Significance
Amino acid sequences are known to be crucial for specifying the folded structure of a protein. Here we show that sequences also play an important role in specifying the properties of a protein’s unfolded ensemble. For an autotransporter protein, rearranging the order of only a few percent of amino acid residues can lead to more collapsed unfolded ensembles which have enhanced aggregation in vitro and degradation in vivo. Sequences that remain expanded, however, are more compatible with proper function. Most significantly, we show that the well-mixed sequence patterns that lead to expanded ensembles are broadly conserved amongst foldable, water-soluble proteins, suggesting expanded disordered states are under selection.
Keywords: unfolded states, autotransporter, IDPs, secretion, protein folding
Abstract
Much attention is being paid to conformational biases in the ensembles of intrinsically disordered proteins. However, it is currently unknown whether or how conformational biases within the disordered ensembles of foldable proteins affect function in vivo. Recently, we demonstrated that water can be a good solvent for unfolded polypeptide chains, even those with a hydrophobic and charged sequence composition typical of folded proteins. These results run counter to the generally accepted model that protein folding begins with hydrophobicity-driven chain collapse. Here we investigate what other features, beyond amino acid composition, govern chain collapse. We found that local clustering of hydrophobic and/or charged residues leads to significant collapse of the unfolded ensemble of pertactin, a secreted autotransporter virulence protein from Bordetella pertussis, as measured by small angle X-ray scattering (SAXS). Sequence patterns that lead to collapse also correlate with increased intermolecular polypeptide chain association and aggregation. Crucially, sequence patterns that support an expanded conformational ensemble enhance pertactin secretion to the bacterial cell surface. Similar sequence pattern features are enriched across the large and diverse family of autotransporter virulence proteins, suggesting sequence patterns that favor an expanded conformational ensemble are under selection for efficient autotransporter protein secretion, a necessary prerequisite for virulence. More broadly, we found that sequence patterns that lead to more expanded conformational ensembles are enriched across water-soluble proteins in general, suggesting protein sequences are under selection to regulate collapse and minimize protein aggregation, in addition to their roles in stabilizing folded protein structures.
In recent years, much attention has been paid to the conformational properties of intrinsically disordered proteins (IDPs) (1–8). Many well-characterized IDPs contain a lower ratio of hydrophobic to charged amino acids than proteins that adopt a stable folded structure (1, 2). However, we recently showed that even polypeptides with sequence compositions typical of foldable proteins can adopt highly expanded ensembles, even under physiological conditions (3, 4). The physical origin of this behavior is currently unclear. Are these sequences merely insufficiently hydrophobic, or do they adopt a highly expanded ensemble due to other factors? Delineating the sequence modifications sufficient to initiate collapse of these expanded ensembles would deepen our understanding of the contributions of the polypeptide sequence to shaping disordered ensembles and the consequences of modifying the extent of collapse on protein function in vivo and illuminate evolutionary pressures on amino acid sequences to specify both the correctly folded and misfolded conformations of proteins.
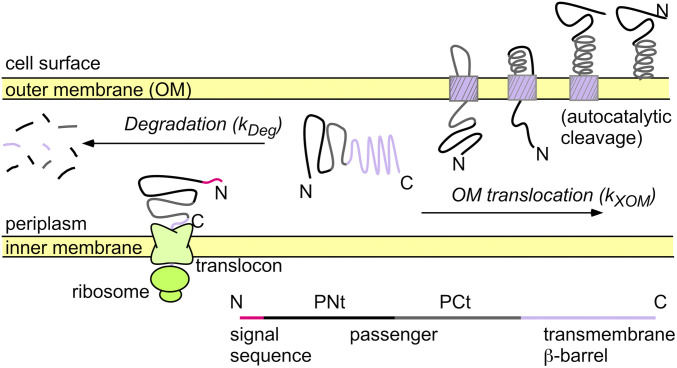
We used our well-characterized PNt system to investigate these questions (3, 5). PNt is the N-terminal 334 amino acids of the passenger domain of Bordetella pertussis pertactin, an archetypal member of the autotransporter (AT) family of virulence proteins from Gram-negative bacterial pathogens (6) (Fig. 1). Autotransporter passenger domains typically possess a β-helical folded structure but must remain unfolded while in the bacterial periplasm, prior to secretion across the outer membrane (OM). Folding of the passenger to a stable structure in the periplasm is sufficient to block its translocation across the OM (7–10). We previously showed that in isolation, PNt adopts a highly expanded conformational ensemble rather than its β-helix structure, despite having an amino acid composition typical of a folded protein (3, 5, 8).
Fig. 1.
Secretion of AT virulence proteins to the bacterial cell surface. The mature AT passenger consists of PNt (black) and PCt (gray) but is synthesized as a larger precursor (see bottom schematic). Efficient translocation of the passenger across the OM requires it remain unfolded while in the periplasm, prior to passage through the narrow AT C-terminal transmembrane β-barrel (purple). PNt swap variants that lead to a more collapsed conformational ensemble impair secretion by 1) increasing the rate of degradation by periplasmic proteases (kDeg; left arrow) and/or 2) decreasing the rate of translocation across the OM (kXOM; right arrow).
These features make pertactin an ideal model system to interrogate how properties of the unfolded state relate to biological function. We found that swapping the positions of just six residues (<2% of 334 total) of PNt, selected to create small hydrophobic clusters, was sufficient to induce significant contraction of the PNt conformational ensemble. This remarkable finding indicates that a well-mixed pattern of hydrophobic residues in a sequence is crucial for adopting an expanded ensemble. Moreover, in the context of the full-length pertactin, sequence-swap mutations that led to contraction were sufficient to impair secretion of the B. pertussis virulence protein to the bacterial cell surface. We discovered that low hydrophobic clustering is a common feature shared by diverse AT passenger sequences, suggesting evolution selects sequences with low hydrophobic clustering in order to enhance secretion of AT virulence proteins to the bacterial cell surface. More broadly, we found that low hydrophobic clustering is a general feature of well-folded protein sequences, indicating amino acid sequences are under selection to yield expanded unfolded states, potentially to reduce aggregation or enhance folding cooperativity.
Results
Swap Variants Maintain Amino Acid Composition but Change Sequence Patterns.
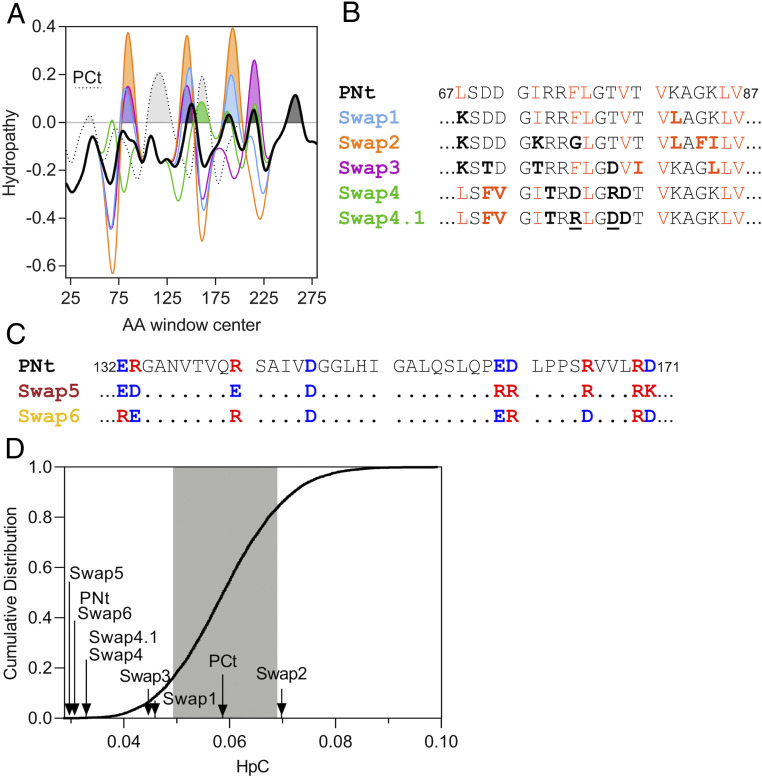
PNt adopts a highly expanded conformational ensemble despite having sequence properties typical of well-folded proteins (3). Specifically, the ratio of hydrophobic to charged residues in PNt (1.7) is higher than the average ratio for proteins in the Protein Data Bank (PDB) (1.3). For this reason, we hypothesized that local amino acid patterns, rather than global amino acid composition, might encode the conformational properties of PNt. In particular, we noted that the hydrophobic residues of PNt, although abundant, are well distributed along the sequence (SI Appendix, Fig. S1). This sequence pattern leads to smaller clusters of hydrophobic groups when compared to the pertactin β-helix C terminus (PCt; Fig. 2A). In contrast to PNt, PCt remains stably folded in isolation (8).
Fig. 2.
Swap variants maintain PNt amino acid composition while changing hydrophobic and charge patterns. (A) Protein sequence hydropathy (hybrid Hopp–Woods/Miyazawa–Jernigan scale; Materials and Methods) averaged over a sliding window of 9 AA. A Gaussian function (σ = 5 AA) was applied to smooth the raw curves. Pertactin C terminus (PCt; dotted) is included as an example of a wild-type sequence with more hydrophobic clustering than PNt (black). Other lines, color-coded as in B, indicate hydropathy of select swap variants. HpC is calculated as the sum of the shaded areas for each construct, divided by sequence length. (B and C) Examples of the sequence swap strategy used here to alter hydrophobic (B) or charge (C) patterns while maintaining amino acid composition. Swapped residues are indicated in bold font. (B) Hydrophobic (orange) and polar/charged (black) residues were swapped to increase hydrophobic clustering (Swap1, Swap2, and Swap3). As controls, in Swap4 and Swap4.1 the positions of a similar number of hydrophobic residues were swapped without significantly altering hydrophobic clustering. Swap4 and Swap4.1 differ only in the distribution of charged residues (underlined); Swap4.1 has a higher κ, more similar to wild-type PNt (Table 1). Hydrophobic and nonhydrophobic residues were defined as those that have Z score >0 and <0, respectively, on the hybrid Hopps–Woods/Miyazawa–Jernigan hydropathy scale (see Materials and Methods for details). (C) In Swap5 and Swap6, the distribution of positive (red) and negative (blue) charges was altered to increase κ (more like-charge segregation; Swap5) or decrease κ (more well-mixed charged residues; Swap6). (D) HpC values of wild-type PNt and swap variants compared to a cumulative distribution of HpC values for 10,000 randomly shuffled PNt sequences. Gray shading indicates 1 SD. The HpC value of PCt is shown for comparison.
To test if local clustering of hydrophobic residues can influence the extent to which a polypeptide chain will collapse in the absence of denaturant, we designed a series of swap variants of PNt, in which the amino acid order was locally rearranged by swapping the positions of a small fraction of hydrophobic residues with polar or charged residues in order to increase local clustering of hydrophobic residues (Fig. 2 A and B). The amino acid composition of all swap variants is identical to PNt, so overall hydrophobicity and charge remain the same. We quantified the extent of hydrophobic clustering using a combination of the well-known Hopp–Woods (11) and Miyazawa–Jernigan (12) hydropathy scales. These scales are based on experimental measures of amino acid partitioning from water to organic solvent and pairwise contact probabilities in proteins, respectively. We calculated the extent of hydrophobic clustering as the positive area under the curve in hydropathy plots, using a sliding window of nine amino acids. We define the sum of these areas divided by sequence length for a given sequence as HpC, for hydrophobic clustering (Materials and Methods and Table 1).
Table 1.
Comparison of PNt and swap variant sequence pattern properties and dimensions in 150 mM KCl (Native) or 2 M Gdn (Gdn) measured by SAXS (3)
| No. mutations | HpC* | κ* | Buffer | Rg (Å) | ν | χ2r | |
| PNt | – | 0.0300 | 0.222 | Native | 51.10 ± 0.13 | 0.542 ± 0.002 | 1.095 |
| Gdn | 58.78 ± 0.11 | 0.583 ± 0.001 | 0.982 | ||||
| Swap1 | 6 (1.8%) | 0.0458 | 0.212 | Native | 49.20 ± 0.59 | 0.514 ± 0.009 | 0.904 |
| Gdn | 59.87 ± 0.20 | 0.580 ± 0.002 | 0.966 | ||||
| Swap2 | 18 (5.4%) | 0.0697 | 0.218 | n.d. | n.d. | n.d. | |
| Swap3 | 18 (5.4%) | 0.0445 | 0.200 | Native | 40.58 ± 1.07 | 0.432 ± 0.017 | 0.909 |
| Gdn | 61.26 ± 0.64 | 0.594 ± 0.006 | 1.023 | ||||
| Swap4 | 21 (6.3%) | 0.0329 | 0.170 | Native | 53.37 ± 0.17 | 0.558 ± 0.002 | 1.069 |
| Gdn | 60.19 ± 0.34 | 0.584 ± 0.003 | 0.975 | ||||
| Swap4.1 | 21 (6.3%) | 0.0296 | 0.213 | Native | 54.45 ± 0.14 | 0.545 ± 0.002 | 0.938 |
| Gdn | 58.97 ± 0.23 | 0.580 ± 0.002 | 1.033 | ||||
| Swap5 | 12 (3.6%) | 0.0302 | 0.291 | Native | 48.71 ± 0.34 | 0.498 ± 0.005 | 0.924 |
| Gdn | 59.95 ± 0.38 | 0.584 ± 0.004 | 1.015 | ||||
| Swap6 | 12 (3.6%) | 0.0694 | 0.137 | Native | 52.61 ± 0.27 | 0.562 ± 0.003 | 1.067 |
| Gdn | 59.25 ± 0.50 | 0.583 ± 0.005 | 0.934 |
See Materials and Methods for explanations of the HpC and κ calculations. n.d., not determined.
Initially, we designed four swap variants (Swap1 to Swap4) to sample a wide range of possible HpC values. Swap1 and Swap3 increase HpC to similar extents but achieve this by rearranging the positions of different sets of residues (6 residues for Swap1, HpC = 0.0458; 18 residues for Swap3, HpC = 0.0445; wild-type PNt, HpC = 0.0300). Swap2 is a more extreme version of Swap1, rearranging 18 residues to yield the highest HpC value tested, 0.0697 (Fig. 2D and Table 1). Swap4 (HpC = 0.0329) serves as a negative control, swapping a similar number of residues as Swap2 and Swap3 but retaining a similar extent of hydrophobic clustering as wild-type PNt.
In some swap variants, clustering hydrophobic residues also changed patterns of charged residues. In highly charged IDPs (>35% charged residues), the distribution of charges along an amino acid chain can influence the conformational ensemble, with larger blocks of like-charges (a concept referred to as “high κ”) leading to collapse (13–15). However, PNt (15% charged residues) is not nearly as highly charged as typical IDP sequences. It is therefore unclear to what extent charge patterning will affect the conformational ensemble of a minimally charged sequence. To test this explicitly, we designed a modification of Swap4 with a charge mixing pattern more similar to PNt (Swap4.1; Fig. 2B, Table 1, and SI Appendix, Fig. S1) and two additional swap variants to test the extent to which altered charge mixing alone affects chain collapse. Swap5 and Swap6 increase (segregate like-charges) and decrease (leading to more mixed charges), respectively, like-charge blocks as much as possible while preserving the remainder of the wild-type PNt sequence pattern, including hydrophobic clustering (Fig. 2C).
All swap variants were expressed and purified from Escherichia coli. After dialysis against 50 mM Tris, pH 7.5, each variant, with the exception of Swap2, was monomeric as judged by size-exclusion chromatography (SEC; SI Appendix, Fig. S2). Swap2 eluted as a monodisperse but multimeric soluble assembly; we were unable to identify conditions where Swap2 behaved as a monomer. As reported previously for wild-type PNt, none of the monomeric swap variants possess regular secondary structure nor a cooperative thermal unfolding transition as measured by far-UV circular dichroism (CD) spectroscopy (SI Appendix, Figs. S3 and S4).
Subtle Changes in Residue Order Affect Polypeptide Chain Dimensions.
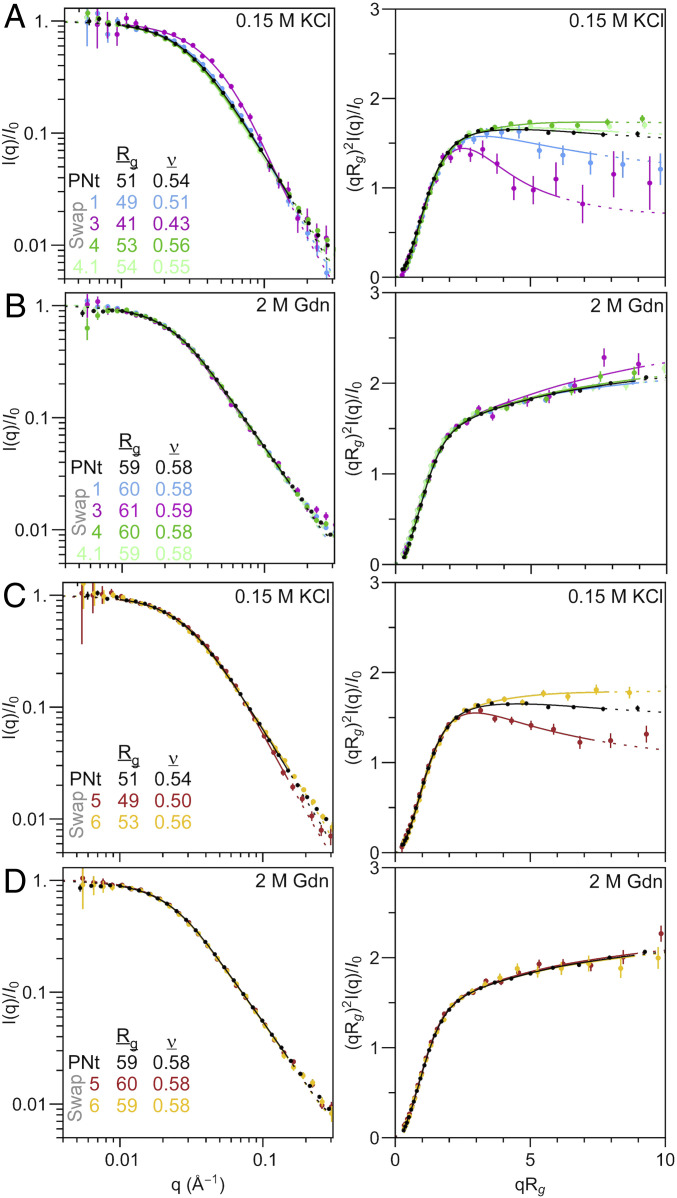
We used SAXS to measure the overall size and shape of the conformational ensemble of each monomeric swap variant (Fig. 3). To ensure that each variant was analyzed as a monodisperse population, each sample was subjected to inline SEC immediately before entering the X-ray beam, as described previously (3). Scattering profiles were analyzed using our molecular form factor (MFF) designed for disordered heteropolymers, to evaluate the overall dimensions (root mean squared radius of gyration, Rg) and solvent quality (Flory exponent, ν), using the scaling relationship Rg = RoNν (3). Under denaturing conditions (2 M guanidine hydrochloride, Gdn), all variants behave at the self-avoiding random walk limit (ν ∼ 0.6), exhibiting the characteristic positive slope in the scattering curve at qRg > 3 in the dimensionless Kratky plots (Fig. 3 B and D). Under native conditions, a range of more collapsed to more expanded ensembles was observed for the swap variants as compared to wild-type PNt. Of note, variants that increase hydrophobic clustering (Swap1 and Swap3) adopted more collapsed ensembles compared to PNt; for Swap1, Rg and ν decreased by 2 Å and 0.03, respectively, relative to wild-type PNt, while for Swap3, the changes were 10 Å and 0.11 (Fig. 3A). The negative control variant Swap4, however, was slightly more expanded (Rg and ν increased by 2 Å and 0.02, respectively; Fig. 3A).
Fig. 3.
Changes in the amino acid pattern alter the conformational ensemble of PNt. Log-log and dimensionless Kratky plots from analysis of SAXS profiles of PNt and swap variants with (A and B) altered hydrophobic clustering or (C and D) altered charge patterning in (A and B) 0 M or (B and D) 2 M Gdn. Rg and ν were extracted from the fit to the MFF (3).
We hypothesized that the slightly more expanded conformational ensemble of Swap4 might be caused by its more highly mixed pattern of charged residues. However, Swap4.1, which preserves the hydrophobic pattern of Swap4 but has a like-charge clustering pattern more similar to wild-type PNt, has a similar expansion as Swap4 (Fig. 3 A and B and Table 1). Yet, charge patterning did have some capacity to affect PNt conformational ensembles, as Swap5 and Swap6, which alter only charged residue pattern in the wild-type PNt background, affected chain collapse in a manner more reminiscent to charge pattern effects in highly charged IDP sequences (13). Specifically, Swap5, with more like-charge segregation, adopts a more collapsed conformational ensemble; the Rg and ν decreased by 2 Å and 0.04, respectively. In contrast, Swap6 with its more highly mixed charge pattern is more expanded compared to PNt (Rg and ν increased by 2 Å and 0.02; Fig. 3C). These variants highlight the complex nature of sequence patterning: polypeptide dimensions are sensitive to charge patterning even when the hydrophobic pattern was held constant, and vice versa. Overall, we found a moderate trend between local amino acid sequence patterns and the extent of polypeptide chain collapse, consistent also with the extent of collapse as measured by SAXS for other IDPs (SI Appendix, Fig. S5).
Amino Acid Sequence Patterns Increase Aggregation Propensity.
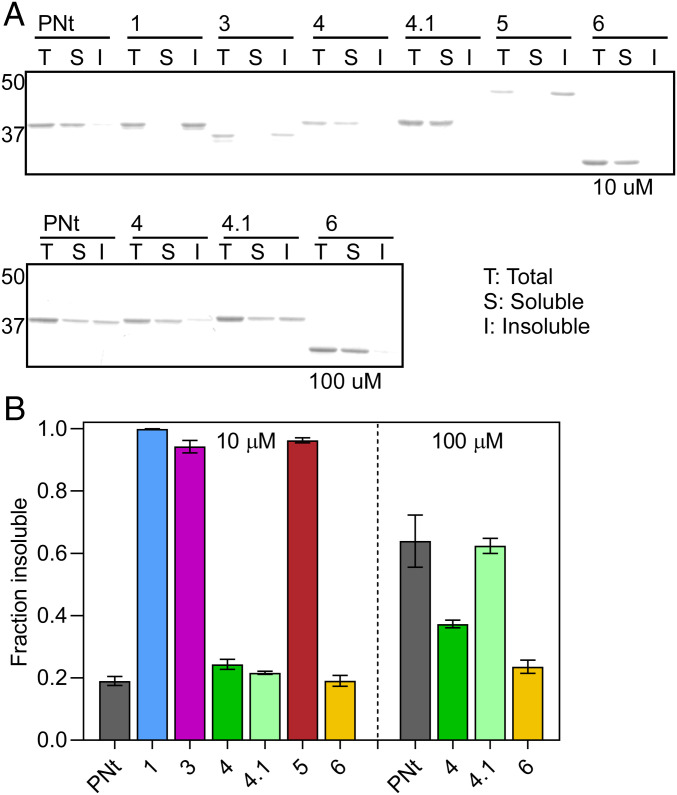
We reasoned that the same forces that lead to intramolecular collapse in swap variants could also lead to intermolecular associations (i.e., aggregation), as noted for IDPs (16). To test this hypothesis, swap variants were incubated overnight under conditions that favor aggregation of foldable proteins (high concentration and/or high temperature; Materials and Methods). The insoluble and soluble fractions were separated by centrifugation, and the fractions of swap variant in the supernatant and pellet were quantified. Each of the more collapsed variants (Swap1, Swap3, and Swap5) was significantly more aggregation prone than PNt (Fig. 4). In contrast, the aggregation propensities of the more expanded variants (Swap4, Swap4.1, and Swap6) were very similar to wild-type PNt. These results indicate that increasing HpC or segregating like charges can stabilize both intramolecular and intermolecular chain interactions. This model is consistent with the behavior of Swap2, which had the highest HpC and formed soluble oligomers (SI Appendix, Fig. S2) under every condition tested (see above).
Fig. 4.
Collapsed swap variants are more prone to aggregation. (A) Proteins were incubated at 37 °C for 16 to 20 h at either 10 or 100 μM. Soluble and insoluble (aggregated) fractions were separated by centrifugation and subjected to SDS/PAGE. Gel mobility differences for some swap variants are attributed to the altered distributions of charged residues. The actual molecular weight of each construct was confirmed by intact mass spectrometry. (B) Quantification of SDS/PAGE bands from A. Each data point represents three replicates; error bars represent the SEM.
Expanded PNt Conformations Facilitate Efficient Pertactin Secretion to the Bacterial Cell Surface.
Pertactin and other members of the Type Va secretion family of virulence proteins are called AT proteins because their secretion to the cell surface is largely autonomous (6). This minimal reliance on other host proteins means that pertactin and many other AT proteins are efficiently secreted to the bacterial cell surface upon expression in standard laboratory strains of E. coli. However, we and others have shown that in order for an AT passenger to be efficiently secreted across the outer OM, it must remain unfolded in the periplasm (7, 9, 10, 17). Further, the N-terminal PNt portion is the first part of the pertactin passenger to enter the periplasm but last to exit, implying that it must maintain an unfolded conformation longer than the passenger C terminus (PCt; Fig. 1). We hypothesized that the expanded conformational ensemble of PNt might prevent premature collapse, folding, and/or aggregation of the pertactin passenger in the periplasm, thereby facilitating efficient secretion of the passenger to the cell surface.
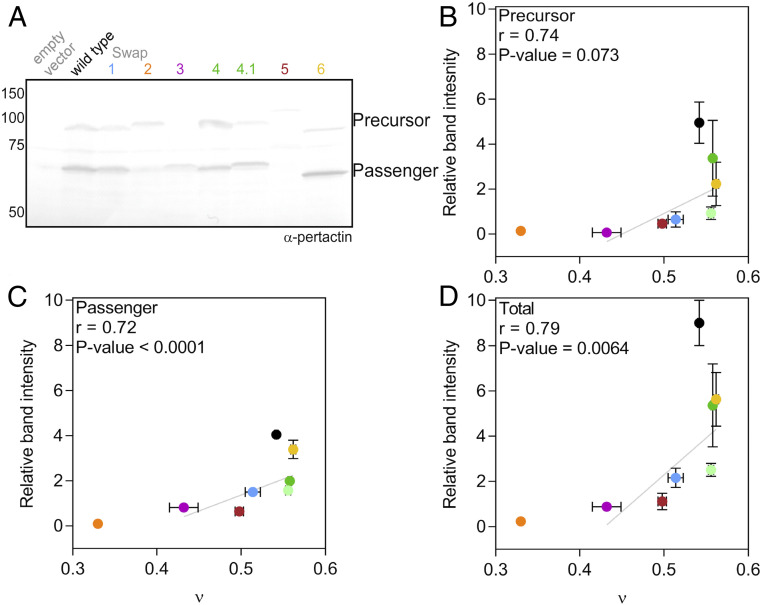
To test this hypothesis, we introduced the swap mutations into the full-length pertactin coding sequence and monitored secretion of the resulting pertactin variants in E. coli, as described previously (7, 8, 17). Amounts of the longer pertactin precursor protein, relative to the postsecretion, cleaved passenger were quantified from Western blots of whole-cell lysates following our established procedures (8, 17) (Fig. 5). We and others have shown that AT precursor cleavage to create the mature passenger occurs only after translocation of the passenger across the OM; passenger cleavage is therefore diagnostic for secretion of the AT passenger to the cell surface (7, 17, 18).
Fig. 5.
Effects of PNt contraction on pertactin secretion in vivo. (A) Secretion of wild-type pertactin and swap variant chimeras, monitored by the intramolecular cleavage of the pertactin precursor (93 kDa apparent MW) to the smaller mature passenger (69 kDa apparent MW) in whole-cell lysate samples, detected using an anti-pertactin passenger antibody. Gel mobility differences for some swap variants are attributed to the altered distributions of charged residues. (B–D) Quantification of (B) precursor, (C) passenger, and (D) total pertactin accumulated (precursor + passenger) as a function of ν. Vertical error bars represent SEM for three replicates; bars not visible are smaller than the data point. Horizontal error bars are the error in the MFF fit (Fig. 2 and refs. 3 and 5). Swap2, which formed soluble oligomers under all conditions tested, is displayed with an estimated ν of 0.33 and was excluded from the statistical analysis. The wild-type protein was also excluded from the correlation analysis due to the separate influence of its folding on secretion efficiency (see fourth paragraph of text section titled Expanded PNt Conformations Facilitate Efficient Pertactin Secretion to the Bacterial Cell Surface). The P values were calculated by parametric bootstrap (Materials and Methods).
We observed a positive correlation between the Flory exponent, ν, and the amount of cleaved pertactin passenger detected for the monomeric swap variants (Pearson correlation coefficient, r = 0.72; P < 0.0001; Fig. 5C), suggesting more collapsed conformational ensembles are less efficiently secreted to the bacterial cell surface than variants with more expanded disordered states, regardless of the specific sequence pattern changes. This result is consistent with studies demonstrating the sensitivity of AT translocation across the OM to the folded status of the passenger while it resides in the periplasm (7–10).
Note, however, that even the most expanded swap variants were secreted less efficiently than wild-type pertactin. This result is expected, as we have previously shown that folding of the passenger to its β-helical structure at the cell surface is a driving force for efficient translocation of pertactin across the OM (8, 17), and in contrast to the wild-type passenger sequence, none of the constructs bearing a swap variant PNt region can stably fold to the pertactin passenger β-helix structure (SI Appendix, Fig. S3). Because wild-type pertactin has the extra driving force of folding to facilitate its secretion, we restricted our comparison of secretion efficiency above to the monomeric swap variants in order to more directly examine the effects of collapse, independent of the influence of folding.
We also observed a positive correlation between ν and the total amount of pertactin detected in vivo (cleaved passenger plus uncleaved precursor) (r = 0.79; P = 0.0064; Fig. 5 B and D). This correlation is unlikely to be due to differences in gene expression, as we observed no significant difference in RNA levels (SI Appendix, Fig. S6), nor have we previously observed expression differences for a wide variety of pertactin variants (7, 8, 17, 19). Differences in the accumulation of the swap variants in vivo are therefore more likely due to increased degradation of more collapsed PNt swap variants. The higher level of hydrophobic clustering in more collapsed swap variants would be expected to lead to enhanced recognition by cellular protein quality control systems, many of which recognize exposed hydrophobic surface area (Fig. 1).
Note that there was no significant correlation between the secreted passenger fraction (passenger divided by total) and ν (SI Appendix, Fig. S7). This result is not surprising, as the extent of degradation in periplasm will likely be affected by the rate of OM translocation. In other words, if kXOM is slow, the precursor will spend more time in the periplasm, leading to more degradation, reducing both numerator and denominator of the fraction secreted (Fig. 1). Therefore, both degradation of the precursor in the periplasm and impaired translocation across the OM are consistent with more collapsed conformations adversely affecting pertactin secretion.
We also explored the alternative hypothesis that swap variants do not disrupt secretion but instead disrupt interactions between the pertactin passenger and the cell surface after secretion, leading to release of the passenger into the growth media. To test this, we measured pertactin passenger accumulation in the spent media. For wild-type pertactin, a very small amount of passenger was detected in the spent media (SI Appendix, Fig. S8). In contrast, no passenger was detected in the spent media for any of the swap chimeras, indicating they disrupt a step prior to secretion of the passenger across the OM. Together, these results support a model where collapse of the pertactin passenger sequence in the periplasm impairs its subsequent secretion to the cell surface, either by a direct blockade of secretion, which leads to precursor degradation in the periplasm, or more rapid degradation in the periplasm, which leads to fewer precursor proteins available for OM translocation (Fig. 1).
Sequence Patterns Leading to Chain Expansion Are Enriched in AT Passengers and Other Water-Soluble Proteins.
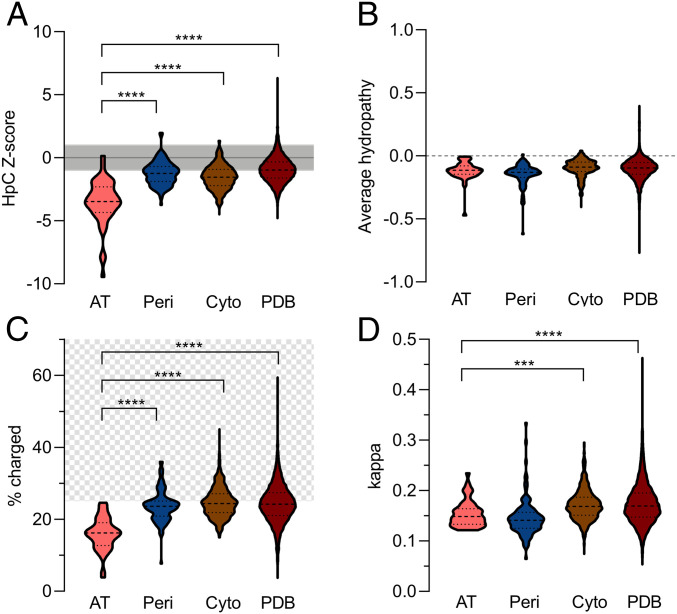
The results presented above led us to hypothesize that AT passenger sequence patterns may be under selection to have a low value of HpC and/or κ in order to enhance secretion efficiency. Consistent with this hypothesis, we noticed that compared to a distribution of 10,000 randomly swapped pertactin passenger sequences, the HpC value for wild-type PNt is nearly 3 SDs below the mean (Z score = −2.84) (Fig. 2D). We detected a similarly low HpC for most AT passengers (Fig. 6A). More broadly, the average Z score for three other protein datasets (E. coli cytoplasmic and periplasmic proteins and a nonredundant PDB dataset of water-soluble proteins; Materials and Methods) was approximately 1 SD below the mean of the randomized distribution (Fig. 6A). These results suggest that amino acid sequences of water-soluble, folded proteins may be broadly under selection in order to avoid hydrophobic clustering, with AT passenger domain sequences under especially stringent selection for low HpC, perhaps to ensure maintenance of an expanded conformation for the passenger during its time in the periplasm (Fig. 1). It is important to note that the average hydropathy (calculated as the average amino acid MJHW Z score for all residues in each sequence) of all sequences analyzed is close to indistinguishable (Fig. 6B), indicating that the strikingly low hydrophobic clustering observed for AT passengers is not due to lower overall hydrophobicity.
Fig. 6.
Properties of amino acid sequences for AT passengers (pink) (20), E. coli periplasmic (blue) or cytoplasmic (brown) proteins (21), and water-soluble proteins from the PDB (red). (A) HpC Z score is the number of SDs of an HpC value for a given naturally occurring protein sequence from the mean of 10,000 randomly shuffled sequences with the same amino acid composition. All protein datasets have a mean Z score well below zero; gray shading indicates 1 SD. Z scores for AT passengers are significantly lower than the other three datasets (Mann–Whitney–Wilcoxon test-based ***P < 0.001; ****P < 0.0001). (B) Average sequence hydropathy, calculated as the average MJHW mean Z score value (SI Appendix, Table S1; Materials and Methods) for all residues in a sequence. While the hydropathy of AT passenger sequences is detectably lower than cytoplasmic protein sequences (P = 0.0262), there is no significant difference between AT passengers and either periplasmic proteins or water-soluble proteins in the PDB. (C) Percent of charged amino acids is significantly lower for AT passengers than both periplasmic and cytoplasmic proteins (P values as for A). Hatched region (>25% charged residues) indicates sequences where κ is predicted to influence polypeptide conformation (13, 14, 22). (D) Autotransporter passengers have significantly lower values of κ than E. coli cytoplasmic proteins and water-soluble proteins in the PDB; P values as for A.
We also investigated the number and distribution of charged residues in amino acid sequences across these protein datasets. We found that AT passenger sequences in general have a significantly lower percentage of charged residues than cytoplasmic and periplasmic proteins (Fig. 6C), despite the frequently observed enrichment of charged residues typically associated with disordered proteins (1, 2, 14) and a prior report suggesting negatively charged residues enhance AT passenger secretion (23). The sparsity of charged residues in AT passengers makes their ability to remain unfolded as they pass through the periplasm even more remarkable, but it is also unclear to what extent charged residue distribution will be predictive of AT protein conformational ensembles, as highlighted by our results with Swap4.1 (Fig. 3A). Indeed, despite the differences we observed in conformational ensemble expansion for our charge-swap PNt variants (Swap5 and Swap6; Fig. 3), we found no significant difference in clustering of like-charge residues between AT passengers and proteins that fold within the periplasm (Fig. 6D). Taken together, the low fraction of charged residues in AT passengers and the modest effects of charge pattern on collapse suggest that the preferred evolutionary path to maintaining an expanded conformational ensemble may be to avoid hydrophobic clustering, rather than specific charge patterns.
Discussion
We and others have shown previously that amino acid sequences typical of foldable proteins adopt highly expanded conformational ensembles under physiological conditions (3, 5, 24, 25). Here we have demonstrated that even subtle changes to sequence patterns, particularly those involving hydrophobic residues, are sufficient to perturb the extent of collapse in one such sequence, PNt. Even a modest increase in local clustering of hydrophobic groups, corresponding to the rearrangement of only six residues (<2% of the 334 AA sequence), yielded a more collapsed PNt conformational ensemble. This suggests that although the PNt sequence composition is sufficiently hydrophobic to engender collapse, its well-mixed sequence pattern results in an expanded conformation.
Crucially, swap variants with more collapsed ensembles disrupted secretion of full-length pertactin in vivo. We found that unusually well-mixed sequence patterns are a common feature across the large and diverse family of AT passenger sequences, despite their low sequence identity. This observation points to a layer of evolutionary selection to stabilize an expanded conformational ensemble and is consistent with studies showing that formation of stable passenger structure in the periplasm impedes transport of the passenger across the OM (7–10). More broadly, we found evidence for selection for unusually well-mixed hydrophobic residues across the amino acid sequences of water-soluble proteins in general. Similar selective pressures may govern the conformational ensembles of intrinsically disordered polypeptides (16, 26).
These results suggest that, particularly within the crowded environment of the cell, protein disorder is not merely the absence of selection for folding but a specific property under selection to avoid aggregation or other inappropriate intermolecular interactions, including those that lead to degradation. We propose that protein sequence patterns are under selection to regulate the collapse properties of the denatured ensemble, balancing (for folded proteins) selection for folding to a stable structure along with selection to avoid misfolding and aggregation. Because the formation of nonnative hydrophobic clusters would stabilize the unfolded state and therefore decrease the net stability of the native state, reducing residual, nonnative structure in the unfolded state generally should increase protein stability while also increasing cooperativity. Hence, selection for well-mixed sequences has multiple benefits.
The bias to be well mixed also influences the allowed folds of naturally occurring proteins. Such structures should not have stretches of hydrophobic residues, for example, in the form of fully buried strands or helices. Consistent with this conjecture, α-helices and β-sheet secondary structures in water-soluble proteins are often amphiphilic (27) and therefore well mixed. Selection for protein sequences that are well mixed may therefore place significant constraints on possible protein architectures, emphasizing those that are compatible with both an expanded unfolded state and a stably folded structure.
By having a well-mixed sequence pattern, water acts as a good solvent for PNt (ν > 0.5), with solvent–protein interactions outweighing protein–protein interactions. This observation appears applicable to many water-soluble proteins, implying that by itself, the hydrophobic effect is insufficient to stabilize the folded structure of a protein. Folding must therefore be due to additional factors, such as improved van der Waals packing, hydrogen bonding, and/or electrostatics, as compared to their values in the unfolded state. Although these findings could be perceived as surprising in light of the long-held notion that folding must begin with a hydrophobic collapse, the preponderance of results now indicates that water typically serves as a good solvent for polypeptide chains, leading to expanded unfolded states. This behavior is consistent with well-known properties of proteins, including their marginal stability, high degree of folding cooperativity, and the presence of a high-energy barrier separating the unfolded and folded states (28).
Materials and Methods
Plasmid Design, Expression, and Purification.
E. coli optimized DNA sequences encoding full-length pertactin (P.93v2) and the pertactin passenger β-helix region alone (P.69Tv2) were cloned into plasmids pET-21b (Invitrogen) and pQE-2 (Qiagen). Pntv2 was constructed by inserting a stop codon starting at nucleotide position 1006 of P.69tv2 in pET21b (29). Swap mutations were introduced to PNtv2 by megaprimer PCR (30) using gBlock (Integrated DNA Technologies) megaprimers. The swap mutations were introduced into P.69tv2 and P.93v2 by megaprimer PCR (30), using megaprimers amplified from PNtv2-Swap.
Plasmids encoding PNt, swap variants, and Swap-PCt composite variants were used to transform E. coli BL21(DE3)pLysS. Proteins were expressed in E. coli and purified from inclusion bodies as previously described (3, 8, 31). Briefly, LB-Amp media was inoculated with a 1:50 dilution of a fresh, saturated overnight culture and shaken at 37 °C to OD600 = 0.35 to 0.45. Expression was induced with the addition of 500 μM IPTG. Cells were grown for 2 h at 37 °C before pelleting and resuspension in sonication buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10 mM benzamidine). Resuspended cells were stored at −80 °C. Cells were lysed by three cycles of freeze/thaw, addition of 1 mg/mL lysozyme (Sigma), and sonication. Inclusion bodies were isolated by centrifugation of the cell lysate and solubilized of the resulting pellet in 50 mM Tris, pH 7.5, plus 6 M Gdn. Denaturant was removed from solubilized inclusion bodies first by diluting to 1.2 M Gdn by slow addition of cold 50 mM Tris, pH 7.5. For all variants except Swap3-PCt, the protein containing solution was dialyzed against 10 L of cold 50 mM Tris, pH 7.5, until the concentration of Gdn was less than 1 mM. The dialyzed solution was centrifuged for 20 min at 15,000 × g and filtered through a 0.22-μm membrane. Solubilized Swap3-PCt was subjected to dropwise refolding from 6 M Gdn to 5 L of cold 50 mM Tris, pH 7.5, and allowed to refold for 72 h before filtration through a 0.22-μM membrane.
PNt, swap, and swap-PCt variants were first purified by strong anion exchange chromatography (Source 15Q; GE Healthcare) and eluted with a 0 to 150 mM NaCl gradient in 50 mM Tris, pH 7.5. The elution peak was pooled and immediately supplemented with 5 mM EDTA. The protein was concentrated (100 to 200 μM) using a 10-kDa molecular weight cutoff centrifugal concentrator (Millipore). At times, this concentration step triggered protein aggregation (especially for the more aggregation-prone variants). If this occurred, Gdn was added from an 8 M stock in 50 mM Tris, pH 7.5, until the aggregates were solubilized. Concentrated protein samples were filtered through a 0.22-μm membrane and further purified using SEC. SEC of PNt and swap variants was carried out in 50 mM Tris, pH 7.5, containing 5 mM EDTA. Monomeric PNt and its variants eluted at ∼60 mL (HiLoad 16/60 Superdex S200; GE Healthcare). The monomer peak was pooled and concentrated to 100 to 200 μM and stored at 4 °C.
Sequence Property Calculations.
Hydropathy was calculated using a hybrid Miyazawa–Jernigan–Hopp–Woods (MJHW) scale (SI Appendix, Table S1). The MJHW scale is the mean Z score of the Hopp–Woods (11) and Miyazawa–Jernigan scales (12), each of which captures a distinct aspect of hydrophobicity expected to impact polypeptide chain collapse (Results). Among several scales tested, MJHW was most predictive of the extent of collapse [as measured by ν, the Flory exponent in the expression Rg = RoNν (3)] of PNt Swap variants. This approach is similar to the development of other hydrophobicity scales. For example, the Hopp–Woods scale was originally adapted from the Levitt-adjusted Tanford hydropathy scale (32, 33) as the scale most predictive of antibody binding sites in proteins (11).
Local clustering of hydrophobic amino acid residues was calculated as the mean hydropathy using the MJHW scale for sliding windows of nine amino acids. Local hydrophobic clustering (HpC) was quantified as a sum of the positive area under the curve (+AUC) in a plot of these sliding window MJHW hydropathy values versus sequence position. For the purposes of visual interpretation only (Fig. 2), plots of hydropathy versus sequence position were smoothed using a Gaussian function with σ = 5 (SI Appendix, Fig. S9). HpC was normalized for chain length by dividing by the number of windows (N):
To generate the cumulative distribution plot of PNt and swap variants, 10,000 randomly shuffled sequences with the amino acid composition of PNt were generated using the “random” package in Python, and the HpC value was calculated for each shuffled sequence. The distribution of HpC values for the PNt shuffled sequences were plotted as a cumulative distribution curve.
Charge patterns, expressed as κ, were calculated using the Classification of Intrinsically Disordered Ensemble Regions (CIDER) (22) webserver at pappulab.wustl.edu/CIDER/ or the module “SequenceParameters” from the Python package “localcider,” available at pappulab.github.io/localCIDER/. In highly charged IDP sequences, lower values of κ are associated with more expanded ensembles (13).
Small-Angle X-Ray Scattering.
Scattering profiles were collected as previously described (3) at the Argonne National Laboratory Advanced Photon Source BioCAT beamline. Purified and concentrated protein in 50 mM Tris, pH 7.5, 5 mM EDTA, 2 to 4 M Gdn was injected onto an S200 gel filtration column equilibrated in 20 mM Hepes, pH 7.5, and 150 mM KCl (native conditions) or 2 M Gdn before entering the X-ray beamline. Scattering profiles were analyzed using the MFF webserver available at sosnick.uchicago.edu/SAXSonIDPs as described previously (3).
Aggregation Propensity Assay.
Purified PNt or swap variant (in 50 mM Tris, pH 7.5, 5 mM EDTA) was diluted to the desired test concentration in 50 mM Tris, pH 7.5, 5 mM EDTA, 150 mM KCl to a final volume of 50 µL in 1.5 mL microcentrifuge tubes. Samples were incubated at 30 °C for 16 to 20 h, then centrifuged at 21,130 × g for 15 min. The supernatant was carefully decanted and placed in a clean microcentrifuge tube. The supernatant and pellet fractions were supplemented with 25 µL of 3× SDS loading dye or 75 µL of 1× SDS loading dye, respectively. Samples were boiled for at least 20 min or until the pellets had solubilized. The supernatant (soluble) and pellet (insoluble) fractions were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. The percent insoluble was quantified using ImageJ using the following equation:
where I is band intensity.
Far-UV CD Spectrophotometry.
Purified PNt and Swap variants were diluted at least 10-fold into 50 mM sodium phosphate, pH 8, to a final concentration of 1 to 5 μM. Far-UV CD spectra were collected using a Jasco J-815 spectropolarimeter at 20 °C. Data points from three spectra (190 to 250 nm) were averaged and buffer subtracted using a buffer only spectrum.
Pertactin Secretion Assay.
P.93v2Swap coding sequences were expressed in E. coli BL21(DE3)pLysS as described previously (7, 8, 17), with minor differences. Briefly, cells were grown at 30 °C in a 12-well plate; optical density at 600 nm was measured using an H1 Synergy plate reader. Cultures consisting of 1.5 mL of LB media supplemented with 100 μg/mL ampicillin, 35 μg/mL chloramphenicol, and 0.1% wt/vol glucose inoculated with fresh overnight culture to a starting OD600 = 0.08 were grown at 30 °C with shaking for 3.5 h to an OD600 of ∼0.3 and induced with 500 μM IPTG. Cultures were grown for three additional hours at 30 °C with shaking.
An aliquot of each culture containing the same number of cells as in 1 mL of culture at an OD600 = 0.5 was removed and centrifuged at 4 °C at 21,130 × g for 5 min. The supernatant was decanted, and the cell pellet was resuspended in 50 μL of 1× SDS loading dye, supplemented with 14 mM BME, and boiled for 20 min. These whole-cell lysate samples were separated by SDS/PAGE and visualized by Western blotting using an anti-pertactin polyclonal antibody (7). The precursor and passenger appear at apparent molecular weights of 93 and 69 kDa, respectively. ImageJ was used to quantify the intensity (I) of the bands.
The significance of the observed correlation coefficients was calculated based on parametric bootstrap. For each mutant variant i, the mean and SD of ƒi and νi are known. Random samples of ƒi and νi are then generated according to normal distributions with the corresponding mean and SD. The drawn samples are then used to calculate one sample correlation coefficient. The procedure is repeated 10,000 times to get an empirical distribution of the correlation coefficient, from which a significant test or confidence interval could be derived. Under 5% critical level, significant positive correlation coefficients are observed for passenger, precursor, and total.
mRNA Quantification.
E. coli were grown as described above. After 3 h of induction with 500 μM IPTG, an aliquot of cells was diluted to OD600 = 0.3, and 500 μL of diluted cells was added to 1,000 μL of RNAprotect Bacteria Reagent (Qiagen). RNA was extracted and purified using the RNeasy Mini Kit (Qiagen). RNA concentration was determined by absorbance at 260 nm, and quality was assessed using an Agilent 2100 Bioanalyzer. All RNA integrity numbers (RIN) were >8.7. Residual DNA (genomic and plasmid) was degraded with RNase-free DNase (Invitrogen). cDNA was generated using the iScript RT kit (BioRad) with the random hexamer primers provided. A control reaction lacking reverse transcriptase was included for all mutants to ensure cDNA amplification of mRNA only. Primers used for cDNA amplification were as follows: PNt and swap variants (forward primer, GGCAGGTAGCGGTCTGTTTC; reverse primer, AACCCACAGACGATGCTGAC) and GAPDH reference gene (forward primer, CGGTACCGTTGAAGTGAAAGAC; reverse primer, ACCAGTTGCTTCAGCGAC). Amplification was performed on a CFX96 Touch Real-Time PCR Detection System (BioRad) using SsoAdvanced SYBR Green Supermix (BioRad) containing either the PNt/Swap or GAPDH primers. Mean fold change was calculated using the 2−ΔΔCT method (34). All qPCR reactions were repeated in biological triplicate, and each biological replicate was analyzed in technical triplicate.
Bioinformatic Analysis of Protein Sequence Properties.
A set of 44 characterized AT gene sequences, including pertactin, was retrieved (see table S1 of ref. 20); three sequences originally included here (GenBank nucleotide accession numbers YP_003516849.1, YP_049087.1, and AP_003226.1) were not included in this study because these sequences no longer return a corresponding protein record in the National Center for Biotechnology Information (NCBI). Each GenBank protein accession number was mapped to its corresponding amino acid sequence. In order to remove contributions from the N-terminal signal sequence to the sequence pattern analysis, the first 60 amino acids were omitted for each sequence. Likewise, the last 300 amino acids were also omitted to remove contributions from the C-terminal transmembrane β-barrel to the AT passenger analysis.
A nonredundant set of PDB IDs were downloaded from NCBI Vector Alignment Search Tool using the nonredundancy P value of 10e-7, resulting in 14,470 PDB IDs. These IDs were mapped to their amino acid sequences; sequences with nonamino acid designations were removed from the dataset. The dataset was trimmed further to remove sequences that were less than 70 amino acids in length and entries that matched the word “membrane” in the sequence description. The resulting water-soluble PDB dataset included 11,894 proteins.
The E. coli cytoplasmic and periplasmic sets of proteins were retrieved from published datasets (607 and 113 proteins, respectively; see ref. 21 and SI Appendix, Table S13). Gene names were mapped to their corresponding E. coli mRNA sequences using an E. coli annotated genome dataset downloaded from ecogene.org (based on ref. 35; now temporarily unavailable) and a custom Python script. All mRNA sequences were translated to their amino acid sequence using Python. The first 60 amino acids were trimmed from the periplasmic proteins to remove signal sequence contributions. Average hydropathy, HpC, and kappa were calculated for all proteins as described above. Percent charge was calculated by counting the number of glutamate (E), aspartate (D), arginine (R), and lysine (K) residues in a given protein sequence, dividing by the total number of residues in that sequence, and multiplying by 100.
To generate HpC distributions, 10,000 shuffled amino acid sequences were generated for each protein across all datasets by randomly shuffling amino acid positions using the “random” package in Python. The mean, SD, and Z score were calculated for each HpC distribution, with Z scores calculated as
Supplementary Material
Acknowledgments
We thank Srinivas Chakravarthy for assistance with the SAXS measurements and Gina Jozwiak for assistance with RNA quantification and protein purification. We thank the members of our groups, K. Plaxco, and other colleagues for stimulating discussions over the past 2 decades. This work was supported by grants from the NIH (GM130122 [T.R.S. and P.L.C.] and GM055694 [T.R.S.]), the W. M. Keck Foundation (P.L.C.), and the NSF (MCB 1516959 [C. R. Matthews]). Use of the Advanced Photon Source, an Office of Science User Facility, operated for the Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the DOE under Contract DEAC02-06CH11357.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003773117/-/DCSupplemental.
Data Availability.
All raw data analyzed in this study are publicly available at the following repository: https://github.com/sosnicklab/SAXSonIDPs/tree/master/Properties_of_unfolded_states_of_proteins_suggest_broad_selection_for_expanded_conformational_ensembles.
References
- 1.van der Lee R. et al., Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uversky V. N., Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 1834, 932–951 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Riback J. A. et al., Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water. Science 358, 238–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riback J. A. et al., Response to Comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 361, eaar7949 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Riback J. A. et al., Commonly used FRET fluorophores promote collapse of an otherwise disordered protein. Proc. Natl. Acad. Sci. U.S.A. 116, 8889–8894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drobnak I. et al., Of linkers and autochaperones: An unambiguous nomenclature to identify common and uncommon themes for autotransporter secretion. Mol. Microbiol. 95, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junker M., Besingi R. N., Clark P. L., Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol. Microbiol. 71, 1323–1332 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Renn J. P., Junker M., Besingi R. N., Braselmann E., Clark P. L., ATP-independent control of autotransporter virulence protein transport via the folding properties of the secreted protein. Chem. Biol. 19, 287–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jong W. S. et al., Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol. Microbiol. 63, 1524–1536 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Leyton D. L. et al., Size and conformation limits to secretion of disulfide-bonded loops in autotransporter proteins. J. Biol. Chem. 286, 42283–42291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopp T. P., Woods K. R., Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U.S.A. 78, 3824–3828 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazawa S., Jernigan R. L., Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 256, 623–644 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Das R. K., Pappu R. V., Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das R. K., Ruff K. M., Pappu R. V., Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 32, 102–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin E. W. et al., Sequence determinants of the conformational properties of an intrinsically disordered protein prior to and upon multisite phosphorylation. J. Am. Chem. Soc. 138, 15323–15335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin E. W. et al., Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drobnak I., Braselmann E., Clark P. L., Multiple driving forces required for efficient secretion of autotransporter virulence proteins. J. Biol. Chem. 290, 10104–10116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnard T. J., Dautin N., Lukacik P., Bernstein H. D., Buchanan S. K., Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat. Struct. Mol. Biol. 14, 1214–1220 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braselmann E., Chaney J. L., Champion M. M., Clark P. L., DegP chaperone suppresses toxic inner membrane translocation intermediates. PLoS One 11, e0162922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celik N. et al., A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One 7, e43245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt A. et al., The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holehouse A. S., Das R. K., Ahad J. N., Richardson M. O., Pappu R. V., CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 112, 16–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang’ethe W., Bernstein H. D., Charge-dependent secretion of an intrinsically disordered protein via the autotransporter pathway. Proc. Natl. Acad. Sci. U.S.A. 110, E4246–E4255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borgia A. et al., Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J. Am. Chem. Soc. 138, 11714–11726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuertes G. et al., Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. U.S.A. 114, E6342–E6351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W., Dignon G., Brown M., Kim Y. C., Mittal J., Hydropathy patterning complements charge patterning to describe conformational preferences of disordered proteins. J. Phys. Chem. Lett. 11, 3408–3415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz R., King J., Frequencies of hydrophobic and hydrophilic runs and alternations in proteins of known structure. Protein Sci. 15, 102–112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark P. L., Plaxco K. W., Sosnick T. R., Water as a good solvent for unfolded proteins: Folding and collapse are fundamentally different. J. Mol. Biol. 432, 2882–2889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C. et al., FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11, 92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazaki K., MEGAWHOP cloning: A method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Methods Enzymol. 498, 399–406 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Junker M. et al., Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitt M., A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 104, 59–107 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Tanford C., Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J. Am. Chem. Soc. 84, 4240–4247 (1962). [Google Scholar]
- 34.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Zhou J., Rudd K. E., EcoGene 3.0. Nucleic Acids Res. 41, D613–D624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data analyzed in this study are publicly available at the following repository: https://github.com/sosnicklab/SAXSonIDPs/tree/master/Properties_of_unfolded_states_of_proteins_suggest_broad_selection_for_expanded_conformational_ensembles.