Significance
Cell division is an essential process in bacteria and an attractive target for antibiotic development. A major activity of the division apparatus is the synthesis of the cell wall peptidoglycan that will ultimately fortify the poles of the daughter cells. How cell wall synthesis by the division machinery is regulated has remained unclear. In this report, we use a combination of genetics and biochemical reconstitution to demonstrate that a complex of the highly conserved FtsQ, FtsL, and FtsB proteins serves as an essential activator of cell wall synthesis within the cytokinetic ring.
Keywords: cell division, peptidoglycan, cell wall, divisome, septal ring
Abstract
Cell division in bacteria is mediated by a multiprotein assembly called the divisome. A major function of this machinery is the synthesis of the peptidoglycan (PG) cell wall that caps the daughter poles and prevents osmotic lysis of the newborn cells. Recent studies have implicated a complex of FtsW and FtsI (FtsWI) as the essential PG synthase within the divisome; however, how PG polymerization by this synthase is regulated and coordinated with other activities within the machinery is not well understood. Previous results have implicated a conserved subcomplex of division proteins composed of FtsQ, FtsL, and FtsB (FtsQLB) in the regulation of FtsWI, but whether these proteins act directly as positive or negative regulators of the synthase has been unclear. To address this question, we purified a five-member Pseudomonas aeruginosa division complex consisting of FtsQLB-FtsWI. The PG polymerase activity of this complex was found to be greatly stimulated relative to FtsWI alone. Purification of complexes lacking individual components indicated that FtsL and FtsB are sufficient for FtsW activation. Furthermore, support for this activity being important for the cellular function of FtsQLB was provided by the identification of two division-defective variants of FtsL that still form normal FtsQLB-FtsWI complexes but fail to activate PG synthesis. Thus, our results indicate that the conserved FtsQLB complex is a direct activator of PG polymerization by the FtsWI synthase and thereby define an essential regulatory step in the process of bacterial cell division.
Bacterial cell division is initiated by polymerization of the tubulin-like FtsZ protein just underneath the cytoplasmic membrane at midcell (1). These polymers associate with membrane-anchored proteins like FtsA and treadmill around the prospective division site, forming what looks like a continuous ring when labeled FtsZ is visualized by epifluorescence microscopy (2–5). Once this so-called “Z-ring” is formed, a host of additional proteins are recruited to midcell to form the mature divisome (septal ring, cytokinetic ring) capable of all of the activities necessary for division, including membrane constriction and the synthesis of a multilayered PG cell wall structure, septal PG, that eventually forms the cap fortifying the daughter cell poles (5).
PG synthesis requires two enzymatic reactions: a PG glycosyltransferase (PGTase) for glycan polymerization and a transpeptidase (TPase) to cross-link adjacent glycans via their attached peptides (6). Most bacteria encode two different types of PG synthases: class A penicillin-binding proteins (aPBPs) and complexes between SEDS (shape, elongation, division, sporulation) proteins and class B PBPs (bPBPs) (6). The aPBPs possess both PGTase and TPase activities in a single polypeptide chain (7), whereas in the SEDS-bPBP complexes the activities are split, with SEDS proteins supplying the glycan polymerase function and the bPBP promoting cross-linking (8–10). Both types of PG synthases are thought to participate in septal PG biogenesis, with a complex between the core divisome components FtsW and FtsI forming the SEDS-bPBP synthase of the division machinery (10). The identity of the aPBP most closely associated with cell division varies from organism to organism. In Escherichia coli, the primary model system for cell division in gram-negative bacteria, PBP1b is thought to perform this function due to its moderate enrichment at division sites and detected interactions with several division proteins (11–14).
Many components of the divisome are thought to have roles in regulating the initiation of septal PG biogenesis. In E. coli, the division protein FtsN has long been thought to be the trigger of cell constriction and PG biogenesis by the division machinery (15). Additional proteins are also thought to participate in divisome activation based on the identification and characterization of so-called “superfission” mutants that bypass the requirement for FtsN and other normally essential components of the machinery for cell division (16–20). The current thinking in the field, therefore, is that PG synthesis at the division site is regulated by an activation pathway involving FtsA; the FtsEX ABC system; a three-protein complex of the division proteins FtsQ, FtsL, and FtsB (FtsQLB); and FtsN (5, 16–18). What has remained unclear and the subject of debate is which PG synthase is the direct regulatory target of this pathway, PBP1b or FtsWI, and how divisome activation is achieved (14, 16, 17, 21). The function of the FtsQLB complex in this pathway and whether it works as an inhibitor or activator of PG biogenesis within the divisome has been particularly controversial (14, 16, 17, 22).
Here we report results supporting a role for the FtsQLB complex in the activation of septal PG biogenesis by the FtsWI synthase. Because of difficulties in overproducing and purifying stable E. coli FtsWI complexes that are active for PG polymerization, a problem similar to those reported by others (13, 14), we chose to study the PG synthase from Pseudomonas aeruginosa, which we previously found to have favorable biochemical properties (10). A five-member complex of FtsQLB-FtsWI was purified and shown to have greatly enhanced PG polymerase activity relative to complexes containing the FtsWI synthase alone. Furthermore, support for this activity being important for the cellular function of FtsQLB was provided by the identification of two division-defective variants of FtsL that still form normal FtsQLB-FtsWI complexes but fail to activate PG synthesis. Thus, our results indicate that the conserved FtsQLB complex is a direct activator of PG polymerization by the FtsWI synthase and thereby define an essential regulatory step in the process of bacterial cell division.
Results
FtsQLB Activates Cell Wall Polymerization by the FtsWI Synthase.
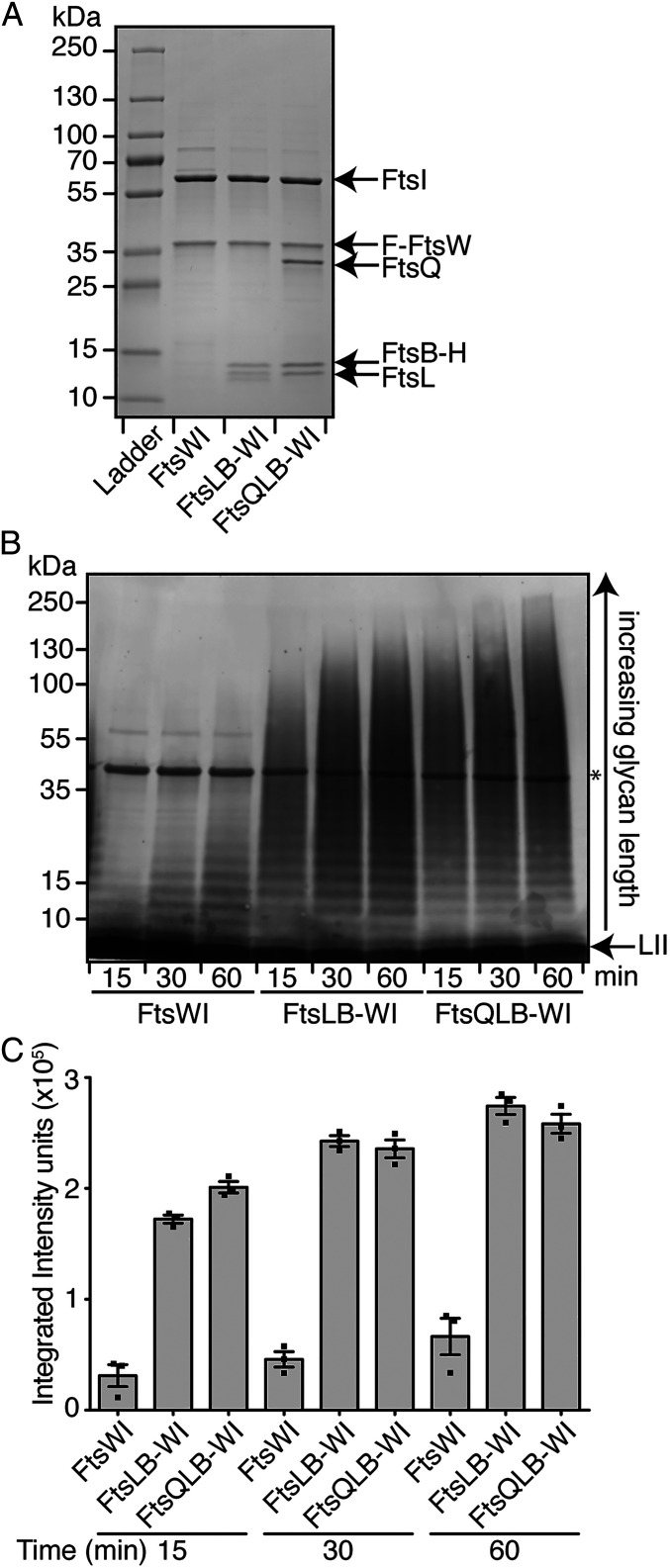
To better understand the potential role of FtsQLB in controlling FtsWI activity within the divisome, a FLAG-tagged variant of FtsW from P. aeruginosa was overproduced in E. coli cells along with FtsI alone or with FtsQ, FtsL, and His-tagged FtsB in addition to FtsI. As we have observed previously (10) the FLAG-FtsW-FtsI complex was readily purified by anti-FLAG affinity chromatography when the two proteins were co-overproduced (Fig. 1A). A modest level of short PG oligomers was formed when the complex was incubated with purified lipid II substrate, indicating that FtsWI alone is a relatively weak and poorly processive PG polymerase (Fig. 1B). When all five proteins were produced, an FtsQLB-FtsWI complex was isolated, demonstrating that the FtsQLB subcomplex interacts directly with the FtsWI synthase (Fig. 1A). Strikingly, the five-member complex produced much longer glycans and roughly five times the amount of total material relative to the FtsWI synthase alone (Fig. 1 B and C).
Fig. 1.
FtsQLB stimulates PG polymerization by the FtsWI synthase. (A) Coomassie- stained gel of purified FLAG(F)-tagged FtsW coexpressed with 1) FtsI alone; 2) FtsI, FtsL, and His(H)-tagged FtsB; or 3) FtsI, FtsQ, FtsL, and FtsB-H. Protein concentrations in each preparation were normalized, and equivalent volumes of each were loaded on the gel. (B) PGTase assays using the same normalized preparations shown in A. Purified complexes (0.5 µM) were incubated with purified E. faecalis lipid II (LII) (10 µM) and cephalexin (200 µM) to block cross-linking. The biotin-labeled glycan polymers thus produced were detected on blots with IRDye-labeled streptavidin. The asterisk marks the enzyme used for biotinylating the glycans, which itself becomes biotin-labeled. In both panels, representative images of three independent experiments are shown. (C) The accumulation of glycan fragments from three independent replicates of polymerization time courses was quantified using densitometry. Error bars represent SEM.
Importantly, the activity of this complex was found to be dependent on FtsW, as complexes purified with an FtsW variant previously shown to be defective for polymerase function displayed dramatically reduced activity (SI Appendix, Fig. S1). When FtsL and FtsB-His were coproduced with FLAG-FtsW and FtsI, a four-protein complex was isolated (Fig. 1A) that displayed robust PG polymerase activity comparable to that of the five-protein complex (Fig. 1 B and C). Thus, the FtsQLB subcomplex of the divisome activates the PG polymerase activity of FtsWI and enhances its processivity in vitro, with the FtsL and FtsB components being sufficient for the observed stimulation.
Identification of Dominant-Negative FtsL Variants.
We reasoned that if the biochemical activation of FtsWI by FtsQLB observed in vitro is physiologically relevant, then it should be possible to identify a subset of FtsL or FtsB variants defective for cell division in vivo that are also impaired for activation of the synthase in vitro. Given the modest sizes of FtsL (97 aa) and FtsB (94 aa), we decided to use site-directed mutagenesis to search for such activation-defective variants. Thus, a collection of 150 expression plasmids was generated to produce derivatives in which every amino acid in the periplasmic region of these proteins was individually substituted with at least one alternative residue (Fig. 2A and SI Appendix, Tables S1 and S2). To maximize our chances of successfully disrupting FtsL and/or FtsB function, the collection was designed such that the majority of the site-directed changes resulted in a charge change or the addition of a charged residue in place of a neutral residue.
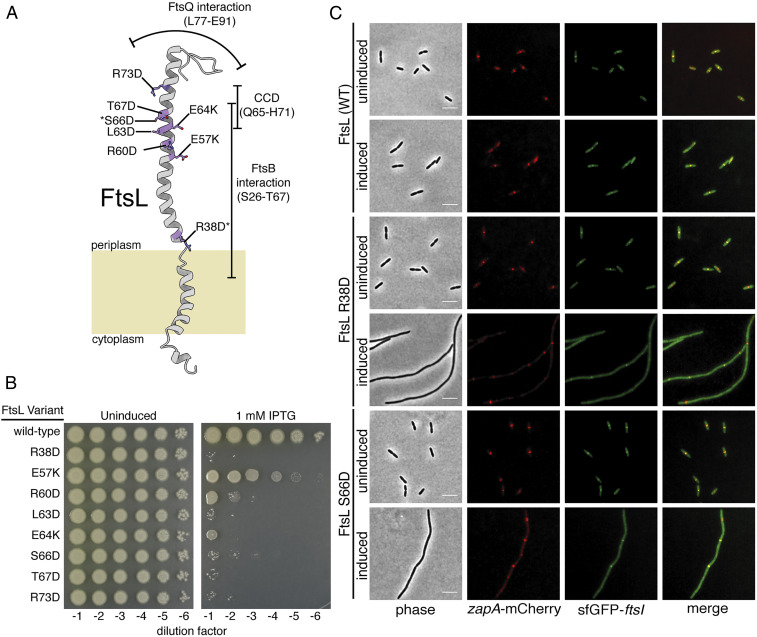
Fig. 2.
Identification of dominant-negative FtsL variants. (A) Structural model of P. aeruginosa FtsL created using Phyre2 (49). Domains of the protein that interact with other division proteins or have been implicated in constriction control (CCD) are highlighted. These assignments were based on the analogous regions of E. coli FtsL implicated in these activities. All periplasmic residues were targeted by site-directed mutagenesis, and the resulting variants were tested for dominant-negative activity. (SI Appendix, Tables S1 and S2 list the FtsL and FtsB variants tested.) Variants of FtsL with dominant-negative activity are highlighted; the residues marked with an asterisk indicate those shown to be defective for FtsWI activation in vitro. (B) Tenfold serial dilutions of P. aeruginosa PAO1 cells harboring expression plasmids producing the indicated FtsL variant. Dilutions were plated on LB agar with or without IPTG to induce the FtsL variants as indicated. (C) Phase-contrast, mCherry, GFP, and merged mCherry/GFP micrographs of P. aeruginosa PAO1 cells grown with or without IPTG to induce the indicated FtsL variants from derivatives of the pLSM11 plasmid. (Scale bar: 5 µm.) Representative images of two independent experiments are shown in B and C.
Plasmids in the mutant collection encoding FtsL or FtsB variants under isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible control were individually transformed into wild-type (WT) P. aeruginosa. The resulting transformants were then tested for inducer sensitivity to identify candidates encoding dominant-negative FtsL or FtsB (DNFtsL or DNFtsB) variants. Surprisingly, no DNFtsB variants were identified, indicating either that the expression plasmid does not overproduce FtsB to high enough levels to induce a dominant-negative phenotype or that it is difficult to generate a functional defect in FtsB with a single amino acid substitution in its periplasmic domain. In contrast, eight DNFtsL variants were identified that, unlike WT FtsL, displayed varying ability to interfere with the growth and division of P. aeruginosa when induced (Fig. 2 A–C and SI Appendix, Figs. S2 and S3). FtsL has been reported to be unstable in some organisms (23, 24), and thus, even though high-level overproduction of WT FtsL was previously shown to be well tolerated in E. coli (25), it is possible that a subset of the DNFtsL variants enhance protein stability to cause lethal hyperaccumulation of FtsL. Therefore, FLAG-tagged versions of the DNFtsL variants were constructed and tested for activity and protein accumulation. All the tagged DNFtsL variants retained lethal activity and accumulated to levels similar to or below that of tagged FtsL(WT), with only one mutant, FtsL(R38D), showing signs of modest processing (SI Appendix, Fig. S4). Therefore, we conclude that instead of acting via an alteration in protein stability, the DNFtsL variants identified are likely to inhibit cell division because they are functionally defective and replace FtsL(WT) in the divisome when they are overproduced.
To further characterize the division defect induced by the DNFtsL variants, divisome formation in the division-inhibited cells was assessed using fluorescent fusions to both ZapA (ZapA-mCherry), an FtsZ-binding protein and marker for the Z-ring (26), and FtsI (sfGFP-FtsI), the TPase component of the FtsWI synthase that is a late recruit to the machinery and a marker for mature divisomes (21). As is often observed in filamentous E. coli cells (27, 28), the frequency of Z-rings appeared to be reduced along the length of division-inhibited P. aeruginosa cells producing the DNFtsL variants relative to normally dividing cells producing FtsL(WT) (Fig. 2C). In the filaments produced by each DNFtsL variant, at least a subset of the Z-rings that formed was also associated with a ring of sfGFP-FtsI, but the frequency of colocalization varied widely in the different filaments (Fig. 2C and SI Appendix, Figs. S3 and S5). Nevertheless, because FtsI is downstream of both FtsW and FtsQLB in the E. coli divisome recruitment hierarchy (21, 28), the fact that the filaments were all capable of recruiting FtsI to the divisome suggested that many, if not all, of the DNFtsL variants retain the ability to associate with FtsWI. Therefore, we considered all eight FtsL derivatives good candidates for in vitro studies to determine whether their defective phenotype is caused by a failure to activate FtsWI.
A Subset of DNFtsL Variants Fail to Activate FtsWI.
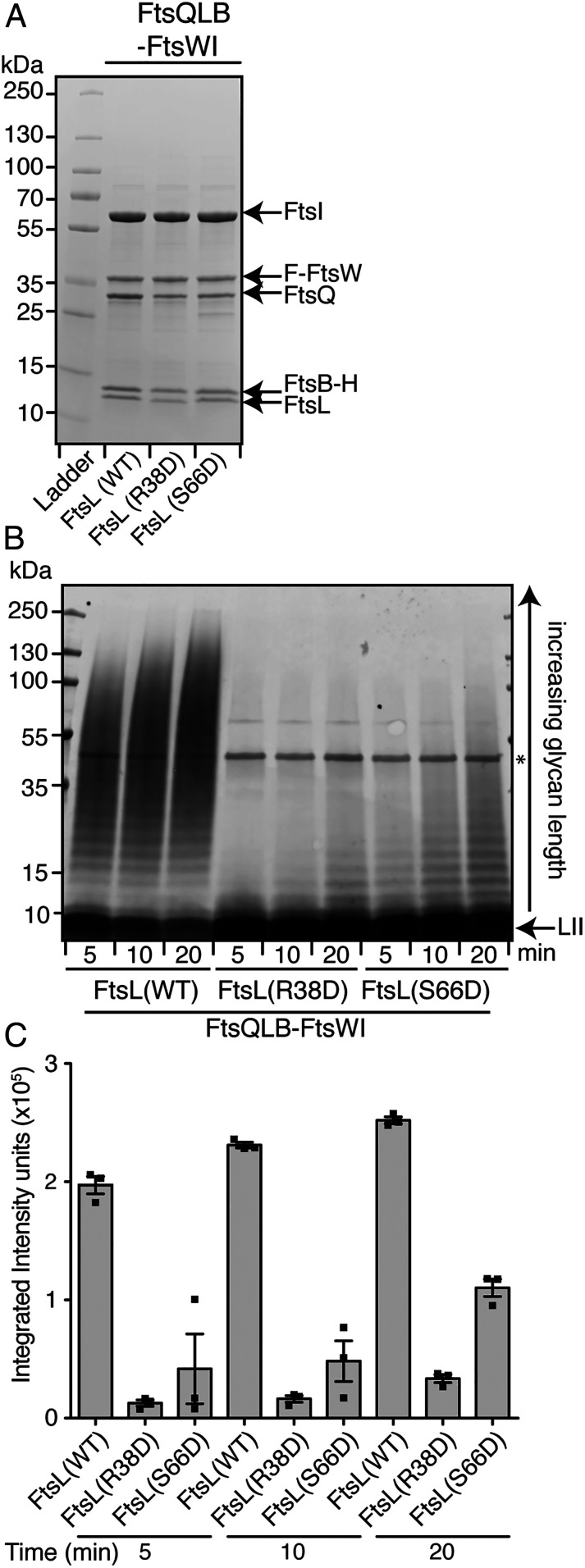
The biochemical function of the DNFtsL variants was tested by purifying FtsQLB-FtsWI complexes containing the altered FtsL proteins. As suggested by the ability of sfGFP-FtsI to be recruited to the division site in cells expressing the DNFtsL variants, they all retained the ability to form five-member FtsQLB-FtsWI complexes (Fig. 3 and SI Appendix, Figs. S6 and S7). Six of the eight DNFtsL variants formed complexes that displayed PG polymerase activity comparable to those with WT FtsL (SI Appendix, Fig. S6), suggesting that they may affect cell division at a step upstream of FtsW activation in vivo that is not captured by the in vitro assay (Discussion). However, FtsQLB-FtsWI complexes that included FtsL(R38D) or FtsL(S66D) had significantly reduced PG polymerase activity relative to WT complexes such that it more closely resembled the activity of the FtsWI synthase alone (Fig. 3 and SI Appendix, Fig. S7). Thus, these derivatives of FtsL retain the ability to form FtsQLB-WI complexes but are functionally defective for cell division in vivo and fail to properly activate PG synthesis by FtsWI in vitro. Although it is possible, based on the GFP-FtsI recruitment results, that FtsL(R38D) and FtsL(S66D) are partially defective for divisome assembly in vivo in addition to the activation of cell constriction, the results with these variants indicate that the biochemical stimulation of FtsWI by FtsQLB is likely to be physiologically relevant, supporting an activation function for FtsQLB in promoting septal PG biogenesis.
Fig. 3.
FtsQLB complexes with DNFtsL variants are defective for FtsWI activation. (A) Coomassie-stained gel of the indicated complexes purified and labeled as shown in Fig. 1A. Protein concentrations in each preparation were normalized and equivalent volumes of each were loaded on the gel. (B) PG polymerization activity of the indicated complexes was assayed as shown in Fig. 1B. Representative images of three independent experiments are shown in A and B. (C) The accumulation of glycan fragments from three independent replicates of polymerization time courses was quantified using densitometry. Error bars represent SEM.
A Superfission Variant of FtsL Does Not Further Activate PG Synthesis In Vitro.
Because FtsN has been implicated in the septal PG synthesis activation pathway in vivo (15–17), we wanted to determine whether it would further activate PG polymerization by the FtsQLB-FtsWI complex in vitro. However, we were unable to copurify FtsN with the complex when it was co-overexpressed in cells with the other proteins. Moreover, when a soluble version of FtsN lacking its transmembrane domain was purified and mixed in excess with the purified FtsQLB-FtsWI or FtsWI complexes, only a minor inhibitory effect on PG polymerase activity was observed (SI Appendix, Fig. S8). Although there are many possible technical reasons for these negative results that cannot be excluded, we considered the possibility that the purified FtsQLB-FtsWI complexes are already in the activated state that normally requires FtsN to form in vivo. If this were true, then one would expect the activity of the synthase to also be insensitive to the presence of a superfission variant of FtsL (SFFtsL) that bypasses the FtsN requirement for cell division in vivo.
One of the best-characterized E. coli SFFtsL variants is FtsL(E88K) (16, 17). The analogous residue in P. aeruginosa FtsL is Q65. To determine whether an FtsL(Q65K) derivative also has superfission activity in P. aeruginosa, we generated the ftsL(Q65K) allele at its native genomic locus. Like its E. coli counterpart (17), P. aeruginosa cells with the ftsL(Q65K) allele were significantly shorter on average than WT (average length, 2.7 µm vs. 3.3 µm) (SI Appendix, Fig. S9A), indicating enhanced cell division activity. To test whether the FtsL(Q65K) variant can bypass a requirement for FtsN function, we first needed to generate an ftsN (PA5052) mutant. A prior transposon sequencing (Tn-Seq) analysis suggested that ftsN may be only conditionally essential in P. aeruginosa, with its inactivation being tolerated in minimal medium, but not in rich medium (29). Therefore, we attempted to construct a ΔftsN strain. In accordance with the Tn-Seq data, FtsN inactivation resulted in a severe growth defect on rich LB medium (SI Appendix, Fig. S9B). The ΔftsN cells also formed long filaments indicative of a major disruption to divisome function (SI Appendix, Fig. S9C).
In the course of mutant construction, we fortuitously observed that the growth defect of ΔftsN cells could be largely alleviated by the addition of 5% sucrose to the medium, which somehow promotes better division in the absence of FtsN (SI Appendix, Fig. S9 B and C), allowing us to more readily propagate the mutant for analysis. Combining the ftsL(Q65K) allele with the ftsN deletion suppressed the LB growth and division defect caused by FtsN inactivation, providing further support for superfission activity of the FtsL(Q65K) variant (SI Appendix, Fig. S9 B and C). Therefore, we purified FtsQLB-FtsWI complexes containing FtsL(Q65K) to assess their activity. The altered complexes were not significantly more active for PG polymerization in vitro than those containing FtsL(WT) (SI Appendix, Fig. S9 D–F). Thus, we infer that the purified FtsQLB-FtsWI complexes that we isolate from membranes are likely to contain the FtsWI synthase in the final activated state achieved on maturation of the divisome in vivo (Discussion).
Discussion
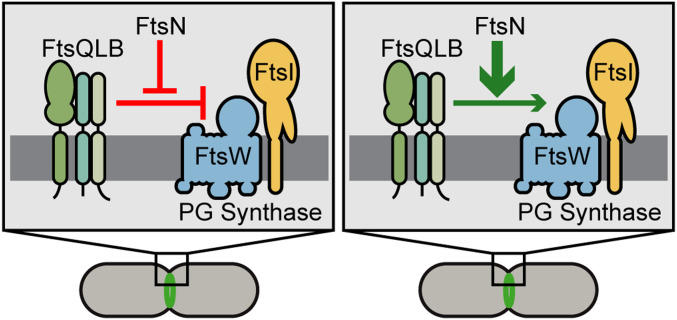
The function of the FtsQLB complex in cell division has been unclear for many years. Recent studies (14, 16, 17) have pointed to a regulatory role in controlling septal PG synthesis, but whether the complex functions as an inhibitor or activator of the process remains the subject of debate. Both possibilities are consistent with the isolation of superfission variants of the FtsL and FtsB components of the complex that bypass the essentiality of FtsN and other divisome proteins (16, 17). According to one proposal, the superfission variants have lost an inhibitory activity such that FtsN is no longer needed to relieve inhibition by FtsQLB to promote the initiation of septal PG biogenesis (16) (Fig. 4). Alternatively, these variants can be interpreted as spontaneously triggering an activation function of the FtsQLB complex that normally requires additional input from or stimulation by other proteins like FtsN to initiate septal PG synthesis and cell division (17) (Fig. 4).
Fig. 4.
Models for the role of FtsQLB in controlling septal PG synthesis by the divisome. Shown are schematics depicting the two current models for the regulation of septal PG synthesis by the FtsQLB complex. Genetic and biochemical results suggest that it is either an inhibitor of PG synthesis by FtsWI, with its inhibition relieved by FtsN (Left), or an activator of FtsWI with an activity enhanced by FtsN (Right). Other division proteins are omitted from the diagram for simplicity. Our results support an activation function for FtsQLB. See the text for details.
Our biochemical results with P. aeruginosa proteins support an activation function for FtsQLB, with the PG polymerase activity of the FtsWI synthase as its regulatory target. However, another recent biochemical analysis found that E. coli FtsQLB inhibits PG polymerization by PBP1b and PG cross-linking by FtsI (14), suggesting an inhibitory function for FtsQLB. Although it is possible that the complex performs each of the detected biochemical activities in vivo, or that the variety of observed functions reflect differences in divisome regulation between E. coli and P. aeruginosa, several lines of reasoning argue that the detected inhibitory activities might not reflect the normal essential function of FtsQLB in cells.
PBP1b is not essential in E. coli and becomes required for growth only when PBP1a, the other major aPBP, is inactivated (30). Moreover, both E. coli aPBPs can be inactivated and viability restored to cells by the production of P. aeruginosa PBP1b and its cognate activator, LpoP (31). Owing to sequence divergence, it is unlikely that E. coli FtsQLB is capable of interacting with and regulating E. coli PBP1a and/or P. aeruginosa PBP1b in the same way as is seen for E. coli PBP1b in vitro. Furthermore, the aPBPs are nonessential for growth or division in Bacillus subtilis (32), yet the equivalent FtsQLB proteins are either essential or very nearly essential in this model organism (33–35). Thus, the inhibition of PG polymerization by aPBPs like PBP1b is unlikely to represent the conserved and essential regulatory function of the FtsQLB complex.
Unlike the polymerase activity of PBP1b, the TPase activity of FtsI is essential (36). Thus, the observed inhibition of FtsI TPase activity by E. coli FtsQ (14) may represent an important activity of the FtsQLB complex, especially since this inhibition also has been observed using Streptococcus pneumoniae proteins (37). However, the cross-linking reaction is not an ideal regulatory target for controlling PG synthesis, because it will merely affect the structure of the final PG product, not whether or not the product is produced (10). Furthermore, it was previously shown that inhibition of the TPase activity of FtsI with cephalexin results in the polymerization and rapid turnover of PG glycans in a futile cycle that contributes to the toxicity of beta-lactam antibiotics (38). Thus, we favor a model in which PG polymerase activity is the key point of regulation for controlling the initiation of septal PG synthesis, not cross-linking.
FtsW is the only PG polymerase known to be essential for cell division such that it is the most attractive target for regulation controlling the onset of PG synthesis by the divisome. Our biochemical system using P. aeruginosa proteins reveals that FtsQLB stimulates PG polymerization by FtsW in vitro when it is part of the FtsWI synthase complex. Evidence that this activity is important for FtsQLB function in cells was provided by the identification of two DNFtsL variants that disrupt cell division in vivo and retain the ability to form stable FtsQLB-WI complexes that can be purified but fail to activate PG polymerase activity by FtsWI in vitro.
Further support for FtsWI PG synthase activity as the conserved target of regulation by the divisome activation pathway involving FtsQLB comes from genetic results in E. coli and Caulobacter crescentus. In C. crescentus, superfission variants of FtsW or FtsI were found to overcome the division block caused by DNA damage-induced inhibitors and to suppress the normally lethal inactivation of an essential upstream regulator of cell division (39–41). Importantly, the amino acid substitutions in the SFFtsI variants are found in the non-TPase domain, a domain in bPBPs implicated in the activation of their partner SEDS proteins (9, 10, 42). Therefore, the effect of these substitutions is likely mediated by stimulating PG polymerase activity, not cross-linking.
In addition, in E. coli, a corresponding activated derivative of FtsW along with superfission variants of FtsL and FtsB were shown to overcome division inhibition by a dominant-negative version of the FtsEX complex that is thought to block the activation of septal PG biogenesis (18). Thus, the genetic studies of septal PG biogenesis regulation largely support FtsW activity, not the PG polymerase activity of PBP1b or the TPase activity of FtsI, as the target of the divisome activation pathway involving FtsL and FtsB. The genetic and biochemical data in P. aeruginosa reported here reinforce this connection and support a model in which the FtsWI synthase is the direct regulatory target of FtsQLB with the complex functioning as an activator of PG polymerization, not an inhibitor.
Although our results indicate that FtsQLB is an activator of PG polymerization by FtsWI, how the formation of the activated complex is normally controlled in cells remains unclear. The finding that the SFFtsL variant did not lead to additional stimulation of PG polymerization by FtsQLB-WI complexes suggests that the complexes that we purify are already in the activated state triggered on divisome formation. One possibility for why other divisome proteins like FtsN are not required to achieve this activated state in vitro is that the P. aeruginosa divisome is regulated differently than E. coli. However, FtsN is required for normal division in both organisms, and in both cases its function can be bypassed by SFFtsL variants (16, 17). Thus, despite the sequence differences between E. coli and P. aeruginosa, the overall process of septal PG synthesis activation likely proceeds by similar mechanisms in the two organisms.
Rather than differences between model organisms, we favor the possibility that the co-overproduction of FtsQ, FtsL, FtsB, FtsW, and FtsI promotes the spontaneous formation of an activated PG synthase complex, and that it does so via a mechanism analogous to the mode of action of the superfission variants of FtsL and FtsB. At their normally low cellular concentrations (low hundreds of molecules per cell), we suspect that FtsN and other divisome proteins are needed to promote septal PG biogenesis because they 1) increase the affinity of the FtsQLB subcomplex for FtsWI, 2) promote a conformational change in FtsQLB that activates it, and/or 3) relieve the inhibitory activity of another divisome component. In any case, it is reasonable to think that increasing the concentrations of FtsQ, FtsL, FtsB, FtsW, and FtsI in the membrane might subvert this regulation, obviating the requirement for other divisome proteins in forming the activated complex. Similarly, we suspect that the DNFtsL variants that retain FtsWI activation activity in vitro are also likely to be defective in these earlier activation events, such that they too are overcome by the overexpression and purification of the FtsQLB-WI complex.
Given how readily the functions of FtsN and other divisome proteins implicated in divisome activation, such as FtsEX, FtsK, and FtsA, can be bypassed by superfission variants of FtsL and FtsB in vivo, it is actually not surprising that parts of the activation pathway can also be easily bypassed in vitro. Thus, although our biochemical system does not report on all the steps of divisome activation, we believe that it faithfully represents the final step of septal PG synthesis initiation by FtsWI. Further work is needed to reconstitute earlier stages of divisome assembly to further dissect the activation pathway, the role played by the other core divisome proteins in the process, and how the substitutions in several of the DNFtsL variants block division. It will also be interesting to determine whether FtsWI is ultimately activated by a conformational change in the non-TPase domain of FtsI similar to that implicated in activation of the analogous RodA-PBP2 synthase of the Rod complex (elongasome) (9, 42). The reconstitution of activated PG synthesis by a subcomplex of the cytokinetic ring paves the way for investigating these and other important mechanistic questions. In addition, the established biochemical system for divisome-mediated PG synthesis will also aid the discovery of small molecule inhibitors of this activity for antibiotic development.
Methods and Materials
Materials.
Unless indicated otherwise, all chemicals and reagents were purchased from Sigma-Aldrich. Restriction enzymes were purchased from New England BioLabs. Oligonucleotide primers were purchased from Integrated DNA Technologies. E. faecalis lipid II was isolated from cells as described previously (43). S. aureus PBP4 was expressed and purified as described previously (43, 44).
Bacterial Strains, Plasmids, Oligonucleotide Primers, and Culture Conditions.
E. coli strains were grown with shaking at 37 °C in Lysogeny broth (LB), in Terrific broth, or on LB agar as indicated. P. aeruginosa strains, all derivatives of PAO1, were grown with shaking at 37 °C in LB, in M9 broth containing 0.2% casamino acids and 0.2% glucose, or on LB agar as indicated. The following concentration of antibiotics were used: ampicillin (Amp), 50 µg/mL; chloramphenicol (Cam), 25 µg/mL; gentamicin (Gent), 15 µg/mL (E. coli); Gent, 30 µg/mL (P. aeruginosa); and kanamycin (Kan), 25 µg/mL Plasmids were maintained in DH5a(λpir). The bacterial strains, plasmids, and oligonucleotide primers used in this study are summarized in SI Appendix, Tables S3–S5. The protocols for plasmid and strain construction are provided in SI Appendix.
Allelic Replacement in P. aeruginosa PAO1.
Chromosomally expressed fluorescent protein fusions in P. aeruginosa were constructed using an established allelic replacement protocol (45). The protocol is described in detail in SI Appendix.
Assessment of Dominant-Negative Phenotypes.
A single colony of P. aeruginosa strain LSM10 [zapA-mCherry sfgfp-ftsI] containing an ftsL or ftsB overexpression plasmid was streaked on an LB Gent agar with or without 1 mM IPTG to induce expression of the cloned ftsL or ftsB allele. Strains that were inducer-sensitive were selected for further analysis.
Protein Expression and Purification.
Expression and purification of P. aeruginosa protein complexes was performed as described previously (10). Details are provided in SI Appendix.
PG Glycosyltransferase Assays.
The protocol for detecting lipid II/peptidoglycan was adapted from previously published methods (44, 46, 47), and that for P. aeruginosa FtsW-FtsI has been described previously (10). More details are provided in SI Appendix.
Microscopy and Image Acquisition.
Cells were grown overnight in LB Gent at 37 °C. In the morning, cultures were diluted (1:500) into M9 minimal medium with 0.2% (wt/vol) glucose and Gent on the day of imaging. Cells were allowed to grow for 2 h at 37 °C before the induction of DNftsL expression using 1 mM IPTG. Cells were allowed to grow for a further 3 h before being immobilized on 1.5% M9 agarose pads and covered with #1.5 coverslips (48). Phase-contrast and fluorescence microscopy images were obtained using a Nikon Ti inverted microscope fitted with a Nikon motorized stage, an Andor Zyla 4.2 Plus sCMOS camera, Lumencore SpectraX LED Illumination, a Plan Apo 100x/1.45 oil Ph3 DM objective lens, and Nikon Elements 4.30 acquisition software. Images in the green and red channels were obtained using Chroma 49002 and 49008 filter cubes, respectively, with 1-s exposures. Colocalization of ZapA-mCherry and sfGFP-FtsI signals was determined by manual examination of each micrograph using Fiji software.
Supplementary Material
Acknowledgments
We thank all the members of the T.G.B. and Rudner labs at Harvard Medical School for their thoughtful discussions and advice throughout this project. Special thanks to Grasiela Torres for providing the ftsN and ftsL mutants of P. aeruginosa. We also thank Andrew Kruse and Megan Sjodt for advice on membrane protein purification; Suzanne Walker and her lab for advice on lipid II purification and the PG polymerization assay setup; and Paula Montero Llopis and the other members of MicRoN (Microscopy Resources on the North Quad) team at Harvard Medical School for help with microscopy. This work was supported by the NIH (AI083365, to T.G.B.) and Investigator funds from the HHMI. L.S.M. was supported in part by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004598117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Bi E. F., Lutkenhaus J., FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Loose M., Mitchison T. J., The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X. et al., GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisson-Filho A. W. et al., Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du S., Lutkenhaus J., At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 27, 781–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H., Patel V., Helmann J. D., Dörr T., Don’t let sleeping dogmas lie: New views of peptidoglycan synthesis and its regulation. Mol. Microbiol. 106, 847–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P., The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Meeske A. J. et al., SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohs P. D. A. et al., A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taguchi A. et al., FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller P. et al., The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J. Biol. Chem. 282, 36394–36402 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Bertsche U. et al., Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61, 675–690 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Leclercq S. et al., Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 7, 43306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boes A., Olatunji S., Breukink E., Terrak M., Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio 10, e01912-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerding M. A. et al., Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B., Persons L., Lee L., de Boer P. A. J., Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945–970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang M.-J., Bernhardt T. G., A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du S., Pichoff S., Lutkenhaus J., FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. U.S.A. 113, E5052–E5061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissler B., Elraheb D., Margolin W., A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 4197–4202 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernard C. S., Sadasivam M., Shiomi D., Margolin W., An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64, 1289–1305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du S., Lutkenhaus J., Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang M.-J., Bernhardt T. G., Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 24, 60–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel R. A., Errington J., Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36, 278–289 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Buddelmeijer N., Judson N., Boyd D., Mekalanos J. J., Beckwith J., YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. U.S.A. 99, 6316–6321 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman L.-M., Barondess J. J., Beckwith J., FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 174, 7716–7728 (1992). [PMC free article] [PubMed] [Google Scholar]
- 26.Gueiros-Filho F. J., Losick R., A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogliano J., Pogliano K., Weiss D. S., Losick R., Beckwith J., Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. U.S.A. 94, 559–564 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss D. S., Chen J. C., Ghigo J. M., Boyd D., Beckwith J., Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181, 508–520 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S. A. et al., General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 5189–5194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato J., Suzuki H., Hirota Y., Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol. Gen. Genet. 200, 272–277 (1985). [DOI] [PubMed] [Google Scholar]
- 31.Greene N. G., Fumeaux C., Bernhardt T. G., Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, 3150–3155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson D. C., Popham D. L., Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185, 1423–1431 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel R. A., Harry E. J., Katis V. L., Wake R. G., Errington J., Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29, 593–604 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Harry E. J., Stewart B. J., Wake R. G., Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol. Microbiol. 7, 611–621 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Levin P. A., Losick R., Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J. Bacteriol. 176, 1451–1459 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spratt B. G., Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 72, 2999–3003 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noirclerc-Savoye M. et al., Reconstitution of membrane protein complexes involved in pneumococcal septal cell wall assembly. PLoS One 8, e75522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho H., Uehara T., Bernhardt T. G., Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modell J. W., Hopkins A. C., Laub M. T., A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25, 1328–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modell J. W., Kambara T. K., Perchuk B. S., Laub M. T., A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, e1001977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lariviere P. J. et al., An essential regulator of bacterial division links FtsZ to cell wall synthase activation. Curr. Biol. 29, 1460–1470.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sjodt M. et al., Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat. Microbiol. 5, 813–820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh M. A. et al., Identification of a functionally unique family of penicillin-binding proteins. J. Am. Chem. Soc. 139, 17727–17730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiao Y. et al., Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hmelo L. R. et al., Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjodt M. et al., Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 556, 118–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett D. et al., Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J. Biol. Chem. 282, 31964–31971 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Llopis P. M., Visualizing bacillus subtilis during vegetative growth and spore formation. Methods Mol. Biol. 1431, 275–287 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.