Significance
Intracellular pathogens utilize specialized mechanisms for replicating within host cells while minimizing cytotoxicity. Misregulation of bacterial virulence factors that are responsible for infection can cause host cell death and the consequent loss of virulence. The results presented here reveal that the translation of a cytolysin that is responsible for host infection is reduced during bacterial growth, due to the formation of an mRNA secondary structure that results in translational inhibition. The coding domain that occludes the ribosome binding site regulates the synthesis and activity of the cytolysin inside the host cytosol, highlighting multiple levels of LLO control from this domain. These findings provide insight into how expression of a virulence factor can be translationally regulated within alternate host cellular compartments.
Keywords: bacteria, cytolysin, pathogenesis, translation
Abstract
Intracellular pathogens are responsible for an enormous amount of worldwide morbidity and mortality, and each has evolved specialized strategies to establish and maintain their replicative niche. Listeria monocytogenes is a facultative intracellular pathogen that secretes a pore-forming cytolysin called listeriolysin O (LLO), which disrupts the phagosomal membrane and, thereby, allows the bacteria access to their replicative niche in the cytosol. Nonsynonymous and synonymous mutations in a PEST-like domain near the LLO N terminus cause enhanced LLO translation during intracellular growth, leading to host cell death and loss of virulence. Here, we explore the mechanism of translational control and show that there is extensive codon restriction within the PEST-encoding region of the LLO messenger RNA (mRNA) (hly). This region has considerable complementarity with the 5′ UTR and is predicted to form an extensive secondary structure that overlaps the ribosome binding site. Analysis of both 5′ UTR and synonymous mutations in the PEST-like domain that are predicted to disrupt the secondary structure resulted in up to a 10,000-fold drop in virulence during mouse infection, while compensatory double mutants restored virulence to WT levels. We showed by dynamic protein radiolabeling that LLO synthesis was growth phase-dependent. These data provide a mechanism to explain how the bacteria regulate translation of LLO to promote translation during starvation in a phagosome while repressing it during growth in the cytosol. These studies also provide a molecular explanation for codon bias at the 5′ end of this essential determinant of pathogenesis.
Intracellular pathogens remain a worldwide public health concern due to the lack of vaccines and their resistance to antimicrobial therapies (1, 2). In order to establish their replicative niche, intracellular pathogens either use secretion systems, which translocate effectors to the cytosol and promote survival within phagosomes or, as is the case of Listeria monocytogenes, produce a pore-forming cytolysin listeriolysin O (LLO) that allows for bacteria to escape the host phagosomal compartment and reach the cytosol where rapid replication ensues (3). LLO is a member of a large family of bacterially encoded toxins called cholesterol-dependent cytolysins (CDCs), but it is unique in that it is the only CDC that is made by an intracellular pathogen, and, thus, its expression and activity must be precisely regulated to promote the intracellular compartmentalization of L. monocytogenes (4). Substitution of LLO with other CDCs results in bacteria that can still escape from a phagosome but subsequently cause cytotoxicity of the host cell due to loss of membrane integrity (5–8), suggesting unique regulatory features that specifically promote the lifecycle of L. monocytogenes. One of these features is a lower optimal pH that maximized pore-forming activity in the acidified phagolysosome, while reducing the cytotoxic effects of cytoplasmic LLO (6, 9–11).
The most distinctive feature of LLO that separates it from other CDCs is contained within its N-terminal PEST-like domain that is essential for L. monocytogenes virulence (5). PEST-like domains were originally described in eukaryotic proteins and were thought to mediate shortened protein half-lives (12, 13); however, it is now appreciated that this region of LLO encodes a polyproline II (PPII) helix, which is involved in intermolecular oligomerization (14). The PEST region also interacts with host proteins of the host endocytic machinery, resulting in intracellular clearance of plasma membrane-associated LLO (15), highlighting its role in the intracellular compartmentalization. Additionally, residues in this region are phosphorylated intracellularly, although the functional consequence of these modifications is unknown (16). While the PEST-like domain is involved in LLO protein regulation, it is also involved in LLO synthesis at the translational level. A synonymous mutation within the domain at S44 (TCT263AGC from the transcription start site) results in increased expression in broth due to elevated translational efficiency, and a 10,000-fold decrease in virulence in mice (17). In this study, we discovered that the messenger RNA (mRNA) transcript of LLO (hly) forms an extensive secondary structure between the PEST-encoding region and the 5' UTR that is responsible for regulating LLO synthesis during growth.
Results
Identification of Nucleotide Complementarity between the hly PEST-Encoding Region and the 5′ UTR.
We wanted to understand how a single synonymous mutation in the coding region of hly could cause such a dramatic virulence defect. Analysis of the codon adaptation index (CAI; a measure of the deviation from codon usage in highly expressed genes) (18) showed that the PEST-encoding sequence has a lower score than hly as a whole (19) (CAI = 0.418 versus 0.534, respectively), but TCT263AGC actually reduced the score of the PEST region (CAI = 0.402), suggesting codon bias does not play a major role. To explore this codon specificity further, we substituted the WT S44 codon with each of the other five serine codons and assessed the effect on protein secretion in broth. To maintain constant transcript levels, the endogenous promoter driving hly transcription was replaced with a constitutive promoter (Phyper) on an integrative plasmid (20) in a Δhly background. Five of the six serine codons resulted in LLO hypersecretion from bacteria grown in broth (Fig. 1B), indicating that the endogenous codon, UCU, is the only codon that supports WT expression. To test if this codon restriction was specific to the S44 codon, 21 noncoding mutations were made at different loci within the PEST-encoding region and many of these led to increased LLO secretion (Fig. 1C).
Fig. 1.
PEST region codon restriction suggests an interaction between the ORF and the 5′ UTR of the mRNA transcript. (A) The LLO PEST-like domain and nucleotides with 5′ complementarity. (B) Representative Western blot and quantification of LLO and P60 proteins from all synonymous S44S mutants. (C) Western blot quantification of LLO/P60 from additional PEST-encoding synonymous mutants in rich media. (D) RNAfold predicted mRNA structures and minimum free energies of WT hly and S44S (UCU263AGC). Red color indicates base-pair probabilities. Error bars represent SDs from three independent replicates. Significance is indicated as compared to WT and was determined by unpaired t test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, NS = not significant (P > 0.05). Unannotated statistical significance in C meant to indicate NS.
We hypothesized that there might be an RNA regulatory element, perhaps a small RNA (sRNA), that could affect translation of LLO. To investigate this possibility, we utilized RNApredator (21) to search for sites of complementarity to the PEST-encoding region of the transcript within the L. monocytogenes genome. This program allows us to treat the input sequence as if it is a sRNA to search for possible targets or regions of complementarity within other genes. Surprisingly, a low probability match was identified within the 5′ UTR of the hly suggesting cis regulation. We used RNAfold (22) to model the structure of the hly transcript starting from the transcription start site to beyond the PEST sequence. The generated structure showed base-pair interactions between the PEST-encoding sequence and the 5′ UTR including the ribosome binding site (RBS) (Fig. 1A), suggesting a potential barrier to the efficient translation initiation of the gene that might result in reduced expression. mRNA folding stability at the 5′ end has been shown to impact gene expression and underlie codon bias in bacteria (23, 24). Folding stabilities of mRNAs are generally estimated by determining the folding free energies of the first L bases (ΔGL,). The predicted ΔG300 of this structure was −53.0 kcal/mol while that of the S44S (TCT263AGC) mRNA structure showed a lower probability interaction at the RBS with a folding free energy of ΔG300 = −49.5 kcal/mol (Fig. 1D). These predictions suggest that hly expression is restricted due to base-pair interactions between the RBS and the PEST-encoding region of the ORF.
Genetic Analysis of the Predicted mRNA Structure.
To interrogate putative interactions between the UTR and the ORF, we performed mutational analysis of the hly mRNA. Our strategy was to introduce synonymous mutations within the PEST-encoding sequence and corresponding compensatory mutations in the 5′ UTR that are predicted to restore the original base pairing. Since the previously characterized S44S mutation is predicted to interact directly with the RBS, we chose to focus on downstream synonymous mutations that should base pair 5′ to the RBS (Fig. 2A). P46 and A47 were selected, and synonymous mutations were made to generate P46P and A47A (T271A and A274T, respectively). Compensatory mutations were also constructed (UTRP46P and UTRA47A). Individual mutations in the 5′ UTR or synonymous mutations in the PEST region led to an increase in protein secretion; however, when combined with their specific compensatory mutation, protein levels were restored to WT levels (Fig. 2B). Importantly, UTR compensatory mutations only complemented the ORF nucleotide to which they were predicted to interact. Transcript levels measured by RT-qPCR showed that RNA abundance did not significantly change between the mutants (Fig. 2C), indicating that the basis of the difference in protein levels was most likely due exclusively to changes in translation efficiency and not RNA stability.
Fig. 2.
Genetic analysis of the predicted mRNA structure. (A) Close-up diagram of the mRNA region to be analyzed. The mutated nucleotides were chosen based on proximity to the RBS and the codon capacity for mutations. (B) Western blot quantification of LLO/P60 from mutants expressing LLO from the Phyper promoter and grown in rich media. (C) mRNA transcript relative quantities (RQ) as a percent of WT. (D) Western blot quantification from mutants P46P* (U271G) and UTRP46P* (A116C). (E) mRNA transcript relative quantities from GC mutants. Error bars represent SDs from three independent replicates. Unless otherwise indicated, significance was determined with respect to WT. Significance was determined by unpaired t test. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001, NS = not significant (P > 0.05).
We also assessed LLO expression of an additional set of synonymous mutants, P46P* and A47A* (T271G and A274C, respectively). These mutants change the GC content of hly and can presumably result in mRNAs with different folding free energies. The LLO expression profile of these strains behaved similarly (Fig. 2D). Although the mRNA abundance of the P46P* mutant was consistently elevated compared to WT, this was not statistically significant and may be due to minor differences in mRNA stability (Fig. 2E).
Effect of hly PEST and UTR Mutants on L. monocytogenes Pathogenesis.
L. monocytogenes with synonymous mutations in the PEST region of LLO (e.g., S44S) escape from a phagosome but, due to misregulation of LLO synthesis, result in cytotoxicity of infected host cells (17). Here, we sought to characterize the consequences of infection with the corresponding UTR and double mutants by introducing mutations into their chromosomal locus to assure native transcriptional regulation, which is critical for in vivo virulence. First, we examined the interaction between the P46P synonymous mutant, its compensatory UTR mutation, and the double mutant using an assay that measures host cell cytotoxicity. Consistently, each of the single mutants was more than twice as cytotoxic as WT (Fig. 3A). We also measured bacterial growth in bone marrow-derived macrophages (BMMs) over 8 h. Not surprisingly, both single mutants were dramatically decreased in colony-forming units (CFUs) after 5 h; which based on previous work, was due to the influx of gentamicin that occurs upon LLO-dependent cytotoxicity (Fig. 3E). In contrast, the double mutant grew like WT in BMMs and was less cytotoxic than either of the single mutants but was still more cytotoxic that WT (Fig. 3 A and E). Strikingly, the double P46P*/UTRP46P* mutant produced less cell death than WT, suggesting that the additional hydrogen bond in the CG interaction reduced expression of hly to less than WT (Fig. 3B). This also indicates that slight differences in LLO expression can result in large variations in host cell death.
Fig. 3.
Characterization of mutants in tissue culture and mouse models of infection. (A and B) LDH release assays in BMMs. PactA-holin-lysin is a strain that expresses holin and lysin genes under the control of the actA promoter, causing Aim2-mediated pyroptosis (25). Cells were washed 30 min postinfection with media containing gentamicin for 10 min, washed three times, and incubated with gent-free media to prevent bacterial death. ND = not detected. (C) Plaque assay to measure cell-to-cell spread of AU mutants in L2 cells. (D) Plaque assay in GC mutants. UTR mutants formed plaques of decreased opacity, indicating less cytotoxicity despite cell-to-cell spread. (E) Growth curve in B6 BMMs. (F) Virulence of the mutant bacteria in CD-1 mice (i.v. dose = 105) for 48 h. Error bars represent SDs from three independent replicates. Significance was determined by unpaired t test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, NS = not significant (P > 0.05). ND = not detected.
We further examined pathogenesis by monitoring the growth and cell to cell spread of the strains over 3 d using a plaque assay in a monolayer of cells. Based on previous work, strains that kill their host cell either fail to form a visible plaque or form smaller plaques. In this study, the PEST mutants were unable to form a visible plaque, while the corresponding UTR single mutant displayed much smaller plaques that WT (Fig. 3C). Unexpectedly, the A116C UTR mutant formed plaques similar in size to the double mutant, but they had decreased opacity, a phenotype we had not previously observed (Fig. 3D and SI Appendix, Fig. S1 A and B). The reduced opacity indicated a lower amount of cell death within the cells comprising the plaque, suggesting that L. monocytogenes is spreading, albeit at a reduced capacity, but fewer cells are killed, which we presume is due to the influx of gentamicin that kills the bacteria before they kill the host cell. The PEST/UTR double mutant was identical to WT in both plaque size and opacity, strongly suggesting that restoration of the WT mRNA structure and translational regulation during infection.
To assess the effect on L. monocytogenes virulence, we infected mice intravenous (i.v.) with the mutant bacteria. The P46P synonymous mutant was 1,000-fold less virulent in spleens and livers, while the UTRP46P mutant was 100-fold reduced in both organs. Strikingly, the double P46P/UTRP46P was as virulent as WT (Fig. 3F and SI Appendix, Fig. S2).
Upstream Mutations that Rescue the Toxic S44S Phenotype Reveal a Trade-Off between Cytotoxicity and Phagosomal Escape.
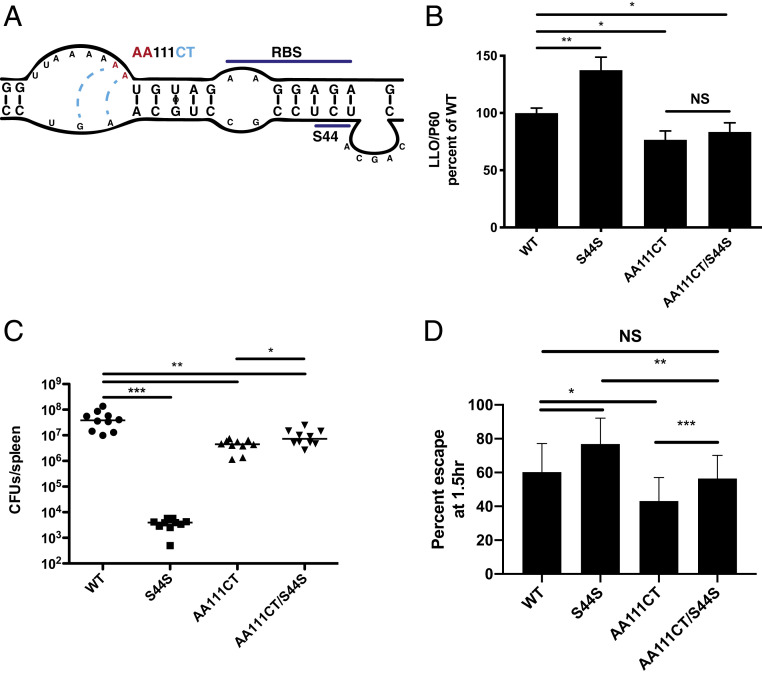
Analysis of the previously characterized S44S mutant was complicated by its predicted interaction with the RBS. Therefore, to determine if WT function could be restored by making another silent mutation elsewhere in the gene, we chose to mutate nucleotides upstream of the predicted interacting site in the 5′ UTR (AA111CT), that is predicted to extend, and therefore stabilize, the stem that is disrupted by S44S (Fig. 4A).
Fig. 4.
Complementation of S44S through restoration of mRNA in a proximal site. (A) The UTR of the endogenous locus was mutated at AA111CT from the TSS so that 5′-CU-3′ would interact with 5′-AG-3′, thereby stabilizing the disrupted hly structure. (B) Quantification of LLO secretion by Western blot. (C) Infection of CD-1 mice (i.v. dose = 105) for 48 h. (D) Phagosomal escape assay using colocalization of bacteria to P62 in the presence of cytochalasin D. Error bars represent SDs from three independent replicates. Significance was determined by unpaired t test for B and C and paired t test for D. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, NS = not significant (P > 0.05).
Quantification of the LLO produced from this strain indicated that this double nucleotide change was sufficient to reduce LLO production (Fig. 4B). To our surprise, the AA111CT mutation restored the S44S strain from more than 10,000-fold less virulent in mice to near WT levels (Fig. 4C). Interestingly, the AA111CT mutant alone was attenuated in virulence by 10-fold, while LLO secretion from this mutant was less than WT. We hypothesized that by stabilizing the mRNA structure, LLO synthesis was reduced, which might result in a phagosomal escape defect. Indeed, the AA111CT mutant had a phagosomal escape defect compared to WT, while the S44S mutant showed enhanced phagosomal escape (Fig. 4D). These results indicate that the hly nucleotide sequence controls the balance between cytotoxicity of host cells and phagosomal escape.
The hly mRNA Translation Inhibition Is Repressed during Stationary Phase.
While it was an intriguing notion that codon bias could be the primary driver of L. monocytogenes virulence by the formation of a highly stable secondary mRNA structure, we suspected the existence of an underlying mechanism that controls a structural change in the mRNA. For example, the mRNA transcript for the L. monocytogenes factor that controls LLO transcription, prfA, contains a thermosensor that alters its structure at 37 °C, allowing for maximal translation efficiency at elevated temperatures (26).
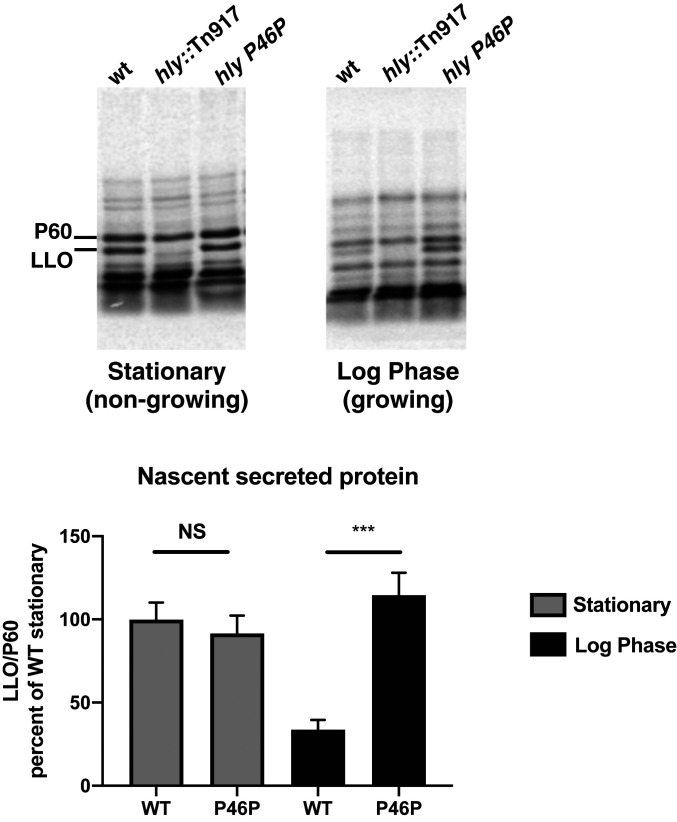
Since LLO is required for L. monocytogenes to escape from a phagosome, but is potentially lethal when expressed in the cytosol, it is reasonable to hypothesize that maximum LLO synthesis is required during starvation in a phagosome while translation is down-regulated during growth in the host cell cytosol to avoid cytotoxicity. Therefore, we examined the differences in LLO secretion from growing versus nongrowing bacteria by pulsing with [35S]methionine for 30 min and quantitating secreted LLO. While growing WT bacteria secreted much less LLO than the PEST mutant, nongrowing bacteria produced equal amounts of LLO (Fig. 5). These data indicated that in WT bacteria LLO translation is reduced during bacteria growth.
Fig. 5.
Translation of LLO is growth phase-dependent. (Upper) Pulse labeling of EGDe Listeria with 35S methionine in met− synthetic media for 30 min in a growing or nongrowing condition. (Lower) Supernatants were TCA precipitated and analyzed for LLO/P60 levels. Error bars represent SDs from three independent experiments. Significance was determined by unpaired t test. ***P ≤ 0.001, NS = not significant (P > 0.05).
Discussion
The results of this study show that translation of LLO is regulated by the formation of an RBS-occluding secondary structure between the 5′ UTR of the mRNA and the PEST-encoding region of the ORF. While disruption of this structure by introduction of noncoding mutations has little effect on transcript levels, the changes in LLO synthesis is enough to result in loss of virulence, highlighting that the formation of this secondary mRNA structure is a critical mechanism of virulence regulation in L. monocytogenes. We showed that LLO expression is precisely controlled at the nucleotide level to provide a balance between maximizing escape from a phagosome but without killing host cells. Although we do not know if this is a general mechanism for intracellular pathogens, an analysis of other Listeria species that encode related cytolysins show similar nucleotide complementarity between the 5′ UTRs and PEST-like sequences (SI Appendix, Fig. S4).
The importance of mRNA secondary structures at the RBS is well-documented to influence protein synthesis by affecting translation initiation (27). Observational and high-throughput experimental studies demonstrate that low mRNA folding stability at the 5′ regions can explain the phenomenon of N-terminal rare codon bias that exists in all domains of life (28, 29). The results of this study suggest that hly translation is governed by the codon usage in the PEST region, which is selective to maintain a mRNA structure that maximizes virulence and cell-to-cell spread. Although low mRNA folding stability can explain N-terminal rare codon bias and is frequently highlighted for its role in promoting efficient translation (24, 28), it is notable that in this study, codons are restricted to ensure a highly stable mRNA molecule that reduces synthesis. We cannot rule out the possibility of an unidentified trans acting factor or factors that could compete with the 5′ UTR and release its translation; however, none of the annotated sRNAs in the L. monocytogenes genome have any nucleotide identity to the 5′ UTR. A search of motifs within the mRNA transcript revealed no potential riboswitches that might regulate mRNA folding as well.
The results suggest that LLO translation is inhibited during growth, but regulation by other means cannot be excluded and we can only speculate as to the mechanism that leads to increased translation at stationary phase. For example, the acidic pH of a phagosome could affect LLO translation (30), although we have no evidence for this (16). Starvation underlies the cessation of growth and accompanies the induction of the stringent response, resulting in rapid reduction of global translation (31–33), although protein synthesis can continue during starvation (34). While growth rate has been shown to affect 5′ UTR-mediated regulation of mRNA stability, as in the case of Escherichia coli ompA (35, 36), in this example growth rate exerted its influence through protection from RNase-mediated degradation, not RBS occlusion. Single molecule fluorescence microscopy studies show that the transcription, translation, and membrane insertion (called transertion) machinery becomes stratified in fast growing cells (37), with RNA polymerase localizing to foci in the nucleoid and ribosomes rich area at the periphery of cells. In Bacillus subtilis, ribosomes in fast-growing cells occupy the poles, while ribosomes at stationary phase are more diffuse (38). Cotranscriptional translation (coupling) is a major piece of the transertion chain that affects 5′ end mRNA-mediated RBS occlusion in E. coli folA (24). In classical experiments the importance of transcription-translation coupling is evident in the mRNA structure-mediated regulation of the biosynthetic operons (39). While cotranscriptional translation is a hallmark of the central dogma in bacteria, mature mRNAs represent the predominant mRNA species translated in the cell. It is possible that the RBS of hly is occluded in either nascent or mature forms, and the relative quantity of these species changes throughout growth phases, an effect that might be dependent on the quantity and spatial distribution of active ribosomes. Understanding the extent of transertion chain reorganization and transcription-translation coupling during nongrowth conditions, such as those seen in the phagosome, may hold answers important for LLO regulation and L. monocytogenes pathogenesis.
The survival of infected host cells is of paramount importance in determining the outcome of infection with intracellular pathogens. While intracellular pathogens secrete virulence factors such as LLO to establish their replicative niche, they must also avoid triggering host cell death pathways designed to prevent pathogen replication. The results of this study show how L. monocytogenes regulates the synthesis of a pore-forming cytolysin to promote escape from a phagosome while repressing its translation once the bacteria reach their intracellular niche in the host cell cytosol. However, to promote its pathogenesis, the bacteria must also avoid triggering host cell pathways of inflammatory cell death called pyroptosis. Indeed, while WT L. monocytogenes induces very low levels of cell death, strains engineered to trigger pyroptosis are highly attenuated for virulence (25, 40). Similarly, other pathogens use mechanisms to avoid pyroptosis to promote their pathogenesis (41). Therefore, to become an intracellular pathogen requires both the intricate regulation of virulence factor expression and the avoidance of endogenous host cell mechanisms that promote cell death. Not surprisingly, the primary mechanism of adaptive immunity to L. monocytogenes is the induction of cell death mediated by antigen-specific cytotoxic T cells (42).
Materials and Methods
Bacterial Strains and Growth Conditions.
L. monocytogenes (WT 10403S and its derivatives) (SI Appendix, Table S1) were grown in Brain Heart Infusion (BHI; BD Biosciences) at 37 °C. EGDe strains, which unlike 10403S do not require methionine, were grown in methionine− Listeria Synthetic Media (LSM). Growth occurred aerobically at an RPM (revolutions per minute) of 220. Growth was measured by optical density at a wavelength of 600 nm (OD600). Frozen bacterial stocks were stored in BHI + 40% glycerol at −80 °C. Antibiotics were used at the following concentrations 200 µg/mL and chloramphenicol at 7.5 µg/mL for L. monocytogenes and 10 µg/mL for E. coli. Derivative strains were made using primers on SI Appendix, Table S2 using pPL2 for knock-ins and pKSV7 for allelic exchange.
In Vitro LLO Quantification.
Overnight cultures were washed and back diluted into fresh media at 1:20 and allowed to grow for 6 h at 37 °C shaking. Cultures were pelleted and supernatants were treated with trichloroacetic acid (TCA) at 10% for 1 h on ice to precipitate protein. Supernatants were centrifuged at 13,000 rpm for 15 min, and pellets were washed with acetone, followed by a 12-min centrifuge. These pellets were washed a final time in acetone and washed for 10 min. Pellets were allowed to dry overnight or in a vacuum and resuspended in 1× lithium dodecyl sulfate buffer (Invitrogen) containing 5% β-mercaptoethanol. Secreted protein was boiled for 5 min and separated on by sodium dodecyl sulphate/ polyacrylamide gel electrophoresis (SDS/PAGE). The primary antibodies, a rabbit polyclonal antibody against LLO and a mouse monoclonal antibody against P60 (Adipogen), were each used at a dilution of 1:5,000. P60 is a constitutively expressed bacterial protein used as a loading control for secreted proteins. Secondary antibodies were goat anti-mouse IRDye 800CW (Licor) and Alexa Fluor 680 goat anti-rabbit (Life Technologies). All immunoblots were visualized using the Odyssey imager and quantified using ImageJ. For labeling experiments, EGDe strains were grown in Met− LSM until stationary and either diluted in fresh Met− LSM, grown for 6 h (5-h doubling time) and pulsed with EasyTag 35S-Methionine (Perkin-Elmer) at 25 μCi/0.1 OD600, or pulsed at stationary. The pulse lasted for 30 min followed by TCA precipitation of supernatants as described above.
RT-qPCR.
Bacteria were grown to midlog (OD600 0.8), and RNA was isolated by RiboPure-Bacteria RNA isolation kit (ambion). RNA was DNase treated, followed by phenol chloroform extraction and cDNA synthesis using iScript reverse transcriptase (Bio-Rad). qPCR was performed with Kapa SYBR fast (Kapa Biosystems) using the manufacturer’s recommended cycle using primers from SI Appendix, Table S3.
Tissue Culture Experiments Using BMMs.
Bacteria were grown at 30 °C shaking overnight. Overnight cultures were washed and resuspended at 1/10 in phosphate buffered saline (PBS). This was used to infect a monolayer of murine bone marrow-derived macrophages at a multiplicity of infection (MOI) of 0.25 that had been seeded the previous day on glass coverslips. Cells were washed three times 30 min postinfection, and media was replaced. At 1 h, gentamicin was added to the tissue culture media to kill the extracellular bacteria (50 μg/mL). Coverslips were collected in water and plated to enumerate CFUs. For cytotoxicity assays, monolayers of PAM3CSK-treated BMMs were infected with bacterial strains for 30 min, washed three times in Dulbecco's phosphate buffered saline (DPBS), followed by incubation with gentamicin-containing media (50 μg/mL) for 10 min. Cells were washed again three times, and fresh media was replaced without gentamicin. At 6 h postinfection (hpi), supernatants were isolated and analyzed as previously described (25). Results are presented as a percentage of 100% cell death as determined by cells treated with 1% Triton X-100.
Phagosome Escape Assay.
Murine BMMs were seeded on glass coverslip within a 24-well plate at 2 × 105 cells per well and allowed to adhere overnight in tissue culture (TC) chamber. Bacteria were grown in BHI at 37 °C overnight with shaking and subcultured 1:20 until reaching OD600 1.0. To target cytosolic bacteria efficiently, 250 ng/mL cytochalasin D (cytD, Sigma) was added to BMMs 30 min before infection to prevent actin polymerization and was maintained in media throughout infection. BMMs were infected at a MOI of 10 for 30 min, washed twice prior to replacing fresh BMM media with cytD, and 50 μg/mL gentamicin was added to kill the extracellular bacteria at 1 hpi. Coverslips were removed and proceeded to immunostaining as previously described (43) at 1.5 hpi. Primary antibodies recognizing L. monocytogenes (1:1,000 dilution; BD Biosciences, no. 223021) and BMM protein p62 (1:200 dilutions; Fitzgerald, no. 20R-PP001) were used and detected with fluorophore-conjugated secondary antibodies, rhodamine red-X goat anti-rabbit IgG (1:2,000 dilution; Invitrogen, R6394), and Alexa Fluor-647 goat anti-guinea pig IgG (1:2,000 dilution; Invitrogen, A21450). Coverslips were mounted in Prolong Gold antifade reagent with DAPI (Invitrogen, P36935), and imaged with BZ-X710 KEYENCE microscope. Bacteria colocalized with p62 were counted as cytosolic ones that successfully escaped from phagosomes. More than 100 bacteria per sample were analyzed. Replicates were performed on different days with different batches of BMMs.
Mouse Infections.
Eight-week-old CD-1 outbred mice (Charles River) were infected i.v. with 1 × 105 CFUs in 200 μL of PBS. Animals were euthanized at 48 h, and spleens and livers were harvested in 5 mL or 10 mL in 0.1% IGEPAL CA-630 (Sigma) in water, respectively, and plated for enumeration of bacterial burdens. Animal work was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council of the National Academy of Sciences (44).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants 1P01 AI063302 (to D.A.P.) and 1R01 AI027655 (to D.A.P). B.N.P. was supported by the NSF Graduate Research Fellowship under Grant DGE 1106400.
Footnotes
Competing interest statement: B.N.P. and J.L.P. are currently employed at Actym Therapeutics (Berkeley, CA 94710) and Affinivax, Inc. (Cambridge, MA 02139), respectively.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004129117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Thakur A., Mikkelsen H., Jungersen G., Intracellular pathogens: Host immunity and microbial persistence strategies. J. Immunol. Res. 2019, 1356540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamaruzzaman N. F., Kendall S., Good L., Targeting the hard to reach: Challenges and novel strategies in the treatment of intracellular bacterial infections. Br. J. Pharmacol. 174, 2225–2236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radoshevich L., Cossart P., Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Tweten R. K., Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 73, 6199–6209 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decatur A. L., Portnoy D. A., A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290, 992–995 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Portnoy D. A., Tweten R. K., Kehoe M., Bielecki J., Capacity of listeriolysin O, streptolysin O, and perfringolysin O to mediate growth of Bacillus subtilis within mammalian cells. Infect. Immun. 60, 2710–2717 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S., Portnoy D. A., Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 62, 5608–5613 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Z. et al., Characterization of Listeria monocytogenes expressing anthrolysin O and phosphatidylinositol-specific phospholipase C from Bacillus anthracis. Infect. Immun. 73, 6639–6646 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoffroy C., Gaillard J.-L., Alouf J. E., Berche P., Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55, 1641–1646 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glomski I. J., Gedde M. M., Tsang A. W., Swanson J. A., Portnoy D. A., The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156, 1029–1038 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuerch D. W., Wilson-Kubalek E. M., Tweten R. K., Molecular basis of listeriolysin O pH dependence. Proc. Natl. Acad. Sci. U.S.A. 102, 12537–12542 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rechsteiner M., Rogers S. W., PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271 (1996). [PubMed] [Google Scholar]
- 13.Rogers S., Wells R., Rechsteiner M., Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234, 364–368 (1986). [DOI] [PubMed] [Google Scholar]
- 14.Köster S. et al., Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat. Commun. 5, 3690 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Chen C. et al., The listeriolysin O PEST-like sequence Co-opts AP-2-mediated endocytosis to prevent plasma membrane damage during Listeria infection. Cell Host Microbe 23, 786–795.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnupf P., Portnoy D. A., Decatur A. L., Phosphorylation, ubiquitination and degradation of listeriolysin O in mammalian cells: Role of the PEST-like sequence. Cell. Microbiol. 8, 353–364 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Schnupf P. et al., Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol. Microbiol. 61, 999–1012 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Sharp P. M., Li W.-H., The codon Adaptation Index–A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grote A. et al., JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, W526–W531 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer P., Chow M. Y. N., Loessner M. J., Portnoy D. A., Calendar R., Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggenhofer F., Tafer H., Stadler P. F., Hofacker I. L., RNApredator: Fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 39, W149–W154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz R. et al., ViennaRNA package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudla G., Murray A. W., Tollervey D., Plotkin J. B., Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharyya S. et al., Accessibility of the shine-dalgarno sequence dictates N-terminal codon bias in E. coli. Mol. Cell 70, 894–905.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer J.-D. et al., Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson J. et al., An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561 (2002). [DOI] [PubMed] [Google Scholar]
- 27.de Smit M. H., van Duin J., Secondary structure of the ribosome binding site determines translational efficiency: A quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 87, 7668–7672 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentele K., Saffert P., Rauscher R., Ignatova Z., Blüthgen N., Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 9, 675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman D. B., Church G. M., Kosuri S., Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Nechooshtan G., Elgrably-Weiss M., Sheaffer A., Westhof E., Altuvia S., A pH-responsive riboregulator. Genes Dev. 23, 2650–2662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaishankar J., Srivastava P., Molecular basis of stationary phase survival and applications. Front. Microbiol. 8, 2000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potrykus K., Cashel M., (p)ppGpp: Still magical? Annu. Rev. Microbiol. 62, 35–51 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Reeve C. A., Amy P. S., Matin A., Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J. Bacteriol. 160, 1041–1046 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gefen O., Fridman O., Ronin I., Balaban N. Q., Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc. Natl. Acad. Sci. U.S.A. 111, 556–561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson G., Belasco J. G., Cohen S. N., von Gabain A., Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature 312, 75–77 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Emory S. A., Belasco J. G., The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 172, 4472–4481 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakshi S., Choi H., Weisshaar J. C., The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 6, 636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis P. J., Thaker S. D., Errington J., Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19, 710–718 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanofsky C., Attenuation in the control of expression of bacterial operons. Nature 289, 751–758 (1981). [DOI] [PubMed] [Google Scholar]
- 40.Sauer J.-D. et al., Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl. Acad. Sci. U.S.A. 108, 12419–12424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen I., Miao E. A., Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 265, 130–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chávez-Arroyo A., Portnoy D. A., Why is Listeria monocytogenes such a potent inducer of CD8+ T‐cells? Cell. Microbiol. 22, 6387–6410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell G. et al., Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 83, 2175–2184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.