Significance
LncRNAs have been identified as regulating antiviral innate responses via different targets through various ways. Whether there is an abundant and highly conserved lncRNA in the nucleus which may regulate antiviral innate signaling through a new mechanism remains to be investigated. Here, we identify that viral infection-reduced expression of Malat1 feedback promotes IRF3-initiated type I IFN production in the innate response against viral infection. Malat1 binds to TDP43 and prevents its cleavage mediated by activated caspase-3. The cleaved TDP35 inhibits IRF3 degradation through promoting the degradation of pre-mRNA of Rbck1, an E3 ubiquitin ligase targeting IRF3. Aberrant MALAT1 reduction and IRF3 activation are found in SLE patients, providing one mechanistic explanation for why SLE patients always have type I interferonopathies.
Keywords: long noncoding RNA, Malat1, TDP43, type I interferon, innate immunity
Abstract
Long noncoding RNAs (lncRNAs) involved in the regulation of antiviral innate immune responses need to be further identified. By functionally screening the lncRNAs in macrophages, here we identified lncRNA Malat1, abundant in the nucleus but significantly down-regulated after viral infection, as a negative regulator of antiviral type I IFN (IFN-I) production. Malat1 directly bound to the transactive response DNA-binding protein (TDP43) in the nucleus and prevented activation of TDP43 by blocking the activated caspase-3-mediated TDP43 cleavage to TDP35. The cleaved TDP35 increased the nuclear IRF3 protein level by binding and degrading Rbck1 pre-mRNA to prevent IRF3 proteasomal degradation upon viral infection, thus selectively promoting antiviral IFN-I production. Deficiency of Malat1 enhanced antiviral innate responses in vivo, accompanying the increased IFN-I production and reduced viral burden. Importantly, the reduced MALAT1, augmented IRF3, and increased IFNA mRNA were found in peripheral blood mononuclear cells (PBMCs) from systemic lupus erythematosus (SLE) patients. Therefore, the down-regulation of MALAT1 in virus-infected cells or in human cells from autoimmune diseases will increase host resistance against viral infection or lead to autoinflammatory interferonopathies via the increased type I IFN production. Our results demonstrate that the nuclear Malat1 suppresses antiviral innate responses by targeting TDP43 activation via RNA-RBP interactive network, adding insight to the molecular regulation of innate responses and autoimmune pathogenesis.
As the first line of defense against viral infection, the production of type I interferons (IFN-I) (IFN-α and IFN-β), plays a central role by activating the expression of hundreds of IFN-stimulated genes (ISGs) for establishing an “antiviral state” to restrict viral replication within infected cells (1). Insufficient production of IFNs causes chronic viral infections, while excessive amounts of IFNs are also harmful to the host, inducing autoimmune inflammation and type I interferonopathies (2–4). Therefore, it is of vital importance to precisely regulate the production of IFNs both in temporal and spatial dimensions to ensure the induction of potent antiviral innate response against viral infection but also avoid the occurrence of autoimmune diseases. The transcription factor IFN regulatory factor 3 (IRF3) is critical for IFNs production and directs expression of diverse genes in the antiviral immune response (5). Activation of IRF3 involves virus infection-induced phosphorylation at several sites in the C terminal, IRF3 dimerization, and translocation to the nucleus. However, how the IRF3 activity for initiating innate gene expression is tightly regulated remains to be fully understood.
Long noncoding RNAs (lncRNAs) are involved in many biological processes, including immunity and inflammation (6, 7). They participate in the battle between host and virus via different mechanisms, including regulating the activity of RNA-binding proteins (RBPs) as well as RNA metabolism, especially the translation and degradation of host mRNAs (8–11). Many viruses induce widespread host RNA decay through virally encoded endonucleases to reduce activation of immune response and provide access to the host’s resources for viral replication. It is estimated that up to two-thirds of total mRNAs are degraded upon expression by these viral endonucleases (12). A significant fraction of lncRNAs are located in the nucleus and nuclear lncRNAs play important roles in physiological, biological, and pathological processes via different mechanisms (13, 14). However, the role of nuclear abundant lncRNAs in regulating RNA metabolism and RBP activity in the antiviral innate immune response remains to be further investigated.
In this study, we functionally screen the differently expressed lncRNAs in macrophages and identify the nuclear abundant lncRNA Malat1 as a negative regulator of antiviral type I IFN production. We find that Malat1 can bind TDP43, inhibit the cleavage of TDP43 to its active form TDP35, then reduce nuclear IRF3 level in resting cells, maintaining immune homeostasis. Upon viral infection, Malat1 expression is down-regulated and releases TDP43 for its activation via cleavage to its active form TDP35, then TDP35 prevents IRF3 proteasomal degradation and thus promotes IRF3-initiated antiviral type I IFNs production, feedback benefitting the host against viral infection. Interestingly, the reduced MALAT1 and increased IRF3 in peripheral blood mononuclear cells (PBMCs) from systemic lupus erythematosus (SLE) patients with type I interferonopathies were confirmed. Thus, our findings provide a potential target for controlling viral infection and IFN-I-related inflammatory autoimmune diseases.
Results
The Expression of Nuclear lncRNA Malat1 Decreases upon Viral Infection.
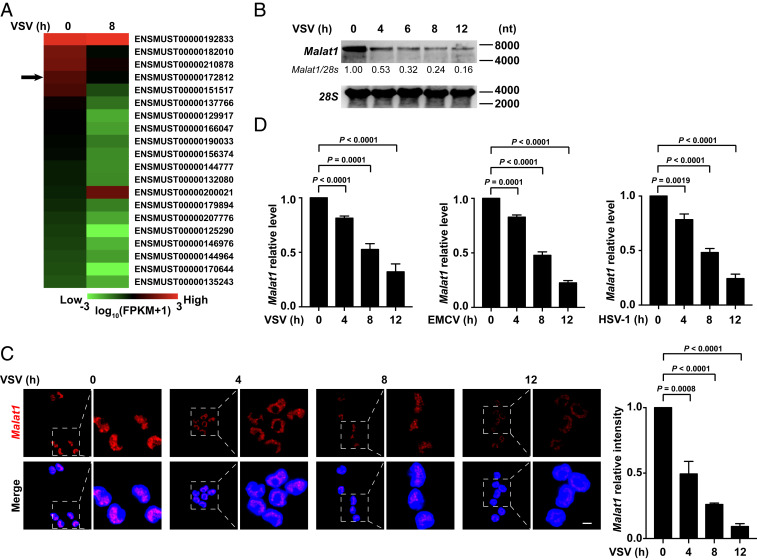
Through RNA sequencing (RNA-Seq), we analyzed nuclear lncRNA profiling in RAW264.7 cells with or without vesicular stomatitis virus (VSV) infection after isolation of nuclear RNAs (SI Appendix, Fig. S1A). The differentially expressed lncRNAs (more than twofold change) are ranked according to their expression abundance in cells without VSV infection (Fig. 1A). We isolated primary peritoneal macrophages from 8-wk-old C57BL/6 male mice and transfected the mixture composition of small interfering RNAs (siRNAs) and antisense oligonucleotide (ASO) targeting the top 20 candidate RNAs, respectively (SI Appendix, Fig. S1B). Then we detected the intracellular GFP intensity with high-content screening (HCS) in cells upon GFP-VSV infection. We found that knockdown of metastasis-associated lung adenocarcinoma transcript 1 (Malat1) robustly decreased virus replication in macrophages upon GFP-VSV infection for 12 h (SI Appendix, Fig. S1C). Notably, Malat1 was the fourth most abundant lncRNA in the uninfected cells and its expression was significantly reduced upon VSV infection (Fig. 1A). Malat1 was universally and highly expressed in immune organs and immune cells including macrophages (SI Appendix, Fig. S1 D and E), suggesting that Malat1 might be involved in the regulation of antiviral innate immune response.
Fig. 1.
The expression of nuclear lncRNA Malat1 decreases upon viral infection. (A) Heat map of top 20 abundant nuclear lncRNAs in RAW264.7 cells at resting state which are changed more than twofold after VSV infection for 8 h. ENSMUST00000172812 (Malat1) is pointed out by black arrow. (B) Northern blot assay of Malat1 level in RAW264.7 cells along with VSV infection for indicated hours. Relative intensity of Malat1 is calculated by the ImageJ program. (C) Confocal microscope images from FISH assay of Malat1 level in RAW264.7 cells along with VSV infection for indicated hours. Relative fluorescence intensity of Malat1 is calculated by the ImageJ program (Right). Red, Malat1; blue, DAPI. (Scale bar, 5 μm.) (D) qRT-PCR analysis of relative Malat1 level in RAW264.7 cells along with VSV, EMCV, and HSV-1 infection for indicated hours, respectively. Data are representative of three independent experiments (B and C) or shown as mean ± SD of n = 3 biological replicates (C and D), two-tailed unpaired Student’s t test (C and D).
Northern blot and fluorescence in situ hybridization (FISH) assay further showed that the expression of Malat1 significantly decreased in macrophages upon VSV infection (Fig. 1 B and C). Fluorescence probes to Malat1 exhibited punctuate staining only in the nuclear compartment of cells (Fig. 1C). In addition, infections with encephalomyocarditis virus (EMCV) (recognized by melanoma differentiation-associated protein 5 [MDA5]) and DNA virus herpes simplex virus type 1 (HSV-1) could also reduce Malat1 expression (Fig. 1D). In contrast, Toll-like receptor (TLR) ligands, such as lipopolysaccharide (LPS) and poly (I:C), could not significantly reduce Malat1 expression (SI Appendix, Fig. S1 F and G), consistent with the findings of the previous study (15). In addition, it has been reported that widespread RNAs are efficiently degraded during apoptosis (16), and then we investigated whether the reduction of Malat1 was also induced by apoptosis. We found the inducer of apoptosis ABT-737 (inhibiting the inhibitors Bcl-xL, Bcl-2, and Bcl-w), could significantly decrease Malat1 level (SI Appendix, Fig. S1H). Furthermore, the reduction of Malat1 mediated by VSV or ABT-737 was reversed by pretreatment of pan caspase inhibitor Z-VAD-FMK or Q-VD-Oph (SI Appendix, Fig. S1H), indicating that host apoptosis might promote the destabilization of Malat1. Taken together, these data above suggest that the expression of Malat1 decreases upon viral infection, indicting its possible role in antiviral innate immune response.
Malat1 Selectively Inhibits Type I IFN Production in Macrophages upon Viral Infection.
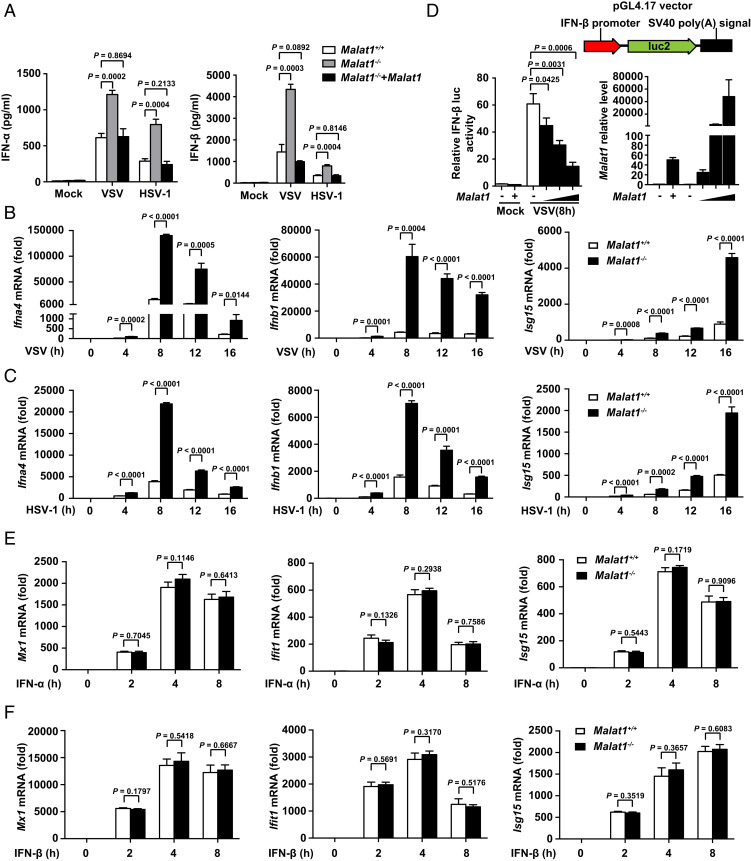
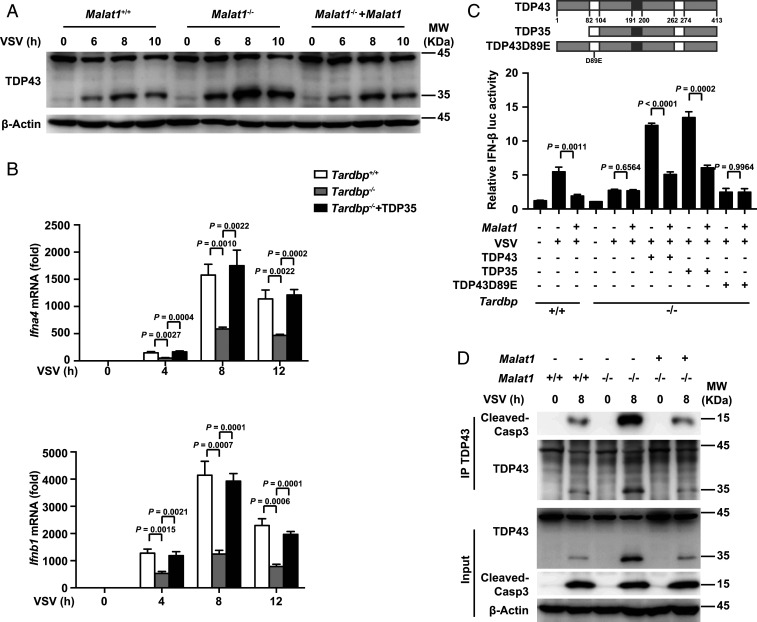
We generated Malat1 knockout RAW264.7 cells (Malat1−/− cells) which lacked a full 7,691-bp genomic region of Malat1 with the CRISPR-Cas9 system (SI Appendix, Fig. S2A). Upon viral infection, Malat1−/− cells produced more IFN-α and IFN-β than Malat1+/+ cells and this effect was reversed by rescue of Malat1 in Malat1−/− cells (Fig. 2A and SI Appendix, Fig. S2B), while the production of proinflammatory cytokines (TNF-α and IL-6) had no difference among these three groups (SI Appendix, Fig. S2C). Correspondingly, the relative mRNA levels of Ifna, Ifnb, and ISG (Isg15) (Fig. 2 B and C), but not Tnfa, Il6, and Il1b (SI Appendix, Fig. S2 D and E), significantly increased in Malat1−/− cells upon viral infection. Furthermore, Malat1 could inhibit IRF3-induced IFN-β promoter activity (Fig. 2D) but not NF-κB-induced IL-6 promoter activity (SI Appendix, Fig. S2F) upon VSV infection. These data indicated that Malat1 selectively inhibits IFN-I production. To investigate whether Malat1 could affect IFN-mediated antiviral function, we treated the Malat1+/+ and Malat1−/− cells with IFN-α or IFN-β and then checked ISG expression. There was no difference on IFN-I effector signaling (p-STAT1 and IFIT1) (SI Appendix, Fig. S2G) and ISG expression (Fig. 2 E and F) between Malat1+/+ and Malat1−/− cells, suggesting that Malat1 does not affect IFN function. So, Malat1 selectively inhibits the production of IFN-I upon viral infection but does not affect IFN-I antiviral function.
Fig. 2.
Malat1 selectively inhibits the production of type I IFNs in macrophages upon viral infection. (A) ELISA analysis of IFN-α and IFN-β in culture supernatants of Malat1+/+, Malat1−/− , and Malat1−/− rescued by Malat1 RAW264.7 cells infected with VSV or HSV-1 for 12 h, respectively. (B) qRT-PCR analysis of relative Ifna4, Ifnb1, and Isg15 mRNA level in Malat1+/+ and Malat1−/− RAW264.7 cells along with VSV infection for indicated hours. (C) qRT-PCR analysis of relative Ifna4, Ifnb1, and Isg15 mRNA level in Malat1+/+ and Malat1−/− RAW264.7 cells along with HSV-1 infection for indicated hours. (D) Luciferase assay of IFN-β promoter activity influenced by gradually enhanced Malat1 in HEK293T cells transfected with RIG-I and Malat1 vectors upon VSV infection for indicated hours (Left) and qRT-PCR analysis of Malat1 relative level in HEK293T cells above transfected with 200, 400, and 800 ng Malat1 vector, respectively (Right). (E) qRT-PCR analysis of relative Mx1, Ifit1, and Isg15 mRNA level in Malat1+/+ and Malat1−/− RAW264.7 cells treated with IFN-α for indicated hours. (F) qRT-PCR analysis of relative Mx1, Ifit1, and Isg15 mRNA level in Malat1+/+ and Malat1−/− RAW264.7 cells treated with IFN-β for indicated hours. All data are shown as mean ± SD of n = 3 biological replicates, two-tailed unpaired Student’s t test.
Deficiency of Malat1 Enhances Antiviral Response In Vivo.
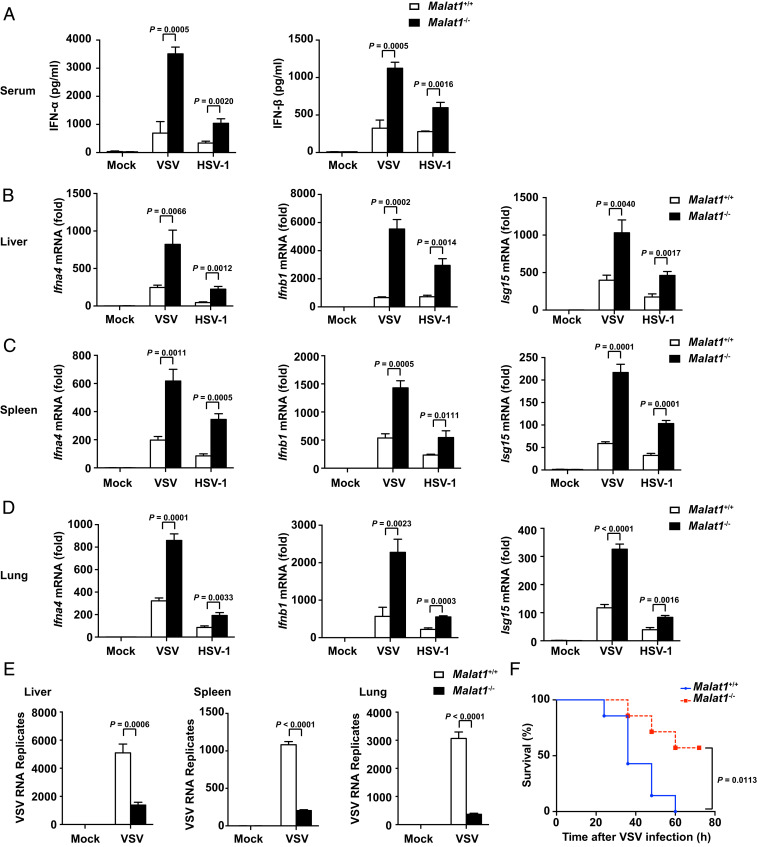
To further evaluate the role of Malat1 in host antiviral innate immune response in vivo, we generated Malat1-deficient mice (Malat1−/− mice, deleting the full 7,691-bp genomic region of Malat1). After i.v. infection with VSV or HSV-1 respectively, Malat1−/− mice produced more IFN-α and IFN-β in serum than Malat1+/+ mice (Fig. 3A). Consistently, the relative mRNA level of Ifna, Ifnb, and Isg15 in organs (liver, spleen, and lung) of Malat1−/− mice was significantly higher than that of their littermates (Fig. 3 B–D). Consequently, the VSV RNA replication of organs above was notably reduced in Malat1−/− mice (Fig. 3E). However, there was no difference of TNF-α or IL-6 expression in serum and organs between two groups infected with VSV or HSV-1 (SI Appendix, Fig. S3 A–D). Consistent with the higher IFNs, Malat1−/− mice showed improved survival compared with Malat1+/+ mice after being challenged with a lethal dose of VSV or HSV-1 infection (Fig. 3F and SI Appendix, Fig. S3E). These data suggest that Malat1 inhibits antiviral innate response in vivo by selectively suppressing IFN-I production.
Fig. 3.
Deficiency of Malat1 enhances antiviral response in vivo. (A) ELISA of IFN-α and IFN-β in serum from Malat1+/+ and Malat1−/− mice infected with VSV (5 × 107 pfu/g) for 18 h or HSV-1 (1 × 107 pfu/g) for 24 h, respectively, via tail i.v. injection. (B) qRT-PCR analysis of relative Ifna4, Ifnb1, and Isg15 mRNA level in liver from Malat1+/+ and Malat1−/− mice corresponding to A. (C) qRT-PCR analysis of relative Ifna4, Ifnb1, and Isg15 mRNA level in spleen from Malat1+/+ and Malat1−/− mice corresponding to A. (D) qRT-PCR analysis of relative Ifna4, Ifnb1, and Isg15 mRNA level in lung from Malat1+/+ and Malat1−/− mice corresponding to A. (E) qRT-PCR analysis of relative VSV RNA replication in liver, spleen, and lung from Malat1+/+ and Malat1−/− mice corresponding to A. (F) Survival curves of 6- to 8-wk-old mice of Malat1+/+ and Malat1−/− infected with VSV (1 × 108 pfu/g) via tail i.v. injection. Data are shown as mean ± SD of n = 3 biological replicates (A–E) or demonstrated as Kaplan–Meier survival curve (F, n = 7), two-tailed unpaired Student’s t test (A–E), or Log-rank (Mantel–Cox) test (F).
Identification of Malat1-Bound TDP43 in Selectively Promoting IFN-I Production.
To analyze the underlying mechanism how Malat1 inhibits the production of type I IFNs, we conducted the chromatin isolation by RNA purification (ChIRP) assay followed by mass spectrography to determine the Malat1-bound proteins in the nucleus. The 10 Malat1-bound proteins with the highest score in nuclear fraction of macrophages without viral infection are shown in SI Appendix, Fig. S4A. Then we used HCS to screen the function of these proteins in antiviral innate response with siRNAs and found that knockdown of Tardbp (siTardbp, encoding TDP43) increased viral replication in macrophages upon GFP-VSV infection compared with negative control (siNC) (SI Appendix, Fig. S4B).
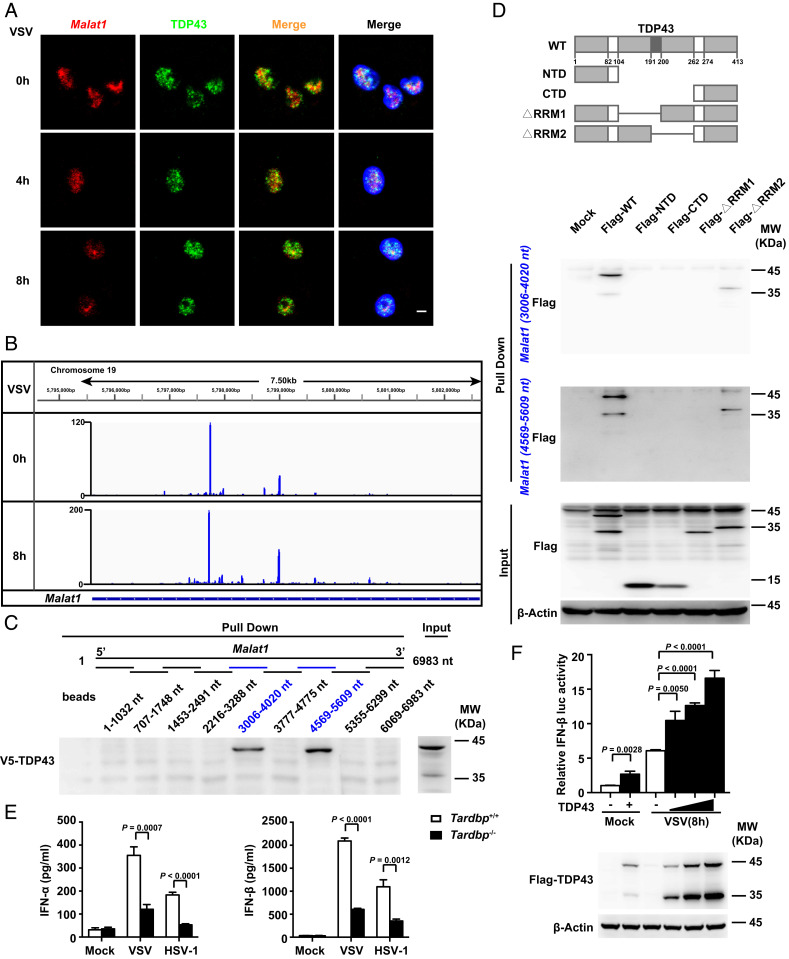
Then we confirmed the interaction of Malat1 with TDP43 by ChIRP assay followed by Western blot (SI Appendix, Fig. S4C). FISH assay further demonstrated that Malat1 colocalized with TDP43 in the nucleus with or without VSV infection (Fig. 4A). To determine the position of Malat1 responsible for interacting with TDP43, we performed individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) in RAW264.7 cells. After RNA digestion, the TDP43-RNA complexes were immunoprecipitated and appeared above the molecular weight of TDP43 (SI Appendix, Fig. S4D). The signal of the TDP43-RNA complex was sharply reduced upon RNase extensive digestion, and no signal was detected by using IgG (SI Appendix, Fig. S4D), demonstrating the high specificity of the immunoprecipitated RNA. Then we analyzed the cross-link-induced truncation positions of Malat1 in the iCLIP library and found that Malat1 cross-linked to TDP43 via 3,618 and 4,887 sites of Malat1 (Fig. 4B). We constructed 9 fragments of Malat1 in a head-to-tail overlap manner, each truncation about 1,000 nt, and found that both fragments of Malat1 (3,006 to 4,020 nt) and Malat1 (4,569 to 5,609 nt), which contained the two cross-link sites, could bind to TDP43, respectively (Fig. 4C). Through RNA pull-down assay, we found that both fragments could bind to the RRM1 domain (Fig. 4D), which was the main RNA-binding domain of TDP43 (17). These data suggest that Malat1 can directly bind to the RRM1 domain of TDP43 with two sites.
Fig. 4.
Identification of Malat1-bound TDP43 in selectively promoting IFN-I production. (A) Confocal microscope images from sequential FISH assay of Malat1 and TDP43 in RAW264.7 cells along with VSV infection for indicated hours. Red, Malat1; green, TDP43; and blue, DAPI. (Scale bar, 5 μm.) (B) iCLIP assay of V5-TDP43 overexpressed in RAW264.7 cells and the likely interaction sites of Malat1 that bind with TDP43. (C) RNA pull-down assay of different truncations of Malat1 with overexpressed V5-TDP43 in RAW264.7 cells. (D) RNA pull-down assay of Malat1 (3,006 to 4,020 nt) and Malat1 (4,569 to 5,609 nt) fragments with HEK293T cell lysates overexpressed different truncations of TDP43, respectively. (E) ELISA analysis of IFN-α and IFN-β in culture supernatants of Tardbp+/+ and Tardbp−/− RAW264.7 cells infected with VSV or HSV-1 for 12 h, respectively. (F) Luciferase assay of IFN-β promoter activity influenced by gradually enhanced TDP43 in HEK293T cells transfected with RIG-I and TDP43 vectors upon VSV infection for indicated hours (Upper) and Western blot analysis of TDP43/TDP35 level in HEK293T cells above transfected with 200, 400, and 800 ng TDP43 vector, respectively (Lower). Data are representative of three independent experiments (A, C, D, and F) or shown as mean ± SD of n = 3 biological replicates (E and F), two-tailed unpaired Student’s t test (E and F).
To further analyze the function of TDP43, we generated TDP43-deficient RAW264.7 cells (Tardbp−/− cells) with the CRISPR-Cas9 system. TDP43 deficiency did not affect the expression of Malat1 in RAW264.7 cells with or without VSV infection (SI Appendix, Fig. S4E). Of note, Tardpb−/− cells produced much less IFN-α and IFN-β than Tardbp+/+ cells (Fig. 4E), but produced almost the same level of TNF-α and IL-6 upon VSV or HSV-1 infection (SI Appendix, Fig. S4F). Overexpression of TDP43 could enhance IFN-β but not IL-6 promoter activity upon VSV infection in a dose-dependent manner (Fig. 4F and SI Appendix, Fig. S4G). These results suggest that Malat1-bound TDP43 selectively promotes the expression of type I IFNs in response to viral infection.
Malat1 Inhibits IFN-I Production by Blocking TDP43 Cleavage to Its Active Form TDP35.
We next investigated how Malat1 inhibited the production of type I IFNs through binding to TDP43. TDP43 was reported to be cleaved to TDP35 mainly by cleaved caspase-3 during apoptosis (18). We found that deficiency of Malat1 did not affect the protein level of TDP43 in RAW264.7 cells in response to VSV infection (Fig. 5A). Unexpectedly, we found that much more TDP43 was cleaved to TDP35 in Malat1−/− cells compared with that in Malat1+/+ cells upon VSV infection (Fig. 5A). Rescue of Malat1 expression in Malat1−/− cells decreased the cleavage of TDP43 to TDP35 (Fig. 5A). To detect the TDP43 protein level without the influence of cleavage by caspase-3 upon viral infection, we pretreated RAW264.7 cells with pan caspase inhibitor Z-VAD-FMK, and found that the activation of caspase-3 and the cleavage of TDP43 were suppressed in both Malat1+/+ and Malat1−/− cells with Z-VAD-FMK treatment and VSV infection (SI Appendix, Fig. S5A). As expected, the protein level of TDP43 was increased in Malat1−/− cells with treatment of Z-VAD-FMK (SI Appendix, Fig. S5A). To determine the function of cleaved TDP35 in innate immunity, we overexpressed TDP35 into Tardbp−/− cells and found that rescue of TDP35 expression could increase the relative mRNA levels of Ifna, Ifnb, and Isg15 (Fig. 5B and SI Appendix, Fig. S5B) but not proinflammatory cytokines (Tnfa, Il6, and Il1b) (SI Appendix, Fig. S5C), suggesting that TDP35 is the key subunit of TDP43 that promoted the production of type I IFNs.
Fig. 5.
Malat1 inhibits IFN-I production by blocking TDP43 cleavage to its active form TDP35. (A) Western blot analysis of Malat1’s effect on TDP43 along with VSV infection for the indicated hours in RAW264.7 cells. (B) qRT-PCR analysis of relative Ifna4 and Ifnb1 mRNA level in Tardbp+/+, Tardbp−/−, and Tardbp−/− rescued by TDP35 RAW264.7 cells upon VSV infection for indicated hours. (C) Luciferase analysis of IFN-β promoter activity in Tardbp+/+ and Tardbp−/− L929 cells transfected with Malat1, RIG-I, as well as wild-type, truncation, or mutant of TDP43 vectors, respectively. (D) Coimmunoprecipitation analysis of interaction between TDP43 and cleaved caspase-3 affected by Malat1 in RAW264.7 cells upon virus infection for indicated hours. Data are representative of three independent experiments (A and D) or shown as mean ± SD of n = 3 biological replicates (B and C), two-tailed unpaired Student’s t test (B and C).
The caspase-3 cleavage in virus-infected cells has been shown recently for HIV, adenovirus, hepatitis C virus, and many other kinds of virus (19). We found that the infection of VSV, EMCV, or HSV-1 in macrophages could induce the activation of caspase-3 (SI Appendix, Fig. S5D), which showed a similar cleavage pattern to TDP43. Moreover, TDP43 mutant (TDP43D89E), in which the consensus motif for caspase-3 cleavage was mutated, failed to promote the IRF3-induced IFN-β promoter activity and STAT1-induced ISRE promoter activity upon VSV infection (Fig. 5C and SI Appendix, Fig. S5E), confirming that TDP43 cleavage into TDP35 subunit by the activated caspase-3 plays an important role in antiviral innate immune response.
Then we further investigated how Malat1 inhibits TDP43 from being cleaved to TDP35 by the activated caspase-3. We found that deficiency of Malat1 increased both the cleavage of TDP43 and the interaction between TDP43/35 (TDP43 antibody targets TDP43 C-terminal domain and fails to distinguish TDP43 from TDP35) and cleaved caspase-3 compared with Malat1+/+ cells, while the opposite results were observed after rescue of Malat1 expression in Malat1−/− cells. However, the activated caspase-3 remained unchanged among three groups (Fig. 5D). Furthermore, immunoprecipitation with cleaved caspase-3 antibody showed that only TDP43 but not TDP35 interacted with the cleaved caspase-3 (SI Appendix, Fig. S5F), indicating that Malat1 inhibits the interaction of the activated caspase-3 and TDP43, and then reduces the cleavage of TDP43 to TDP35 during viral infection. Of note, Malat1 inhibited the IFN-β promoter activity with rescue of TDP43 or TDP35 but not TDP43D89E in Tardbp−/− L929 cells (Fig. 5C). Taken together, these data demonstrate that Malat1 inhibits the production of type I IFNs through inhibiting the activated caspase-3-mediated cleavage of TDP43 to TDP35 upon viral infection.
The Cleaved TDP35 Increases the Nuclear IRF3 Protein Level by Degrading Pre-mRNA of Rbck1.
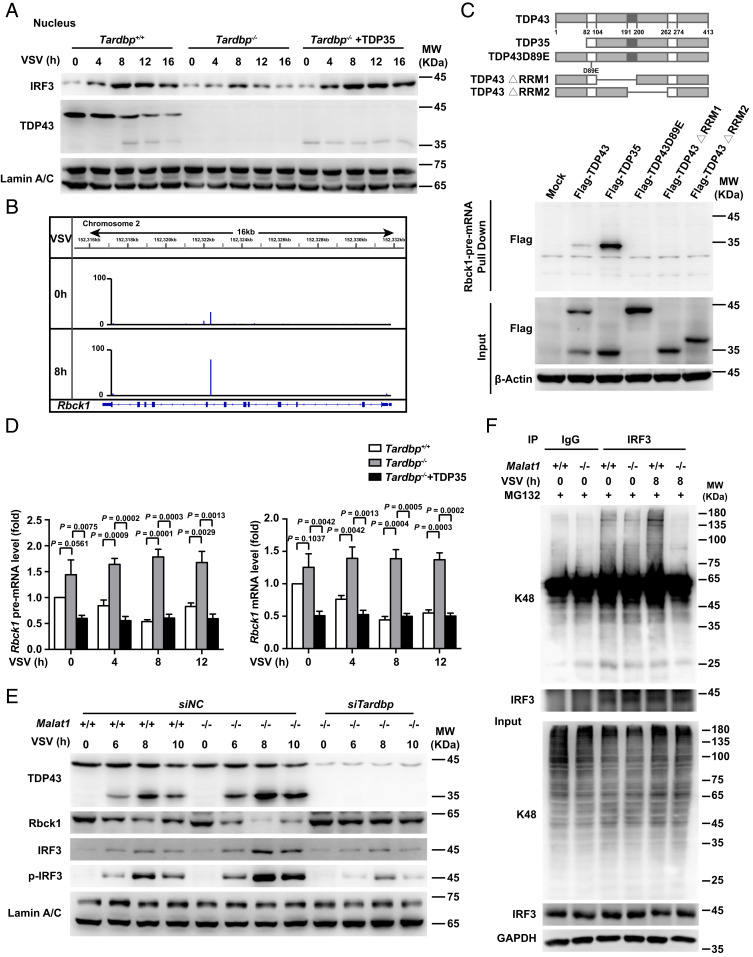
We next investigated how the cleaved TDP35 promotes the production of type I IFNs upon viral infection. It is demonstrated that the intracellular localization of TDP43 is important for its function in the central nervous system (20). We found that both TDP43 and TDP35 mainly located in the nucleus after viral infection, and the cleavage pattern of TDP43 was positively associated with the level of IRF3 in the nucleus upon VSV infection (SI Appendix, Fig. S6A). Furthermore, deficiency of the Tardbp gene in macrophages significantly decreased the level of nuclear IRF3, and rescue of TDP35 expression significantly increased the level (Fig. 6A). Since TDP43 as a RNA-binding protein participates in different RNA processes, including alternative splicing, mRNA stability, and transcriptional regulation, we wondered whether the cleaved TDP35 increases IRF3 protein level through regulating Irf3 mRNA. However, TDP35 was not bound to Irf3 mRNA and there was no difference in the level of mRNA of Irf3 in Tardbp+/+ and Tardbp−/− cells with or without TDP35 rescue upon VSV infection (SI Appendix, Fig. S6B).
Fig. 6.
The cleaved TDP35 increases nuclear IRF3 protein level by binding and degrading Rbck1 pre-mRNA to prevent IRF3 proteasomal degradation upon viral infection. (A) Western blot analysis of TDP35’s effect on protein level of IRF3 in the nucleus of Tardbp+/+, Tardbp−/−, and Tardbp−/− rescued by TDP35 RAW264.7 cells upon VSV infection for indicated hours. (B) iCLIP assay of V5-TDP43 overexpressed RAW264.7 cells and the likely interaction sites of Rbck1 pre-mRNA that binding with TDP43/TDP35. (C) RNA pull-down assay of Rbck1 pre-mRNA and the different truncations or mutant of TDP43 overexpressed in HEK293T cells, respectively. (D) qRT-PCR analysis of relative pre-mRNA and mRNA level of Rbck1 in Tardbp+/+, Tardbp−/−, and Tardbp−/− rescued by TDP35 RAW264.7 cells upon virus infection for indicated hours. (E) Western blot analysis of the necessity of TDP35 for Malat1 regulating the expression of Rbck1 and phosphorylation of IRF3 in nucleus of Malat1+/+ and Malat1−/− RAW264.7 cells transfected with siRNA targeting negative control (siNC) or Tardbp (siTardbp), respectively, upon VSV infection for indicated hours. (F) Coimmunoprecipitation analysis of the K48-linked polyubiquitination on IRF3 in Malat1+/+ and Malat1−/− RAW264.7 cells infected with VSV for indicated hours. Data are representative of three independent experiments (A, C, E, and F) or shown as mean ± SD of n = 3 biological replicates (D), two-tailed unpaired Student’s t test (D).
Then, we wondered whether TDP35 directly targets the mRNA of genes associated with IRF3 protein degradation. We analyzed the interaction of TDP35 with genes encoding the proteins which can regulate IRF3 protein level, including Rbck1 (21), Pin1 (22), Cullin1 (23), and PP2A (24), and found that TDP43/35 robustly cross-linked to intron 7 of Rbck1 transcript upon VSV infection but not the other three genes in iCLIP libraries of TDP43/35 (Fig. 6B). RNA pull-down confirmed that intron 7 of Rbck1 pre-mRNA could interact with TDP35 but not TDP43 or its mutants (Fig. 6C). We then constructed 20 fragments of Rbck1 pre-mRNA each of which was about 1,000 nt in a head-to-tail overlap manner. Through RNA pull-down assay, we found that only one fragment (10,801 to 11,777 nt), which contained the cross-link site, was bound to TDP35 (SI Appendix, Fig. S6C), suggesting that Rbck1 pre-mRNA can directly bind to TDP35 through its seventh intron.
Furthermore, the level of Rbck1, both pre-mRNA and mRNA, increased robustly in Tardbp−/− cells compared with that in Tardbp+/+ cells, and rescue of TDP35 expression in Tardbp−/− cells decreased both levels (Fig. 6D). Correspondingly, the change of Rbck1 protein expression was similar to Rbck1 mRNA among these cells (SI Appendix, Fig. S6D). To investigate whether TDP43 or Malat1 directly regulates Rbck1 pre-mRNA stability, we examined the half-life time of Rbck1 pre-mRNA with actinomycin D (ActD) which can inhibit RNA polymerase II activity. We demonstrated that the level of Rbck1 pre-mRNA decreased much more quickly with ActD treatment in Malat1−/− cells upon VSV infection. Rescue of the Malat1 level in Malat1−/− cells significantly retarded the reduction of Rbck1 pre-mRNA (SI Appendix, Fig. S6E). Additionally, overexpressing TDP35 in Malat1−/− cells robustly accelerated the decrease of Rbck1 pre-mRNA (SI Appendix, Fig. S6E). Consistently, the level of Rbck1 pre-mRNA decreased much more slowly along with the treatment of ActD in Tardbp−/− cells. Rescue of TDP35 expression in Tardbp−/− cells significantly promoted the reduction of Rbck1 pre-mRNA, while rescue of Malat1 in Tardbp−/− cells had little effect on its level (SI Appendix, Fig. S6F), indicating that TDP43/TDP35 directly regulates Rbck1 pre-mRNA stability, while Malat1 depends on TDP43/TDP35 to achieve its regulation on Rbck1 pre-mRNA stability.
The Cleaved TDP35 Degrades Pre-mRNA of E3 Ligase Rbck1 for Preventing IRF3 Degradation.
Next we detected whether Rbck1 acts as the E3 ubiquitin ligase targeting IRF3 for degradation in macrophages upon viral infection. We demonstrated that overexpression of Rbck1 reduced the protein level of IRF3 in a dose-dependent manner and this effect was significantly diminished in the presence of proteasome inhibitor MG132. And both TDP43 and TDP35 but not TDP43D89E could decrease Rbck1 and increase the IRF3 level as well (SI Appendix, Fig. S7A). Exposure of cells under the same culture conditions resulted in the corresponding changes of K48-linked polyubiquitination of IRF3 (SI Appendix, Fig. S7B). Furthermore, deficiency of Tardbp showed increased Rbck1 but decreased IRF3 levels in the nucleus of cells upon VSV infection, whereas knockdown of Rbck1 by siRNA in Tardbp−/− cells significantly increased the level of nuclear IRF3 (SI Appendix, Fig. S7C). Taken together, these data demonstrate that virus infection promotes the cleavage of TDP43 to TDP35, which increases the nuclear IRF3 protein level through preventing IRF3 protein degradation by inhibiting expression of its ubiquitin ligase Rbck1 in an antiviral innate immune response.
Next we demonstrated that deficiency of Malat1 significantly increased the cleavage of TDP43 to TDP35, decreased the expression of Rbck1, and increased the nuclear IRF3 protein compared with that in control cells upon VSV infection, while knockdown of Tardbp in Malat1−/− cells increased Rbck1 expression and decreased the IRF3 level, suggesting that Malat1 regulates the expression of Rbck1 and IRF3 depending on TDP35 (Fig. 6E). Furthermore, deficiency of Malat1 reduced the K48-linked polyubiquitination on IRF3 compared with that in control cells (Fig. 6F), indicating that Malat1 promotes IRF3 degradation through the proteasome pathway upon VSV infection.
The Decreased Expression of MALAT1 Is Associated with Increased TDP43 Cleavage, IRF3 Protein, and IFNA Expression in PBMCs of SLE Patients.
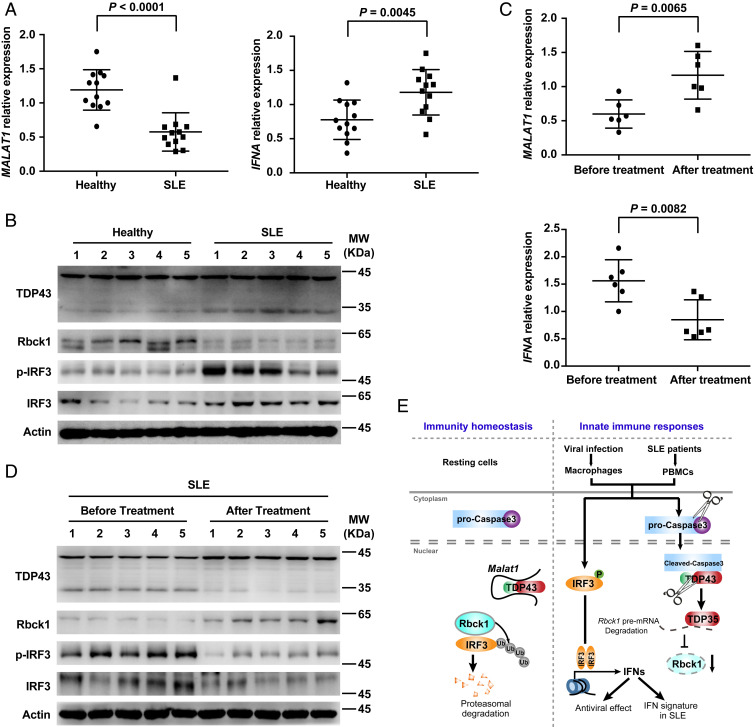
Dysregulation of innate immune response plays a critical role in the development of many autoimmune diseases. SLE, a common and potentially fatal autoimmune disorder, has been characterized by autoantibody production and higher type I IFN signature (25). Early studies indicated that higher expression of p-IRF3 is found in PBMCs from patients with SLE (26). Inspired by these indications, we analyzed the expression of MALAT1 in PBMCs isolated from SLE patients and control normal donors. As expected, MALAT1 expression was drastically reduced and IFNA mRNA level significantly increased in SLE PBMCs compared with that in control cells (Fig. 7A). Furthermore, the key molecules, including increased TDP43 cleavage, reduced Rbck1, and enhanced IRF3 and p-IRF3, were observed in PBMCs from SLE patients (Fig. 7B).
Fig. 7.
The decreased expression of MALAT1 is associated with increased TDP43 cleavage, IRF3 protein, and IFNA expression in PBMCs of SLE patients. (A) qRT-PCR analysis of relative MALAT1 and IFNA in PBMCs from healthy donors and SLE patients. (B) Western blot analysis of key molecules in MALAT1-regulated antiviral response in PBMCs from healthy donors and SLE patients. (C) qRT-PCR analysis of relative MALAT1 and IFNA in PBMCs from SLE patients before and after effective treatment. (D) Western blot analysis of key molecules in MALAT1-regulated antiviral response in PBMCs from SLE patients before and after effective treatment. (E) Proposed working model for Malat1 as a negative regulator of antiviral innate response by targeting TDP43 activation via an RNA-RBP interactive network. Data are shown as mean ± SD of n = 12 (A) or n = 6 (C) biological replicates, two-tailed unpaired Student’s t test (A and C).
The glucocorticoids and antimalarial drugs (hydroxychloroquine and chloroquine) have been a standard and prominent treatment for SLE patients for a long time (27). The treatment can achieve rapid immune suppression and alleviated inflammation through inhibiting TLR signaling and cGAS stimulator of IFN genes (STING) signaling and reducing the production of proinflammatory cytokines, including type I interferons (27–29). We demonstrated that effective treatment not only rescues the IFNA expression but also MALAT1 expression in PBMCs from SLE patients (Fig. 7C). Furthermore, a series of molecular events, from reduced TDP43 cleavage to decreased IRF3, were also achieved in PBMCs from SLE patients after effective treatment (Fig. 7D)
Discussion
Here, we identified Malat1 as a negative regulator of antiviral IFN-I production. The viral infection-induced reduction of Malat1 promotes IRF3 activation and type I IFNs production. The results of this study demonstrated that, in resting cells, Malat1 binds to TDP43, inhibits IRF3-induced IFNs expression, and maintains immune homeostasis. Upon viral infection, Malat1 expression is reduced, and without the binding of Malat1, TDP43 is cleaved into TDP35 by activated caspase-3. Then cleaved TDP35 increases nuclear IRF3 level by inhibiting its ubiquitination through degradation of Rbck1 pre-mRNA, and the increased production of IFNs participate in antiviral effects. A series of molecular events, from MALAT1 reduction to IFN production, spontaneously exist in the PBMCs of SLE patients. In sum, we found a nuclear lncRNA inhibits the production of type I IFNs, decreasing the antiviral capacity and inhibiting the progress of autoimmune diseases (Fig. 7E).
LncRNAs have been reported closely associated with a repertoire of physiological and pathological processes, including antiviral innate immune responses (30). Generally, the level of most lncRNAs is much lower than that of mRNAs. Consistently, the abundance of lncRNAs is much lower than that of the proteins which results in the limitation of the interaction between lncRNAs and proteins. The great function of lncRNAs (link-EPS and lnc13) with few copy numbers is debatable (31). Most lncRNAs have a 5′ cap and a poly(A) tail, yet some of the most abundant long noncoding RNAs are processed in unexpected ways and lack these canonical structures. It has been reported that widespread RNA decay during apoptosis is initiated by the mitochondrial exoribonuclease PNPT1 (16). RNAs with 3′ end structures, like many nonpoly(A) ncRNAs, block PNPT1 degradation and likely contribute to the relative resistance of nonpoly(A) ncRNAs to accelerated decay during apoptosis. It is possible that structured sequences near the 3′ end of imported RNAs protect them from being degraded during import. The Malat1 is not polyadenylated as the tRNA biogenesis machinery generates its mature 3′ end. In place of a poly(A) tail, Malat1 is stabilized by highly conserved triple helical structures, which accumulates to high levels in the nucleus (32). Upon viral infection, however, Malat1 was reduced. As Malat1 used noncanonical strategies to ensure its stability, Malat1 may be regulated by unique posttranscriptional control mechanisms. The study by Bhattacharyya and Vrati detected an increased Malat1 level by flaviviral infection in Neuro2a but not in macrophages (BV2 cells) in the nervous system (33). The study by Yao et al. showed Malat1 KO had no defects following lymphocytic choriomeningitis virus (LCMV) infection in T cells (34). These studies indicate that the function of Malat1 varied in different kinds of cells which respond diversely to viral infection. In addition, we found the reduction of Malat1 was induced by apoptosis caused by viral infection; therefore, we speculated that various degrees of apoptosis induced by different types of virus lead to specific changes in Malat1 expression. Malat1 has been reported closely associated with many tumors, ranging from breast cancer to prostate cancer. It is involved in tumorigenesis, progression, and metastasis, which has been generally studied since it was discovered as metastasis-associated lung adenocarcinoma transcript 1 (35–39). Our findings identify that Malat1 reduction contributes to the production of type I IFNs upon viral infection, which uncovered a mechanism that host cells can actively decrease the expression of certain types of suppressive lncRNAs in response to viral infection and then promote the induction of type I IFNs to resist viral infection.
The most popular role of nuclear lncRNAs is regulating the genes’ expression through epigenetic regulation. They could promote or suppress the transcription of certain genes directly or by transcription factors (30). Our study provides a mechanism for nuclear lncRNA in reducing the cleavage of TDP43 to TDP35 by blocking the interaction of TDP43 and cleaved caspase-3. Malat1 was directly bound to TDP43 with two fragments around the cross-link sites of Malat1 transcript at 3,618 and 4,887 nt in different sequences. It has been previously demonstrated that Malat1 with a short interspersed nuclear element (SINE) deletion form creates more available TDP43-binding sites, suggesting that Malat1 has a specific secondary structure which lays the foundation for the binding to TDP43 (40). However, it needs more profound investigation of secondary structures of RNAs to fully explain the properties of RNA-binding proteins.
The function of TDP43, especially as a RBP, in antiviral response is rarely investigated. Here, we discovered that TDP35 was a positive regulator of type I IFN production by binding with pre-mRNA of Rbck1 and facilitating its degradation. In a previous study, TDP43 binds GU-rich distal intronic sites (41). However, the two sites where TDP43 binds to Malat1 are not classical GU-rich sites which enlarge the TDP43-binding RNA sequences. Moreover, TDP35 likely differs from TDP43 in secondary structure which provides the basis that full-length TDP43 could bind to Malat1 but not Rbck1 pre-mRNA, while cleaved TDP35 could interact with Rbck1 pre-mRNA. It’s possible that secondary structure changes of TDP35 caused by cleavage exposes the sites for binding Rbck1 pre-mRNA.
Notably, it has been widely accepted that persistent type I IFN exposure and subsequent type I IFN signaling leading to ISG expression (referred to as the IFN-I signature) are characteristic features of SLE pathogenesis (42). In animal models, reduced IFN-I expression or IFN-I signaling showed clear beneficial effects for lupus treatment (43). Clinically, anti-IFN-I therapies attracted much attention for treatment of SLE patients (44). However, the mechanisms governing negative regulation of IFNs is incomplete and requires further investigation and clarification. Malat1 is one of the most conserved lncRNAs between human and mouse (about 70% conserved between human and mouse) (45). We found SLE patients displayed reduced MALAT1 expression, coupled with enhanced cleavage of TDP43, decreased Rbck1 expression, increased IRF3 activation, and finally increased IFNA production. Furthermore, posttreatment of SLE patients could robustly increase MALAT1 expression and alleviate the aberrant IRF3 activation cascade in patient-derived PBMCs, consistent with the available data in the Gene Expression Omnibus that MALAT1 expression is decreased dramatically in monocytes and lymphocytes (accession no. GSE51997) of SLE patients, further supporting the potential role of MALAT1 in SLE patients. Our findings uncover an unexpected pathophysiological connection between lncRNA and type I IFNs, which allows us to gain insight into therapeutic targets and disease biomarkers to provide potential support for clinical treatment in autoimmune diseases.
Materials and Methods
Mice and Cells.
Malat1-deficient mice were generated via the CRISPR-Cas9 genomic editing system based on C57BL/6J background. Primary peritoneal macrophages were acquired from the peritoneal lavage fluids of mice that were intraperitoneally injected with 3% thioglycollate 72 to 96 h in advance. All animal experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and with approval of the Scientific Investigation Board of the Chinese Academy of Medical Sciences. Malat1-deficient RAW264.7, TDP43-deficient RAW264.7, and L929 cell lines were produced through the CRISPR-Cas9 system with pGL3-U6 plasmid containing guide RNA sequence. Sequences for sgRNAs and genotype identification are listed in SI Appendix, Table S1. HEK293T, RAW264.7, L929, BHK-21, and Vero cell lines were purchased from American Type Culture Collection (ATCC) company. Malat1−/− mice and cells were generated as described in SI Appendix, Materials and Methods.
Control Normal Donors, SLE Patients, and Treated SLE Patients.
Healthy normal donors had no history of autoimmune diseases. Patients with concurrent infection were excluded from the study. All SLE patients (ages ranging from 17 to 55, mainly female, SI Appendix, Table S2) fulfilled the American College of Rheumatology (ACR) classification criteria for SLE (46). The Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) score was determined for each patient at the time of the blood draw. Patients were categorized as having active disease (scores >4) or inactive disease (scores ≤4) based on the SLEDAI results. The study was approved by the Research Ethics Board of Peking Union Medical College Hospital (JS-1239). Written informed consent was signed before sample collection. Information of healthy donors, SLE patients, and treated patients are listed in SI Appendix, Table S2.
High-Content Screening.
Primary peritoneal macrophages from wild-type mice were seeded into 96-well plates. Then the lncRNA Smart Silencer (a mixture of three ASOs and three siRNAs with equal proportion; RiboBio) (47–49) were transfected into the macrophages at the optimal final concentration of 50 nM to silence target genes, respectively. Forty-eight hours later, the transfected macrophages were infected with or without GFP-VSV for 12 h. Cells were then fixed by 4% paraformaldehyde for 15 mins at room temperature (RT) and stained with DAPI for 10 min at RT to dye the nucleus. GFP intensity which represented the titer of VSV was measure by the ArrayScan High-Content System (Thermo Scientific). The siRNA- and ASO-targeted sequences for knockdown of 20 lncRNAs are outlined in SI Appendix, Table S1.
ChIRP Assay.
Cells infected with or without VSV were washed three times with PBS. The assay was performed with Magna ChIRP RNA Interactome Kits (Merck Millipore) according to the manufacturer’s protocols and followed by Western blot analysis or mass spectrography.
Statistical Analysis.
Two-tailed unpaired Student’s t test was applied to analyze the statistical significance of data from two groups with GraphPad Prism software. Mice survival curve data were demonstrated as Kaplan–Meier curves and analyzed with Log-rank (Mantel–Cox) test. P values less than 0.05 were regarded as statistically significant.
Supplementary Material
Acknowledgments
This work is supported by grants from the National Natural Science Foundation of China (81788101), National 135 Mega Program of China (2017ZX10102032-001, 2017ZX10202203-002, and 2017ZX10203206-001), and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016-12M-1-003).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003932117/-/DCSupplemental.
Data Availability.
The sequencing data for RNA-Seq and iCLIP have been deposited in the Gene Expression Omnibus with accession no. GSE134032 (50). All study data are included in the article and SI Appendix.
References
- 1.Yan N., Chen Z. J., Intrinsic antiviral immunity. Nat. Immunol. 13, 214–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kretschmer S., Lee-Kirsch M. A., Type I interferon-mediated autoinflammation and autoimmunity. Curr. Opin. Immunol. 49, 96–102 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lee-Kirsch M. A., The type I interferonopathies. Annu. Rev. Med. 68, 297–315 (2017). [DOI] [PubMed] [Google Scholar]
- 4.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A., Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S. et al., Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Atianand M. K., Caffrey D. R., Fitzgerald K. A., Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 35, 177–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satpathy A. T., Chang H. Y., Long noncoding RNA in hematopoiesis and immunity. Immunity 42, 792–804 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Jiang M. et al., Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Wang P., Xu J., Wang Y., Cao X., An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358, 1051–1055 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Lin H. et al., The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20, 812–823 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Xu H. et al., Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 20, 1621–1630 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Muller M., Glaunsinger B. A., Nuclease escape elements protect messenger RNA against cleavage by multiple viral endonucleases. PLoS Pathog. 13, e1006593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engreitz J. M., Ollikainen N., Guttman M., Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Quinodoz S., Guttman M., Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao G., Su Z., Song D., Mao Y., Mao X., The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 590, 2884–2895 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Liu X. et al., PNPT1 release from mitochondria during apoptosis triggers decay of Poly(A) RNAs. Cell 174, 187–201.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Buratti E., Baralle F. E., Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Lee E. B., Lee V. M., Trojanowski J. Q., Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 13, 38–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang J. Y. et al., HIV infection increases HCV-induced hepatocyte apoptosis. J. Hepatol. 54, 612–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann M. et al., Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Zhang M. et al., Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 18, 1096–1104 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Saitoh T. et al., Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7, 598–605 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Bibeau-Poirier A. et al., Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 177, 5059–5067 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Long L. et al., Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40, 515–529 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Tsokos G. C., Lo M. S., Costa Reis P., Sullivan K. E., New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12, 716–730 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Santana-de Anda K. et al., Interferon regulatory factor 3 as key element of the interferon signature in plasmacytoid dendritic cells from systemic lupus erythematosus patients: Novel genetic associations in the Mexican mestizo population. Clin. Exp. Immunol. 178, 428–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatto M., Zen M., Iaccarino L., Doria A., New therapeutic strategies in systemic lupus erythematosus management. Nat. Rev. Rheumatol. 15, 30–48 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Luijten R. K., Fritsch-Stork R. D., Bijlsma J. W., Derksen R. H., The use of glucocorticoids in systemic lupus erythematosus. After 60 years still more an art than science. Autoimmun. Rev. 12, 617–628 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Schrezenmeier E., Dörner T., Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q., Cao X., Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 19, 417–432 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Cabili M. N. et al., Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Hamblin M. H., Yin K. J., The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 14, 1705–1714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya S., Vrati S., The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Sci. Rep. 5, 17794 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao Y. et al., Long noncoding RNA Malat1 is not essential for T cell development and response to LCMV infection. RNA Biol. 15, 1477–1486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendell J. T., Targeting a long noncoding RNA in breast cancer. N. Engl. J. Med. 374, 2287–2289 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Malakar P. et al., Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 77, 1155–1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin D. et al., m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 12, 135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhang Y. et al., MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Differ. 26, 2061–2073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R. et al., Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9((R)) to suppress enzalutamide-resistant prostate cancer progression. Eur. Urol. 72, 835–844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen T. M. et al., The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 48, 2621–2642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polymenidou M. et al., Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crow M. K., Type I interferon in the pathogenesis of lupus. J. Immunol. 192, 5459–5468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nacionales D. C. et al., Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 56, 3770–3783 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morand E. F. et al.; TULIP-2 Trial Investigators , Trial of anifrolumab in active systemic lupus erythematosus. N. Engl. J. Med. 382, 211–221 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Zhang B. et al., The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aringer M., EULAR/ACR classification criteria for SLE. Semin. Arthritis Rheum. 49, S14–S17 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Lennox K. A., Behlke M. A., Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Q. et al., A new long noncoding RNA ALB regulates autophagy by enhancing the transformation of LC3BI to LC3BII during human lens development. Mol. Ther. Nucleic Acids 9, 207–217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng Q. et al., The DGCR5 long noncoding RNA may regulate expression of several schizophrenia-related genes. Sci. Transl. Med. 10, eaat6912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W., et al. , LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134032. Deposited 9 July 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for RNA-Seq and iCLIP have been deposited in the Gene Expression Omnibus with accession no. GSE134032 (50). All study data are included in the article and SI Appendix.