Abstract
Autophagy is a conserved catabolic process characterized by degradation and recycling of cytosolic components or organelles through a lysosome-dependent pathway. It has a complex and close relationship to drug resistance in breast cancer. MicroRNAs (miRNAs) are small noncoding molecules that can influence numerous cellular processes including autophagy, through the posttranscriptional regulation of gene expression. Autophagy is regulated by many proteins and pathways, some of which in turn have been found to be regulated by miRNAs. These miRNAs may affect the drug resistance of breast cancer. Drug resistance is the main cause of distant recurrence, metastasis and death in breast cancer patients. In this review, we summarize the causative relationship between autophagy and drug resistance of breast cancer. The roles of autophagy-related proteins and pathways and their associated miRNAs in drug resistance of breast cancer are also discussed.
Keywords: Autophagy, MicroRNA, Breast cancer, Drug resistance
1. Introduction
Breast cancer is one of the most prevalent malignant tumors and the leading cause of cancer morbidity and mortality in women worldwide (Dai et al., 2015). Systemic therapies, including chemotherapy, endocrine therapy, and targeted therapy, are the standard methods for treating breast cancer, and have made a significant progress in the past decade. However, distant recurrence and metastasis, caused mainly by the resistance of breast cancer to systematic treatment, are the leading causes of death in breast cancer patients (Fahad Ullah, 2019). So understanding the mechanisms of drug resistance has important value and clinical significance in improving the prognosis of breast cancer.
Given that the systemic treatments for different subtypes of breast cancer are diverse, the corresponding mechanisms of drug resistance differ. For example, the resistance of estrogen receptor (ER)-positive breast cancer to endocrine drugs may be related to regulation of the receptor and changes to the downstream signal pathway (Clarke et al., 2015); the resistance of human epidermal growth factor receptor-2 (HER-2)-positive breast cancer to trastuzumab could be related to the antibody-dependent cell-mediated cytotoxicity (ADCC) effect (Musolino et al., 2008); and the resistance of triple-negative breast cancer to chemotherapy may be due to changes in various pathways (Guestini et al., 2016). However, autophagy is one of the drug resistance mechanisms shared by different breast cancer cells and their treatment. Autophagy (macroautophagy) is a catabolic process whereby intracellular cytosolic components or organelles are enveloped in double-membraned vesicles, known as autophagosomes, which ultimately fuse with lysosomes where the contents are degraded and recycled into the cytosol (Levine and Klionsky, 2004; Zhou et al., 2018). The correlation between autophagy and drug resistance has been confirmed in many other tumors, such as osteosarcoma, ovarian cancer, strong malignant glioma, multiple myeloma, colorectal cancer, prostate cancer, bladder cancer, and leukemia (Pan et al., 2013; Li et al., 2017).
Autophagy is a complex process involving regulation by a large number of proteins and related enzymes. MicroRNAs (miRNAs) are a class of endogenously expressed, short non-coding RNAs, which play important gene-regulatory roles by pairing to the target messenger RNAs (mRNAs) of protein-coding genes to direct their posttranscriptional repression. This results in mRNA degradation and/or translational inhibition and then a reduction in the synthesis of the corresponding proteins (Bartel, 2009). So it can be inferred that miRNAs can regulate autophagy via targeting the mRNAs of autophagy-related proteins, thereby affecting the sensitivity of breast cancer cells to drugs (Pan et al., 2013). Indeed, numerous recent studies summarized below have demonstrated several relationships between miRNAs, autophagy, and drug resistance of breast cancer.
2. Relationship between autophagy and drug resistance
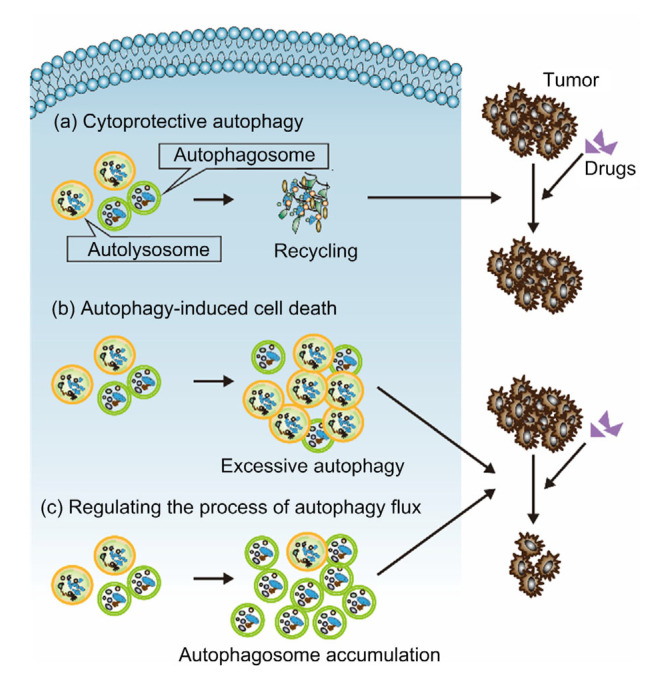
In general, autophagy mediates both tumor cell survival and death (Hanahan and Weinberg, 2011). Traditionally, the relationship between autophagy and drug resistance has been divided in terms of two contrasting mechanisms and their associated effects: a protective mechanism which induces tumor drug resistance, or autophagy-induced cell death which increases the sensitivity of tumors to killers (Hanahan and Weinberg, 2011; Li et al., 2017). However, more and more studies have found that the relationship between autophagy and drug resistance is not so simple, and the regulation of the process of autophagy seems to be the key to this relationship (Periyasamy-Thandavan et al., 2010; Li et al., 2012; Schwartz-Roberts et al., 2013). In summary, in studies of breast cancer, the relationship between autophagy and drug resistance is reflected in three ways: (1) autophagy induces breast cancer drug resistance; (2) autophagy-induced cell death increases the sensitivity of breast cancer to drugs; and, (3) regulating the process of autophagy flux affects the sensitivity of breast cancer to drugs (Fig. 1). These aspects are introduced in detail below.
Fig. 1.
Relationship between autophagy and drug resistance of breast cancer
(a) Cytoprotective autophagy leads to drug resistance of breast cancer; (b) Autophagy-induced cell death increases the sensitivity of breast cancer cells to drugs; (c) Regulating the process of autophagy flux affects the sensitivity of breast cancer to drugs
2.1. Autophagy induces breast cancer drug resistance (cytoprotective autophagy)
Epirubicin-resistant MCF-7 cells, tamoxifen-resistant ER-positive breast cancer cells, and trastuzumab-resistant SKBR3 cells (HER-2-positive) have all been shown to have a higher autophagy level than their corresponding drug-sensitive cells. Also, by inhibiting autophagy, the drug sensitivity of these three kinds of breast cancer cells can be improved (Vazquez-Martin et al., 2009; Sun et al., 2011; Kim et al., 2015; Gu et al., 2017). It seems that relatively high levels of autophagy are closely related to resistance to systemic treatments of breast cancer.
Autophagy is a highly conserved catabolic process which occurs in response to unfavorable conditions, including starvation, caspase inhibition, and the presence of cytokines or chemical reagents, to ensure cell survival and homeostasis (Kim et al., 2013). In the treatment of breast cancer, the uses of endocrine drugs, chemotherapy drugs, and targeted drugs prevent cell growth and cause cell death by having a huge impact on the DNA and key proteins of breast cancer cells. Under these conditions, autophagy can degrade macromolecular waste, long-lived proteins, and damaged organelles in lysosomes, and some residual cells even enter into a dormant state. This affords time and nutritional support for tumor cells and provides opportunities for cell repair, which leads to tumor drug resistance, recurrence, and progression (Rabinowitz and White, 2010; Guo et al., 2011; Kim et al., 2013). This kind of autophagy, which protects breast cancer cells and reduces their sensitivity to drugs, is called cytoprotective autophagy.
Cytoprotective autophagy may also lead to the occurrence of “dual drug resistance.” Everolimus/RAD001 (Afinitor®), a phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway inhibitor, can enhance 4-hydroxy-tamoxifen (4-OHT) sensitivity in both MCF-7:2A cells (cloned from parental MCF-7 cells, partially sensitive to 4-OHT) and MCF-7:5C cells (cloned from parental MCF-7 cells, unresponsive to 4-OHT). However, it increases the number of autophagosomes as well. Combined inhibition of autophagy with chloroquine (CQ) significantly improves the efficacy of everolimus treatment on cell proliferation, indicating that autophagy is a mechanism of everolimus insensitivity (Lui et al., 2016). The PI3K/AKT/mTOR pathway plays an important role in the regulation of autophagy (Yang and Klionsky, 2010). Yang and Klionsky (2010) showed that the inhibition of the PI3K/AKT/mTOR pathway often leads to an increase in intracellular autophagy (which will be introduced in the context of autophagy-related proteins/pathways later in this review). Therefore, it is not difficult to understand why everolimus leads to an increase in autophagy. The above research suggests that “anti-drug resistance drugs” may fail due to autophagy, which is a barrier we cannot ignore.
2.2. Autophagy-induced cell death increases the sensitivity of breast cancer to drugs
Autophagy does not have only a single effect on breast cancer resistance. In some studies, we found that increased autophagy flux induced cell death and increased the sensitivity of breast cancer cells to drugs, which is opposite to the effect of the above-mentioned cytoprotective autophagy (Gonzalez-Malerva et al., 2011; Wu et al., 2012; Abdel-Mohsen et al., 2019). Autophagy-induced cell death can be divided into two categories. The first is autophagy-dependent cell death (ADCD), which was defined by the Nomenclature Committee on Cell Death in 2018 as regulated cell death that depends on the autophagy machinery without involving alternative death pathways (Galluzzi et al., 2018). That definition was consistent with previous definition of autophagic cell death (Shen and Codogno, 2011). The second category is autophagic related cell death, in which increased autophagy induces other death pathways, and so is quite different from ADCD. Autophagy can be associated with necrosis-like cell death induced by caspase inhibition; autophagy and apoptosis can occur simultaneously or inversely, depending on the experimental conditions and the signaling pathways shared by both; and autophagy may regulate some other types of cell death such as necrosis (Zhong et al., 2017; Yan et al., 2019).
Using high-throughput cell-based screens, researchers have identified 31 kinases, including small heat shock protein β-8 (HSPB8), that confer drug resistance on sensitive breast cancer cells. Silencing HSPB8 increases the sensitivity of MCF-7 cells to 4-OHT by inducing ADCD, whereas ectopic expression of HSPB8 blocks autophagy and provides a proliferation advantage with 4-OHT treatment compared with a luciferase control (Gonzalez-Malerva et al., 2011). Macrophage migration inhibitory factor (MIF) is of great importance to cell survival. Overexpression of MIF promotes cell survival and proliferation and reduces chemosensitivity to doxorubicin (DOX) and etoposide through decreasing ADCD. In contrast, knockdown of MIF promotes autophagy-specific light chain 3-II (LC3-II) and enhances the cytotoxicity of DOX and etoposide in MCF-7 breast cancer cells compared to controls (Wu et al., 2012). In a recent study, Torin-1 (autophagy inducer) in combination with DOX caused a statistically significant increase of autophagy and a more cytotoxic effect on cell growth than the same dose of DOX used alone, which indicated that the combination therapy increased the sensitivity of breast cancer cells to DOX via ADCD (Abdel-Mohsen et al., 2019). This is an example of autophagic related cell death increasing the drug sensitivity of breast cancer cells. Endoplasmic reticulum stress activated by tunicamycin could enhance chemosensitivity of MCF-7 cells by promoting autophagy and apoptosis at the same time via inhibiting the PI3K/AKT/mTOR signaling pathway (Zhong et al., 2017).
The different effects of autophagy on cell survival have puzzled researchers. When does a rise of autophagy lead to cell death or play a role in cell protection? Bialik et al. (2018) have shown that lethality may result from extreme levels of autophagy flux that lead to overconsumption of cellular organelles and rerouting of cellular membrane sources to support autophagosome generation, to the point where cellular membrane homeostasis is disturbed. Or excessive autophagy of cells damages important organelles, such as mitochondria, and induces other forms of cell death (Bialik et al., 2018). Autophagy associated with other cell death patterns may result from pathway crosstalk among different phenotypes (Green and Llambi, 2015).
2.3. Regulating the process of autophagy flux affects the sensitivity of breast cancer to drugs
The regulation of drug sensitivity to breast cancer cells is achieved by up-or down-regulating intact autophagy flux. Detailed studies of autophagy found that regulating the intermediate process of autophagy flux seems to have a great influence on its final outcome.
Periyasamy-Thandavan et al. (2010) demonstrated crosstalk between the autophagy and proteasome-mediated pathways of protein degradation. The proteasome inhibitor, bortezomib, blocks the catabolic process of autophagy. Cells showed high levels of LC3-II and p62, which suggests that autolysosomal turnover of proteins was not occurring (Periyasamy-Thandavan et al., 2010). GX15-070 (obatoclax) inhibits autophagosomal degradation. This causes breast cancer cells to lose their ability to recycle subcellular components through autophagy and restore metabolic homeostasis (Schwartz-Roberts et al., 2013). Both bortezomib and GX15-070 increased the sensitivity of antiestrogen-resistant breast cancer cells to tamoxifen by inhibiting autolysosomal function without inhibiting autophagosome formation (Periyasamy-Thandavan et al., 2010; Schwartz-Roberts et al., 2013).
Lysosomal-associated protein transmembrane 4 β (LAPTM4B) plays an important role in lysosomal functions and is critical for autophagic maturation. Knockdown of LAPTM4B by small interfering RNA (siRNA) increases the sensitivity of breast cancer to chemotherapy, and overexpression of LAPTM4B induces drug resistance (Li et al., 2010). There seem to be three reasons why inhibition of LAPTM4B increases the sensitivity of breast cancer cells to chemotherapy drugs: (1) LAPTM4B acts on anthracycline trafficking by reducing drug entry into the nucleus and decreasing drug-induced DNA damage, and the loss of LAPTM4B will increase the permeability of the lysosomal (autolysosomal) membrane, which leads to a large number of drugs entering the nucleus; (2) the increase of lysosomal (autolysosomal) membrane permeability also results in the release of cathepsin, which leads to lysosomal-mediated programmed cell death; (3) the failure of the fusion of autophagic bodies and lysosomes leads to the accumulation of autophagosomes, thereby inducing cell death (Li et al., 2010, 2011, 2012).
Further research has proved that increased accumulation of autophagosomes through diverse targets widely induces cytotoxicity. Accumulation of autophagosomes causes loss of cell viability independent of apoptosis and necroptosis. Lowering accumulation of autophagosomes by partial depletion of autophagosome synthesis provides a rescue from aggregation-prone protein toxicity (Button et al., 2017). Furthermore, combining chemotherapeutic drugs with autophagosome clearance inhibitors has been proven to increase treatment potency in a variety of tumors (Heng et al., 2010; Decressac et al., 2013; Kimura et al., 2013; Li et al., 2017). So even if there is no direct research confirmation, it is probable that the accumulation of autophagosomes regulates the sensitivity of breast cancer to drugs.
Therefore, regulation of breast cancer resistance could be achieved not only through simple up-or down-regulation of autophagy, but also by finding out how to affect the sensitivity of breast cancers to drugs by regulating the specific process of autophagy.
3. Effects of miRNAs on autophagy-related proteins and pathways involved in breast cancer drug resistance
As shown above, autophagy is closely related to drug resistance of breast cancer. With further research on autophagy, the major proteins and related pathways that regulate it have been studied in depth. The up- or down-regulation of mTOR1, Beclin 1, autophagy-related gene protein (ATG), unc-51-like autophagy activating kinase 1 (ULK1), adenosine 5'-monophosphate (AMP)-activated protein kinase (AMPK), high mobility group box 1 protein (HMGB1), and other proteins can affect autophagy and thus tumor drug resistance.
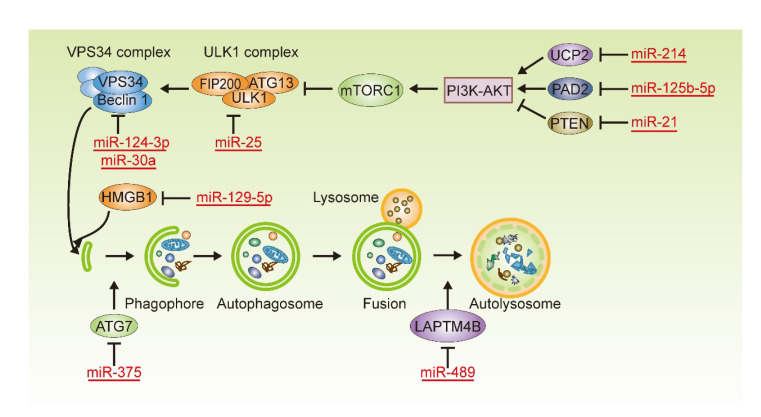
In most previous studies, autophagy was inhibited or induced by drugs and chemicals which were not specific. However, according to the mechanism of action of miRNAs, they precisely regulate the expression of autophagic related proteins by acting on mRNA complementary to their base sequences, thereby affecting autophagy and breast cancer cell resistance. miRNAs affecting drug sensitivity in breast cancer cells in this way are summarized in Table 1 and Fig. 2, and their regulatory relationships are described in detail below.
Table 1.
miRNAs involved in the regulation of autophagy which affects drug sensitivity in breast cancer cells
| miRNA | Target | Autophagy protein/pathway | Autophagy | Sensitivity | Cell type | Reference |
| miR-214 | UCP2 | AKT/mTOR | ↓ | ↑ (Endo) | MCF7/LCC9 cells | Yu et al., 2015 |
| miR-125b-5p | PAD2 | AKT/mTOR | ↑ | ↑ (Endo) | MCF7/TamR cellsa | Li et al., 2019 |
| miR-21 | PTEN | PI3K-AKT-mTOR | ↓ | ↓ (Endo) | MCF-7 cells | Yu et al., 2016 |
| miR-25 | ULK1 | ULK1 | ↓ | ↓ (Chemo) | MCF-7/ADR cellsb | Wang et al., 2014 |
| miR-124-3p | Beclin 1 | Beclin 1 | ↓ | ↑ (Endo) | MCF-7 cells | Zhang et al., 2016 |
| miR-30a | Beclin 1 | Beclin 1 | ↓ | ↑ (Chemo) | MCF-7 cells | Zou et al., 2012 |
| miR-375 | ATG7 | ATG7 | ↓ | ↑ (lapatinib and imatinib) | FRBC cellsc | Liu et al., 2018 |
| miR-129-5p | HMGB1 | HMGB1 | ↓ | ↑ (Chemo) | MCF-7 cells | Shi et al., 2019 |
| miR-489 | LAPTM4B | LAPTM4B | Block maturation | ↑ (Chemo) | Multiple breast cancer cells | Soni et al., 2018 |
UCP2: uncoupling protein 2; PAD2: peptidylarginine deiminases 2; PTEN: phosphatase and tensin homologue; ULK1: unc-51-like autophagy activating kinase 1; ATG7: autophagy-related gene protein 7; HMGB1: high mobility group box 1 protein; LAPTM4B: lysosomal protein transmembrane 4 β; AKT: protein kinase B; mTOR: mammalian target of rapamycin; PI3K: phosphoinositide 3-kinase; Endo: endocrine drug; Chemo: chemotherapeutic drug.
Tamoxifen-resistant MCF7 cells;
Adriamycin-resistant MCF7 cells;
Fulvestrant-resistant MCF7 cells
Fig. 2.
miRNAs and their targets involved in the regulation of autophagy which affects drug sensitivity in breast cancer cells
miRNAs affect autophagy by precisely regulating the expression of autophagic related proteins, thereby affecting the drug resistance of breast cancer cells. PI3K: phosphoinositide 3-kinase; VPS34: the class III PI3K vacuolar protein sorting 34; ULK1: unc-51-like autophagy activating kinase 1; FIP200: FAK family kinase-interacting protein of 200 kDa; ATG: autophagy-related gene; mTORC1: mammalian target of rapamycin complex 1; AKT: protein kinase B; UCP2: uncoupling protein 2; PAD2: peptidylarginine deiminases 2; PTEN: phosphatase and tensin homologue; HMGB1: high mobility group box 1 protein; LAPTM4B: lysosomal protein transmembrane 4 β
3.1. mTOR and its related miRNAs
mTOR is a master regulator of autophagy. It inhibits the ULK1-Atg13-FIP200 complex, which is crucial for the beginning of autophagosome formation (Romero et al., 2019). There are several ways to regulate mTOR. In breast cancer cells, the PI3K/AKT pathway is an upstream major modulator (Yang and Klionsky, 2010).
Overexpression of uncoupling protein 2 (UCP2) contributes to endocrine resistance to 4-OHT by the induction of autophagy in MCF7 cells accompanied by activation of AKT and mTOR, whereas it shows the opposite results in UCP2-knockdown cells. In addition, UCP2 is a direct target of miR-214, and an increase of miR-214 improves the sensitivity of breast cancer cells to the 4-OHT/fulvestrant-induced apoptosis via inhibition of autophagy through down-regulation of UCP2 (Yu et al., 2015).
Glutathione S-transferase P1 (GSTP1) is a phase II detoxifying enzyme. Its level is very low in human breast cancer cell line MCF-7, but is high in adriamycin (ADR)-resistant MCF-7/ADR cells (Dong XL et al., 2019). A high level of GSTP1 maintains breast cancer cell resistance to ADR by enhancing the autophagy level through interacting with PI3K, and then inhibits PI3K/AKT/mTOR activity (Dong XL et al., 2019). Although the regulation of miRNA on GSTP1 and autophagy was not included in that study, other studies have shown that miR-513a-3p can sensitize human lung adenocarcinoma cells to cisplatin by targeting GSTP1 (Zhang et al., 2012), and miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells (Lin et al., 2018). Therefore, it is very likely that breast cancer sensitivity to drugs can also be regulated by miRNAs targeting GSTP1 and then affecting autophagy. E twenty-six (ETS)-like transcription factor 3 (ELK3) normally functions as a transcriptional repressor of gene expression. Suppression of ELK3 causes much lower autophagy activity than control cells by activating PI3K/AKT/mTOR, and increases the killing effect of DOX on MDA-MB-231 cells (Park et al., 2016). ELK3 can be regulated by miR-155-5p, miRNA-135a, and miR-378 (Chan et al., 2014; Robertson et al., 2014; Ahmad et al., 2018), which might have potential for regulating breast cancer drug resistance. Promoting autophagy via the PI3K/AKT pathway causes chemoresistance to pharmorubicin in breast cancer cells. The process can be mediated by heme oxygenase-1 (HO-1) (Pei et al., 2018), which can be regulated by miRNAs including miR-1254, miR-29a, miR-218-5p, and miR-92a (Pu et al., 2017; Fan et al., 2018; Gou et al., 2018; Zhang and Xiang, 2019).
The above describes the manifestation of protective autophagy in breast cancer cell drug resistance. The following shows that autophagy increases the sensitivity of breast cancer to drugs through the PI3K/AKT/mTOR pathway. Cyclovirobuxine D, an alkaloid component in a traditional Chinese herb, induces autophagic related cell death by suppressing the phosphorylation of AKT and mTOR in MCF-7 breast cancer cells (Lu et al., 2014).
miR-21 was found to be overexpressed in breast cancers (Yu et al., 2016). Silencing of miR-21 increased the sensitivity of ER-positive breast cancer cells to tamoxifen or fulvestrant by increasing ADCD through inhibition of the PI3K/AKT/mTOR pathway (Yu et al., 2016). Peptidylarginine deiminases 2 (PAD2) was dramatically up-regulated in MCF7/TamR cells (tamoxifen-resistant breast cancer cells). Decreasing PAD2 expression by Cl-amidine or depletion via a lentivirus-based approach accelerated autophagy processes and increased apoptosis by synergistically decreasing the activation of AKT/mTOR signaling, thereby enhancing the efficacy of tamoxifen and even docetaxel. More importantly, miR-125b-5p also improved the sensitivity of MCF7/TamR cells to drugs through targeting PAD2 and produced the same effect (Li et al., 2019).
In addition to the PI3K/AKT pathway, AMPK is also an important upstream regulator of the mTOR protein. AMPK is an evolutionarily conserved energy-sensing kinase that is activated by metabolic stress or adenosine triphosphate (ATP) consumption. It globally promotes catabolic processes linked to the regulation of autophagy, and calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK)-mediated enhancement of AMPK activity induces autophagy (Alers et al., 2012). Human canonical transient receptor potential channel 5 (TRPC5) is a Ca2+-permeable cation channel which is associated with cancer chemotherapy (Ma et al., 2014). It regulates cytosolic Ca2+ concentration to gradually activate CaMKKβ and AMPKα, inhibiting mTOR, thereby inducing autophagy and increasing the sensitivity of breast cancer cells to drugs (Zhang et al., 2017). At present, there are no reports of the regulation of the AMPK/mTOR pathway by miRNA to affect autophagy and change the drug resistance of breast cancer cells.
3.2. ULK1 and miR-25
ULK1 is in a large complex that includes Atg13 and the scaffold protein FIP200. Under certain conditions, upstream proteins dephosphorylate mTOR complex 1 (mTORC1) and dissociate it from the ULK1-Atg13-FIP200 complex, so as to induce ULK1 activity and autophagy; conversely, phosphorylated mTORC1 is bound to the complex, thereby inhibiting autophagy (Hosokawa et al., 2009; Jung et al., 2009; Yang and Klionsky, 2010; Sun DJ et al., 2018). Studies have shown that fluoxetine and flavonoids activate ULK and induce ADCD through the eukaryotic elongation factor-2 kinase (eEF2K)/AMPK/mTOR and PI3K/AKT/mTOR pathways in human breast cancer cells individually (Sun DJ et al., 2018; Zhang et al., 2018).
Isoliquiritigenin (ISL) chemosensitizes MCF-7/ADR cells primarily by inducing ADCD. A miRNA 3.0 array analysis revealed that miR-25 is the primary target of ISL and a novel autophagy-related miRNA, and this was confirmed by a functional study (Wang et al., 2014). ISL dose-dependently inhibited miR-25 expression, and miR-25 overexpression blocked ISL-induced autophagy and chemosensitization by targeting ULK1. MCF-7/ADR cells transfected with miR-25 inhibitors also showed increased autophagy and ADCD from epirubicin (EPI), which indicates that inhibiting miR-25 has the potential to help breast cancer cells overcome drug resistance (Wang et al., 2014).
3.3. Beclin 1, miR-30a, and miR-124-3p
Beclin 1 is a well-known key protein regulating autophagy. It functions not only during autophagosome formation, but also during autophagosome and endosome maturation (Xu and Qin, 2019). Beclin 1 regulates autophagy by forming complexes with different proteins, among which the B cell lymphoma 2 (Bcl-2)–Beclin 1 complex is the most important and has been most thoroughly studied (Yang and Klionsky, 2010; Xu and Qin, 2019)
miR-30a is significantly reduced and autophagy activity is promoted in breast cancer cells by treatment with cis-dichloro-diamine platinum (cis-DDP). Forced expression of miR-30a enhances cis-DDP-induced cancer cell apoptosis by repressing autophagy, which in this case plays a cell protection role. Cells transfected with pre-miR-30a and treated with only 5 g/mL cis-DDP achieved an effect equivalent to that of 20 g/mL cis-DDP in a control group, which means that elevation of miR-30a significantly increases sensitivity to cis-DDP. Beclin 1 was proved to be the target of miR-30a and links it to autophagy, enabling miR-30a to regulate the drug sensitivity of breast cancer cells (Zou et al., 2012).
miR-124-3p is reduced in breast cancer tissues and Beclin 1 is its target gene. In breast cancer cells, overexpression of miR-124-3p can decrease the expression of Beclin 1 and partially reverse 4-OHT-induced autophagy and ADCD, thereby increasing the sensitivity of cells to 4-OHT (Zhang et al., 2016).
3.4. ATG proteins and miR-375
More than 35 ATG proteins have been identified in yeast, and the 14 core Atg genes (Atg1–10, 12, 14, 16, and 18) required for starvation-induced autophagy and selective autophagy are highly conserved in mammals. These proteins are involved in multiple processes of autophagosome formation (Wen and Klionsky, 2016; Li and Zhang, 2019). The following summarizes studies that affect the autophagy and drug resistance of breast cancer cells through regulating ATG proteins with or without miRNA. These studies currently all involve cytoprotective autophagy.
When autophagy was compromised by ATG5-siRNA or ATG7-siRNA before treatment with tamoxifen in the ER-positive T47D breast cancer cell line, which is known to exhibit reduced sensitivity to tamoxifen, the reduction in viable cell numbers was much greater than that with tamoxifen alone or tamoxifen with a scramble-siRNA sequence treatment (Qadir et al., 2008). Atg gene silencing alone did not produce any significant decrease in cell viability. Similar results were found in the tamoxifen-resistant MCF7-HER2 cell line when ATG5 and ATG7 were separately knocked down (Qadir et al., 2008). A recent study found that treatment of MDA-MB-231 cells with EPI was accompanied by an increase in autophagy levels and the expression of autophagy/Beclin 1 regulator 1 (AMBRA1). The autophagy level was negatively correlated with EPI sensitivity (Sun WL et al., 2018). Sun WL et al. (2018) explored the relationship between autophagy and drug resistance. AMBRA1 regulated autophagy by targeting ATG12, and their expression was positively correlated. Overexpression of ATG12 dramatically increased the cell viability following EPI treatment and decreased cell death. Sun WL et al. (2018) concluded that the regulatory effect of AMBRA1 on EPI sensitivity is achieved through the regulation of autophagy by targeting ATG12. Icariin, a major component isolated from Epimedium brevicornum Maxim, was shown to significantly suppress autophagy and increase sensitivity to tamoxifen in MCF-7/TamR cells. Enhanced autophagy via ATG5 overexpression can partially reverse the effects of icariin (Cheng et al., 2019).
Expression of miR-375 regulates autophagy by targeting ATG7 and affects the sensitivity of fulvestrant-resistant breast cancer (FRBC) cells to lapatinib and imatinib treatment (dual treatment). When FRBC cells were transfected with miR-375, cell viability after dual treatment significantly decreased compared with the control group, and the effect was attenuated when miR-375 was inhibited (Liu et al., 2018). Liu et al. (2018) showed that for breast cancer cells with resistance to systemic therapy, other non-systemic therapy drugs can be considered, and their sensitivity may also be regulated by the influence of miRNAs on autophagy.
3.5. Histone deacetylase
Histone deacetylases (HDACs) are enzymes involved in the remodeling of chromatin, and have a key role in the epigenetic regulation of gene expression (Bolden et al., 2006). They have gained growing attention for their application in disease treatment, mainly through research using HDAC-inhibiting compounds. One of the effects of HDAC inhibition is induction of autophagy (Trüe and Matthias, 2012).
HDAC inhibitors were proved to induce autophagic-related cell death in ER-positive breast cancer cells (Thomas et al., 2011; Lee et al., 2012; Park et al., 2012). Valproic acid (VPA), an HDAC inhibitor, increased cell autophagy and apoptosis in combination with tamoxifen, beyond the effects seen with tamoxifen alone. Autophagy and apoptosis were both induced by LC3 in the study, indicating that the cell death was associated with autophagy (Thomas et al., 2011). The results demonstrated that VPA can increase the sensitivity of ER-positive breast cancer cells to tamoxifen by increasing autophagic-related cell death.
Although there are no reports of the relationship between miRNAs, HDAC, autophagy, and drug resistance in breast cancer, some studies show that HDAC inhibitor regulates the tumorigenicity, proliferation, invasion, and drug resistance of cancer via miRNAs. miRNAs also target HDAC and affect the growth of tumors (Noonan et al., 2009; Jeon et al., 2012; Hsieh et al., 2015; Lai et al., 2016; Napoli et al., 2016; Jung et al., 2017; Bamodu et al., 2018; Bian et al., 2018).
3.6. Other autophagy-related proteins
miRNAs may regulate other autophagy-related proteins or pathways in addition to those mentioned above to regulate autophagy, thereby affecting the drug resistance of breast cancer cells. For example, up-regulation of miR-129-5p enhanced the chemosensitivity of Taxol by inhibiting autophagy through down-regulation of HMGB1 in MCF-7 cells (Shi et al., 2019). miR-489 acts as a therapeutic sensitizer in MDA-MB-231 cells, mainly by directly targeting LAPTM4B (Soni et al., 2018). Decreased expression of LAPTM4B inhibits the fusion of autophagosomes and lysosomes, the mature step of autophagy. This causes DOX gathered in the lysosome to be released into the nucleus, thereby enhancing the sensitivity of breast cancer cells to chemotherapy (Soni et al., 2018). This shows that regulating the process of autophagy flux affects the sensitivity of breast cancer cells to drugs.
4. Other miRNAs related to drug resistance of breast cancer
Other miRNAs that regulate the sensitivity of breast cancer cells to drugs have been identified, but their association with autophagy has not been reported. miR-105-2, miR-877, let-7f, miR-125a, miR-342, miR-221/222, and miR-574-3p are associated with a loss of inhibition by tamoxifen in MCF-7 cells (Miller et al., 2008; Cittelly et al., 2010; Ujihira et al., 2015). miR-30c, miR-21, miR-203, miR-134, miR-218, miR-638, and miR-141 affect breast cancer chemotherapy resistance in a variety of ways (Ru et al., 2011; Bockhorn et al., 2013; Chen and Bourguignon, 2014; Tan et al., 2014; He et al., 2015; O'Brien et al., 2015; Yao et al., 2015). miR-125b, miR-200c, and miR-18a can also affect the sensitivity of breast cancer cells to targeted drugs through non-autophagy pathways (Shi et al., 2015; Zhu et al., 2018; Dong HY et al., 2019).
5. Conclusions and perspectives
In this review, the role of autophagy in drug resistance of breast cancer is described in detail. The autophagy-related proteins and pathways related to drug resistance, and the relationships among miRNAs, autophagy, and drug resistance are summarized. Specific miRNAs inhibit the synthesis of autophagy-related proteins by binding to target mRNAs, thereby regulating autophagy and then affecting the sensitivity of breast cancer to endocrine drugs, chemotherapy drugs, and targeted drugs.
However, due to the complexity of the relationship between autophagy and breast cancer resistance, miRNAs also have multiple manifestations in regulating drug resistance through autophagy. So it is essential to determine when to increase or inhibit autophagy, and when to regulate the specific process of autophagy to increase the sensitivity of breast cancer cells to drugs. Based on this premise, using miRNAs to regulate target proteins may be a way to overcome breast cancer drug resistance.
Footnotes
Project supported by the Department of Science and Technology of Sichuan Province, China (No. 2019YFH0146) and the Health Commission of Sichuan Province, China (Research Project on Healthcare in Sichuan Province, No. 2019-107)
Contributors: Zheng-gui DU contributed to the conception of the article. Nan WEN wrote and edited the manuscript and created the figures. Qing LV edited and checked the final version. All authors participated in manuscript revision, and have read and approved the final version.
Compliance with ethics guidelines: Nan WEN, Qing LV, and Zheng-gui DU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Abdel-Mohsen MA, Abdel Malak CA, El-Shafey ES. Influence of copper (I) nicotinate complex and autophagy modulation on doxorubicin-induced cytotoxicity in HCC1806 breast cancer cells. Adv Med Sci. 2019;64(1):202–209. doi: 10.1016/j.advms.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad A, Zhang WJ, Wu MM, et al. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genomics. 2018;40(3):243–251. doi: 10.1007/s13258-017-0624-6. [DOI] [PubMed] [Google Scholar]
- 3.Alers S, Löffler AS, Wesselborg S, et al. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/mcb.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamodu OA, Kuo KT, Yuan LP, et al. HDAC inhibitor suppresses proliferation and tumorigenicity of drug-resistant chronic myeloid leukemia stem cells through regulation of hsa-miR-196a targeting BCR/ABL1. Exp Cell Res. 2018;370(2):519–530. doi: 10.1016/j.yexcr.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialik S, Dasari SK, Kimchi A. Autophagy-dependent cell death–where, how and why a cell eats itself to death. J Cell Sci. 2018;131(18):jcs215152. doi: 10.1242/jcs.215152. [DOI] [PubMed] [Google Scholar]
- 7.Bian XH, Liang ZX, Feng A, et al. HDAC inhibitor suppresses proliferation and invasion of breast cancer cells through regulation of miR-200c targeting CRKL. Biochem Pharmacol. 2018;147:30–37. doi: 10.1016/j.bcp.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bockhorn J, Dalton R, Nwachukwu C, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun, 4:1393. 2013 doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 10.Button RW, Roberts SL, Willis TL, et al. Accumulation of autophagosomes confers cytotoxicity. J Biol Chem. 2017;292(33):13599–13614. doi: 10.1074/jbc.M117.782276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JK, Kiet TK, Blansit K, et al. MiR-378 as a biomarker for response to anti-angiogenic treatment in ovarian cancer. Gynecol Oncol. 2014;133(3):568–574. doi: 10.1016/j.ygyno.2014.03.564. [DOI] [PubMed] [Google Scholar]
- 12.Chen LQ, Bourguignon LYW. Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol Cancer, 13:52. 2014 doi: 10.1186/1476-4598-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Tan SR, Duan FF, et al. Icariin induces apoptosis by suppressing autophagy in tamoxifen-resistant breast cancer cell line MCF-7/TAM. Breast Cancer. 2019;26(6):766–775. doi: 10.1007/s12282-019-00980-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cittelly DM, Das PM, Spoelstra NS, et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer, 9:317. 2010 doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–an overview and update. Mol Cell Endocrinol. 2015;418:220–234. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai XF, Li T, Bai ZH, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5(10):2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 17.Decressac M, Mattsson B, Weikop P, et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci USA. 2013;110(19):E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong HY, Hu JG, Zou KJ, et al. Activation of lncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting microRNA-125b in breast cancer. Mol Cancer. 2019;18(1):3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Dong XL, Yang Y, Zhou Y, et al. Glutathione S-transferases P1 protects breast cancer cell from adriamycin-induced cell death through promoting autophagy. Cell Death Differ. 2019;26(10):2086–2099. doi: 10.1038/s41418-019-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fahad Ullah M. Breast cancer: current perspectives on the disease status. In: Ahmad A , editor. Breast Cancer Metastasis and Drug Resistance. Springer, Cham; 2019. pp. 51–64. [DOI] [Google Scholar]
- 21.Fan WX, Wen XL, Xiao H, et al. MicroRNA-29a enhances autophagy in podocytes as a protective mechanism against high glucose-induced apoptosis by targeting heme oxygenase-1. Eur Rev Med Pharmacol Sci. 2018;22(24):8909–8917. doi: 10.26355/eurrev_201812_16660. [DOI] [PubMed] [Google Scholar]
- 22.Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Malerva L, Park J, Zou LH, et al. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci USA. 2011;108(5):2058–2063. doi: 10.1073/pnas.1018157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gou LS, Zhao L, Song WC, et al. Inhibition of miR-92a suppresses oxidative stress and improves endothelial function by upregulating heme oxygenase-1 in db/db mice. Antioxid Redox Signal. 2018;28(5):358–370. doi: 10.1089/ars.2017.7005. [DOI] [PubMed] [Google Scholar]
- 25.Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7(12):a006080. doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, Chen TX, Li GL, et al. Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget. 2017;8(32):52156–52177. doi: 10.18632/oncotarget.11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guestini F, Mcnamara KM, Ishida T, et al. Triple negative breast cancer chemosensitivity and chemoresistance: current advances in biomarkers indentification. Expert Opin Ther Targets. 2016;20(6):705–720. doi: 10.1517/14728222.2016.1125469. [DOI] [PubMed] [Google Scholar]
- 28.Guo JY, Chen HY, Mathew R, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.He X, Xiao X, Dong L, et al. miR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1. Tumour Biol. 2015;36(3):2065–2075. doi: 10.1007/s13277-014-2814-z. [DOI] [PubMed] [Google Scholar]
- 31.Heng MY, Detloff PJ, Paulson HL, et al. Early alterations of autophagy in huntington disease-like mice. Autophagy. 2010;6(8):1206–1208. doi: 10.4161/auto.6.8.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh TH, Hsu CY, Tsai CF, et al. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther. 2015;23(4):656–666. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeon HS, Lee SY, Lee EJ, et al. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012;76(2):171–176. doi: 10.1016/j.lungcan.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung DE, Park SB, Kim K, et al. CG200745, an HDAC inhibitor, induces anti-tumour effects in cholangiocarcinoma cell lines via miRNAs targeting the Hippo pathway. Sci Rep. 2017;7(1):10921. doi: 10.1038/s41598-017-11094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HJ, Lee SG, Kim YJ, et al. Cytoprotective role of autophagy during paclitaxel-induced apoptosis in Saos-2 osteosarcoma cells. Int J Oncol. 2013;42(6):1985–1992. doi: 10.3892/ijo.2013.1884. [DOI] [PubMed] [Google Scholar]
- 38.Kim HS, Tian LJ, Jung M, et al. Downregulation of choline kinase-alpha enhances autophagy in tamoxifen-resistant breast cancer cells. PLoS ONE. 2015;10(10):e0141110. doi: 10.1371/journal.pone.0141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura T, Takabatake Y, Takahashi A, et al. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 40.Lai TH, Ewald B, Zecevic A, et al. HDAC inhibition induces microRNA-182, which targets RAD51 and impairs HR repair to sensitize cells to sapacitabine in acute myelogenous leukemia. Clin Cancer Res. 2016;22(14):3537–3549. doi: 10.1158/1078-0432.Ccr-15-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YJ, Won AJ, Lee J, et al. Molecular mechanism of SAHA on regulation of autophagic cell death in tamoxifen-resistant MCF-7 breast cancer cells. Int J Med Sci. 2012;9(10):881–893. doi: 10.7150/ijms.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 43.Li FJ, Miao LX, Xue T, et al. Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells. J Exp Clin Cancer Res. 2019;38(1):414. doi: 10.1186/s13046-019-1404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Zhang LN. Regulation of ATG and autophagy initiation. In: Qin ZH , editor. Autophagy: Biology and Diseases. Springer, Singapore; 2019. pp. 41–65. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Zou LH, Li QY, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16(2):214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Zhang Q, Tian RY, et al. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. 2011;71(24):7481–7489. doi: 10.1158/0008-5472.Can-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Iglehart JD, Richardson AL, et al. The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy. 2012;8(2):273–274. doi: 10.4161/auto.8.2.18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer, 36:52. 2017 doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C, Xie LY, Lu Y, et al. miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells. Int J Mol Med. 2018;41(4):2050–2058. doi: 10.3892/ijmm.2018.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu LM, Shen WF, Zhu ZH, et al. Combined inhibition of EGFR and c-ABL suppresses the growth of fulvestrant-resistant breast cancer cells through miR-375-autophagy axis. Biochem Biophys Res Commun. 2018;498(3):559–565. doi: 10.1016/j.bbrc.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Sun DP, Gao S, et al. Cyclovirobuxine D induces autophagy-associated cell death via the Akt/mTOR pathway in MCF-7 human breast cancer cells. J Pharmacol Sci. 2014;125(1):74–82. doi: 10.1254/jphs.14013fp. [DOI] [PubMed] [Google Scholar]
- 52.Lui A, New J, Ogony J, et al. Everolimus downregulates estrogen receptor and induces autophagy in aromatase inhibitor-resistant breast cancer cells. BMC Cancer, 16:487. 2016 doi: 10.1186/s12885-016-2490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X, Chen Z, Hua D, et al. Essential role for trpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc Natl Acad Sci USA. 2014;111(17):6389–6394. doi: 10.1073/pnas.1400272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27kip1 . J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Musolino A, Naldi N, Bortesi B, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26(11):1789–1796. doi: 10.1200/jco.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 56.Napoli M, Venkatanarayan A, Raulji P, et al. ΔNp63/DGCR8-dependent microRNAs mediate therapeutic efficacy of HDAC inhibitors in cancer. Cancer Cell. 2016;29(6):874–888. doi: 10.1016/j.ccell.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien K, Lowry MC, Corcoran C, et al. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget. 2015;6(32):32774–32789. doi: 10.18632/oncotarget.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan BZ, Yi J, Song HZ. MicroRNA-mediated autophagic signaling networks and cancer chemoresistance. Cancer Biother Radiopharm. 2013;28(8):573–578. doi: 10.1089/cbr.2012.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JH, Ahn MY, Kim TH, et al. A new synthetic HDAC inhibitor, MHY218, induces apoptosis or autophagy-related cell death in tamoxifen-resistant MCF-7 breast cancer cells. Invest New Drugs. 2012;30(5):1887–1898. doi: 10.1007/s10637-011-9752-z. [DOI] [PubMed] [Google Scholar]
- 61.Park JH, Kim KP, Ko JJ, et al. PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem Biophys Res Commun. 2016;477(2):277–282. doi: 10.1016/j.bbrc.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 62.Pei L, Kong YR, Shao CF, et al. Heme oxygenase-1 induction mediates chemoresistance of breast cancer cells to pharmorubicin by promoting autophagy via PI3K/Akt pathway. J Cell Mol Med. 2018;22(11):5311–5321. doi: 10.1111/jcmm.13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Periyasamy-Thandavan S, Jackson WH, Samaddar JS, et al. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress, and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy. 2010;6(1):19–35. doi: 10.4161/auto.6.1.10323. [DOI] [PubMed] [Google Scholar]
- 64.Pu MF, Li CG, Qi XM, et al. miR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth. PLoS Genet. 2017;13(7):e1006896. doi: 10.1371/journal.pgen.1006896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qadir MA, Kwok B, Dragowska WH, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112(3):389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 66.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robertson ED, Wasylyk C, Ye T, et al. The oncogenic microRNA hsa-miR-155-5p targets the transcription factor ELK3 and links it to the hypoxia response. PLoS ONE. 2014;9(11):e113050. doi: 10.1371/journal.pone.0113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Romero MA, Bayraktar Ekmekcigil O, Bagca BG, et al. Role of autophagy in breast cancer development and progression: opposite sides of the same coin. In: Ahmad A , editor. Breast Cancer Metastasis and Drug Resistance. Springer, Cham; 2019. pp. 65–73. [DOI] [PubMed] [Google Scholar]
- 69.Ru P, Steele R, Hsueh EC, et al. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2011;2(7):720–727. doi: 10.1177/1947601911425832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz-Roberts JL, Shajahan AN, Cook KL, et al. GX15-070 (obatoclax) induces apoptosis and inhibits cathepsin d- and L-mediated autophagosomal lysis in antiestrogen-resistant breast cancer cells. Mol Cancer Ther. 2013;12(4):448–459. doi: 10.1158/1535-7163.Mct-12-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy. 2011;7(5):457–465. doi: 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- 72.Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi Y, Gong WH, Lu L, et al. Upregulation of miR-129-5p increases the sensitivity to taxol through inhibiting HMGB1-mediated cell autophagy in breast cancer MCF-7 cells. Braz J Med Biol Res. 2019;52(11):e8657. doi: 10.1590/1414-431x20198657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soni M, Patel Y, Markoutsa E, et al. Autophagy, cell viability, and chemoresistance are regulated by miR-489 in breast cancer. Mol Cancer Res. 2018;16(9):1348–1360. doi: 10.1158/1541-7786.Mcr-17-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun DJ, Zhu LJ, Zhao YQ, et al. Fluoxetine induces autophagic cell death via eEF2K-AMPK-mTOR-ULK complex axis in triple negative breast cancer. Cell Prolif. 2018;51(2):e12402. doi: 10.1111/cpr.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun WL, Chen J, Wang YP, et al. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7(9):1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 77.Sun WL, Wang L, Luo J, et al. Ambra1 modulates the sensitivity of breast cancer cells to epirubicin by regulating autophagy via ATG12. Cancer Sci. 2018;109(10):3129–3138. doi: 10.1111/cas.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan X, Peng J, Fu Y, et al. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res, 16:435. 2014 doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas S, Thurn KT, Biçaku E, et al. Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res Treat. 2011;130(2):437–447. doi: 10.1007/s10549-011-1364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trüe O, Matthias P. Interplay between histone deacetylases and autophagy–from cancer therapy to neurodegeneration. Immunol Cell Biol. 2012;90(1):78–84. doi: 10.1038/icb.2011.103. [DOI] [PubMed] [Google Scholar]
- 81.Ujihira T, Ikeda K, Suzuki T, et al. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci Rep, 5:7641. 2015 doi: 10.1038/srep07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the Anti-HER2 monoclonal antibody trastuzumab. PLoS ONE. 2009;4(7):e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang ZY, Wang N, Liu PX, et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5(16):7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428(9):1681–1699. doi: 10.1016/j.jmb.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu MY, Fu JJ, Xu JM, et al. Steroid receptor coactivator 3 regulates autophagy in breast cancer cells through macrophage migration inhibitory factor. Cell Res. 2012;22(6):1003–1021. doi: 10.1038/cr.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu HD, Qin ZH. Beclin 1, Bcl-2 and autophagy. In: Qin ZH , editor. Autophagy: Biology and Diseases. Springer, Singapore; 2019. pp. 109–126. [DOI] [PubMed] [Google Scholar]
- 87.Yan XJ, Zhou RM, Ma ZY. Autophagy–cell survival and death. In: Qin ZH , editor. Autophagy: Biology and Diseases. Springer, Singapore; 2019. pp. 667–696. [DOI] [PubMed] [Google Scholar]
- 88.Yang ZF, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yao YS, Qiu WS, Yao RY, et al. miR-141 confers docetaxel chemoresistance of breast cancer cells via regulation of EIF4E expression. Oncol Rep. 2015;33(5):2504–2512. doi: 10.3892/or.2015.3866. [DOI] [PubMed] [Google Scholar]
- 90.Yu XF, Luo AP, Liu YC, et al. miR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol Cancer, 14:208. 2015 doi: 10.1186/s12943-015-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu XF, Li RL, Shi WN, et al. Silencing of microRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed Pharmacother. 2016;77:37–44. doi: 10.1016/j.biopha.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 92.Zhang FF, Wang BB, Long HL, et al. Decreased miR-124-3p expression prompted breast cancer cell progression mainly by targeting Beclin-1. Clin Lab. 2016;62(6):1139–1145. doi: 10.7754/clin.lab.2015.151111. [DOI] [PubMed] [Google Scholar]
- 93.Zhang HW, Hu JJ, Fu RQ, et al. Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70s6K/ULK signaling pathway in human breast cancer cells. Sci Rep, 8:11255. 2018 doi: 10.1038/s41598-018-29308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang P, Liu XY, Li HJ, et al. TRPC5-induced autophagy promotes drug resistance in breast carcinoma via CaMKKβ/AMPKα/mTOR pathway. Sci Rep. 2017;7(1):3158. doi: 10.1038/s41598-017-03230-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang T, Xiang L. Honokiol alleviates sepsis-induced acute kidney injury in mice by targeting the miR-218-5p/heme oxygenase-1 signaling pathway. Cell Mol Biol Lett, 24:15. 2019 doi: 10.1186/s11658-019-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang XL, Zhu J, Xing RY, et al. miR-513a-3p sensitizes human lung adenocarcinoma cells to chemotherapy by targeting GSTP1. Lung Cancer. 2012;77(3):488–494. doi: 10.1016/j.lungcan.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 97.Zhong JT, Yu J, Wang HJ, et al. Effects of endoplasmic reticulum stress on the autophagy, apoptosis, and chemotherapy resistance of human breast cancer cells by regulating the PI3K/AKT/mTOR signaling pathway. Tumour Biol. 2017;39(5):1010428317697562. doi: 10.1177/1010428317697562. [DOI] [PubMed] [Google Scholar]
- 98.Zhou H, Chen Y, Huang SW, et al. Regulation of autophagy by tea polyphenols in diabetic cardiomyopathy. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(5):333–341. doi: 10.1631/jzus.B1700415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu HY, Bai WD, Ye XM, et al. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem Biophys Res Commun. 2018;496(4):1308–1313. doi: 10.1016/j.bbrc.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 100.Zou ZY, Wu LP, Ding HY, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing Beclin 1-mediated autophagy. J Biol Chem. 2012;287(6):4148–4156. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]