdvanced Universal SYBR Green Supermix and the following primers: for common-F CTCCATTCACGGGCTCGGAT, for common-R ATCTCGCTGATTCCCCCACG, for pr1-F TTTGTCGCGATCGTGCTTGG, for pr1-R ACCACCGAACTAAATGGCGATG, for pr2-F GGATGTCGCTAGCACGCAAAC, for pr2-R TAACACACGCACAAGCACACC, for pr1/3-F TCTGGTGGGTGGCATTGTGA, for pr1/3-R TGTGGGTCAGCACATCCGAG, for pr4-F GCGCTCGGTTGCGAAAAGTG, for pr4-R CCAAACACGAAGTGGGGGAGT, 1433ε-F ACCAACACACCCCATCCGTT, 1433ε-R ATGGCATCATCGAAAGCGGC, act5c-F GCTGAGCGTGAAATCGTCCG, act5c-R GATGCCGCAAGCCTCCATTC, tub-F GGACGTCAACGCCGCTATTG, and tub-R TTGGACAACATGCACACGGC. Since pr3 is embedded in an exon that is shared by the P1 (made by pr1 and pr2) and P3 (made by pr3) protein isoforms, and thus cannot be amplified separately, the primer pair designed for pr3 was designated P1/3. Primer efficiency ranged between 98% and 104%. Target gene expression was standardized to three reference genes (α-tub, act5c, and 1433ε) with robust stability values (mean coefficient variance < 0.05, mean M value < 0.1), and fold change cycle threshold values (2-ΔΔCt) were determined to quantify relative expression differences between genotypes.
Optogenetics.
A single first, second, or third instar larva was placed on a small Petri dish with distilled water to clean off remnants from the culture medium. The larva was then transferred to a 2% agar plate (100 × 15 mm) and allowed to acclimate for 20 s. Following acclimatization, the larva was stimulated three times for 5 s by a red light-emitting diode (LED) light source (M617F1; Thorlabs) controlled by an LED driver (LED1B; Thorlabs), with 5-s pauses between each pulse of light. An overhead camera (Canon Vixia HFR400) recorded larval behavior.
The videos were reviewed and scored to determine the number of curling and rolling occurrences. Curling was defined as the larval anterior and posterior regions moving simultaneously toward each other. Rolling was defined as a corkscrew-like rotation along the larval body axis (10). Each 5-s light stimulation was defined as a trial, for a total of three sequential trials of stimulation for each larva. Each trial was scored as 0 if the larva did not curl and as 1 if the larva did curl. Rolling behavior was scored in the same manner. The scores of the three trials were totaled, and a score out of three was determined for each behavioral response.
Thermal Nociception.
Thermal nociception was tested as described previously (10, 59). A single larva was removed from its food vial and placed on a small Petri dish with distilled water to wash off food remnants. The larva was then transferred to a Petri dish (100 × 15 mm) with a drop of water shallow enough to allow contact of the larva’s ventral cuticle with the surface of the dish. A soldering iron with a 0.6-mm-wide tip was used as the probe, and the voltage of the probe was adjusted using a variable autotransformer (VARIAC). The temperature of the probe was calibrated with a thermocouple (Physitemp BAT-12) and was determined using a temperature sensor (Physitemp IT-23) wrapped around the tip of the probe. The probe was heated to 42 ± 1 °C and was used to gently touch each larva in segments 4–6 of the abdominal region. An overhead camera (Canon Vixia HFR400) recorded larval behavior.
Larval behavior was analyzed and scored as described above in the optogenetics experiments. Larval response latency was measured as the time from stimulation of the probe to the onset of the curling or rolling response.
Developmental Assay.
Larvae were reared in the dark in small Petri dishes with a thin layer of fly medium supplemented with all-trans retinal. Larvae were stimulated for 5 s every 5 min using a red LED light source (Thorlabs M617F1) controlled by a LED driver (Thorlabs LED1B) for 16 h (at either 79–95 or 103–119 h AEL) or 1 h (at either 103 or 118 h AEL). Control larvae were reared in the same conditions but were not exposed to red light.
Immunohistochemistry.
Immunohistochemistry was performed as previously described (60). Briefly, preparations were fixed in Bouin’s solution (Sigma) for 5 min, washed, and then incubated overnight at 4 °C with a mouse anti-GFP monoclonal antibody (mAb) 3E6 (1:500; Molecular Probes) or a rabbit polyclonal anti-dilp7 antibody (1:5,000) (61). Primary antibodies were diluted in blocking solution. Preparations were mounted in Permafluor (Immunon) on a glass slide with a coverslip and viewed under a Leica TCS SP5 confocal laser-scanning microscope with a 20× air objective [0.7 numerical aperture (NA)] or 63× oil-immersion objective (1.4 NA).
GRASP.
Activity-dependent GRASP was performed as previously described (49). Larval CNSs were dissected and stimulated by high K+ depolarization for 20 min using the following high K+ saline: 25 mM NaCl, 90 mM KCl, 10 mM NaHCO3, 5 mM Hepes, 30 mM sucrose, 5 mM trehalose, 10 mM MgCl2, and 2 mM CaCl2 (pH 7.2) (62). Preparations were subsequently fixed in Bouin’s solution (Sigma) for 5 min. Preparations were washed and then incubated overnight at 4 °C with a monoclonal anti‐GFP mAb 3E6 (1:500; Molecular Probes) diluted in blocking solution. Preparations were mounted in Permafluor (Immunon) on a glass slide with a coverslip and viewed under a Leica TCS SP5 confocal laser-scanning microscope with a 20× air objective (0.7 NA) or 63× oil-immersion objective (1.4 NA).
Statistical Analysis.
SigmaPlot (version 11.0; Systat Software) was used for statistical analysis. Unpaired t tests or Mann–Whitney rank sum test were used for comparing datasets of two groups, and one-way ANOVA tests (with a Kruskal–Wallis or Holm–Sidak post hoc test) were used for comparing datasets of more than two groups. Error bars in all figures represent ± SEM.
Supplementary Material
Acknowledgments
We thank Scott Douglas for generating the UAS-forRNAi construct and Robert Lee for conducting preliminary experiments for the optogenetic assay. We thank Stefan Pulver for advice on optogenetic experiments and Dan Tracey for advice on the thermal nociception assay. We thank Gero Miesenböck, Julie Simpson, and Dan Tracey for fly stocks and Irene Miguel-Aliaga for anti-dilp7 antibody. This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) (Grants RGPIN 3397-11 and RGPIN-2016-06185 to M.B.S. and Grant RGPIN 06582 to J.S.D.), a Heart and Stroke Fellowship (to J.S.D.), an Ontario Graduate Scholarship (to I.A.), and an NSERC Undergraduate Student Research Award (to V.A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820840116/-/DCSupplemental.
References
- 1.Tracey W. D., Jr, Nociception. Curr. Biol. 27, R129–R133 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Eaton R. C., Bombardieri R. A., Meyer D. L., The Mauthner-initiated startle response in teleost fish. J. Exp. Biol. 66, 65–81 (1977). [DOI] [PubMed] [Google Scholar]
- 3.Edwards D. H., Heitler W. J., Krasne F. B., Fifty years of a command neuron: The neurobiology of escape behavior in the crayfish. Trends Neurosci. 22, 153–161 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Card G. M., Escape behaviors in insects. Curr. Opin. Neurobiol. 22, 180–186 (2012). [DOI] [PubMed] [Google Scholar]
- 5.De Franceschi G., Vivattanasarn T., Saleem A. B., Solomon S. G., Vision guides selection of freeze or flight defense strategies in mice. Curr. Biol. 26, 2150–2154 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Janssen A., Driessen G., Haan M. D., Roodbol N., The impact of parasitoids on natural populations of temperate woodland Drosophila. Neth. J. Zool. 38, 61–73 (1988). [Google Scholar]
- 7.Fleury F., et al. , Ecological and genetic interactions in Drosophila-parasitoids communities: A case study with D. melanogaster, D. simulans and their common Leptopilina parasitoids in south-eastern France. Genetica 120, 181–194 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Hwang R. Y., et al. , Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 17, 2105–2116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson J. L., Tsubouchi A., Tracey W. D., Larval defense against attack from parasitoid wasps requires nociceptive neurons. PLoS One 8, e78704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tracey W. D. Jr, Wilson R. I., Laurent G., Benzer S., painless, a Drosophila gene essential for nociception. Cell 113, 261–273 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Zhong L., Hwang R. Y., Tracey W. D., Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr. Biol. 20, 429–434 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong L., et al. , Thermosensory and nonthermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat-sensor domains of a thermoTRP Channel. Cell Rep. 1, 43–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauthner S. E., et al. , Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr. Biol. 24, 2920–2925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honjo K., Mauthner S. E., Wang Y., Skene J. H. P., Tracey W. D. Jr, Nociceptor-enriched genes required for normal thermal nociception. Cell Rep. 16, 295–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honjo K., Tracey W. D. Jr, BMP signaling downstream of the Highwire E3 ligase sensitizes nociceptors. PLoS Genet. 14, e1007464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walcott K. C. E., Mauthner S. E., Tsubouchi A., Robertson J., Tracey W. D., The Drosophila small conductance calcium-activated potassium channel negatively regulates nociception. Cell Rep. 24, 3125–3132.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honjo K., Hwang R. Y., Tracey W. D. Jr, Optogenetic manipulation of neural circuits and behavior in Drosophila larvae. Nat. Protoc. 7, 1470–1478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohyama T., et al. , A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kaneko T., et al. , Serotonergic modulation enables pathway-specific plasticity in a developing sensory circuit in Drosophila. Neuron 95, 623–638.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne K. A., et al. , Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277, 834–836 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Anreiter I., Kramer J. M., Sokolowski M. B., Epigenetic mechanisms modulate differences in Drosophila foraging behavior. Proc. Natl. Acad. Sci. U.S.A. 114, 12518–12523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen A. M., Anreiter I., Neville M. C., Sokolowski M. B., Feeding-related traits are affected by dosage of the foraging gene in Drosophila melanogaster. Genetics 205, 761–773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaun K. R., Hendel T., Gerber B., Sokolowski M. B., Natural variation in Drosophila larval reward learning and memory due to a cGMP-dependent protein kinase. Learn. Mem. 14, 342–349 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mery F., Belay A. T., So A. K., Sokolowski M. B., Kawecki T. J., Natural polymorphism affecting learning and memory in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 13051–13055 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen A. M., Anreiter I., Vesterberg A., Douglas S. J., Sokolowski M. B., Pleiotropy of the Drosophila melanogaster foraging gene on larval feeding-related traits. J. Neurogenet. 32, 256–266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel J. E., Xie X. J., Sokolowski M. B., Wu C. F., A cGMP-dependent protein kinase gene, foraging, modifies habituation-like response decrement of the giant fiber escape circuit in Drosophila. Learn. Mem. 7, 341–352 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaun K. R., et al. , Natural variation in food acquisition mediated via a Drosophila cGMP-dependent protein kinase. J. Exp. Biol. 210, 3547–3558 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Kaun K. R., Chakaborty-Chatterjee M., Sokolowski M. B., Natural variation in plasticity of glucose homeostasis and food intake. J. Exp. Biol. 211, 3160–3166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent C. F., Daskalchuk T., Cook L., Sokolowski M. B., Greenspan R. J., The Drosophila foraging gene mediates adult plasticity and gene-environment interactions in behaviour, metabolites, and gene expression in response to food deprivation. PLoS Genet. 5, e1000609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renger J. J., Yao W. D., Sokolowski M. B., Wu C. F., Neuronal polymorphism among natural alleles of a cGMP-dependent kinase gene, foraging, in Drosophila. J. Neurosci. 19, RC28 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Q., et al. , cGMP-dependent protein kinase encoded by foraging regulates motor axon guidance in Drosophila by suppressing Lola function. J. Neurosci. 36, 4635–4646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dason J. S., Allen A. M., Vasquez O. E., Sokolowski M. B., Distinct functions of a cGMP-dependent protein kinase in nerve terminal growth and synaptic vesicle cycling. J. Cell Sci. 132, jcs227165 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Sokolowski M. B., Turlings T., Drosophila parasitoid-host interactions: Vibrotaxis and ovipositor searching from the host’s perspective. Can. J. Zool. 65, 461–464 (1987). [Google Scholar]
- 34.Carton Y., Sokolowski M. B., Interactions between searching strategies of Drosophila parasitoids and the polymorphic behavior of their hosts. J. Insect Behav. 5, 161–175 (1992). [Google Scholar]
- 35.Hughes K., Sokolowski M. B., Natural selection in the laboratory for a change in resistance by Drosophila melanogaster to the parasitoid wasp Asobara tabida. J. Insect Behav. 9, 477–491 (1996). [Google Scholar]
- 36.Lewin M. R., Walters E. T., Cyclic GMP pathway is critical for inducing long-term sensitization of nociceptive sensory neurons. Nat. Neurosci. 2, 18–23 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Luo C., et al. , Presynaptically localized cyclic GMP-dependent protein kinase 1 is a key determinant of spinal synaptic potentiation and pain hypersensitivity. PLoS Biol. 10, e1001283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krzyzanowski M. C., et al. , Aversive behavior in the nematode C. elegans is modulated by cGMP and a neuronal gap junction network. PLoS Genet. 12, e1006153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Klapoetke N. C., et al. , Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grueber W. B., Ye B., Moore A. W., Jan L. Y., Jan Y. N., Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol. 13, 618–626 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Lee T., Luo L., Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Suster M. L., Seugnet L., Bate M., Sokolowski M. B., Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis 39, 240–245 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Harris R. M., Pfeiffer B. D., Rubin G. M., Truman J. W., Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. elife 4, e04493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grueber W. B., et al. , Projections of Drosophila multidendritic neurons in the central nervous system: Links with peripheral dendrite morphology. Development 134, 55–64 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Yoshino J., Morikawa R. K., Hasegawa E., Emoto K., Neural circuitry that evokes escape behavior upon activation of nociceptive sensory neurons in Drosophila Larvae. Curr. Biol. 27, 2499–2504.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Burgos A., et al. , Nociceptive interneurons control modular motor pathways to promote escape behavior in Drosophila. eLife 7, e26016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clyne J. D., Miesenböck G., Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell 133, 354–363 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Macpherson L. J., et al. , Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 10024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eguchi K., Nakanishi S., Takagi H., Taoufiq Z., Takahashi T., Maturation of a PKG-dependent retrograde mechanism for exoendocytic coupling of synaptic vesicles. Neuron 74, 517–529 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Taoufiq Z., Eguchi K., Takahashi T., Rho-kinase accelerates synaptic vesicle endocytosis by linking cyclic GMP-dependent protein kinase activity to phosphatidylinositol-4,5-bisphosphate synthesis. J. Neurosci. 33, 12099–12104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anreiter I., Sokolowski M. B., Deciphering pleiotropy: How complex genes regulate behavior. Commun. Integr. Biol. 11, 1–4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu C., et al. , Sensory integration and neuromodulatory feedback facilitate Drosophila mechanonociceptive behavior. Nat. Neurosci. 20, 1085–1095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anreiter I., Vasquez O. E., Allen A. M., Sokolowski M. B., Foraging path-length protocol for Drosophila melanogaster larvae. J. Vis. Exp., e53980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai S. L., Lee T., Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9, 703–709 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Wodarz A., Hinz U., Engelbert M., Knust E., Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82, 67–76 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Vogelstein J. T., et al. , Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344, 386–392 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Lee Y. S., Carthew R. W., Making a better RNAi vector for Drosophila: Use of intron spacers. Methods 30, 322–329 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Caldwell J. C., Tracey W. D. Jr, Alternatives to mammalian pain models 2: Using Drosophila to identify novel genes involved in nociception. Methods Mol. Biol. 617, 19–29 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Dason J. S., Smith A. J., Marin L., Charlton M. P., Cholesterol and F-actin are required for clustering of recycling synaptic vesicle proteins in the presynaptic plasma membrane. J. Physiol. 592, 621–633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miguel-Aliaga I., Thor S., Gould A. P., Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol. 6, e58 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dason J. S., Smith A. J., Marin L., Charlton M. P., Vesicular sterols are essential for synaptic vesicle cycling. J. Neurosci. 30, 15856–15865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.