Significance
This study describes the discovery of a class of drug that can help the immune system eliminate hard-to-kill cells infected with HIV. HIV establishes a persistent infection for which there is no cure, necessitating the development of new approaches to enhance the clearance of HIV-infected cells. HIV encodes Nef, which down-regulates MHC-I expression in infected cells to impair immune-mediated clearance by cytotoxic T lymphocytes. We identified the plecomacrolide family of natural products as potent inhibitors of Nef, and concanamycin A restored MHC-I and enhanced the clearance of HIV-infected primary cells by cytotoxic T lymphocytes. Concanamycin A counteracted Nef from diverse clades of HIV targeting multiple allotypes of MHC-I, indicating the potential for broad therapeutic utility.
Keywords: HIV, MHC-I, Nef, cytotoxic T lymphocytes, concanamycin A
Abstract
Nef is an HIV-encoded accessory protein that enhances pathogenicity by down-regulating major histocompatibility class I (MHC-I) expression to evade killing by cytotoxic T lymphocytes (CTLs). A potent Nef inhibitor that restores MHC-I is needed to promote immune-mediated clearance of HIV-infected cells. We discovered that the plecomacrolide family of natural products restored MHC-I to the surface of Nef-expressing primary cells with variable potency. Concanamycin A (CMA) counteracted Nef at subnanomolar concentrations that did not interfere with lysosomal acidification or degradation and were nontoxic in primary cell cultures. CMA specifically reversed Nef-mediated down-regulation of MHC-I, but not CD4, and cells treated with CMA showed reduced formation of the Nef:MHC-I:AP-1 complex required for MHC-I down-regulation. CMA restored expression of diverse allotypes of MHC-I in Nef-expressing cells and inhibited Nef alleles from divergent clades of HIV and simian immunodeficiency virus, including from primary patient isolates. Lastly, we found that restoration of MHC-I in HIV-infected cells was accompanied by enhanced CTL-mediated clearance of infected cells comparable to genetic deletion of Nef. Thus, we propose CMA as a lead compound for therapeutic inhibition of Nef to enhance immune-mediated clearance of HIV-infected cells.
The development of combination antiretroviral therapy (ART) has drastically altered the course of the HIV epidemic, yet HIV infection remains a lifelong condition for which there is no cure. The virus persists despite the presence of HIV-specific cytotoxic T lymphocytes (CTLs), the main effectors of cellular adaptive immunity responsible for clearing viral infections (1, 2). While rare elite controllers with particularly potent CTLs or CTLs targeting vulnerable antigens can spontaneously suppress the virus, even these individuals fail to clear the infection (3–5). In controllers or ART-treated patients with suppressed viral loads, HIV persists in long-lived latent reservoirs of virus. Approaches to clear these reservoirs by reactivating latent viruses have provided evidence that latency can be reversed in vivo, but this alone is unlikely to eliminate infected cells, and the cells harboring latent HIV may be resistant to CTL killing (6–10). Thus, new strategies are needed to enhance the clearance of infected cells following latency reversal.
CTLs recognize peptide antigens presented in the context of major histocompatibility complex class-I (MHC-I) on the surface of infected cells, mediating death of the target cell through perforin and Fas lytic pathways (11). MHC-I is both polygenic, with genes encoding HLA-A, -B, -C, -E, -F, and -G, and polymorphic, with remarkable allelic variation particularly in HLA-A, -B, and -C (12). Polygeny allows for functional separation as HLA-A and -B, and to a lesser extent -C, are responsible for presenting peptides to CTLs, which recognize non-self antigens expressed by intracellular pathogens. HLA-C, -E, and -G are predominantly responsible for inhibiting the responsiveness of natural killer (NK) cells, which recognize cells with low MHC-I (13) and elevated natural killer cell activating ligands (14). Allelic variation in the antigen-presenting forms of MHC-I yields alleles that are optimized for presentation of diverse peptides (15). Some alleles of HLA-B, in particular, are associated with rapid or delayed progression of HIV disease, and this may be attributable to whether the optimal peptide repertoire for an allele includes vulnerable regions in the HIV genome (5, 16, 17).
MHC-I is loaded with peptides in the endoplasmic reticulum (ER) and proceeds through the secretory pathway to reach the cell surface (18). The HIV accessory protein Nef alters MHC-I trafficking by binding to the cytoplasmic tail of MHC-I early in the secretory pathway, stabilizing an interaction between a tyrosine residue in the MHC-I cytoplasmic tail and the tyrosine-binding pocket in the μ subunit of clathrin adaptor protein 1 (AP-1) (19, 20). Formation of the AP-1:Nef:MHC-I complex mediates the redirection of MHC-I into the endolysosomal trafficking pathway in an ADP ribosylation factor-1 (ARF-1)–dependent manner where it is degraded in the lysosome (21, 22). Lysosomal acidification, which is required for the function of lysosomal proteases responsible for this degradation, is maintained by the vacuolar H+-ATPase (V-ATPase), a rotary proton-pumping motor (23). X-ray crystallography (24) and cryo-electron microscopy analyses (25) have confirmed the direct contacts between Nef, MHC-I, AP-1, and ARF-1 and described the structural basis for these interactions (26).
As a result of these interactions, HIV-infected cells expressing Nef experience a loss of cell-surface MHC-I, which protects them from being killed by HIV-specific CTLs (27). Nef binds specifically to the cytoplasmic domains of HLA-A and -B, but not HLA-C and -E. Because of their different functional roles, this differentiation optimizes evasion of both CTL and NK cell responses and is conserved across primate lentiviruses (28–30). The identification of a potent inhibitor of Nef that restores MHC-I to the surface of HIV-infected cells therefore represents an important and perhaps essential step toward the goal of efficiently clearing HIV reservoirs following therapeutic latency reversal. Here, we describe that the plecomacrolide family of natural products, in particular concanamycin A (CMA), potently counteracts Nef down-regulation of MHC-I to enhance CTL-mediated clearance of HIV-infected primary lymphocytes.
Results
Screening for Natural Product Inhibitors of HIV Nef.
To identify a Nef inhibitor capable of reversing MHC-I downmodulation in HIV-infected cells, we performed a high-throughput flow cytometric screen for compounds that increased expression of recombinant HLA-A2 in a CEM T cell line (CEM-A2) expressing Nef from a gutted adenoviral vector (SI Appendix, Fig. S1A) (31). HLA-A2 was chosen as a representative allele of MHC-I because HLA-A allotypes are strongly targeted by Nef for down-regulation (28), HLA-A2 is abundant in diverse ethnic groups (32), and a high-affinity monoclonal antibody selective for HLA-A2 (BB7.2) is available (33). Initial screening of over 200,000 small molecules failed to identify convincing hits. Screening of over 20,000 natural product extracts (34) identified 37 that were positive in the primary screen and negative in the counter screen. A secondary screen using CEM-A2 cells infected with a VSV-G pseudotyped single-round reporter virus derived from the HIV NL4-3 molecular clone (NL4-3-ΔGPE-EGFP [ΔGPE]) (Fig. 1A) confirmed that extracts from 11 strains inhibit Nef in the context of HIV infection (SI Appendix, Fig. S1 B and C).
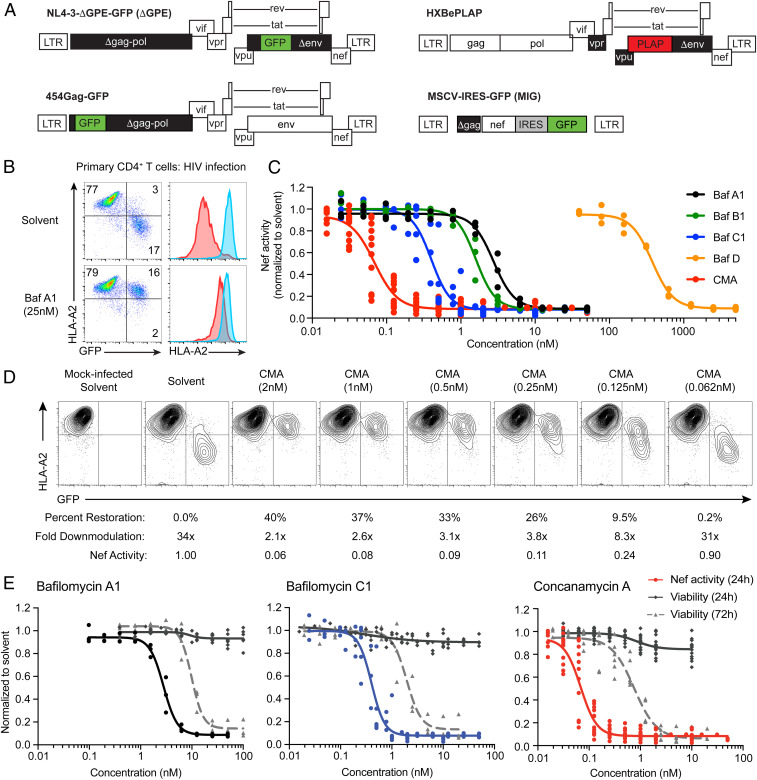
Fig. 1.
Plecomacrolides possess distinct potencies for Nef inhibition and cellular toxicity, which are separable for CMA. (A) Schematic representations of viral genomes used throughout the manuscript. Deleted genes are in black, HIV genes are in white, reporter genes are in red or green. (B) Representative flow cytometry plots (n = 3) from primary CD4+ T cells infected with HIV ΔGPE and treated with Baf A1 as in SI Appendix, Fig. S1B. Blue histograms are from GFP− cells; red histograms are from infected GFP+ cells. Compiled data are in C. (C) Summary graph of flow cytometric data from primary CD4+ T cells infected as in B and treated with the indicated plecomacrolides (n = 3 for Baf A1, B1, and D; n = 8 for Baf C1; n = 12 for CMA). Nef activity is the fold downmodulation of Nef normalized to solvent control as shown in D. (D) Representative flow cytometric plots from experiments summarized in C, shown from the donor with results closest to the mean among 12 donors tested. Percent restoration, fold downmodulation, and Nef activity were calculated as described in Materials and Methods. (E) Summary graph comparing Nef activity as in D after 24 h (n = 3 for Baf A1, n = 8 for BafC1, n = 12 for CMA) and viability as in SI Appendix, Fig. S8A after 24 h (n = 7 for Baf A1, n = 8 for Baf C1, n = 15 for CMA) and 72 h of plecomacrolide exposure (n = 3 for Baf A1 and Baf C1, n = 6 for CMA). Solvent control is dimethyl sulfoxide (DMSO). Extra sum-of-squares F test was used to compare IC50 values of curves.
Natural Product Nef Inhibitors Are Plecomacrolides.
Natural product metabolites with anti-Nef activity analyzed by NMR and mass spectrometry were identified as members of the bafilomycin (Baf) plecomacrolide family of natural products (SI Appendix, Figs. S2 A–C and S3A) (35). Baf A1 and C1 purified from our natural product extracts by standard fractionation procedures had near identical activity to commercial sources of these compounds (SI Appendix, Fig. S3 B and C). Baf A1 dramatically increased cell-surface HLA-A2 in CEM-A2 cells expressing Nef from the cytomegalovirus promoter in the context of an adenoviral delivery vector (SI Appendix, Fig. S4 A and B) and the HIV long terminal repeat (LTR) using ΔGPE (SI Appendix, Fig. S4 C and D). Nef-dependent HLA-A2 downmodulation was reduced by over 10-fold in CEM-A2 cells (SI Appendix, Fig. S4 C and D) and by 18-fold in primary T cells transduced with ΔGPE (Fig. 1B).
Plecomacrolides Have Diverse Nef Inhibitory Potencies and Achieve Superior Restoration of MHC-I Compared to Recently Identified Nef Inhibitors B9 and Lovastatin.
The plecomacrolide family includes the bafilomycins, which have a characteristic 16-member ring, and the concanamycins, which have an 18-member ring (SI Appendix, Fig. S3A). We exposed HIV-infected primary CD4+ T cells to plecomacrolide family members and determined that each restored MHC-I to similar levels, but with variable potencies (Fig. 1 C and D). CMA counteracted Nef at the lowest concentrations (50% effective concentration [EC50] = 0.07 nM) while Baf C1 was most potent among the bafilomycins (EC50 = 0.4 nM, Baf B1 = 1.6 nM, Baf A1 = 2.8 nM, Baf D = 380 nM). We further confirmed that the effects of CMA on Nef activity could not be attributed to a reduction in viral gene expression (SI Appendix, Fig. S5).
For comparison, we also tested recently identified Nef inhibitors B9 and lovastatin. Both B9 (36, 37) and lovastatin (38) have been reported to impair multiple Nef functions, with the effects of lovastatin evident only at supratherapeutic concentrations. We observed no effect of B9 on Nef-dependent MHC-I downmodulation across a wide range of concentrations, including those that had been previously reported to inhibit Nef in cells (SI Appendix, Fig. S4 A–F) (36, 37). For lovastatin, we observed little restoration of MHC-I at 24 h posttreatment but did confirm that lovastatin partially restored MHC-I to the surface of Nef-expressing cells after 48 h of exposure. However, restoration of MHC-I by lovastatin required 2,000-fold higher concentrations than CMA and failed to achieve comparable levels (SI Appendix, Fig. S4 G and H). To confirm that the negative results achieved with B9 were not due to receipt of the wrong compound, we independently validated that the purchased material matched the published structure of B9 by 1H NMR (SI Appendix, Fig. S6) and mass spectrometry (SI Appendix, Fig. S7) analysis. Based on these results, we conclude that CMA achieves the greatest magnitude of MHC-I restoration and is the most potent Nef inhibitor yet described.
CMA Restores MHC-I at Concentrations That Are Nontoxic to Primary Cells.
High-dose plecomacrolide treatment is toxic to cells, and questions remain over the safety and utility of plecomacrolides in clinical applications targeting V-ATPase activity (39–41). We did not observe any notable toxicity with 24-h exposure to plecomacrolides in the above or any subsequent experiments (Fig. 1E). However, in agreement with published reports, we observed toxicity when primary cells were exposed to plecomacrolides at high concentrations for extended periods (42, 43). Nevertheless, based on MTT assays and flow cytometric viability assays (SI Appendix, Fig. S8 A and B), inhibition of Nef in CD4+ primary T cells occurred at concentrations that were nontoxic even with 72 h of direct exposure (Fig. 1E). For CMA, there was an 11-fold difference between the 50% effective and toxic concentrations (EC50 and TC50). This compared with 3.5-fold and 4.8-fold differences for Baf A1 and C1, respectively (Fig. 1E). For subsequent experiments in primary CD4+ T cells, we utilized 24-h incubations with 0.5 nM CMA, which suppressed Nef activity by 10-fold and maintained 95% viability (Fig. 1E).
We also found that G0/G1 cell cycle arrest, a reported effect of plecomacrolide exposure, was minimal even at concentrations well above the EC50 for Nef inhibition (SI Appendix, Fig. S8C) (44). CD4+ T cells treated with 0.5 nM CMA showed a 1.15-fold increase in the proportion of cells in S phase compared to the solvent control, but the corresponding decrease in cells in G2/M was not statistically significant. Given the small magnitude of these changes, it is unlikely that cell cycle arrest meaningfully contributes to toxicity in cells treated with 0.5 nM CMA. Thus, plecomacrolides, and particularly CMA, may be promising lead compounds for therapeutic Nef inhibition.
Plecomacrolides Have Distinct Nef Inhibitory and Lysosome Neutralization Potencies.
The toxicity associated with plecomacrolide treatment likely results from their inhibition of V-ATPase, which is responsible for many cellular processes, including lysosomal acidification (45). To determine whether inhibition of lysosomal pH might be responsible for reversal of MHC-I downmodulation in Nef-expressing cells, we adapted a previously described method to measure the pH of the lysosome of primary human monocyte-derived macrophages (MDMs) by measuring ratiometric fluorescence of an endocytosed dextran (46) (SI Appendix, Fig. S9A). We first confirmed that Baf A1 completely neutralized lysosomal pH (SI Appendix, Fig. S9 B and C). We then tested each of the plecomacrolides over a range of concentrations. Interestingly, the most potent inhibitor of Nef, CMA, was not the most potent inhibitor of V-ATPase. Instead, Baf C1 (EC50 = 7.3 nM) neutralized lysosomes more potently than CMA (EC50 = 12.7 nM, P < 0.0001), which had comparable potency to Baf A1 (EC50 = 18.5 nM, P = 0.06) (Fig. 2A). This evidence indicated a qualitative separation between plecomacrolide inhibition of Nef in primary T cells and V-ATPase–mediated acidification in MDMs.
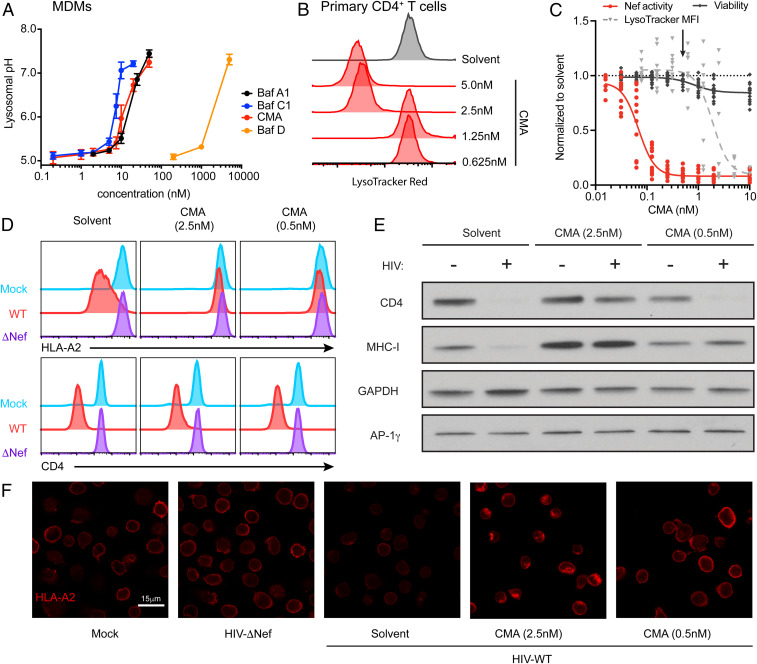
Fig. 2.
Lysosome function and acidification remain intact at concentrations of CMA that restore MHC-I. (A) Summary graph of flow cytometric data from MDMs as in S9A-C, treated with plecomacrolides for 1 h as indicated (n = 8 for Baf A1 and Baf C1, n = 2 for Baf D, n = 6 for CMA). (B) Representative flow cytometry histograms of primary activated CD4+ T cells treated for 24 h with CMA as indicated and incubated with Lysotracker Red for 1 h. (C) Summary graph of flow cytometric data from B comparing the normalized median fluorescence intensity (MFI) of Lysotracker Red (n = 9) with the normalized Nef activity (n = 12) and viability (n = 15) from Fig. 1E in primary CD4+ T cells treated with CMA at the indicated concentrations. The arrow indicates the concentration of CMA used in primary cells in Figs 4 and 5. (D) Representative flow cytometry histograms from primary activated CD4+ T cells infected with HxBePLAP (Fig. 1A) for 72 h, sorted for PLAP+ cells, and treated with CMA as indicated for 24 h. Blue histograms represent mock-infected cells, red histograms represent sorted PLAP+ cells infected with HXBePLAP (wild type [WT]), and purple histograms represent sorted PLAP+ cells infected with HXBePLAP in which Nef was deleted (ΔNef, representative of three independent experiments). (E) Western blot of whole cell lysates from CD4+ T cells prepared as described for D (representative of four independent experiments). (F) Representative confocal microscopy images of primary activated CD4+ T cells prepared as described in D, stained for HLA-A2. Mock cells are uninfected. All images were captured with identical microscope settings. Solvent control is DMSO. An extra sum-of-squares F test was used to compare IC50 values of curves.
Because CD4+ T cells did not efficiently endocytose dextran, we assessed lysosomal neutralization in these cells with Lysotracker Red dye, which freely crosses cell membranes until it reaches an acidic compartment, where it is protonated and retained. As measured by flow cytometry (Fig. 2 B and C) and confocal microscopy (SI Appendix, Fig. S9D), the EC50 for lysosome neutralization by CMA in primary CD4+ T cells (EC50 = 1.9 nM) was significantly higher than the EC50 for Nef inhibition (EC50 = 0.07 nM, 27-fold difference, P < 0.0001). Taken together, these results indicate that CMA counteracted Nef in primary human CD4+ T cells at concentrations that were nontoxic and did not alter lysosomal pH.
CMA Restores Cell-Surface MHC-I, but Not CD4, in Nef-Expressing Cells.
Both MHC-I and CD4 are targeted to the lysosome by Nef, but by distinct mechanisms. MHC-I is redirected from the trans-Golgi network to the lysosome via the AP-1 adaptor complex (19, 20) while CD4 is internalized from the cell surface and trafficked to the lysosome in an AP-2–dependent manner (47). Based on the above data, we hypothesized that surface restoration of MHC-I was not simply secondary to lysosome dysregulation. To explore this hypothesis, we treated pure populations of HIV-infected primary cells with a high dose of CMA that neutralizes the lysosome (2.5 nM) or a low dose that leaves acidic compartments intact (0.5 nM). We found that neither dose of CMA reversed Nef-dependent down-regulation of cell-surface CD4 while down-regulation of surface MHC-I was reversed equally in both conditions (Fig. 2D). This indicated that CMA specifically redirects MHC-I, and not all proteins targeted to the lysosome by Nef, to the cell surface.
CMA Restores MHC-I in Nef-Expressing Cells with Functional Lysosomal Protein Degradation.
We then sought to confirm that CMA restores MHC-I by a mechanism independent of its effects on lysosomal degradation by directly observing Nef-mediated lysosomal degradation. As expected, both MHC-I and CD4 were degraded in Nef-expressing cells (Fig. 2E, leftmost lanes). A high dose of CMA, which neutralized the lysosome (Fig. 2 B and C and SI Appendix, S9D), inhibited Nef-mediated degradation of both HLA-A2 and CD4 (Fig. 2E, centermost lanes). Notably, high-dose CMA also increased MHC-I expression in uninfected cells, consistent with disruption of the steady-state turnover of MHC-I in the lysosome. Low-dose CMA, however, did not prevent degradation of CD4 and did not increase steady-state levels of MHC-I, indicating that the lysosome was functional for protein degradation. Despite this, MHC-I was not degraded in Nef-expressing cells (Fig. 2E, rightmost lanes). Thus, low-dose CMA treatment selectively alters the transport of MHC-I in HIV-infected primary cells, preventing redirection to the lysosome and restoring MHC-I to the cell surface.
To validate these results, we performed immunofluorescence microscopy on pure populations of HIV-infected primary CD4+ T cells. Cells infected with Nef-expressing HIV had a dramatic reduction in cell surface and total expression of HLA-A2 compared to uninfected cells or cells infected with Nef-deficient HIV (Fig. 2F). Exposure to 0.5 nM CMA restored the appearance of MHC-I staining to that observed in the absence of Nef (Fig. 2F). High doses (2.5 nM) of CMA caused accumulation of HLA-A2 in intracellular compartments, consistent with inhibition of lysosomal degradation. Taken together, these experiments confirmed that low doses of CMA that do not disrupt lysosomal function specifically restore MHC-I to the surface of Nef-expressing HIV-infected CD4+ T cells. These results strongly suggest a role for CMA in this process that is independent of its effects on lysosomal pH through its known target, V-ATPase.
CMA Reduces the Association of Nef and AP-1 with MHC-I.
The observation that CMA selectively affects MHC-I and not CD4 degradation suggests that CMA disrupts the formation of the AP-1:Nef:MHC-I complex. To test this directly, we used CEM-A2 cells transduced with an adenoviral vector expressing Nef. This system has been used to study the formation of the AP-1:Nef:MHC-I complex under conditions where ammonium chloride (NH4Cl) prevents lysosomal degradation (19). Higher concentrations of CMA were required for reversal of Nef activity in CEM cells than in primary CD4+ T cells. However, we identified 1.25 nM CMA as a nontoxic concentration that inhibited Nef without significantly altering intracellular acidification (Fig. 3A). Thus, we consider this to be functionally similar to 0.5 nM CMA treatment in primary CD4+ T cells.
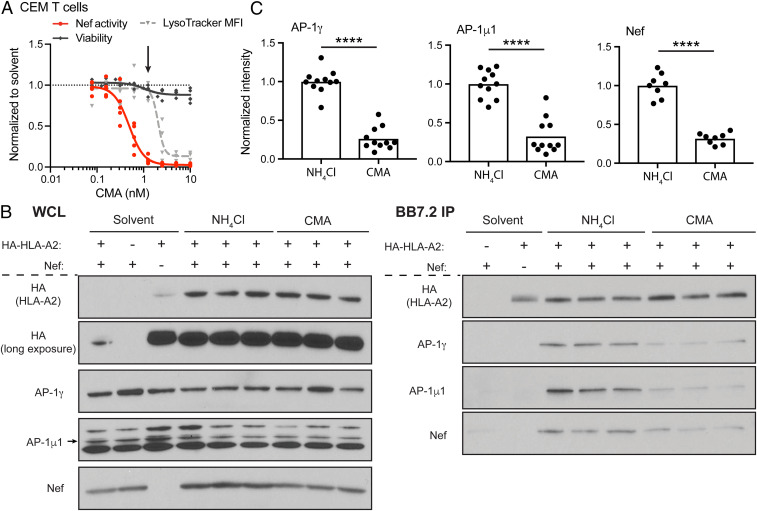
Fig. 3.
CMA reduces the abundance of Nef:MHC-I:AP-1 complexes. (A) Summary graph of flow cytometric data comparing the normalized median fluorescence intensity (MFI) of Lysotracker Red (n =4) with the normalized Nef activity following ΔGPE infection as in SI Appendix, Fig. S1B (n = 5) and viability as in SI Appendix, Fig. S8A (n = 5) in CEM-A2 cells treated with a range of CMA concentrations for 24 h. The arrow indicates the concentration of CMA used in remaining experiments with CEM cells. (B) Representative Western blot depicting three experimental replicates (of 11 total replicates) of whole cell lysates (WCL) before (Left) or matched samples after (Right) immunoprecipitation using BB7.2-conjugated beads (specific for HLA-A2) from CEM-SS or CEM-A2 cells transduced with Nef-expressing adenoviral vector construct or the control vector lacking Nef. NH4Cl, 35 mM NH4Cl; CMA, 1.25 nM CMA. (C) Summary graphs quantifying experimental replicates of Western blots for AP-1 subunits (n = 11) and Nef (n = 8) as in B. Band intensities were recorded for each protein from a single exposure in which all bands were visible but none were saturated. Band intensity was normalized to the intensity of HLA-A2 for each sample to account for differences in HLA-A2 recovery. Results were normalized to NH4Cl, and the mean of NH4Cl values was used where multiple replicates were run simultaneously as in B. ****P < 0.0001, unpaired two-tailed t tests.
Because both NH4Cl and CMA stabilized HLA-A2 expression to similar levels, we were able to compare whether CMA specifically resulted in a reduction in the abundance of the AP-1:Nef:MHC-I relative to what is observed under conditions of lysosomal neutralization. We found that CMA treatment led to a reproducible reduction in the abundance of Nef, AP-1γ, and AP-1μ1 subunits associating with HLA-A2 compared to cells treated with NH4Cl (Fig. 3B). These effects were highly significant when compiled across multiple experiments (Fig. 3C) (P < 0.0001). Control experiments confirmed the specificity of the assay, as detection of AP-1γ and AP-1μ1 in pulldowns required both HLA-A2 and Nef (Fig. 3B, leftmost lanes). In addition, we confirmed that the complex could not reliably be observed in the absence of NH4Cl due to robust HLA-A2 degradation and minimal HLA-A2 recovery in the presence of Nef (Fig. 3B, leftmost lanes).
To determine whether CMA directly binds to components of the AP-1:Nef:MHC-I complex, we performed differential scanning fluorimetry thermal stability assays using a comprehensive panel of purified components of the AP-1:Nef:MHC-I complex (SI Appendix, Fig. S10A), including the AP-1µ1–C-terminal domain (CTD), the MHC-I tail fused with HIV-1 NL4-3 Nef, the µ1-CTD:MHC-NL4-3 Nef complex, the AP-1 core, the AP-1 trimer containing AP-1 core:Arf1-GTP: MHC-NL4-3 Nef complex, and NL4-3 Nef alone (SI Appendix, Fig. S10 B–G). No significant changes in melting temperature (Tm) were observed with 1-h incubation of any of the samples with CMA compared to solvent control, indicating that CMA does not bind directly to any of these members of the ARF-1:AP-1:Nef:MHC-I complex in vitro. Furthermore, when the GST-tagged MHC-I cytoplasmic tail was immobilized on resin, the presence of CMA did not alter pulldown of NL4-3 Nef, simian immunodeficiency virus (SIV) Nef, or the AP-1μ1-CTD (SI Appendix, Fig. S10H. In summary, these results demonstrate that CMA impairs the formation of the AP-1:Nef:MHC-I complex in cells. However, we did not observe direct binding of CMA to known protein components of the complex, implicating the existence of an alternative target necessary for Nef-specific MHC-I trafficking.
CMA Enhances CTL-Mediated Clearance of HIV-Infected Cells Comparably to Genetic Deletion of Nef.
There is a large body of literature indicating that increases in cell-surface MHC-I on target cells yield proportional increases in CTL-mediated clearance of target cells (48–51). Given that Nef-expressing CD4+ T cells treated with 0.5 nM CMA have near normal surface expression of HLA-A2 (Fig. 2D), we hypothesized that CMA would eliminate Nef-mediated protection of HIV-infected cells from HIV-specific CTLs. To test this directly, we performed in vitro flow cytometric CTL killing assays (SI Appendix, Fig. S11A) with two HLA-A2–restricted CTL clones expressing T cell receptors specific for HLA-A2 presenting the Gag SL9 epitope, which is expressed in the HIV molecular clone HXBePLAP with placental alkaline phosphatase (PLAP) as the viral reporter (Fig. 1A) (27).
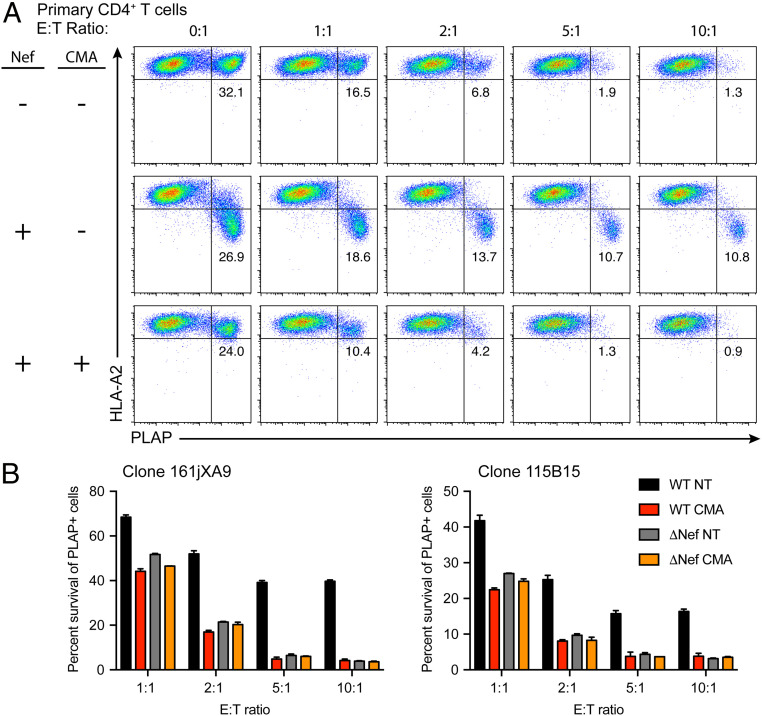
As previously observed (27), PLAP+ primary cells infected with a Nef-deleted virus were efficiently eliminated by CTLs (Fig. 4 A, Top Row). In contrast, CTL-mediated elimination of cells infected with a Nef-competent virus was notably reduced at every effector:target (E:T) ratio. Importantly, there was no further elimination of PLAP+ cells when the E:T ratio was increased from 5:1 to 10:1, indicating that there was a residual population of Nef-expressing cells that were highly resistant to clearance even by a large excess of potent HIV-specific CTLs (Fig. 4 A, Middle Row). Cells infected with Nef-competent virus and treated with 0.5 nM CMA, however, had restored HLA-A2 expression, and the PLAP+ subset was efficiently eliminated by CTLs (Fig. 4 A, Bottom Row). The effect of CMA on CTL killing of HIV-infected cells was indistinguishable from genetic deletion of Nef and was Nef-dependent as there was no increase in clearance of cells infected with Nef-deleted virus (Fig. 4B). Importantly, when target cells from a donor lacking HLA-A2 were cocultured with CTLs, there was no reduction in PLAP+ target cells regardless of whether they were treated with CMA (SI Appendix, Fig. S11B), validating the specificity of the CTLs and demonstrating that CMA only enhances clearance of HIV-infected cells in the presence of both Nef and specific anti-HIV CTL responses. Nef-dependent increases in killing of CMA-treated cells were statistically significant for both CTL clones tested (P < 0.0001) (SI Appendix, Fig. S11C). These observations confirm that low-dose CMA treatment of Nef-expressing cells restores HLA-A2 that is properly loaded with an HIV-derived peptide that can be successfully presented to CTLs without impairing responsiveness to CTL-derived lytic signals.
Fig. 4.
CMA enhances clearance of HIV-infected cells by HIV-specific CTLs. (A) Representative flow cytometry plots depicting CTL-mediated killing of primary CD4+ T cells infected with HXBePLAP plus or minus Nef (Fig. 1A) for 72 h and treated for 24 h with 0.5 nM CMA or matched DMSO solvent control prior to 4-h coculture with CTLs. Cells were gated for CD4+ T cell targets as in SI Appendix, Fig. S11A. (B) Summary graph of results from A in two independent experiments using two distinct CTL clones. Each condition was performed in duplicate, and survival of PLAP+ cells was determined by normalizing to the mean of quadruplicate 0:1 samples. Error bars represent SD. WT, HXBePLAP wild-type; ΔNef, HXBePLAP in which Nef was deleted; E:T, effector:target ratio, indicates the number of anti-HIV CTLs present in the 4-h coculture per CD4+ T cell target cell; NT, no treatment (matched DMSO solvent control); CMA, 0.5nM CMA.
Previous reports have indicated that concentrations of CMA greater than 1 nM could inhibit the effector functions of CTLs, but no such effect was observed at 0.5 nM CMA (52, 53). We confirmed these published results using our anti-HIV CTLs. We observed no change in the clearance of SL9 peptide-pulsed target cells in the presence of 0.5 nM CMA (SI Appendix, Fig. S11D).
CMA Reverses Nef-Mediated Down-Regulation of HLA-B in Primary Cells.
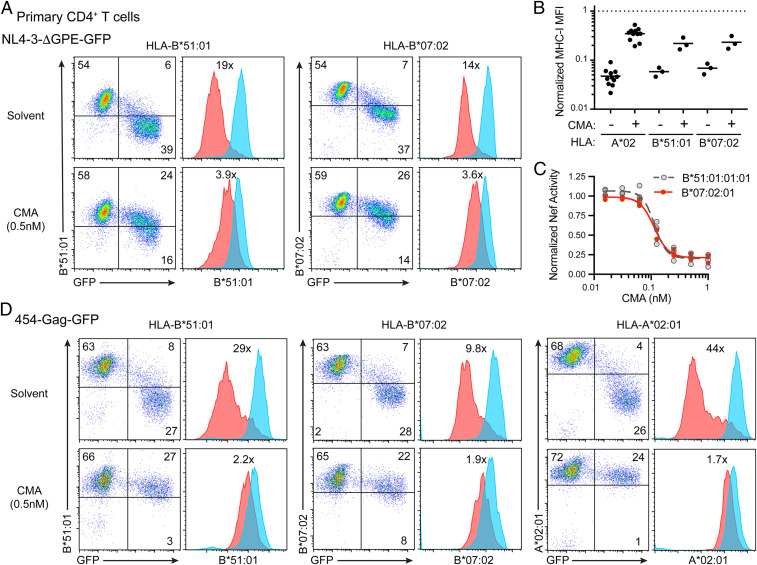
Nef down-regulates both HLA-A and HLA-B allotypes, and many patients possess robust HLA-B–restricted, HIV-specific CTLs (54–56). Thus, we sought to determine whether CMA would restore HLA-B expression in HIV-infected cells. Sequence differences in HLA-B allotypes classify them as either HLA-Bw4 or HLA-Bw6 serotypes. Each serotype can be detected with monoclonal antibodies, but these antibodies are cross-reactive with some HLA-A (Bw4) and HLA-C (Bw6) allotypes (57). As previously reported, we identified a donor that was heterozygous for Bw4 (B*51:01) and Bw6 (B*07:02) with no cross-reactive HLA-A alleles and minimal cross-reactivity from HLA-C, allowing us to reliably measure expression of two HLA-B alleles (SI Appendix, Fig. S12A) (57). We observed significant downmodulation of both HLA-B*51:01 and HLA-B*07:02 in cells infected with ΔGPE, which was consistently counteracted by CMA (Fig. 5 A–C). The effects of Nef and CMA on both HLA-B alleles in this donor were similar in magnitude to those observed for HLA-A*02 in an array of donors (Fig. 5B). Thus, we conclude that CMA can potently counteract Nef-mediated down-regulation of both HLA-A and HLA-B allotypes in primary CD4+ T cells.
Fig. 5.
CMA counteracts Nef-mediated HLA-B down-regulation in primary cells, including those expressing Nef from a primary isolate of HIV. (A) Representative flow cytometry plots (n = 3 independent replicates from a single donor) from primary CD4+ T cells infected with NL4-3-ΔGPE for 48 h, treated with 0.5 nM CMA for 24 h, and stained with monoclonal antibodies to Bw4 (B*51:01) and Bw6 (B*07:02). Blue histograms are from GFP− cells; red histograms are from infected GFP+ cells. (B) Summary graph of data from A plotting the MHC-I MFI from infected GFP+ cells normalized to that in uninfected cells treated with solvent control (dotted line). Data for HLA-A*02 are from independent experiments with 11 different donors, data for HLA-B*51:01 and HLAB*07:02 are from three independent experiments with a single donor. (C) Summary graph of data from A depicting the relative Nef activity against the indicated HLA-B allotypes in cells treated with a range of CMA concentrations (n = 3). (D) Flow cytometry plots from CD4+ T cells infected with 454-Gag-GFP (Fig. 1A) and treated and stained as in A (n = 1 for HLA-B allotypes, n = 3 for HLA-A*02). Solvent control is DMSO. Numbers indicate the proportion of live cells in each quadrant gate or the fold change in MHC-I MFI between infected and uninfected cells.
A Primary HIV Isolate from an Optimally Treated Patient Down-Regulates MHC-I and Is Inhibited by CMA.
Enhancing the clearance of latent reservoirs of virus that persist in optimally treated patients, likely following therapeutic reactivation from latency, is an important clinical application for a Nef inhibitor. We previously isolated a full-length provirus that was expressed as residual plasma virus in an optimally treated patient and was further shown to be infectious (58). We deleted Gag-Pol and introduced GFP, allowing identification of infected cells in a single round infection while preserving Nef from the original isolate (454-Gag-GFP) (Fig. 1A). Primary CD4+ T cells infected with 454-Gag-GFP demonstrated down-regulation of HLA-A*02:01, HLA-B*51:01, and HLA-B*07:02, which was Nef-dependent (Fig. 5D and SI Appendix, S12B). CMA restored expression of all three allotypes of MHC-I in the context of infection with the primary isolate virus (Fig. 5D), and this result was confirmed in two additional donors for HLA-A*02 (SI Appendix, Fig. S12C). Thus, we have demonstrated that subnanomolar concentrations of CMA can potently restore MHC-I to the surface of primary cells expressing Nef from a virus isolated from an optimally treated patient.
CMA Broadly Inhibits Nef Alleles from Diverse Clades of HIV and SIV Targeting a Range of MHC-I Alleles.
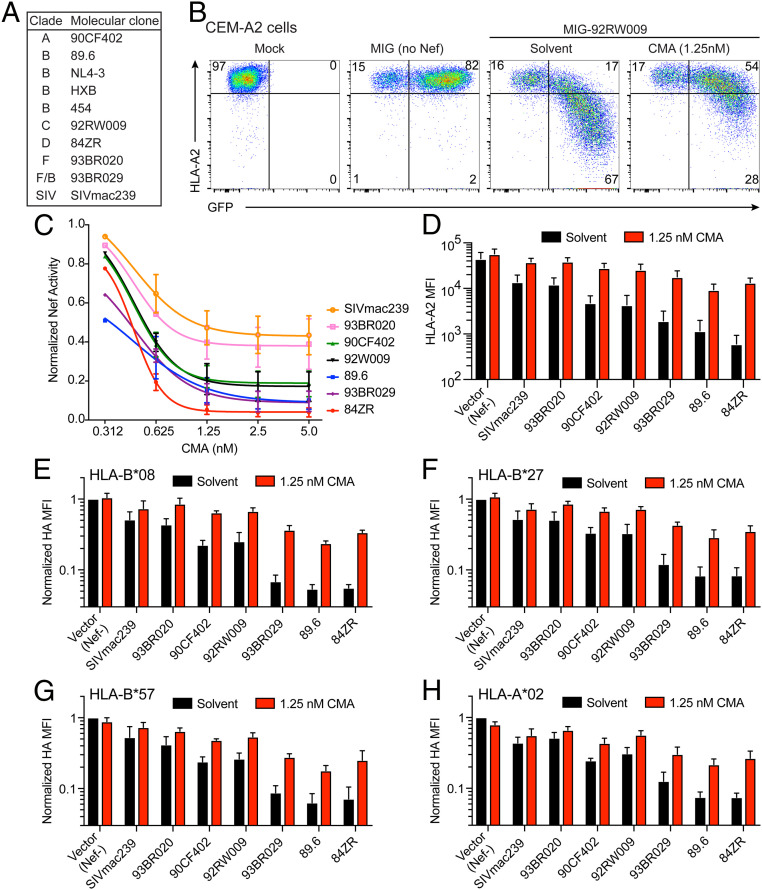
Globally, HIV possesses remarkable genetic diversity. To this point, we had only investigated the inhibitory activity of plecomacrolides against Nef alleles from NL4-3, HXB, and the 454 patient molecular clones, all of which are clade B viruses. To determine whether plecomacrolides offer broad therapeutic promise against a diverse range of Nef sequences, we tested nef alleles from HIV clades A, B, C, D, F, and F/B, as well as one from SIV (Fig. 6A) cloned into the murine stem cell virus internal ribosome entry site GFP (MSCV-IRES-GFP, MIG) vector (Fig. 1A) (59). We observed that CMA restored expression of HLA-A2 in cells expressing each nef allele, indicating that plecomacrolides broadly inhibit nef alleles from genetically diverse HIV isolates (Fig. 6 B–D), and the potency of CMA was comparable for each allele (Fig. 6C). CMA was able to restore HLA-A2 expression more completely for nef alleles that down-regulate HLA-A2 to a lesser extent but had the most dramatic effect on HLA-A2 expression in cells expressing the most potent nef alleles (Fig. 6D). Similarly, CMA restored surface expression of HA-tagged MHC-I allotypes HLA-A*02, HLA-B*08, HLA-B*27, and HLA-B*57 expressed in CEM cells. Each allele of Nef down-regulated each allele of MHC-I, with varying magnitudes, and CMA restored MHC-I surface expression in every context (Fig. 6 E–H). These observations support the hypothesis that CMA can enhance cellular adaptive immunity regardless of the initial degree of impairment and may provide therapeutic benefit despite HLA and viral diversity.
Fig. 6.
CMA inhibits Nef alleles from diverse clades of HIV and SIV targeting diverse alleles of MHC-I. (A) Summary of Nef alleles tested and clade of HIV or SIV to which the isolate belongs. (B) Representative flow cytometry plots depicting CEM-A2 cells infected with MSCV-IRES-GFP (MIG) alone or expressing the Nef allele from clade C HIV isolate 92RW009, the median Nef allele from C, and treated for 24 h with 1.25 nM CMA. (C) Summary graph of data from B, showing the relative Nef activity of each Nef allele after treatment with varying concentrations of CMA (n = 3). (D) Summary graph of HLA-A2 MFI from experiments shown in B and C. (E–H) Summary graphs of flow cytometric data from CEM cells expressing the indicated HA-tagged MHC-I alleles treated as in D. Cell-surface MHC-I expression was assessed by staining for HA, and the median fluorescence intensity in GFP+ cells was normalized to vector control for each cell line. (E and F) n = 4; (G and H) n = 3.
Discussion
In summary, we have identified plecomacrolides as potent inhibitors of HIV Nef-mediated down-regulation of MHC-I. Inhibition of Nef by CMA, in particular, occurs at concentrations that are nontoxic to primary T cells and that do not inhibit lysosome function. Restoration of cell-surface MHC-I in HIV-infected cells enhances their clearance by CTLs comparably to genetic deletion of Nef, confirming that the restored MHC-I is functional for presentation of viral antigens. We further demonstrated that CMA inhibits nef alleles isolated from diverse clades of HIV, an allele of SIV nef, and an allele of HIV nef from an optimally treated patient. Additionally, we found that CMA restored diverse allotypes of MHC-I in HIV-infected cells and that CMA treatment enhanced antigen presentation for all combinations of nef alleles and MHC-I alleles tested. These results provide evidence that inhibition of Nef by this mechanism could have broad clinical utility.
The identification of plecomacrolides as inhibitors of HIV Nef may seem intuitive, given that Nef redirects many of its targets for lysosomal degradation and plecomacrolides lead to potent lysosomal neutralization and loss of degradative capacities (19, 22, 60–63). Yet, we determined that CMA restored MHC-I in Nef-expressing primary CD4+ T cells at concentrations that were nontoxic and did not alter the function of the lysosome or reduce the abundance of acidified intracellular compartments. Thus, we propose that CMA specifically alters a step prior to lysosomal degradation. This may be an unrecognized activity of the known target, V-ATPase, or the activity of a novel non–V-ATPase target, although our data suggest this target is unlikely to be a component of the ARF-1:AP-1:Nef:MHC-I complex itself. Inhibition of this putative target activity reduces the capacity of Nef to interact with MHC-I and AP-1 in CMA-treated cells but does not alter the ability of Nef to down-regulate and degrade CD4. The fact that AP-1:Nef:MHC-I complex formation in cells is CMA-sensitive suggests that there may be more subtle pH-dependent steps that are unrelated to lysosomal function. Plecomacrolides have previously been shown to alter intracellular trafficking, but generally with the effect of reducing the expression of cell-surface markers rather than increasing them (64, 65). A subunit of V-ATPase has been reported to form a complex with Nef and CD4 to promote endocytosis (66–68). However, this subunit has not been implicated in Nef-dependent MHC-I trafficking, which occurs by a different mechanism that begins in the Golgi rather than at the cell surface (19, 31). Other explanations are also possible. For example, there may be an as yet unidentified target of CMA that is required for Nef activity against MHC-I.
In addition, we demonstrated that the potency of CMA is greater than previously published Nef inhibitors, including B9 and lovastatin. B9 failed to restore MHC-I to the surface of Nef-expressing cells in any of the assays employed, while lovastatin was able to restore MHC-I to a fraction of the levels achieved by CMA with prolonged incubations and supratherapeutic concentrations. The explanation for the negative result obtained with B9 is unclear. A recent paper showed small but statistically significant effects of B9 on CTL clearance, and the authors claimed these effects were mediated through reversal of Nef-dependent MHC-I down-regulation (37). Because of these reports, we confirmed the structure of the commercially obtained B9 to rule out the possibility that we had received the wrong compound. Our results clearly demonstrated that any potential effect of B9 on MHC-I down-regulation in cells is not biologically meaningful under the conditions of our assays.
In short-term cocultures with HIV-specific CTLs, primary CD4+ T cells infected with Nef-expressing HIV revealed a residual population of infected cells that could not be cleared from the culture, regardless of how many CTLs were present. If the cells were treated with CMA or Nef was genetically removed from the virus, this population was virtually nonexistent. This raises the possibility that Nef activity in a subset of HIV-infected cells in vivo renders those cells refractory to killing even by highly responsive CTLs. Following therapeutic reactivation in a “shock and kill” effort to eliminate the HIV reservoir, such cells could escape CTL killing long enough to proliferate and return to latency, reseeding the reservoir with clonally expanded sequences expressing potent alleles of nef. Our coculture assays demonstrate proof of concept that therapeutic Nef inhibition with low-dose CMA is sufficient to dramatically enhance the clearance of previously hard-to-kill cells when effective CTLs are present. Cells harboring reactivated HIV may be particularly resistant to clearance by even potent anti-HIV CTLs (10), inviting further investigation to determine whether CMA enhances the clearance of these cells.
The CTL response in vivo is polyclonal, with CTLs responding to a diverse array of HIV antigens presented predominantly by HLA-A and HLA-B. Furthermore, MHC-I is remarkably polymorphic, and HIV sequences are tremendously diverse both within and between infected individuals. A CTL-based therapeutic intervention will therefore need to function in a wide range of immune contexts. CTL responses restricted to HLA-B are predominant in HIV infection, and many MHC-I genes associated with HIV control are HLA-B alleles (54–56). We observed reversal of Nef down-regulation of HLA-A and HLA-B allotypes following CMA treatment. Restoration was particularly dramatic when MHC-I was targeted most strongly by Nef. Therefore, CMA could facilitate the elimination of resistant reservoirs of virus by enhancing the efficiency of both already-effective HLA-B-restricted and previously suboptimal HLA-A–restricted CTL responses. In the absence of Nef-mediated protection and in combination with vaccination strategies to increase the abundance and breadth of HIV-specific CTLs, antigens that had not experienced strong selective pressure to generate CTL-escape mutants prior to the initiation of ART could become vulnerable targets for immune-mediated clearance of HIV reservoirs.
Despite the promising nature of these results, several limitations warrant further investigation. CMA restores expression of diverse MHC-I allotypes in HIV-infected primary T lymphocytes, but CTL killing was only confirmed for a single MHC-I allotype. In addition, we report an 11-fold window between EC50 and TC50 for CMA in primary cells, which is encouraging for a lead compound. However, chemical modifications that further reduce toxicity and negative effects on CTLs or enhance activity will likely be necessary for clinical utility. Moreover, additional research using animal models is needed to determine the toxicity, accessibility, and efficacy of CMA in vivo. Lastly, Nef inhibition alone is unlikely to achieve a cure for HIV as latent reservoirs of virus express neither Nef nor viral-derived peptide antigens. Thus, the parallel development of improved latency-reversal agents (LRAs) will be needed to achieve a cure. LRAs that achieve sufficient reactivation from latency to sensitize infected cells to CTLs also cause T cell activation (10, 69, 70). The identification of LRAs that induce sufficient HIV gene expression for CTL recognition while maintaining CD4+ T cells in a resting state is a critical need. It is important to note that resting CD4+ T cells may harbor additional mechanisms to resist CTL killing that will need to be addressed (10, 70). Our data indicating that CMA is effective at enhancing CTL-mediated clearance is thus far limited to actively infected primary T cells and may not translate to clearance of reactivated resting cells in vivo.
In summary, we demonstrated that CMA potently counteracts HIV Nef at subnanomolar concentrations to restore immune-mediated clearance of HIV-infected cells. This approach has the potential to broadly enhance anti-HIV immunity in diverse immune contexts. Thus, we propose CMA as a lead compound for further development as a therapeutic inhibitor of Nef, a crucial addition to current efforts to cure HIV by eradicating residual viral reservoirs.
Materials and Methods
Detailed methodology can be found in SI Appendix.
Ethics Statement.
Anonymized leukocytes isolated by apheresis from healthy donors were obtained from the New York Blood Center.
For experiments for which the HLA genotype was determined, blood was collected in Ann Arbor, MI, with informed consent from healthy donors. This study was approved by the University of Michigan Institutional Review Board (HUM00071750).
Nef Inhibitory Compounds.
The following compounds were used as described below: B9 (500653; Calbiochem, MilliporeSigma), lovastatin (PHR1285; MilliporeSigma), Baf A1 (11038; Cayman Chemical), Baf B1 (14005; Cayman), Baf C1 (19624; Cayman), Baf D (19438; Cayman), and CMA (Fermentek, 80890-47-7; Cayman, 11050)
Preparation of Primary CD4+ T Lymphocytes and MDMs.
Anonymized leukocytes isolated by apheresis were obtained from the New York Blood Center, and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque Plus (17144002; GE Healthcare) centrifugation using SepMate tubes (85450; Stemcell Technologies) according to the manufacturer’s protocol. CD8+ lymphocytes were depleted with Dynabeads according to the manufacturer’s protocol (11147D; Invitrogen), and the remaining cells were incubated at a density of 2 × 106 cells per milliliter in R10 medium and stimulated with 10 μg/mL phytohemagglutinin (PHA) (431784; EMD/MilliporeSigma). Then, 16 to 24 h post-PHA activation, cells were cultured in R10-50. Primary CD4+ T cells were infected via spinoculation or treated for other experiments 48 h after IL-2 addition.
Genotyping of donor PBMCs was performed as previously described (57).
Primary MDMs were isolated with a CD14-positive isolation kit (17858; StemCell Technologies), stimulated with 50 ng/mL each of M-CSF and GM-CSF (216-MC-025/CF and 215-GM-050; R&D Systems), and cultured as previously described (71). MDMs were used for lysosomal pH measurements 7 to 10 d postisolation.
Viral Constructs and Infections.
Infectious supernatants for HIV constructs were prepared by cotransfection of 293T cells using polyethylenimine (PEI) as previously described (72) with each viral construct, the HIV packaging plasmid pCMV-HIV, and pHCMV-G at a mass ratio of 1:1:1. Infections were performed by spinoculation. Murine stem cell virus internal ribosome entry site GFP (pMIG) constructs containing various nef alleles were generated as previously described (59). Nef-expressing and control adenoviral vectors were obtained from the University of Michigan Gene Vector Core as previously described (31).
Flow Cytometry Surface Staining.
Detailed methodology can be found in SI Appendix. In all experiments, cells were gated sequentially by forward scatter vs. side scatter for cells, doublet exclusion (forward scatter area vs. height) for singlets, and exclusion of viability dye for viable cells.
Lysosensor Yellow/Blue Dextran Analysis of Lysosomal pH.
To measure the lysosomal pH in human MDMs, MDMs adhered to 24-well plates were exposed to 500 μg/mL Lysosensor Yellow/Blue dextran, 10,000 molecular weight (MW) (L22460; Invitrogen) in R10 for 24 h. MDMs were then exposed to plecomacrolides for 1 h and harvested with 0.05% Trypsin-(ethylenedinitrilo)tetraacetic acid (EDTA) (25300054; Gibco). Cells were washed twice in FACS buffer, and analyzed on a MoFlo Astrios flow cytometer, with blue signal excited from a 354-nm laser and yellow signal excited from a 405-nm laser. A standard curve was generated by resuspending MDMs in equilibration buffers of known pH as previously described (46). The ratio of blue:yellow fluorescence intensity was calculated for each cell, the median blue:yellow ratio for the cell population for each condition was obtained, and the lysosomal pH in MDMs was calculated for each condition using the standard curve.
Lysotracker Flow Cytometry Assay.
Cells were treated with plecomacrolides at a density of 1 × 106 cells per milliliter for 24 h, and then treated with 100 nM LysoTracker Red DND-99 (L7528; Invitrogen) in phosphate-buffered saline (PBS) at a density of 1 × 106 cells per milliliter for 1 h at 37 °C, washed twice in PBS, and fixed in 2% paraformaldehyde (PFA) before flow cytometric analysis on a Bio-Rad Ze5 flow cytometer.
Western Blotting.
Detailed methodology can be found in SI Appendix. Briefly, sorted PLAP+ CD4+ T cells isolated as previously described (73) were pelleted and lysed, sonicated, separated by gel electrophoresis, and transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked in 5% milk prior to probing with target-specific antibodies. Western blotting results were quantified using Photoshop by determining the mean pixel density in a box of equal size over each band from a single, unedited film displaying a single gel. Background pixel density was subtracted. No quantification comparisons were made from bands on different films or gels at any point.
Confocal Immunofluorescence Microscopy.
For HLA-A2 staining, sorted PLAP+ primary CD4+ T cells isolated as previously described (73) were attached to poly-l-lysine (P4707; Sigma Aldrich) coated chambered slides (154534; Fisher), fixed in PBS plus 2% PFA and permeabilized in PBS plus 0.2% Tween 20. Staining was performed as previously described (19) with primary antibody against HLA-A2 (BB7.2, 2 μg/mL) and secondary goat anti-mouse IgG2b-AF546 (A21143, 1:250; Invitrogen). Slides were coated with ProLong Gold Antifade Mountant (P36930; Invitrogen), coverslips were added, and images were collected on a Leica SP5 confocal microscope using identical instrument settings for each sample.
HLA-A2 Coimmunoprecipitation.
Immunoprecipitation (IP) of CEM cell lysates with BB7.2-conjugated beads was performed as previously described (21). Briefly, 25 × 106 CEM-A2 cells were transduced with Nef-expressing or control adenoviral vectors. Then, 48 h postinfection, cells were counted and resuspended at a density of 1 × 106 cells per milliliter R10 supplemented with 35 mM NH4Cl, 1.25 nM CMA, or solvent control for 24 h. Cells were pelleted, washed twice in PBS, and lysed in 1% digitonin lysis buffer (1% digitonin [043-21371; Wako], 100 mM NaCl, 50 mM Tris, pH 7.0, 1 mM CaCl2, and 1 mM MgCl2) as previously described (20). Then, 1% of the lysate was removed for input controls. After overnight preclear with isotype control antibody and protein A/G agarose (IP-10; EMD Bioscience), lysates were immunoprecipitated overnight with protein A/G agarose cross-linked to BB7.2. After pulldown, resin was washed five times in 0.1% digitonin wash buffer, and proteins were eluted by incubating in 150 mM dithiothreitol (DTT) (Invitrogen) for 30 min at 37 °C and analyzed by Western blot.
Flow Cytometric CTL Killing Assays.
CTL elimination assays were performed as previously described (27) with the following modifications: 72 h postinfection, primary CD4+ T cells (target cells) were stained with CellTracker Green CMFDA (C7025; Fisher) according to the manufacturer’s protocol and treated with 0.5 nM CMA or solvent control for 24 h. For each condition, 50,000 target cells were resuspended in fresh R10/50 without CMA with the corresponding number of effector CTLs to achieve the desired E:T ratio. Following the 4 h of coculture, the cells were stained with DAPI as a viability dye in addition to anti-PLAP and BB7.2 antibodies. Viable target cells were separated by gating for cells that were CellTracker Green-positive and excluded DAPI. The proportion of PLAP+ cells present in each condition was divided by that in the mean of target cells-only conditions (E:T = 0:1) to report the proportion of PLAP+ cells surviving in the presence of CTLs. All samples were performed in experimental duplicates, except the target cells-only conditions (E:T = 0:1), which were performed in quadruplicate. Flow cytometry data were collected on a Bio-Rad Ze5 cytometer.
Calculations.
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism software as described in the figure legends for each experiment. Curves were generated using GraphPad Prism software using [Inhibitor] vs. response with variable slope (four parameters), and the extra sum-of-squares F test was used to compare the EC50 for different curves.
Supplementary Material
Acknowledgments
We thank the University of Michigan Gene Vector Core facility for producing Nef-expressing adenoviral vectors, and the University of Michigan Center for Chemical Genomics for providing the equipment and chemical libraries used in the initial screening. The University of Michigan Flow Cytometry Core and the University of Michigan Microscopy Core provided access to instruments and technical support. The University of Michigan Advanced Genomics Core sequenced recombinant DNA constructs. We thank S.-J.-K. Yee (City of Hope National Medical Center) for providing pCMV-HIV-1. This work was funded by NIH Grants R21/R33 AI116158, R01 AI148383, and F31 AI131957; The University of Michigan Life Sciences Institute; the Dorothy and Herman Miller Award for Innovative Immunology Research; the Margolies Fund for the Center for Chemical Genomics at the Life Sciences Institute; the Hans W. Vahlteich Professorship; and the Taubman Institute. Additional funding was provided by Grants T32GM007315 (to J.A.L.), T32AI007413 (to M.M.P.), T32GM008353 (to J.L.), R01 AI044115 (to M.R.), R01 AI120691 (to X.R.), and P50 AI150476 (to J.H.H.).
Footnotes
Competing interest statement: A provisional patent relating to this study has been filed.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008615117/-/DCSupplemental.
Data and Materials Availability.
All data associated with this study are available in the main text or SI Appendix.
References
- 1.Borrow P. et al., Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3, 205–211 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Ndhlovu Z. M. et al., Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity 43, 591–604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migueles S. A. et al., HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3, 1061–1068 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Sáez-Cirión A. et al.; Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group , HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. U.S.A. 104, 6776–6781 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaiha G. D. et al., Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science 364, 480–484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrman G. et al., Depletion of latent HIV-1 infection in vivo: A proof-of-concept study. Lancet 366, 549–555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan L. et al., Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Archin N. M. et al., Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng K. et al., Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S.-H. et al., Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J. Clin. Invest. 128, 876–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowin B., Hahne M., Mattmann C., Tschopp J., Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature 370, 650–652 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Robinson J. et al., The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 43, D423–D431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parham P., MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5, 201–214 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Champsaur M., Lanier L. L., Effect of NKG2D ligand expression on host immune responses. Immunol. Rev. 235, 267–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parham P., Ohta T., Population biology of antigen presentation by MHC class I molecules. Science 272, 67–74 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Pereyra F. et al.; International HIV Controllers Study , The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaslow R. A. et al., Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2, 405–411 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Donaldson J. G., Williams D. B., Intracellular assembly and trafficking of MHC class I molecules. Traffic 10, 1745–1752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roeth J. F., Williams M., Kasper M. R., Filzen T. M., Collins K. L., HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 167, 903–913 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wonderlich E. R., Williams M., Collins K. L., The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J. Biol. Chem. 283, 3011–3022 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Wonderlich E. R. et al., ADP ribosylation factor 1 activity is required to recruit AP-1 to the major histocompatibility complex class I (MHC-I) cytoplasmic tail and disrupt MHC-I trafficking in HIV-1-infected primary T cells. J. Virol. 85, 12216–12226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer M. R., Wonderlich E. R., Roeth J. F., Leonard J. A., Collins K. L., HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLoS Pathog. 4, e1000131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishi T., Forgac M., The vacuolar (H+)-ATPases–Nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Jia X. et al., Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat. Struct. Mol. Biol. 19, 701–706 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Q. T., Ren X., Zhang R., Lee I. H., Hurley J. H., HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 350, aac5137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffalo C. Z., Iwamoto Y., Hurley J. H., Ren X., How HIV Nef proteins hijack membrane traffic to promote infection. J. Virol. 93, e01322-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins K. L., Chen B. K., Kalams S. A., Walker B. D., Baltimore D., HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391, 397–401 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Cohen G. B. et al., The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10, 661–671 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Williams M. et al., Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J. Virol. 76, 12173–12184 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Specht A. et al., Selective downmodulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 373, 229–237 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kasper M. R., Collins K. L., Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J. Virol. 77, 3041–3049 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao K. et al., Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62, 1009–1030 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Hilton H. G., Parham P., Direct binding to antigen-coated beads refines the specificity and cross-reactivity of four monoclonal antibodies that recognize polymorphic epitopes of HLA class I molecules. Tissue Antigens 81, 212–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacob R. T. et al., MScreen: An integrated compound management and high-throughput screening data storage and analysis system. J. Biomol. Screen. 17, 1080–1087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M. et al., Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emert-Sedlak L. A. et al., Effector kinase coupling enables high-throughput screens for direct HIV-1 Nef antagonists with antiretroviral activity. Chem. Biol. 20, 82–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mujib S. et al., Pharmacologic HIV-1 Nef blockade promotes CD8 T cell-mediated elimination of latently HIV-1-infected cells in vitro. JCI Insight 2, e93684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B. et al., Lovastatin inhibits HIV-1-induced MHC-I downregulation by targeting Nef–AP-1 complex formation: A new strategy to boost immune eradication of HIV-1 infected cells. Front. Immunol. 10, 2151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dröse S., Altendorf K., Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200, 1–8 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Kinashi H., Someno K., Sakaguchi K., Isolation and characterization of concanamycins A, B and C. J. Antibiot. (Tokyo) 37, 1333–1343 (1984). [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Sayáns M., Somoza-Martín J. M., Barros-Angueira F., Rey J. M. G., García-García A., V-ATPase inhibitors and implication in cancer treatment. Cancer Treat. Rev. 35, 707–713 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Hall T. J., Cytotoxicity of vacuolar H(+)-ATPase inhibitors to UMR-106 rat osteoblasts: An effect on iron uptake into cells? Cell Biol. Int. 18, 189–193 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Manabe T., Yoshimori T., Henomatsu N., Tashiro Y., Inhibitors of vacuolar-type H(+)-ATPase suppresses proliferation of cultured cells. J. Cell. Physiol. 157, 445–452 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Gao X. et al., Bafilomycin C1 induces G0/G1 cell-cycle arrest and mitochondrial-mediated apoptosis in human hepatocellular cancer SMMC7721 cells. J. Antibiot. (Tokyo) 71, 808–817 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Beyenbach K. W., Wieczorek H., The V-type H+ ATPase: Molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209, 577–589 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Yanagawa M. et al., Cathepsin E deficiency induces a novel form of lysosomal storage disorder showing the accumulation of lysosomal membrane sialoglycoproteins and the elevation of lysosomal pH in macrophages. J. Biol. Chem. 282, 1851–1862 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Jin Y. J. et al., HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J. Immunol. 175, 3157–3164 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Sykulev Y., Joo M., Vturina I., Tsomides T. J., Eisen H. N., Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity 4, 565–571 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Anmole G. et al., A robust and scalable TCR-based reporter cell assay to measure HIV-1 Nef-mediated T cell immune evasion. J. Immunol. Methods 426, 104–113 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Christinck E. R., Luscher M. A., Barber B. H., Williams D. B., Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature 352, 67–70 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Valitutti S., Müller S., Cella M., Padovan E., Lanzavecchia A., Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375, 148–151 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Kataoka T. et al., Concanamycin A, a powerful tool for characterization and estimation of contribution of perforin- and Fas-based lytic pathways in cell-mediated cytotoxicity. J. Immunol. 156, 3678–3686 (1996). [PubMed] [Google Scholar]
- 53.Togashi K., Kataoka T., Nagai K., Characterization of a series of vacuolar type H(+)-ATPase inhibitors on CTL-mediated cytotoxicity. Immunol. Lett. 55, 139–144 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Kiepiela P. et al., Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432, 769–775 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Apps R. et al., Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J. Immunol. 194, 3594–3600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mwimanzi F. et al., Resistance of major histocompatibility complex class B (MHC-B) to Nef-mediated downregulation relative to that of MHC-A is conserved among primate lentiviruses and influences antiviral T cell responses in HIV-1-infected individuals. J. Virol. 92, e01409-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yarzabek B. et al., Variations in HLA-B cell surface expression, half-life and extracellular antigen receptivity. eLife 7, e34961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaikos T. D. et al., Hematopoietic stem and progenitor cells are a distinct HIV reservoir that contributes to persistent viremia in suppressed patients. Cell Rep. 25, 3759–3773.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Leonard J. A., Filzen T., Carter C. C., Schaefer M., Collins K. L., HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements. J. Virol. 85, 6867–6881 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J., et al. , HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J. Virol. 92, e00196-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo T., Anderson S. J., Garcia J. V., Inhibition of Nef- and phorbol ester-induced CD4 degradation by macrolide antibiotics. J. Virol. 70, 1527–1534 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El-Far M. et al., Down-regulation of CTLA-4 by HIV-1 nef protein. PLoS One 8, e54295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pawlak E. N., Dirk B. S., Jacob R. A., Johnson A. L., Dikeakos J. D., The HIV-1 accessory proteins Nef and Vpu downregulate total and cell surface CD28 in CD4+ T cells. Retrovirology 15, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson L. S., Dunn K. W., Pytowski B., McGraw T. E., Endosome acidification and receptor trafficking: Bafilomycin A1 slows receptor externalization by a mechanism involving the receptor’s internalization motif. Mol. Biol. Cell 4, 1251–1266 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marshansky V., Futai M., The V-type H+-ATPase in vesicular trafficking: Targeting, regulation and function. Curr. Opin. Cell Biol. 20, 415–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geyer M. et al., Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J. Biol. Chem. 277, 28521–28529 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Lu X., Yu H., Liu S.-H., Brodsky F. M., Peterlin B. M., Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8, 647–656 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Mandic R. et al., Negative factor from SIV binds to the catalytic subunit of the V-ATPase to internalize CD4 and to increase viral infectivity. Mol. Biol. Cell 12, 463–473 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mota T. M. et al., Integrated assessment of viral transcription, antigen presentation, and CD8+ T cell function reveals multiple limitations of class I-selective histone deacetylase inhibitors during HIV-1 latency reversal. J. Virol. 94, e01845-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren Y. et al., BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo. J. Clin. Invest. 130, 2542–2559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mashiba M., Collins D. R., Terry V. H., Collins K. L., Vpr overcomes macrophage-specific restriction of HIV-1 Env expression and virion production. Cell Host Microbe 16, 722–735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuroda H., Kutner R. H., Bazan N. G., Reiser J., Simplified lentivirus vector production in protein-free media using polyethylenimine-mediated transfection. J. Virol. Methods 157, 113–121 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Zaikos T. D., Painter M. M., Sebastian Kettinger N. T., Terry V. H., Collins K. L., Class 1-selective histone deacetylase (HDAC) inhibitors enhance HIV latency reversal while preserving the activity of HDAC isoforms necessary for maximal HIV gene expression. J. Virol. 92, e02110-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are available in the main text or SI Appendix.