Highlights
-
•
BLACAT1 promotes cell proliferation, migration and invasion, and dampens cell apoptosis in OS.
-
•
BLACAT1 sponges miR-608 in OS.
-
•
SOX12 is the target of miR-608.
-
•
BLACA1 promotes OS cell growth and migration via targeting miR-608/SOX12 axis.
Abbreviations: SOX12, sex determining region Y-box protein 12; OS, osteosarcoma; SOX, sex-determining region Y (SRY)-box; lncRNAs, long non-coding RNAs; ceRNAs, competing endogenous RNAs; miRNAs, microRNAs; mRNA, messenger RNA; ATCC, American type culture collection; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; RIPA, radioimmunoprecipitation assay; SDS-PAGE, sulphate-polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride; RT-qPCR, RNA extraction and quantitative real-time polymerase chain reaction; HRP, horseradish peroxidase; FISH, Fluorescence in situ hybridization; WT, wild-type; Mut, mutant; SD, standard deviation; ANOVA, analysis of variance
Keywords: Osteosarcoma, BLACAT1, miR-608, SOX12
Abstract
Background
Osteosarcoma is the most common type of bone malignancy. Increasing evidence indicated that long non-coding RNAs (lncRNAs) possess multiple functions in the development of cancer and can be used as indicators of prognosis and diagnosis. LncRNA BLACAT1 has been found to promote the proliferation of breast cancer cells. However, the role of BLACAT1 in osteosarcoma remains largely unknown.
Methods
QRT-PCR analysis was employed to evaluate mRNA expressions. Western blot was performed to measure relevant protein level. Colony formation and EdU assays were conducted to certify proliferative ability. TUNEL assay was finalized to assess apoptotic cells. Wound-healing and transwell assays were utilized for the exploration of migrating and invasive abilities. The subcellular distribution of BLACAT1 was studied by nucleus-cytoplasm separation assay. Relevant mechanical experiments were combined to elucidate molecular relationship between molecules.
Results
BLACAT1 was highly expressed in osteosarcoma. BLACAT1 promoted the proliferation and migration of osteosarcoma cells. BLACAT1 acted as a sponge for miR-608 to augment the expression of Sex determining region Y-box protein 12 (SOX12), the direct target of miR-608. Further, inhibiting miR-608 recovered the repressive effect of silenced BLACAT1 on the malignant behaviors of osteosarcoma cells.
Conclusion
This study highlighted the contribution of BLACAT1/miR-608/SOX12 axis to the progression of osteosarcoma, suggesting novel targets for osteosarcoma therapy.
1. Introduction
Usually diagnosed in children and adolescents, osteosarcoma (OS) is one of the most common malignant bone tumors originating from mesenchymal cells [1], [2]. Combined therapy strategies, including surgery and neoadjuvant chemotherapy, have been applied for treating OS. Nevertheless, the overall survival rate of OS patients is still low, particularly for those with distant metastasis [3]. Tumor resection or standard adjuvant chemotherapy is futile for patients tolerated with severe metastasis [4], [5]. Hence, uncovering the molecular mechanism that regulates the carcinogenic behaviors in OS is required for identifying novel biomarkers and targets.
Featured by >200 nucleotides in length, long noncoding RNAs (lncRNAs) are RNA transcripts with very finite or no protein-coding ability [6]. In the tumorigenesis of OS, lncRNAs were found to elicited profound impacts on diverse genetic processes, including transcription activity, messenger RNA (mRNA) stability and chromosome structure [7]. Over the last decade, lncRNAs have emerged as potential biomarkers and targets for their intricate mechanism in cancers. Similarly, some lncRNAs have been verified to possess therapeutic target values for OS. For example, lncRNA TUG1 was manifested to promote OS metastasis via modulating HIF-1α through sponging miR-143-5p [8]. BLACAT1 was a relatively novel lncRNA, which was initially found overexpressed in human cervical cancer and facilitated cell proliferation and invasion [9]. Previously, BLACAT1 has been detected to be up-regulated in OS tissues and regulate the phosphorylation of STAT3 to promote OS progression [10]. However, in-depth research needs to be conducted for better elucidation of the underlying mechanism of BLACAT1 in OS.
Enriched documents have revealed the essential epigenetic regulatory role of lncRNAs in biological performance. Increasing lncRNAs were verified to act as miRNA sponges to post-transcriptionally regulate the expression of particular mRNAs, and such crosstalk among above molecules was termed as competing endogenous RNA (ceRNA) network [11]. MiR-608 was lowly expressed in melanoma and played a progression-inhibitory role in melanoma [12]. Besides, miR-608 was discovered to be engaged in ceRNA network in malignancies. For instance, miR-608 was found to be sponged by lncRNA NORAD to suppress the growth of gastric cancer cells [13].
Sex determining region Y-box protein 12 (SOX12) is a member of Sry-related HMG box protein family. It is expressed in the mesenchymal cells of many human organs. It was found to be associated with the progression of acute leukaemia via modulating β-catenin [14]. However, the connection between SOX12 and the development of OS has never been investigated.
This study aimed to probe into the possible molecular mechanism of BLACAT1 in osteosarcoma. It was discovered that BLACAT1 was up-regulated in OS cell lines. Mechanically, BLACAT1 sponged to miR-608 to elevate SOX12 level leading to deterioration of OS in vitro.
2. Materials and methods
2.1. Cell lines and culture
Human osteoblast cell line hFOB1.19 and human osteosarcoma cell lines (143B, U2OS, MG63 and Saos2), available from the Chinese Academy of Sciences (Shanghai, China), were cultured under standard condition of 5% CO2 and 37 °C. DMEM culture medium (Hyclone, Logan, UT) was acquired for cell culture with fetal bovine serum (10%, FBS; Hyclone) and Pen/Strep mixture (1%).
2.2. Total RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNAs were isolated from the confluent cells and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA synthesis was performed by reverse transcription using PrimeScript® strand cDNA Synthesis Kit (TaKaRa, Shiga, Japan). Gene expression was quantified by PCR amplification and calculated by the 2-ΔΔCt method, with GAPDH or U6 as the internal control.
2.3. Cell transfection
To stably silence BLACAT1, the duplex short hairpin RNAs (shRNAs) specifically targeted to BLACAT1 and control-shRNAs were produced by Genepharma Company (Shanghai, China). The miR-608 mimics/inhibitor and NC mimics/inhibitor, also from Genepharma, were transfected into 143B and MG63 cells applying Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 48 h.
2.4. Colony formation assay
48 h after transfection, 143B and MG63 cells were subjected to the 14-day incubation process in 6-well plates (500 cells/well). Then, clones were dyed by 0.5% crystal violet after fixing in 4% paraformaldehyde. After washing in PBS, colonies were counted manually.
2.5. EdU incorporation assay
143B and MG63 cells were reaped after transfection and cultured for 2 h with EdU medium diluent in 96-well plates. Then, cell proliferation was tested by EdU kit (Ribobio) based on the protocol. Followed by washing in PBS, cells were incubated with DAPI solution for 30 min, followed by cell observation under the fluorescence microscope (Olympus, Tokyo, Japan).
2.6. TUNEL assay
Transfected 143B and MG63 cells were cultured with 2% formaldehyde for 1 h, with 0.1% Triton X-100 for 2 min, and then washed in PBS. Cells were incubated in the dark at 37℃ with 50 μl of TUNEL reaction buffer for 1 h. Apoptotic cells were analyzed under Olympus microscope.
2.7. Wound-healing assay
The sub-confluent monolayers of 143B and MG63 cells were treated with P-200 pipette tips. After removing the detached cells, medium was replaced with 1% FBS-medium. Images were captured at 0 and 24 h, the percentage of wound-healing was calculated.
2.8. Transwell invasion analysis
Cell invasion was investigated by transwell chamber-coated with Matrigel (Corning Co, Corning, NY). 50,000 cells cultured in serum-free medium were added to upper chamber, with lower chamber supplied with complete medium. Cells on the lower chamber were fixed for crystal violet staining after 24 h of incubation. Invading cells were counted in 5 random fields and also imaged under a microscope (Olympus).
2.9. Western blot
Transfected cells were collected for preparing total proteins using RIPA lysis buffer on ice, followed by separation on 10% SDS-PAGE. Proteins were shifted to PVDF membranes, which were then cultured with 5% skimmed milk. Primary antibodies against MMP9, MMP2, Bcl2, Bax, SOX12 and the loading control GAPDH, along with the HRP-marked secondary antibodies were all available from Abcam (Cambridge, MA) and employed after dilution. At last, protein bands were quantified by ECL Prime Western Blotting Detection reagent (GE Healthcare, Chicago, IL).
2.10. Nucleus-cytoplasm separation
PARIS™ Kit (Ambion, Austin, TX) was acquired for nucleus-cytoplasm separation as per instruction. 1 × 107 collected cells were placed in cell fraction buffer and treated on ice for 10 min. After centrifugation, supernatant was discarded and the nuclear pellet was kept for culturing in cell disruption buffer, followed by RNA detection via qRT-PCR.
2.11. RNA immunoprecipitation (RIP)
After 48 of transfection, cells were reaped for RIP assay with Magna RIP™ Kit (Millipore, Bellerica, MA). Cells suspended in complete RIP lysis buffer were then prepared to incubate with magnetic beads and anti-Ago2 antibody overnight. Normal rabbit IgG acted as the control. RNAs bound in beads were extracted for qRT-PCR analysis.
2.12. RNA pull down assay
RNAs were biotin-labeled using Biotin RNA Labeling Mix (Roche, Basel, Switzerland), and then in vitro transcribed for RNA pull down assay. After purification, the biotinylated miR-608-WT/MUT were mixed with cell lysates and streptavidin-conjugated magnetic beads. The biotin-labeled nonsense sequence (Bio-NC) was used as the negative control. Relative RNA enrichment was estimated by qRT-PCR.
2.13. Luciferase reporter assay
The fragments of BLACAT1 or SOX12 3′UTR covering the wild-type or mutated miR-608 binding sites were acquired and then inserted at downstream of luciferase reporter pmirGLO vectors (Promega, Madison, WI). After co-transfecting with miR-608 mimics or NC mimics, luciferase activities normalized to Renilla activity were detected utilizing Luciferase Reporter Assay System (Promega).
2.14. Statistical analysis
GraphPad PRISM 6 (GraphPad, San Diego, CA) was used for statistical analysis. Data were represented as the mean ± SD from three independent bio-repeats. Group difference was compared by t-test or one-way analysis of variance (ANOVA), with significance defined as p < 0.05.
3. Results
3.1. BLACAT1 promotes cell proliferation, migration and invasion, yet dampens cell apoptosis in OS
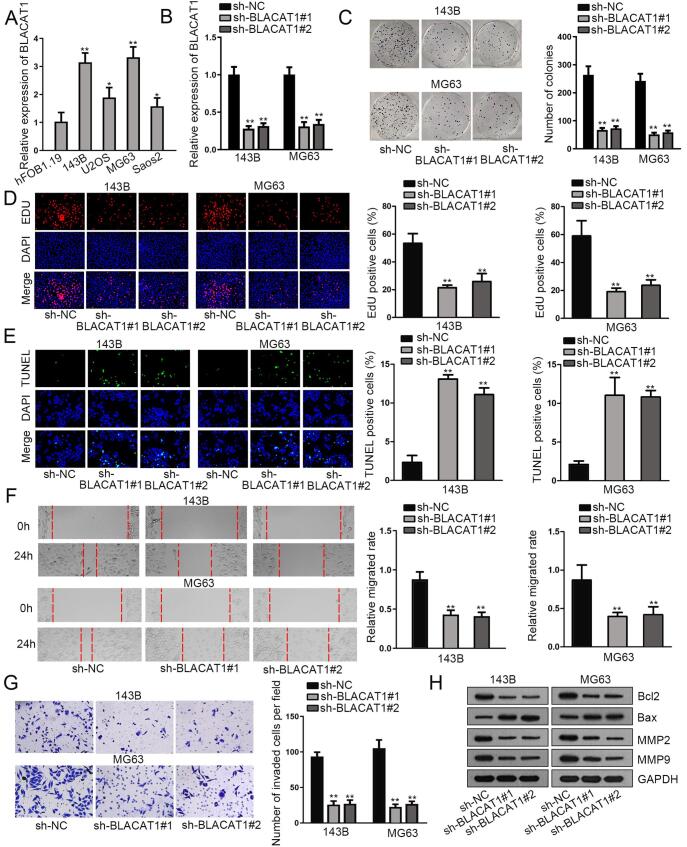
To determine the expression status of BLACAT1 in OS, qRT-PCR analysis was utilized to detect its expression in OS cell lines. The results showed that BLACAT1 expression was notably higher in four OS cell lines (143B, U2OS, MG63, Saos2) than the normal osteoblasts hFOB1.19 (Fig. 1A). Then, loss-of-function experiments were performed to evaluate the role of BLACAT1 in OS. Firstly, we constructed shRNAs targeting BLACAT1 (sh-BLACAT1#1/2) and corresponding negative control (sh-NC) and transfected them into selected 143B and MG63 cells, as BLACAT1 was most abundantly expressed in these two cells. As a result, BLACAT1 expression was declined by sh-BLACAT1#1/2 in the above cells (Fig. 1B). Colony formation and EdU assay were performed to study the role of BLACAT1 on OS cell proliferation. The results manifested that BLACAT1 silencing remarkably suppressed the proliferative ability of OS cells (Fig. 1C, D). TUNEL assay revealed that BLACAT1 silencing notably enhanced OS cell apoptotic capacity (Fig. 1E). Wound-healing assay demonstrated a reduction of migrated cell number in BLACAT1-inhibited 143B and MG63 (Fig. 1F). Later, transwell assay was performed to assess the impact of BLACAT1 depletion on OS cell invasion. As expected, BLACAT1 silencing greatly decreased the number of invaded cells (Fig. 1G). Last but not least, western blot was conducted to measure the level of proteins related to cell apoptosis and migration. It showed an increase in Bax, while a decrease in Bcl-2, MMP2 and MMP9 in response to BLACAT1 deficiency (Fig. 1H). Together, BLACAT1 facilitated cell proliferation, migration and invasion, yet dampened cell apoptosis in OS.
Fig. 1.
BLACAT1 promotes cell proliferation, migration and invasion, yet dampens cell apoptosis in OS. BLACAT expression level was measured by qRT-PCR analysis in OS cell lines (143B, U2OS, MG63, Saos2) versus normal osteoblast hFOB1.19. B. The inhibitory efficiency of sh-BLACAT1#1/2 was detected by qRT-PCR analysis. C-D. Cell proliferation ability was evaluated by EdU and colony formation assays after transfecting sh-BLACAT1 #1/2 into selected OS cells. E. Apoptotic cells were evaluated by TUNEL assay after silencing BLACAT1. F-G. Wound-healing and transwell invasion assays were performed to assess cell migration and invasion ability after silencing BLACAT1. H. western blot was conducted to measure the level of proteins related to cell apoptosis and migration. *P < 0.05; **P < 0.01.
3.2. BLACAT1 sponges miR-608 in OS
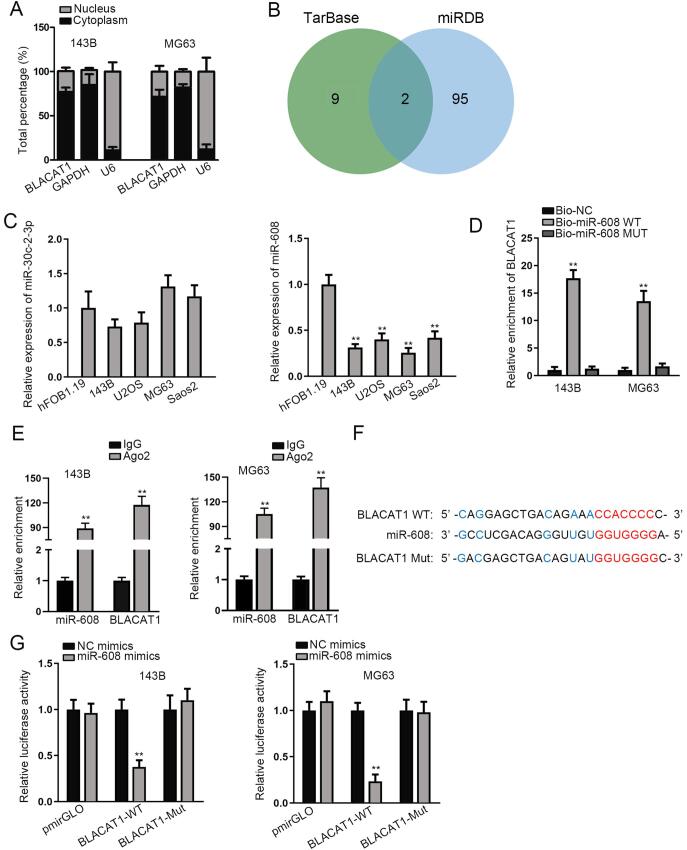
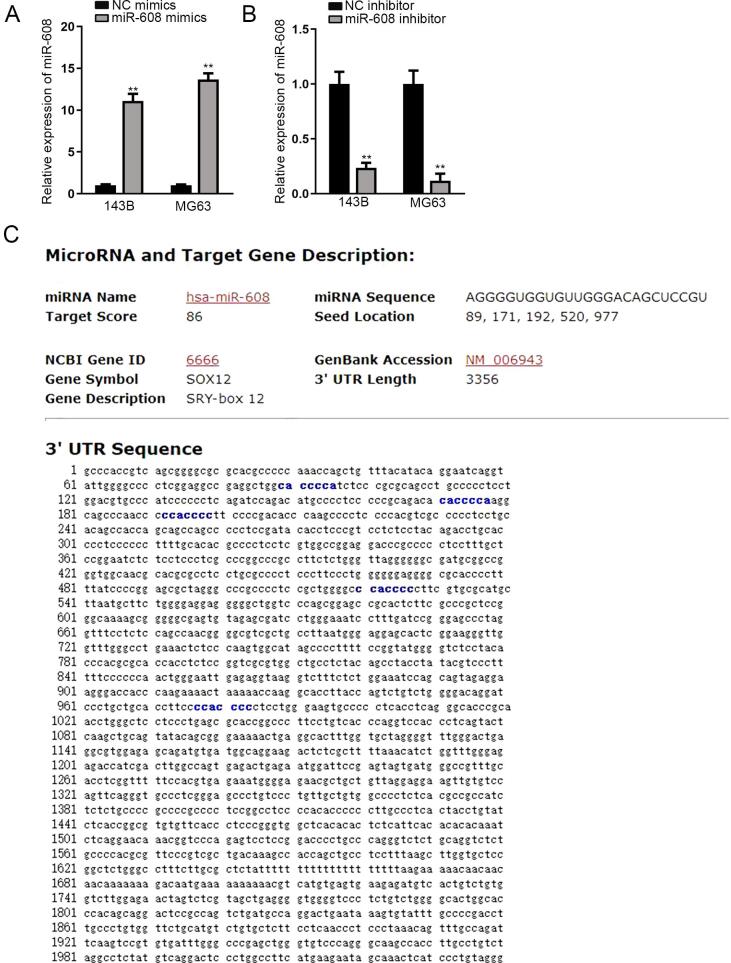
Growing lncRNAs have been discovered to serve as a sponge for specific miRNAs [15]. To examine whether BLACAT1 could act as a miRNA sponge in OS, we performed nucleus-cytoplasm separation assay. The results manifested that BLACAT1 was mainly distributed in the cytoplasm of 143B and MG63 cells (Fig. 2A), suggesting the post-transcriptional mediation of BLACAT1 plausible. By using RegRNA (http://regrna.mbc.nctu.edu.tw/html/prediction.html) and miRDB (http://mirdb.org/) tools, two candidate miRNAs (miR-30c-2-3p and miR-608) that might bind to BLACAT1 were screened out (Fig. 2B). In addition, we observed an aberrant down-regulation of miR-608 in OS cells, while revealed no significant change of miR-30c-2-3p expression in OS cells (Fig. 2C). Therefore, miR-608 was chosen for further mechanical study. RNA pull down assay delineated that BLACAT1 was noticeably enriched by Bio-miR-608- wt (Fig. 2D). Moreover, RIP assay manifested that both miR-608 and BLACAT1 were enriched by antibody targeting Ago2 in 143B and MG63 cells (Fig. 2E). Furthermore, the wild and mutated binding sites of miR-608 in the sequence of BLACAT1 were displayed in Fig. 2F. Besides, it was proved that miR-608 expression was strongly increased by the transfection of miR-608 mimics in OS cells (Fig. S1A). The results of luciferase reporter assay delineated that miR-608 overexpression evidently repressed the luciferase activity of BLACA1-WT in 143B and MG63 cells, while exerted no evident effect on the activity of BLACA1-MUT (Fig. 2G). Above data proved that BLACA1 sponged miR-608 in OS.
Fig. 2.
BLACAT1 sponges miR-608 in OS. Nucleus-cytoplasm separation assay was used to determine the subcellular distribution of BLACAT1 in 143B and MG63 cells. B. RegRNA and miRDB bioinformatics tools predicted the candidate miRNAs for BLACAT1. C. Candidate miRNA expressions were measured by qRT-PCR in OS cell lines and normal hFOB1.19 cells. D. RNA pull down was conducted to testify the physical combination between BLACAT1 and miR-608. E. RIP assay showed the abundance of BLACAT1 and miR-608 in antibody targeting Ago2. F. Putative miR-608 binding site in BLACAT1 predicted by. G. Luciferase reporter assays was performed in 143B and MG63 to verify the interaction between BLACAT1 and miR-608. **P < 0.01.
3.3. SOX12 is the target of miR-608
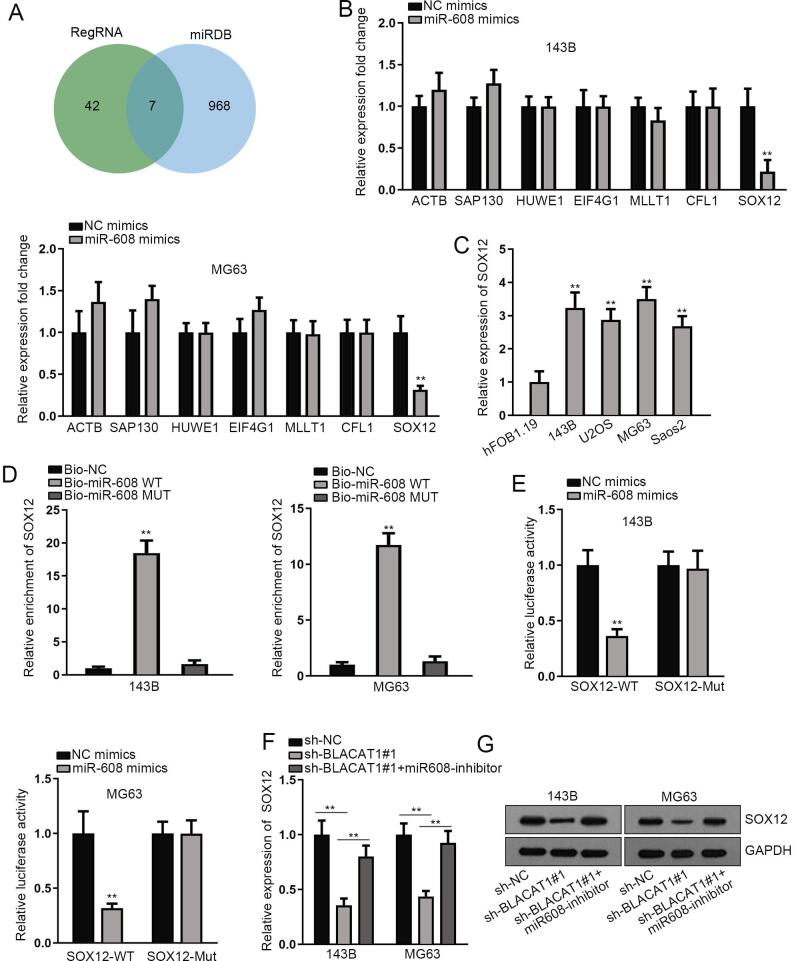
To further probe the potential ceRNA regulatory mechanism involving BLACA1 and miR-608, we searched possible targets of miR-608. After browsing miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and miRDB (http://mirdb.org/) databases, 7 potential targets, including ACTB, SAP130, HUWE1, EIF4G1, MLLT1, CFL1 and SOX12, were screened out for miR-608 (Fig. 3A). Later, we adopted qRT-PCR analysis to study the impact of miR-608 overexpression on these targets. Outcomes manifested that the expression level of SOX12, instead of balance candidates, was decreased most dramatically by miR-608 mimics (Fig. 3B). Also, a higher expression pattern of SOX12 in OS cells than normal control was also observed subsequently (Fig. 3C). RNA pull down assay delineated that SOX12 possessed a preference to be pulled down by Bio-miR-608- wt (Fig. 3D). Furthermore, the results of luciferase reporter assays showed obvious attenuation in the luciferase activity of SOX12-WT by miR-608 mimics, while no distinct change in that of SOX12-Mut (Fig. 3E). We then testified the expression level of SOX12 after co-transfecting miR-608 inhibitor into BLACA1-depleted OS cells. Before that, the interfering efficiency of miR-608 inhibitor in 143B and MG63 were ensured (Fig. S1B). Besides, the description of miR-608 and SOX12 was presented according to miRDB (Fig. S1C). As expected, both the mRNA and protein levels of SOX12 were down-regulated by silencing BLACA1, and restored again by co-transfection of miR-608 inhibitor (Fig. 3F, 3G). In a word, SOX12 targeted by miR-608 was the downstream of BLACAT1/miR-608 in OS.
Fig. 3.
SOX12 is the target of miR-608. A. MiRTarBase and miRDB databases predicted potential targets of miR-608. B. QRT-PCR analysis was conducted to study the expression of above mRNA candidates after transfecting miR-608 mimics into 143B and MG63 cells. C. The expression pattern of SOX12 was detected in OS cells versus normal controls by qRT-PCR. D. RNA pull down assay verified the binding relationship between SOX12 and miR-608 in 143B and MG63 cells. E. Luciferase reporter assays were conducted to determine the relationship between SOX12 and miR-608. F-G. QRT-PCR and western blot analyses were utilized to study the expression of SOX12 mRNA and protein after co-transfecting miR-608 inhibitor into BLACAT1-silenced 143B and MG63 cells. **P < 0.01.
3.4. BLACA1 promotes OS cell growth and motility via targeting miR-608/SOX12 axis
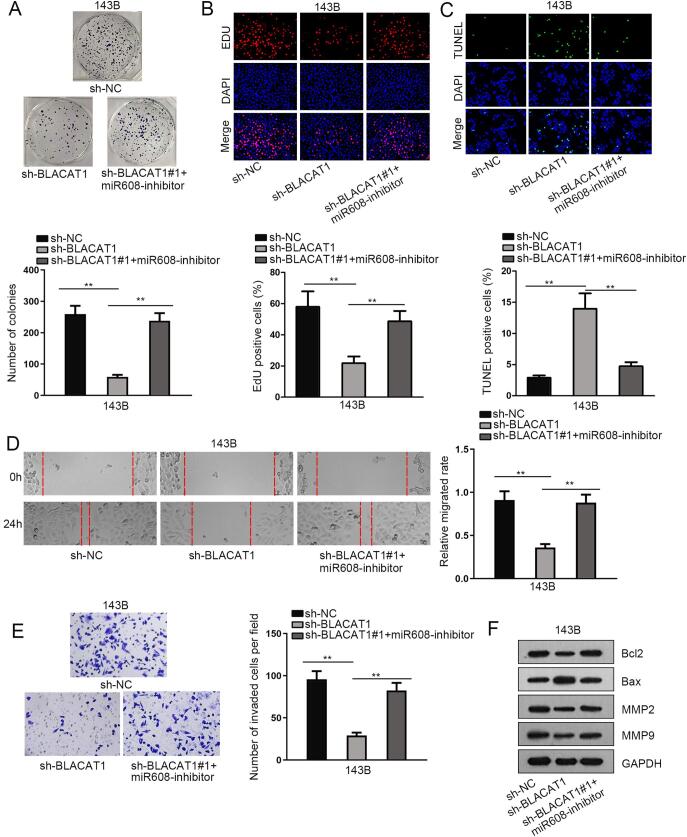
To confirm that BLACA1 could up-regulate SOX12 via sponging miR-608 in the regulation of OS progression, we conducted rescue assays. We noticed that the repressing effect of inhibited BLACA1 on cell proliferation was abolished by miR-608 inhibitor (Fig. 4A, B). Meanwhile, the apoptosis ability was strengthened by silencing BLACA1, while dampened again by miR-608 inhibitor (Fig. 4C). Additionally, the migrating and invasive abilities of OS cells were impeded by BLACA1 deficiency, yet enhanced by miR-608 inhibitor (Fig. 4D, E). Consistently, the results of western blot further proved the counteracting effect of miR-608 inhibition on BLACA1 knockdown-affected biological activities in OS cells (Fig. 4F). Collectively, our data revealed that miR-608 inhibition rescued the anti-tumor role of silenced BLACA1 in OS. Our work elucidated that BLACA1 promotes OS cell growth and motility via targeting miR-608/SOX12 axis.
Fig. 4.
MiR-608 inhibitor countervailed the biological function of BLACA1 silence. MiR-608 inhibitor countervailed the inhibitory function of BLACA1 silence on OS cell proliferation (A-B), apoptosis (C), migration (D) and invasion (E). F. Western blot analysis was carried out to measure the level of proteins relevant to cell apoptosis and migration after co-transfecting miR-608 inhibitor into BLACA1-silenced 143B cells. **P < 0.01.
4. Discussion
OS accounts for almost 20% of all bone malignancies cases, and it emerges as the second leading cause amongst adolescents. To improve the unsatisfactory overall survival rate, more exploration needs to be made in discovering the etiology of OS.
In recent years, accumulating evidence revealed the critical biological role of lncRNAs in pathologic progression. For instance, LINC00470 boosted the degradation of PTEN mRNA to promote cell proliferation, migration and invasion in gastric cancer [16]. Among the multifold regulatory mechanism of lncRNAs, the post-transcriptional ceRNA network has gained increasing attention [17]. For example, lncRNA H19 up-regulated the expression of BRD4 via sponging miR-152-3p, promoting the malignant behaviors of multiple myeloma cells [18]. LncRNA served as a ceRNA in targeting miR-194-5p-mediated PCED1B to promote cell proliferation and inhibit apoptosis in glioma [19]. To date, a large body of research established close linkages between abnormally expressed lncRNAs and OS progression. Up-regulated SNHG16 enhanced the proliferative ability of OS cells by elevating ZEB1 expression through sponging miR-205 [20]. Previous study showed that BLACAT1 was up-regulated in OS tissues. Similarly, we also identified an aberrant overexpression of BLACAT1 in OS cells. Similar to previous research [10], we validated that BLACAT1 could stimulate cell proliferation, migration and invasion, but restrained cell apoptosis in OS. This carcinogenic property of BLACAT1 was also confirmed in breast cancer [21], papillary thyroid carcinoma [22] and cervical cancer [23].
MiRNAs are short sing-standard RNA transcripts that couldn’t be translated into proteins. However, they harbor sequences complementary to the 3′UTR of target messenger RNAs (mRNAs), which provides the possibility for regulating gene expression at post-transcriptional stage in human etiology. MiR-92b was found to negatively regulate DAB2IP and indirectly activate PI3K/AKT signaling pathway in gastric cancer [24]. MiR-27a-5p was found to bind to SMAD4 and negatively modulateSMAD4 in endometrial cancer [25]. Interestingly, miRNAs could be sponged by lncRNAs and their suppressive function in target genes could be lost by such sponge. In this study, miR-608 was sponged by BLACAT1. Furthermore, SOX12 was confirmed as a target gene of miR-608 in OS. SOX12 belongs to sex-determining region Y (SRY)-box (SOX) family that has been revealed to play a crucial role in tumorigenesis. Here, we discovered that SOX12 expression could be elevated by BLACAT1 via sequestering miR-608 in OS. Moreover, miR-608 inhibition reversed the biological effects of BLACAT1 absence on the behaviors of OS cells.
5. Conclusion
In short, this study revealed that BLACAT1 contributed to OS development. Mechanically speaking, we found that BLACAT1 facilitated OS cell growth and motility by up-regulating SOX12 through sponging miR-608. This regulatory network might provide some theoretical foundations for opening novel targets for OS treatment.
CRediT authorship contribution statement
Xiaotao Chen: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Funding acquisition, Writing - original draft, Writing - review & editing. Yubao Cui: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Funding acquisition, Writing - original draft, Writing - review & editing. Yanming Ma: Resources, Software, Supervision, Validation, Visualization, Funding acquisition, Writing - original draft, Writing - review & editing.
Acknowledgments
Acknowledgement
We appreciate our experimenters.
Conflicts of interests
The authors declare no relevant conflicts of interests.
Funding
This research was supported by the Xining Health Commission project (XPF-2020-163).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2020.100314.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
References
- 1.Al-Mendalawi M.D. Spontaneous conventional osteosarcoma transformation of a chondroblastoma: A case report and literature review. Indian J. Orthop. 2018;52(6):682. doi: 10.4103/ortho.IJOrtho_107_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L., Troisi R.J., Savage S.A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer. 2009;125(1):229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruksakorn D., Kongthavonskul J., Teeyakasem P., Phanphaisarn A., Chaiyawat P., Klangjorhor J., Arpornchayanon O. Surgical outcomes of extracorporeal irradiation and re-implantation in extremities for high grade osteosarcoma: A retrospective cohort study and a systematic review of the literature. J. Bone Oncol. 2019;14 doi: 10.1016/j.jbo.2018.100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luetke A., Meyers P.A., Lewis I., Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat. Rev. 2014;40(4):523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y., Han L., He Z., Li X., Yang S., Yang J., Zhang Y., Li D., Yang Y., Yang Z. Advances in limb salvage treatment of osteosarcoma. J. Bone Oncol. 2018;10:36–40. doi: 10.1016/j.jbo.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung J., Miscianinov V., Sluimer J.C., Newby D.E., Baker A.H. Targeting non-coding RNA in vascular biology and disease. Front. Physiol. 2018;9:1655. doi: 10.3389/fphys.2018.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X., Wei Y., Wang Z., Liu W., Yang Y., Yu X., He J. LncRNA LINC00163 upregulation suppresses lung cancer development though transcriptionally increasing TCF21 expression. Am. J. Cancer Res. 2018;8(12):2494–2506. [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X., Hu L., Li S., Shen J., Wang D., Xu R., Yang H. Long non-coding RNA Taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1alpha via miR-143-5p. Cell Death Dis. 2019;10(4):280. doi: 10.1038/s41419-019-1509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan D., Shang Y., Hu T. Long noncoding RNA BLACAT1 promotes cell proliferation and invasion in human cervical cancer. Oncol. Lett. 2018;15(3):3490–3495. doi: 10.3892/ol.2018.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z., Wang Y. LncRNA BLACAT1 accelerates the proliferation and migration of osteosarcoma cells through regulating STAT3. Pathol. Res. Pract. 2019;215(3):571–579. doi: 10.1016/j.prp.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao H., Jiang S., Wang H., Li Y., Zhang W. Upregulation of LINC00963 facilitates melanoma progression through miR-608/NACC1 pathway and predicts poor prognosis. Biochem. Biophys. Res. Commun. 2018;504(1):34–39. doi: 10.1016/j.bbrc.2018.08.115. [DOI] [PubMed] [Google Scholar]
- 13.Miao Z., Guo X., Tian L. The long noncoding RNA NORAD promotes the growth of gastric cancer cells by sponging miR-608. Gene. 2019;687:116–124. doi: 10.1016/j.gene.2018.11.052. [DOI] [PubMed] [Google Scholar]
- 14.Wan H., Cai J., Chen F., Zhu J., Zhong J., Zhong H. SOX12: a novel potential target for acute myeloid leukaemia. Br. J. Haematol. 2017;176(3):421–430. doi: 10.1111/bjh.14425. [DOI] [PubMed] [Google Scholar]
- 15.Feng Z., Li X.u., Qiu M., Luo R., Lin J., Liu B.o. LncRNA EGFR-AS1 upregulates ROCK1 by sponging miR-145 to promote esophageal squamous cell carcinoma cell invasion and migration. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2019.2926. [DOI] [PubMed] [Google Scholar]
- 16.Yan J., Huang X., Zhang X., Chen Z., Ye C., Xiang W., Huang Z. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem. Biophys. Res. Commun. 2019 doi: 10.1016/j.bbrc.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Qi X., Zhang D.H., Wu N., Xiao J.H., Wang X., Ma W. ceRNA in cancer: possible functions and clinical implications. J. Med. Genet. 2015;52(10):710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 18.Zheng J.F., Guo N.H., Zi F.M., Cheng J. Long non-coding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting miR-152-3p. Mol. Cell. Biol. 2019 doi: 10.1128/MCB.00382-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Yu D., Liu X., Changyong E., Yu S. LncRNA PCED1B-AS1 activates the proliferation and restricts the apoptosis of glioma through cooperating with miR-194-5p/PCED1B axis. J. Cell. Biochem. 2019 doi: 10.1002/jcb.29417. [DOI] [PubMed] [Google Scholar]
- 20.Zhu C., Cheng D., Qiu X., Zhuang M., Liu Z. Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell. Physiol. Biochem. 2018;51(1):429–440. doi: 10.1159/000495239. [DOI] [PubMed] [Google Scholar]
- 21.Hu X., Liu Y., Du Y., Cheng T., Xia W. Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 2019;9:14. doi: 10.1186/s13578-019-0274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao D., Lv G., Wang T., Min J., Wang Y., Liu S. Prognostic value of long non-coding RNA BLACAT1 in patients with papillary thyroid carcinoma. Cancer Cell Int. 2018;18:47. doi: 10.1186/s12935-018-0544-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C.H., Li Y.H., Tian H.L., Bao X.X., Wang Z.M. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22(10):3002–3009. doi: 10.26355/eurrev_201805_15057. [DOI] [PubMed] [Google Scholar]
- 24.H.Q. Mu, Y.H. He, S.B. Wang, S. Yang, Y.J. Wang, C.J. Nan, Y.F. Bao, Q.P. Xie, Y.H. Chen, MiR-130b/TNF-alpha/NF-kappaB/VEGFA loop inhibits prostate cancer angiogenesis, Clin. Transl. Oncolo. (2019). [DOI] [PubMed]
- 25.Che X., Jian F., Chen C., Liu C., Liu G., Feng W. PCOS serum-derived exosomal miR-27a-5p stimulates endometrial cancer cells migration and invasion. J. Mol. Endocrinol. 2019 doi: 10.1530/JME-19-0159. [DOI] [PubMed] [Google Scholar]