Significance
DNA methylation is the most studied modification in human population epigenetics. Its information content can be explored in 2 principal ways—epigenome-wide association studies and epigenetic age. The latter likely reflects cellular/biological age and works with impressive accuracy across most tissues. In adults, it associates with various environments and health. However, current epigenetic clocks are not very accurate in the pediatric age range perhaps because DNA methylation changes much faster in children. Addressing this crucial gap, we created a precise tool to estimate DNA methylation age specific to pediatric buccal epithelial cells. This tool has the potential to become the standard reference for epigenetic studies broadly relevant to child development across the spectrum from health to disease.
Keywords: DNA methylation, age, development, epigenetic clock, adolescence
Abstract
The development of biological markers of aging has primarily focused on adult samples. Epigenetic clocks are a promising tool for measuring biological age that show impressive accuracy across most tissues and age ranges. In adults, deviations from the DNA methylation (DNAm) age prediction are correlated with several age-related phenotypes, such as mortality and frailty. In children, however, fewer such associations have been made, possibly because DNAm changes are more dynamic in pediatric populations as compared to adults. To address this gap, we aimed to develop a highly accurate, noninvasive, biological measure of age specific to pediatric samples using buccal epithelial cell DNAm. We gathered 1,721 genome-wide DNAm profiles from 11 different cohorts of typically developing individuals aged 0 to 20 y old. Elastic net penalized regression was used to select 94 CpG sites from a training dataset (n = 1,032), with performance assessed in a separate test dataset (n = 689). DNAm at these 94 CpG sites was highly predictive of age in the test cohort (median absolute error = 0.35 y). The Pediatric-Buccal-Epigenetic (PedBE) clock was characterized in additional cohorts, showcasing the accuracy in longitudinal data, the performance in nonbuccal tissues and adult age ranges, and the association with obstetric outcomes. The PedBE tool for measuring biological age in children might help in understanding the environmental and contextual factors that shape the DNA methylome during child development, and how it, in turn, might relate to child health and disease.
Epigenetic age, based on CpG methylation and often referred to as DNA methylation (DNAm) age, has emerged as a highly accurate estimator of chronological age (1). A widely used pan-tissue age estimator based on 353 CpG sites (hereon referred to as the pan-tissue Horvath DNAm clock) was developed on DNAm data of over 8,000 samples from 51 healthy tissues (1). This epigenetic clock has been applied to many independent datasets, each showing strong correlations with chronological age. Deviations between DNAm age and chronological age, referred to as DNAm age acceleration, are associated with several age-related health variables, with higher epigenetic age associated with an increase in mortality, cognitive decline, and a decrease in time until death (2).
Although correlations between chronological age and DNAm age, as measured by the Horvath clock, have been reported in pediatric samples, a high degree of variability from chronological age has been observed (3). The inaccuracy in predicting age in pediatric versus adult populations is not surprising, as challenges have been reported when extrapolating several adult-based biomarkers to children (4). Furthermore, the rate of DNAm change is greater in the pediatric age range compared to adulthood (5). Thus, there is a need for an epigenetic predictor of age, specific to pediatrics, to accurately detect deviations across populations that may reflect developmental trajectories, risk for pediatric disease, or certain environmental conditions that may accelerate or decelerate biological development in children.
Taking advantage of a large collection of pediatric DNAm profiles from buccal epithelial cells (BECs), we generated a tool to estimate age using DNAm at 94 CpG sites specific to pediatric buccal swab samples, referred to as the Pediatric-Buccal-Epigenetic (PedBE) clock. As a predictor of age, we focused on BECs because collection of this tissue is noninvasive, and thus more feasible in pediatric populations, contains less cellular heterogeneity as compared to other accessible tissues, such as blood, and has a high degree of DNAm stability (6–8). The utility of this highly precise pediatric molecular biomarker has yet to be fully explored; however, we anticipate deviations between pediatric DNAm age and chronological age to be representative of developmental processes and/or other pediatric diseases, as they are in adults.
Methods
Cohort Descriptions.
Training and test dataset inclusion criteria consisted of BEC Illumina Infinium450 (450K) or BEC Illumina InfiniumEPIC (EPIC) microarray DNAm data derived from typically developing individuals ranging from birth to 20 y old. For the training and test datasets, samples were excluded if exact age in days (collection date − date of birth) was not available or if predicted biological sex did not match with reported sex. We obtained DNAm profiles of 2,778 samples from 16 independent cohorts for our analyses (SI Appendix, Table S1); samples for datasets 1–8, 10–12, 15, and 16 were collected by our group, whereas datasets 9, 13, and 14 (9–25) were downloaded from the Gene Expression Omnibus (GEO) online database (26). All experimental procedures were conducted in accordance with institutional review board policies at the University of British Columbia and Children’s & Women’s Health Centre of British Columbia Research Ethics Board. Written informed consent was obtained from a parent/legal guardian and assent, where possible, was obtained from each child before study participation. Further details regarding each cohort’s ethics, informed consent, and sample processing can be found in SI Appendix.
We divided these data into a training dataset (datasets 1 through 7, n = 1,032, age range = 0.17 to 19.47 y) to generate the PedBE model and an independent test dataset (datasets 8 through 11, n = 689, age range = 0.01 to 19.96 y) in order to report unbiased performance metrics. Dataset 9B was an autism spectrum disorder (ASD) cohort and was used to evaluate whether deviations from the predicted age may associate with a pediatric disorder, and therefore these data were processed independently. We also included 3 non-BEC datasets, none of which were in the training or test data, to assess the predictor accuracy in saliva (dataset 12, n = 65) and blood (datasets 13 and 14, n = 134 and n = 19, respectively). Finally, 2 datasets, not included in our training or test analyses, were used to examine: 1) the accuracy of the pedBE clock in adults (dataset 15, n = 248, age range = 25.5 to 51.4 y), and 2) the association between obstetric outcomes and PedBE age acceleration in infancy (dataset 16, n = 510, age range = 2.8 to 10.3 mo). Further dataset specific details, including genomic DNA extraction methods, are provided in SI Appendix.
DNA Methylation Data Processing.
For all datasets, approximately 750 ng of genomic DNA was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA). Next, ∼160 ng of bisulfite converted DNA was processed using the 450K or EPIC array, according to manufacturer’s instructions (Illumina). Beta values (ranging from 0 to 1) were background subtracted and color corrected using GenomeStudio software. Data were subsequently processed using R statistical software (version 3.2.3). Cross-hybridizing probes, probes that target polymorphic CpGs, and XY probes were removed (27, 28). Additionally, we reduced our dataset to only probes that are represented on both the 450K and EPIC array. Probes with a bead count of <3 in 5% of samples as well as probes having a detection P value greater than 0.01 in 1% of samples were removed. Nonvariable probes, defined as those with an interquantile range of ≤0.05, were also removed (29). Missing DNAm data were imputed for remaining missing beta values (<1% of probes) with the “impute.knn” function based on nearest neighbor averaging (30). Data were normalized using a modified beta-mixture quantile (BMIQ) method to adjust for the microarray probe-type design differences (1, 31). We estimated BEC proportions using a previously described DNAm-based method (7). For blood-derived DNAm datasets (datasets 13 and 14), cell proportions were accounted for by predicting proportions using a commonly applied reference-based method (32, 33), where the top principal components of these estimates were regressed out from the DNAm data. For both the training and test data, ethnicity was not controlled for as availability of this variable was limited; however, for datasets 9B and 16, genotyping information were available and used to control for genetic differences in the ASD and obstetric longitudinal analyses, respectively (see SI Appendix for additional information).
Pan-Tissue Horvath DNAm Age.
For all test (datasets 8 through 11) and non-BEC datasets (datasets 12 through 14), data were processed using methods as described above. The previously established pan-tissue Horvath DNAm clock was performed using R statistical software with code supplied from https://dnamage.genetics.ucla.edu/home.
PedBE Clock.
Methods similar to the development of the Horvath DNAm clock were used to create the PedBE clock (1). We employed an elastic net approach with 10-fold cross-validation in the training dataset to empirically select age-informative CpG sites. An independent test dataset was used to evaluate and report accuracy metrics of the selected model. R code to generate PedBE age is available online: https://github.com/kobor-lab/Public-Scripts/.
Results
Cohort Characteristics.
We separated the 1,721 samples into 2 datasets: 1) a training dataset (n = 1,032, age range = 0.17 to 19.47 y), containing DNA profiles derived from BECs of typically developing individuals, evenly distributed across our selected age range of 0 to 20 y old; and 2) a test dataset (n = 689, age range = 0.01 to 19.96 y) also including typically developing individuals, constructed for the purpose of independently validating the predictor (SI Appendix, Table S1). We had a balanced sample of males and females in both datasets (training: 48% male, test: 53.3% male) but we note that due to a lack of appropriate information, we were not able to account for possible ethnic differences. The training and test datasets, along with all subsequently analyzed datasets, were processed independently during all quality control, filtering, and normalization steps.
A Precise Tool to Measure Pediatric DNAm Age in BECs: The PedBE Clock.
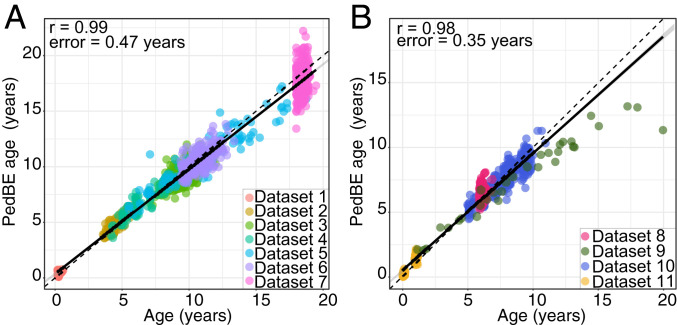
Using the training dataset, we generated an unbiased measure of pediatric DNAm age using elastic net regression, which empirically selected 94 informative age-related CpG sites (the PedBE clock). To validate this tool, we applied the PedBE clock to the test dataset, revealing a correlation between chronological age and pediatric DNAm age of r = 0.98 (P value ≤ 2.2 × 10−16), with a test error (defined as median absolute difference between DNAm age and chronological age) of 0.35 y (Fig. 1). The difference between PedBE age and chronological age in the test dataset was significantly correlated with that of the pan-tissue Horvath DNAm age difference (r = 0.54, P value ≤ 2.2 × 10−16); however, the PedBE clock had reduced variation as compared to the Horvath clock (Horvath DNAm age − age, median absolute difference = 1.73 y) (SI Appendix, Fig. S1). Of the 94 CpGs, DNAm at 50 CpG sites increased and 44 decreased with chronological age. We found that CpGs included in the PedBE clock were significantly depleted in open sea regions of the genome (Monte Carlo simulations, false discovery rate [FDR] ≤ 0.01), trended nonsignificantly toward enrichment in CpG islands (Monte Carlo simulations, FDR = 0.17), and showed no significant enrichment in other annotated gene features (SI Appendix, Fig. S2).
Fig. 1.
Pediatric buccal DNA methylation age accurately predicted chronological age. (A) The training data (datasets 1 through 7) was used in an elastic net regression for selection of 94 age-related CpG sites. Estimated age (PedBE age) (y axis) versus chronological age (x axis). (B) The test data (datasets 8 through 11) was to independently assess the PedBE clock (r = 0.98, test error = 0.35 y). Each data point represents an individual, with the color indicating the corresponding dataset. Solid black and dashed lines denote the linear regression and perfect correlation lines, respectively.
Notably, it was possible to obtain an equally performing predictor based on an entirely different set of CpGs in these data; for example, by using the same elastic-net feature selection approach as above, we found that a set of 392 CpG sites (none of which overlapped with the 94 CpGs of the PedBE clock) to have comparable accuracy in the test dataset (test error = 0.36, r = 0.98; SI Appendix, Fig. S3A). Furthermore, the 392 CpG sites were not found to be significantly enriched in any genomic features tested; however, there was a trend toward depletion in open sea regions (FDR = 0.12), similar to what we observed for the PedBE model, as well as a slight trend toward enrichment for intragenic regions (FDR = 0.18) (SI Appendix, Fig. S3B).
PedBE Clock Age Prediction Was Highly Accurate across Longitudinal Sampling.
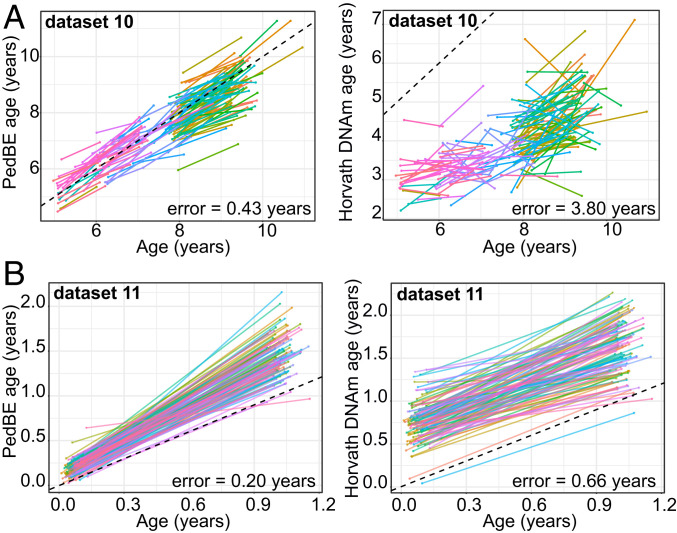
To further investigate the accuracy of the PedBE clock, we took advantage of the longitudinal nature of test datasets 10 and 11, which had repeated measures separated by 6 mo to 2 y, depending on the individual and study. As expected, after predicting PedBE age at each time point, all samples at time point 2 were estimated as being older than at time point 1. Additionally, when estimating Horvath DNAm age in these samples, a larger error was observed across both time points compared to the PedBE clock (dataset 10: PedBE clock error = 0.43 y, Horvath DNAm clock error = 3.8 y; dataset 11: PedBE clock error = 0.20 y, Horvath DNAm clock error = 0.66 y), highlighting the precision of the PedBE clock for the pediatric age range tested here (Fig. 2).
Fig. 2.
Longitudinal data demonstrated higher accuracy of the PedBE clock as compared to the pan-tissue Horvath DNAm clock. (A, Left) Dataset 10 PedBE clock versus chronological age across 2 time points (follow-up ranged from 6 mo to 2 y, depending on the individual). Each point color represents an individual separated by a line indicating the time between sampling. (Right) Dataset 10 pan-tissue Horvath DNAm clock versus chronological age. (B, Left) Dataset 11 PedBE clock versus age across 2 times points separated by 1 y between sampling. (Right) Dataset 11 pan-tissue Horvath DNAm clock versus chronological age. Each colored line represents an individual and the time between sampling is denoted by the beginning and end of each line. Time gaps between sample collection varied across individuals ranging from 6 mo to 2 y. Additionally, for dataset 10, individuals at time point one varied in age between 4 and 12 y. PedBE age was calculated for each individual at both time points.
PedBE Age Deviation Associated with Obstetric Outcomes.
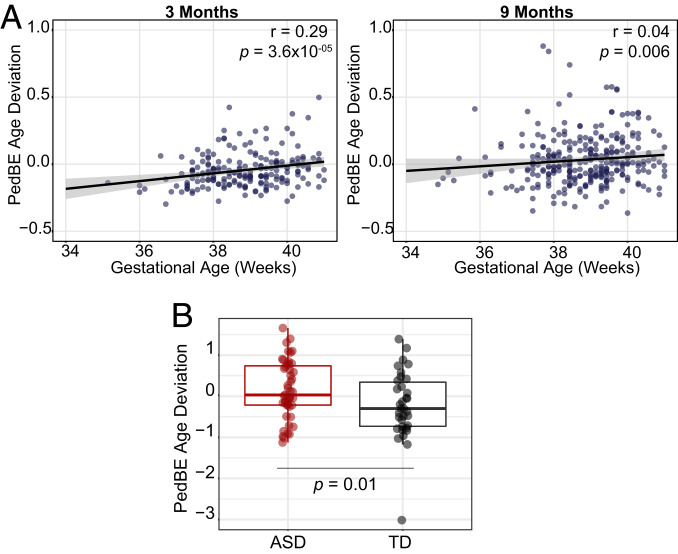
The PedBE clock was trained and tested on typically developing children to best represent general developmental patterns. As such, we expected that deviations from this estimate might serve as a biomarker for altered developmental trajectories. Similar to how the Horvath epigenetic clock has been investigated in adult samples, we extracted the residuals from a linear model of PedBE age regressed on chronological age to obtain the “age acceleration residual” (referred to here as “PedBE age deviation”). In dataset 16 (n = 339, a longitudinal cohort with sampling at 3 mo and 9 mo), which was not included in either the training or test datasets, we assessed whether PedBE age deviation was associated with obstetric outcomes, including gestational age and birthweight adjusted for gestational age. We had a similar observation to longitudinal test dataset 10, where the PedBE clock was more accurate than the Horvath clock in reflecting differences in chronological time between the sampling intervals (SI Appendix, Fig. S5). We found a significant positive association between length of gestation and age deviation at 3 mo (Pearson’s r = 0.29, P value = 4.0 × 10−5), which was relatively unchanged following adjustment for estimated BEC proportions, biological sex, and a composite score for genetic background represented by the top 2 principal components of the same samples run on the OmniExpress genotyping array (P value = 3.6 × 10−5) (Fig. 3 A, Left). Furthermore, this effect was also observed at 9 mo, although to a lesser extent and only statistically significant when controlling for the covariates (P value = 0.006, r = 0.04) (Fig. 3 B, Right). A similar, yet much weaker trend was observed using the pan-tissue Horvath DNAm clock at 3 mo (Pearson’s r = 0.14, P value = 0.05) but not 9 mo (Pearson’s r = 0.08, P value = 0.10).
Fig. 3.
PedBE deviation was associated with gestational age at 3 and 9 mo and individuals diagnosed with ASD in independent cohorts. (A) Dataset 16 is a longitudinal cohort with sampling at 3 mo (Left) and 9 mo (Right) of age in the same individuals. (B) In dataset 9B (n = 81), PedBE age deviation equals PedBE regressed onto chronological age, while controlling for sex, batch, predicted buccal proportion, and ethnicity. A nonparametric propensity score-matching method was applied to ensure the groups were balanced regarding covariate measures.
We also tested whether birthweight (adjusted for gestational age) was associated with PedBE age deviation but did not find any significant associations at either 3 mo (Pearson’s r = 0.05, P value = 0.50) or 9 mo (Pearson’s r = 0.11, P value = 0.07), and neither association was significant following adjustment for covariates.
Positive PedBE Age Deviation Was Associated with Autism Spectrum Disorder.
While the focus of this work was on developing a tool to carefully assess BEC DNAm age in children, we also wanted to begin exploring whether childhood disorders might associate with deviations in pediatric DNAm age. Unfortunately, public availability of BEC DNAm data for these valuable cohorts is rare. However, dataset 9 is one such cohort that has publicly available BEC DNAm from children affected with ASD. This cohort included individuals with ASD and their non-ASD affected siblings as controls (9). ASD has been characterized as a pediatric disorder with an altered development trajectory and has also been shown to have differential DNAm patterns as compared to the typically developing group (TD) individuals. We tested whether PedBE age differed in ASD-affected children as compared to TD children in dataset 9B (subset of GSE50759, age range: 1.2 to 20 y) (26). We assessed PedBE age deviation in a cohort of 47 ASD cases and 34 TD individuals (dataset 9B) while controlling for self-reported ethnicity, experimental batch, and estimated cell proportions (7). We observed a significant difference in PedBE age between ASD and controls (P value = 0.01), with ASD cases having a mean deviation of 0.37 y higher than the TD group (Fig. 3B). We performed a sensitivity analysis by retesting this association after removing the outlier in the TD group and observed a stronger association in the same direction (P value = 0.005, mean deviation = 0.38 y; SI Appendix, Fig. S5). To further verify this association, we employed a nonparametric propensity score-matching method (34) to attempt to reduce any bias by ensuring the groups were balanced in terms of covariate measures; specifically, estimated buccal cell proportion, experimental batch, biological sex, and percent HapMap Central European ancestry (see original paper for details) (26). Using this approach, a sample of 17 ASD cases and 22 TD cases was obtained and the difference remained significant (P value = 0.02, mean deviation = 0.56 y). Furthermore, to address any concern that familial status was influencing this result, we randomly removed 1 individual from each sibling pair from our analyses. The ASD and TD groups remained significantly different in terms of age deviation (P value = 0.03, ASD = 47, TD = 21, median deviation = 0.40 y). To account for unbalanced sample sizes between ASD and TD, we again performed propensity score matching on this subset and observed a moderate difference between ASD and TD age deviation (P value = 0.04, ASD = 12, TD = 16, median deviation = 0.56 y). Given the small group sizes of these analyses, we emphasize cautionary interpretation of this result and present these findings as strictly exploratory requiring independent validation in additional samples.
PedBE Clock Age Prediction in Saliva, Blood, and Adult BECs.
Although we trained the PedBE clock in samples obtained exclusively from BECs, we explored the performance in saliva and blood, both of which are minimally invasive, commonly used tissues for epigenetic interrogations in human populations.
Saliva is a heterogeneous mixture of varying proportions of BECs, white blood cells, amongst other cell types (7). We assessed PedBE age in saliva samples from dataset 12 (n = 65, age range = 6 to 13 y), which resulted in a moderate association between predicted age using the PedBE clock and chronological age (r = 0.50, error = 1.31 y, P value = 2.0 × 10−5; SI Appendix, Fig. S6). However, it should be noted that these data reported age in years rather than days, therefore reported predictor performance was not as precise, potentially deviating by up to 1 y.
In 2 (datasets 13 and 14) publicly available blood DNAm datasets we assessed the performance of the PedBE clock with and without correcting for blood cell type heterogeneity. In dataset 13 (n = 134, age range = 1 to 17 y restricted to data from control TD individuals 20 y and younger), PedBE age without correcting for blood cell type variance led to a correlation of r = 0.79 and a median absolute error of 3.26 y, which was much larger than the test BEC datasets. After statistically regressing out the variance associated with blood cell type heterogeneity, the correlation between predicted age and chronological age was lower with a comparable degree of error (r = 0.60, error = 3.02) (SI Appendix, Fig. S7 A and C). In dataset 14 (n = 19, age range = 2.3 to 10.8 y), the PedBE clock correlated strongly with age on uncorrected data (r = 0.88, error = 1.89 y), but when applied to cell type corrected DNAm data the correlation was considerably lower (r = −0.27, error = 2.82 y; SI Appendix, Fig. S8 A and C). Collectively, these findings suggested that DNAm variance associated with blood cell type proportions improved the accuracy of the PedBE clock when used in blood. For both blood datasets 13 and 14, the pan-tissue Horvath DNAm clock performed well (r = 0.96, error = 0.57 y; r = 0.95, error = 1.66 y, respectively) on data prior to cell composition adjustment (SI Appendix, Figs. S7B and S8B); however, similar to the PedBE clock, a reduction in accuracy was observed when the pan-tissue Horvath DNAm clock was applied after cell type correction (dataset 13: r = 0.82, error = 1.29 y; dataset 14: r = −0.23, error = 5.69 y; SI Appendix, Figs. S7D and S8D). These observations highlighted the effect of blood cellular heterogeneity on age prediction in both the pan-tissue Horvath DNAm clock and PedBE clock.
We also assessed the performance of the PedBE clock in adult buccal samples. In dataset 15 (adult females, age range: 25.5 to 51.4 y), we observed a significant correlation between PedBE age and chronological age (r = 0.72, P value < 2.0 × 10−16) with a median absolute error of 1.40 y (SI Appendix, Fig. S9A). In contrast, the pan-tissue Horvath clock consistently underestimated DNAm age, as compared to chronological age, but had a similar correlation value with chronological age (r = 0.66, P value < 2.0 × 10−16) with a higher median absolute error (3.10 y) (SI Appendix, Fig. S9B). Finally, the estimated PedBE ages and Horvath DNAm ages were significantly correlated (r = 0.44, P value = 2 × 10−13; SI Appendix, Fig. S9C).
A Pediatric BEC DNAm Predictor Generated from All Available Samples.
Following a recently suggested approach to increase the rigor of epigenetic clocks (35), we also investigated a model based on the entirety of our samples (n = 1,721) (SI Appendix, Fig. S10A). The correlation between the “all data model” and the PedBE clock model was very high (r > 0.99; SI Appendix, Fig. S10B). With this approach, we cannot report test accuracy but aimed to compare the PedBE clock to a model established on all samples to ensure our predictor was not compromised by the reduced training dataset sample size. We employed the same parameters as the initial predictor from the n = 1,032 training data, but with all 1,721 samples included, and the number of probes in this model was also 94 CpGs (SI Appendix, Table S2); however, the overlap with this predictor and the PedBE clock was only 64 CpGs (SI Appendix, Table S3 and Fig. S10C). The genomic enrichment of the 94 CpG sites based on all data did not perfectly reflect the enrichment of the PedBE sites: the 94 CpGs (all) were not significantly enriched or depleted for most annotated gene features except for a nonsignificant trend toward enrichment in north shores (SI Appendix, Fig. S10D).
Lastly, we examined whether the association between gestational age and PedBE age deviation was robust in the “all-sample” 94-CpG model. We observed nearly identical results as the training model, where we found a significant association between gestational age and PedBE age deviation at 3 mo (r = 0.27, P value of correlation test: 1.0 × 10−4, P value obtained from linear model while adjusting for covariates = 1.4 × 10−4) and a moderate association at 9 mo (r = 0.08, P value of correlation test: 0.2, P value obtained from linear model while adjusting for covariates = 6.2 × 10−4). Additionally, no significant association was observed with birthweight adjusted for gestational age.
Discussion
Birth to late adolescence is a tremendously dynamic period of development and growth, and an accurate molecular marker of development or age, specific to this age, range has yet to be established. We assessed DNAm profiles in BECs from 1,721 healthy individuals, ranging in age from 0 to 20 y old, and generated a predictor of age, specific to pediatric populations, using weighted DNAm values at 94 CpG sites (the PedBE clock). We characterized the PedBE clock in longitudinal data, different tissues, an adult cohort, and importantly, in the context of obstetric outcomes, finding that infants with a higher gestational age had an older PedBE age. We also assessed PedBE age in children with a neurodevelopmental disorder, ASD, which showed a higher PedBE age than those considered to be typically developing.
Although in adults, positive DNAm age acceleration from the Horvath pan-tissue clock has been associated with increased risk for certain diseases, mortality, frailty, and other negative outcomes (2, 36, 37), DNAm age acceleration in children may not follow a similar pattern, in that accelerated age deviation could potentially reflect positive outcomes. While definite evaluation of these relationships awaits larger surveys, it is tempting to speculate that age acceleration in pediatric samples may be an indicator of progressive development through milestones, whereas deceleration might be an indicator of delays in maturation.
The previously established pan-tissue Horvath DNAm clock was created from prenatal samples to supercentenarians (1). However, the variation in age estimates makes its application to pediatric populations somewhat challenging. More accurate age estimators can be constructed by focusing on a more limited age range. For example, more accurate epigenetic age estimators have been developed for gestational age based on cord blood samples (38, 39). Our study similarly demonstrated that the PedBE clock easily outperforms the pan-tissue estimator in BEC samples from a pediatric population. It is not surprising that previous predictors of age do not perform exceptionally well in children, as this is a unique period of rapid change in DNAm that is unlikely to mimic adult methylome dynamics (5). Given the differences in the pace of developmental and age-related changes across the life course, applying adult-based markers to pediatric populations may not be an appropriate approach.
In addition, the Horvath DNAm clock is agnostic to tissue type, which inevitably sacrifices some precision when estimating age within any single tissue. Part of the reason for the increased accuracy of the PedBE clock stems from building the predictor in a single target tissue within a focused age range of 0 to 20 y old. The specificity of the sample target may be viewed as a limitation, as we have shown, making the predictor less robust to other tissues. However, the loss of applicability to other tissues was necessary as it allowed our predictor to reach the highest accuracy of any epigenetic clock to date. We believe that since age-related DNAm has differential rates across tissues, the focus of a single tissue type is needed for obtaining the highest estimate accuracy possible. Additionally, BECs are very commonly used in pediatric biomarker research, as well as populations from remote communities, due to the fact that they can be obtained by a noninvasive collection protocol (40, 41). Furthermore, BEC swabs are less heterogeneous in terms of cell type population as compared to saliva (6), further illustrating their utility as a tissue of choice for pediatric DNAm studies.
Heterogeneous tissues, such as blood, can change in cell proportion with age (42) and focusing on a more homogeneous tissue, such as BECs, would reduce this source of confounding when constructing a DNAm-based age predictor. The association between PedBE age and chronological age was considerably lower in the blood test datasets, understandably so, as this epigenetic clock was trained specifically on BEC samples. Interestingly, when blood cell type composition was adjusted for in the DNAm data prior to calculating PedBE age, the accuracy was considerably lower than estimated age on cell type uncorrected data for both the PedBE clock and the pan-tissue Horvath DNAm clock. This is most likely because cell type proportions change over time and therefore this variance is captured by the DNAm-based age clocks, underscoring the intended purposes of the present tool to be primarily applied in BEC samples.
Having established an epigenetic clock with great accuracy for the pediatric age range from a single tissue, it will be interesting to compare its associations with chronological age, environmental variables, and developmental outcomes to that of epigenetic clocks derived for the same age range from other single tissues. We note that it is not only feasible that different tissues age at different rates in children, but also that unaccounted factors likely unrelated to the developmental process might contribute noise to such predictors.
We also investigated the PedBE clock in adult BEC samples and found that the PedBE clock and pan-tissue clock had similar correlation values with age; however, the PedBE clock had a lower median absolute error. Perhaps not unexpectedly, this finding suggests that tissue-specific epigenetic clocks might have a higher overall accuracy for the tissue they were trained in than clocks trained on a tissue compendium, even if the training was done for a different stage of the human life course. In our case, even though the PedBE model was only trained on pediatric BEC samples, it was still able to predict age in adult BEC samples with a lower error than the pan-tissue Horvath DNAm clock.
As DNAm is strongly associated with age and tissue type, it is not surprising that we note the overlap of only 1 CpG site (cg06144905) when comparing the Horvath pan-tissue clock sites and the CpG sites used in the PedBE clock. This CpG site is within the promoter region of the PIPOX gene which encodes an enzyme that metabolizes sarcosine, l-pipecolic acid, and l-proline; interestingly, circulating sarcosine decreases with age and is increased with dietary restriction (43).
Since this epigenetic clock is unique to children, and the pan-tissue Horvath clock has been extensively correlated to later life age-related measures, we expect the PedBE clock may be capturing developmental phenotypes related to growth. Furthermore, we note that the performance of both the pan-tissue Horvath and PedBE clock was more accurate closer to birth than in adolescence. While it is possible that this feature was due to slight imbalances in the adolescent age range in our datasets, it is tempting to speculate that in part, it might be due to the dynamic nature of the developing DNA methylome during adolescent stages, emphasizing the unique relationship between DNAm and development over childhood.
The post hoc investigation into whether the PedBE CpGs were of unique importance as compared to the remaining CpG sites measured, was particularly interesting as we were able to generate an equally accurate clock using 392 CpGs of which none overlapped with the PedBE 94 sites. With this distinct 392-CpG model, we also observed less accurate estimates in the adolescent age range, perhaps suggesting that interindividual differences in DNAm age may become more pronounced as children get older; however, the smaller sample size in this age range is an important limitation to note and additional cohorts are required to further explore this. Nonetheless, the lack of the overlap in CpG sites between the 392 model and the PedBE clock was insightful, highlighting that the age-associated nature of the DNA methylome across sets of CpGs is sufficient to accurately predict age. We note that both models had significant or close-to significant depletion in open sea genomic areas but no strong significant enrichment in any particular CpG-island features. Future work comparing the specific nature of these CpGs and other epigenetic clocks would be important for gaining a comprehensive understanding of the methylome landscape in the context of the human life course.
Our results investigating the ASD cohort suggested that deviations between PedBE age and chronological age might be associated with altered developmental trajectories and potentially pediatric disorders. ASD is associated with altered developmentally related phenotypes, such as increased body growth, head growth, and body weight, as well as accelerated postnatal cortical development (44). We showed that individuals with ASD had increased PedBE age deviation compared to controls, consistent with advanced biological development. While the exact mechanisms underlying this “acceleration” remain to be determined, previous research indicates DNAm differences in individuals affected by ASD (26, 45, 46), thus further emphasizing the potential utility for DNAm as a biomarker in ASD. We note that while we would have preferred to add additional independently ascertained datasets and conditions to our analysis, the vast majority of published BEC association studies are not publicly accessible, which constitutes a major limitation in the field (47).
The association between PedBE age deviation with gestational age was of interest, as it might intersect with other long-term measures of child development. Length of gestation is a well-established predictor of a range of child health outcomes, including structural variation in the neonatal brain (48). Gestational age was positively associated with PedBE age acceleration making it tempting to speculate that this is related to positive association between gestational age and brain maturation previously reported for this cohort (dataset 16), that was independent of neonatal birthweight. Thus, our study further supported the role of the length of gestation in shaping variation in the neonatal DNA methylome (39). In contrast, perhaps somewhat unexpectedly, birthweight was not consistently associated with PedBE age deviation. Our findings thus strongly hinted at a specificity of the PedBE clock, which may reflect the distinct genetic and environmental factors that influence gestational age and birthweight (49).
In conclusion, this study described a highly accurate molecular measure of chronological age using DNAm obtained from BECs in pediatric samples. To maximize the rigor, accuracy, and objectivity of this tool we followed recent recommendations (35) and compared a model based on all available data with the PedBE clock. Overall, we found significant correlations between the estimates, and demonstrated that both predictors had an association with gestational age in a completely separate cohort not contained in our test sample. While the utility of this tool as a developmental metric remains to be explored, we envision that by testing additional pediatric datasets, as they become available, this tool will become important for evaluating the environmental and contextual factors shaping child development, chiefly through the DNA methylome, and how this in turn associates with health and disease.
Supplementary Material
Acknowledgments
We would like to acknowledge all study participants. We also thank the researchers who generously uploaded their data to the public repository GEO, especially Dr. John Greally and Dr. Masako Suzuki for kindly sharing further details regarding the ASD cohort; without their contributions this work would not be possible. M.S.K. is the Canada Research Chair in Social Epigenetics, Senior Fellow of the Canadian Institute for Advanced Research, and Sunny Hill British Columbia Leadership Chair in Child Development. L.M.M. is supported by a Frederick Banting and Charles Best Canadian Institutes of Health Research Doctoral Research Award (F15-04283). S.H. acknowledges support by NIH/National Institute on Aging (U34AG051425-01).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.B.S. is a guest editor invited by the Editorial Board.
Data deposition: Unpublished data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers: GSE137495 (APrON, dataset 1), GSE137884 (C3ARE, dataset 2), GSE137903 (GECKO, dataset 3), GSE80261 (NDN, dataset 4), GSE137894 (MAVAN I, dataset 5), GSE94734 (PAWS, dataset 6), GSE137502 (WSFW, dataset 7), GSE137682 (BIBO, dataset 8), GSE50759 (GSE50759, dataset 9), GSE137898 (MAVAN II, dataset 10), GSE137841 (UCI, dataset 11), GSE138279 (Grady [saliva], dataset 12), GSE36054 (GSE36054 [blood], dataset 13), GSE64495 (GSE64495 [blood], dataset 14), GSE137688 (MAVAN [mothers], dataset 15), and GSE137904 (GUSTO, dataset 16). These data have been deposited under the GEO superseries GSE137503.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820843116/-/DCSupplemental.
References
- 1.Horvath S., DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath S., Raj K., DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 23, 223 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Simpkin A. J., et al. , Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum. Mol. Genet. 25, 191–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman J., Becker M. L., Jones B., Clements M., Leeder J. S., Development of biomarkers to optimize pediatric patient management: What makes children different? Biomark Med 5, 781–794 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alisch R. S., et al. , Age-associated DNA methylation in pediatric populations. Genome Res. 22, 623–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theda C., et al. , Quantitation of the cellular content of saliva and buccal swab samples. Sci. Rep. 8, 6944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A. K., et al. , DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168B, 36–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dongen J., et al. , Genome-wide analysis of DNA methylation in buccal cells: a study of monozygotic twins and mQTLs. Epigenetics Chromatin 11, 54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [APrON]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137495. Deposited 18 September 2019.
- 11.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [C3ARE]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137884. Deposited 26 September 2019.
- 12.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [GECKO]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137903. Deposited 25 September 2019.
- 13.Portales-Casamar E., et al. , DNA methylation signature of human fetal alcohol spectrum disorder. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80261. Deposited 22 March 2019. [DOI] [PMC free article] [PubMed]
- 14.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [MAVAN I]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137894. Deposited 26 September 2019.
- 15.Bush N. R., et al. , The biological embedding of early life socioeconomic and family adversity in children’s genome-wide DNA methylation. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94734. Deposited 23 April 2019. [DOI] [PMC free article] [PubMed]
- 16.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [WSFW]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137502. Deposited 23 September 2019.
- 17.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [BIBO cohort]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137682. Deposited 23 September 2019.
- 18.Berko E. R., Greally J. M, Epigenetic Analysis of ASD in AMA. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50759. Deposited 22 March 2019.
- 19.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [MAVAN II]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137898. Deposited 26 September 2019.
- 20.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [UCI]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137841. Deposited 26 September 2019.
- 21.Ressler K. J., Binder E. B., Smith A. K., Jovanovic T., Grady kids saliva methylation. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138279. Deposited 2 October 2019.
- 22.Barwick B. G., Alisch R. S., Chopra P., Warren S. T., Methylation Profiling of Blood DNA from Healthy Children. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36054. Deposited 22 March 2019.
- 23.Horvath S., R. Walker, DNA methylation profiles of human blood samples from a severe developmental disorder and controls. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64495. Deposited 22 March 2019.
- 24.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [MAVAN mothers]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137688. Deposited 29 September 2019.
- 25.McEwen L. M., et al. , The PedBE clock estimates DNA methylation age in pediatric buccal cells [GUSTO]. GEO Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137904. Deposited 25 September 2019.
- 26.Berko E. R., et al. , Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genet. 10, e1004402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y.-A., et al. , Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price M. E., et al. , Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R. D., Jones M. J., Robinson W. P., Kobor M. S., An empirically driven data reduction method on the human 450K methylation array to remove tissue specific non-variable CpGs. Clin. Epigenetics 9, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troyanskaya O., et al. , Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Teschendorff A. E., et al. , A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman E. A., et al. , DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koestler D. C., et al. , Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics 8, 816–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho D. E., Imai K., King G., Stuart E. A., Matching as Nonparametric Preprocessing for Reducing Model Dependence in Parametric Causal Inference. Polit. Anal. 15, 199–236 (2017). [Google Scholar]
- 35.Simpkin A. J., Suderman M., Howe L. D., Epigenetic clocks for gestational age: statistical and study design considerations. Clin. Epigenetics 9, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marioni R. E., et al. , DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitling L. P., et al. , Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenetics 8, 21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight A. K., et al. , An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 17, 206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohlin J., et al. , Prediction of gestational age based on genome-wide differentially methylated regions. Genome Biol. 17, 207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witsø E., Stene L. C., Paltiel L., Joner G., Rønningen K. S., DNA extraction and HLA genotyping using mailed mouth brushes from children. Pediatr. Diabetes 3, 89–94 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Le Marchand L., et al. , Feasibility of collecting buccal cell DNA by mail in a cohort study. Cancer Epidemiol. Biomarkers Prev. 10, 701–703 (2001). [PubMed] [Google Scholar]
- 42.Jaffe A. E., Irizarry R. A., Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walters R. O., et al. , Sarcosine Is Uniquely Modulated by Aging and Dietary Restriction in Rodents and Humans. Cell Reports 25, 663–676.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surén P., et al. , Early growth patterns in children with autism. Epidemiology 24, 660–670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews S. V., et al. , Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat. Commun. 8, 1011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feinberg J. I., et al. , Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. Int. J. Epidemiol. 44, 1199–1210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones M. J., Moore S. R., Kobor M. S., Principles and Challenges of Applying Epigenetic Epidemiology to Psychology. Annu. Rev. Psychol. 69, 459–485 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Broekman B. F. P., et al. ; GUSTO Study Group , Gestational age and neonatal brain microstructure in term born infants: a birth cohort study. PLoS One 9, e115229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lunde A., Melve K. K., Gjessing H. K., Skjaerven R., Irgens L. M., Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am. J. Epidemiol. 165, 734–741 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.