Abstract
Ginseng rusty root (GRR) symptom is one of the primary diseases of ginseng. There has been a problem of ginseng rusty root, leading to a severe decline in the quality of ginseng. To clarify the relationship between root symptoms of ginseng rust and soil, the physical and chemical properties, enzyme activity, community structure and microbial diversity of GRR and healthy ginseng (HG) rhizosphere soil were analyzed and compared. The pH and redox potential (Eh) of GRR soil decreased, and the contents of total phosphorus (TP), available phosphorus (AP), and available potassium (AK) decreased. The activity of catalase and phosphatase and invertase was lower than that of HG groups. Besides, the microbial community of GRR rhizosphere soil changes much, and its abundance and diversity are significantly reduced. The community structure of GRR rhizosphere soil also shows apparent differences, and the samples of the HG group gathered together, and the samples of the GRR group were dispersed. In general, GRR was closely associated with decreases in soil pH and Eh; decreases in TP, AP, and AK; decreases in the activity of several enzymes. Additionally, it is strongly associated with an increase in pathogenic microorganisms such as Ilyonectria and a reduction of beneficial microorganisms such as Tremellomycetes Acidobacteria subgroup 6 and Gemmatimonadetes.
Subject terms: Ecology, Microbiology, Plant sciences
Introduction
Ginseng (Panax ginseng Mayer) is an essential medicinal material in China. Because of the relatively high planting benefits of ginseng and the limitation of planting soil, ginseng production areas are relatively concentrated. Moreover, in recent years, the scale of cultivated ginseng in the cutting forests has been shrinking; it is increasingly essential to produce high-quality ginseng. One of the significant factors affecting the ginseng quality is the various diseases that occur during the growth of ginseng.
Rusty root symptom is one of the central diseases in cultivation and production. It produces reddish-brown spots on the periderm of ginseng roots. With the increase of cultivation years, the patch may gradually expand, which will lead to the decline of commodity-grade and ginseng quality. At present, the mechanism of rust root is not precise. Whether ginseng rusty root is an infectious disease or a non-infectious physiological disease is still controversial, so some scholars have distinguished it1. Previous studies have shown that American ginseng (Panax quinquefolius L.) rusty root is the defense mechanism of ginseng itself, which is due to the invasion of some fungi that stimulates the production of phenolic compounds in ginseng2,3. Besides, some research results suggest that the induction of rusty root by fungal complex is the cause of rust roots in ginseng4. A variety of enzymes were used to hydrolyze plant structural materials and make ginseng roots rusty. Fe3+, combined with pectinase, had a synergistic effect on the formation of rust roots in ginseng5. Current studies suggest that ginseng rusty root symptom is related to soil acidity and alkalinity, water content, and metal element content, as well as the accumulation and oxidation of phenolic compounds in roots.
Soil microorganism is the most active part of the soil, driving force of soil material transformation and nutrient cycling. It participates in various soil processes, such as decomposition of soil organic matter, the formation of humus, transformation, and circulation of soil nutrients. Therefore, the composition and diversity of soil communities are closely related to soil function and plant health. Microorganisms in the soil are often the leading cause of plant diseases6,7. These previous studies focused on the soil enzyme activity and nutrient characteristics of different tree species and their relationship with the incidence index of GRR. Besides, some scholars explored the soil characteristics of several ginseng farms with GRR to explore the relationship between GRR and soil characteristics8,9. No data, however, are available on healthy ginseng and ginseng of rusty root on the same land on microbial community structure and enzyme activity in the plant rhizosphere.
In previous studies, the roots of GRR were collected, and fungi were isolated from the lesions to reveal the pathogen causing GRR4. Based on this phenomenon, it is hypothesized that some microbial communities should be closely related to GRR. By comparing to the rhizosphere soil of HG, any changes in the microbial communities of GRR may be explored.
In this study, we investigated the differences in nutrient content, enzyme activity, and other physical and chemical properties between GRR and HG soils. pH, redox potential, catalase, phosphatase, urease, invertase activities, and soil phenolic acids were measured. In addition, the 16S and ITS rRNA gene fragment obtained directly from soil samples were sequenced to study the microbial community structure and diversity in soil samples.
Results
Chemical properties of soil
Count the degree of disease of 100 ginsengs and calculate the rusty root index based on the formula (Table S1). Calculated by formula, the rusty root index is 0.7725, and this is a ginseng farm with very severe ginseng rusty root (Fig. S1).
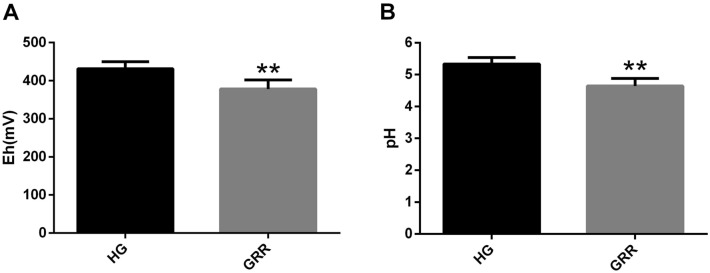
In soil pH measurement, we found that the ginseng planting of soil with dangerous rusty root phenomenon decreased significantly (Fig. 1A). In soil Eh measurements, GRR soils decreased compared considerably to soils of healthy ginseng (Fig. 1B).
Figure 1.
Chemical properties of soil. (A) Soil redox potential; (B) pH. HG: healthy ginseng soil groups; GRR: ginseng rusty root symptom soil groups. Letters indicate significant differences among different soil samples by ANOVA at P ** < 0.01 versus HG group (n = 6 in each group).
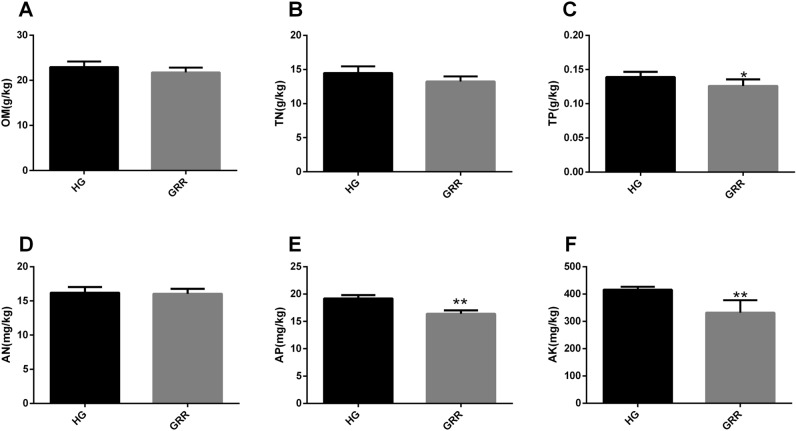
The nutrient content of the soil was also analyzed. The results showed that there was no significant difference in organic matter (OM), total nitrogen (TN), and available nitrogen (AN) between GRR soil and HG soil (Fig. 2A–C). Interestingly, however, tests of total phosphorus (TP), available phosphorus (AP), and available potassium (AK) showed a significant reduction in GRR soil compared to HG soil (Fig. 2D–F).
Figure 2.
Nutrient content of soil. (A) Soil organic matter; (B) total nitrogen; (C) total phosphorus; (D) available nitrogen; (E) available phosphorus; (F) available potassium. HG: healthy ginseng soil groups; GRR: ginseng rusty root symptom soil groups. Letters indicate significant differences among different soil samples by ANOVA at P * < 0.05, P ** < 0.01 versus HG group (n = 6 in each group).
Enzyme activity of soil
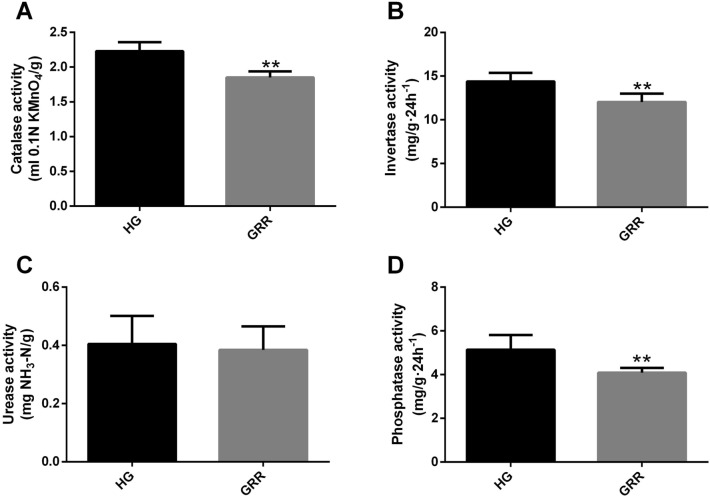
Catalase, invertase, urease, and phosphatase of the two groups were compared. Among them, the activity of invertase, catalase and phosphatase in group GRR were significantly lower than those in the soil of healthy ginseng (Fig. 3A, B, D). However, there was no significant difference in urease activity among the two groups (Fig. 3C).
Figure 3.
Soil enzyme activity. (A) Catalase activity; (B) invertase activity; (C) urease activity; (D) phosphatase activity. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom-high soil group. Letters indicate significant differences among different soil samples by ANOVA at P ** < 0.01 versus HG group (n = 6 in each group).
Microbial diversity and richness in GRR and HG rhizosphere soils
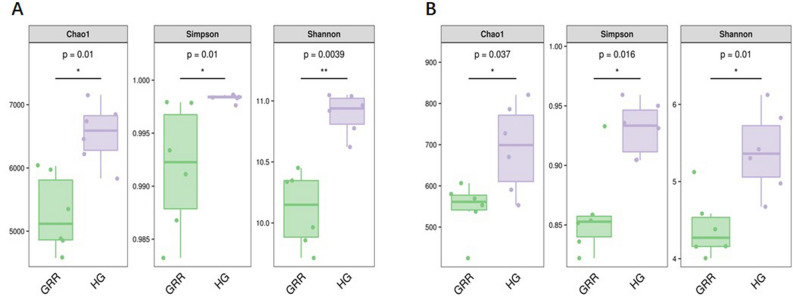
The GRR and HG’s microbial diversity was characterized by partial 16S and ITS rRNA gene sequencing obtained from DNA directly extracted from soil samples. Rarefaction curves of observed amplicon sequence variants (ASVs) for each sample were produced at the ASV level (Fig. S2). The curves of most of the samples reached close to the plateau, indicating that our sequencing depth was able to capture most bacterial species of ginseng rhizosphere soil microbiota. Compared with the rhizosphere soil of HG, the alpha diversity (including Chao1, Simpson, and Shannon diversity) of bacteria and fungi community in rhizosphere soil of GRR decreased significantly (Fig. 4). In other words, compared with the healthy ginseng rhizosphere soil, the microbial richness of GRR rhizosphere soil decreased significantly, whether it was bacteria (P < 0.05) or fungi (P < 0.05). Besides, the comparison between the two groups of samples on Simpson and Shannon index showed that the diversity of bacteria and fungi in the rhizosphere soil of GRR also decreased significantly (P < 0.05).
Figure 4.
Microbial richness and diversity. (A) Bacteria; (B) fungi. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group. Letters indicate significant differences among different soil samples by ANOVA at P * < 0.05, P ** < 0.01 versus HG group (n = 6 in each group).
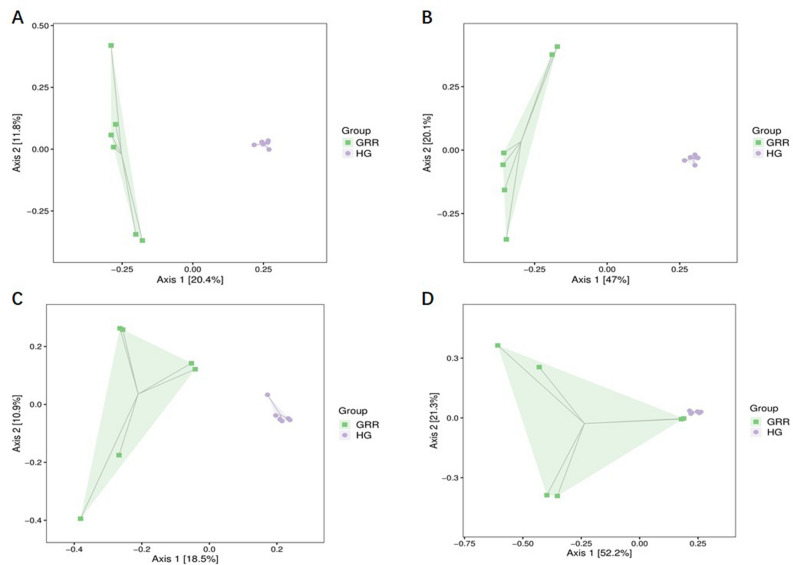
According to the principal coordinate analysis (PCoA) based on Jaccard and Bray–Curtis matrix, the microbial community structure of rhizosphere soil is significantly different in HG and GRR (Fig. 5). Analysis of HG and GRR rhizosphere soil bacteria revealed a significant separation between the Jaccard (Fig. 5A) and Bray Curtis (Fig. 5B) matrices, indicating substantial differences in bacterial composition between the two groups. In the fungi analysis, except that two samples in the GRR group in Fig. 5D are close to HG, other samples also show apparent separation. The above results show that the composition of the two groups of fungi is also different.
Figure 5.
Microbial principal coordinate analysis (PCoA). (A) Bacteria Jaccard distance; (B) bacteria Bray–Curtis distance; (C) fungi Jaccard distance; (D) fungi Bray–Curtis distance. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group (n = 6 in each group).
Interestingly, based on the Jaccard or Bray–Curtis difference matrix, the samples of the HG group were highly clustered, while the samples of the GRR group were highly separated. In other words, the rhizosphere soil microbial community of HG is relatively stable, and the microorganism of GRR rhizosphere soil showed even differences.
Species difference analysis and biomarkers of GRR and HG rhizosphere soil
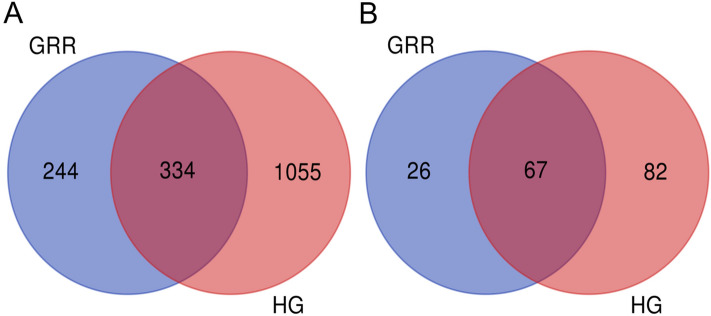
By sequencing, we obtained 33,798 ASVs in bacteria and 3,225 ASVs in fungi. We carried out the Venn map analysis of identify the potential biomarkers (taxa with stable significant differences between groups) of bacteria and fungi in two groups of soil samples. As shown in Fig. 6A, in bacteria, the number of ASVs present in all six samples in the GRR group was 578, and the number of ASVs present in all samples in the HG group was 1,389. Two groups of common ASVs are 334. Compared with HG rhizosphere soil, the total ASVs of GRR rhizosphere soil decreased, and the specific ASVs increased. In fungi, the 67 ASVs are common to all the samples, 26 specific ASVs in the GRR group, and 82 specific ASVs in the HG group (Fig. 6B). Thus, there was also a large difference in fungi ASVs between the rhizosphere soils of GRR and HG.
Figure 6.
Venn diagrams of ASVs among soil samples. (A) Bacteria; (B) fungi. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group.
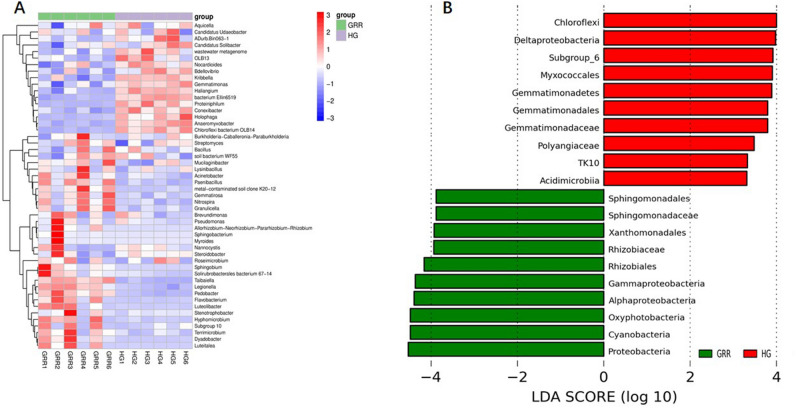
In this study, we used the abundance data of the top 50 genera of the average abundance of bacteria to draw a heatmap (Fig. 7A). Then, we use the histogram of LDA effect value of biomarker species based on linear discriminant analysis (LDA) effect size (Lefse) analysis to show the bacterial taxon with a significant difference between groups, and Fig. 7B shows the 20 taxa with the most significant differences between the two groups (All significantly different taxa are provided in Fig. S3). Compared with HG rhizosphere soil, the abundance of Proteobacteria, Cyanobacteria, Oxyphotobacteria, Alphaproteobacterial, Gammaproteobacterial, Rhizobiales, Xanthomonadales, and Sphingomonadales in GRR rhizosphere soil significantly increased. On the other hand, the Deltaproteobacteria; Chloroflexi; Myxococcales; Gemmatimonadetes; Gemmatimonadales; Gemmatimonadaceae; Polyangiaceae; TK10; Acidimicrobiia is more abundant in HG groups compared to GRR groups. Besides, the cladogram of intergroup difference taxa based on Lefse analysis is provided in Figure S4.
Figure 7.
Bacterial abundance and biomarkers. (A) Heatmap of the 50 most abundant genera in soil samples; (B) histogram of LDA effect value of the histogram of LDA effect value of differentially abundant taxa. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group.
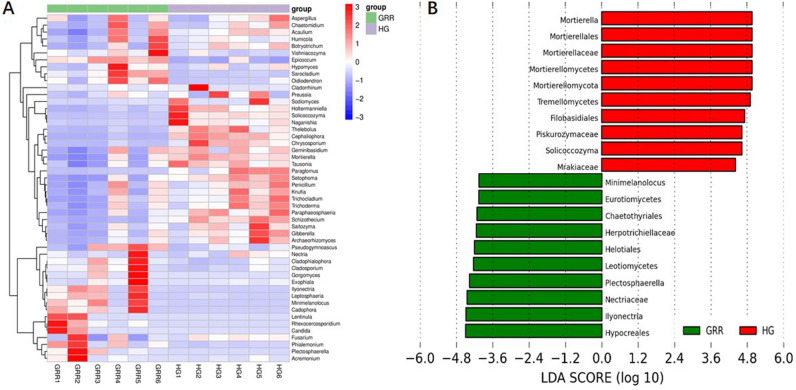
In the fungi analysis, we also used the abundance data of the first 50 genera of average abundance to make the heatmap (Fig. 8A). The histogram of the LDA effect value of 20 biomarkers, with the most significant difference between the two groups, is shown in Fig. 8B. Among them, the 20 most significant taxa of the GRR group are Minimelanolocus, Eurotiomycetes, Chaetothyriales, Herpotrichiellaceae, Helotiales, Leotiomycetes, Plectosphaerella, Nectriaceae, Ilyonectria and Hypocreales. The most apparent ten taxa in the HG group are Mortierellaceae, Mortierellomycota, Mortierellomycetes, Tremellomycetes, Filobasidiales, Piskurozymaceae, Solicoccozyma, and Mrakiaceae (all significantly different taxa are provided in Fig. S3). The cladogram of the intergroup differences in fungi is presented in Figure S4.
Figure 8.
Fungi abundance and biomarkers. (A) Heatmap of the 50 most abundant genera in soil samples; (B) histogram of LDA effect value of differentially abundant taxa. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group.
Relationship of soil physicochemical properties and enzyme activity to microbial abundance
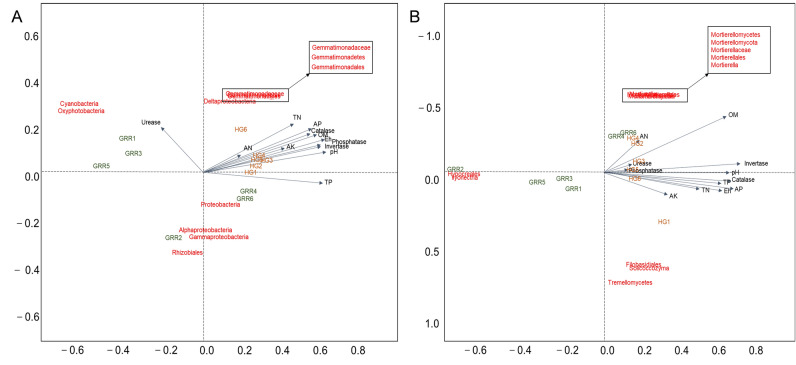
In redundancy analysis (RDA), we used measured soil physicochemical properties and multiple enzyme activities as environmental variables to investigate their correlation with high relative abundance taxa in soil samples. The analysis results show that bacteria and fungi in two rhizosphere soils are greatly influenced by physicochemical properties and enzyme activities. Furthermore, RDA plots reflect the relationship between several beneficial or pathogenic microorganisms and environmental variables. In the analysis results of bacteria, only Cyanobacteria and Oxyphotobacteria showed a positive correlation with Urease. Gemmatimonadaceae, Gemmatimonadetes and Gemmatimonadales are positively correlated with a variety of environmental variables. Proteobacteria, Alphaproteobacteria, Rhizobiales, Gammaproteobacteria and Deltaproteobacteria have not shown a strong correlation with various environmental variables (Fig. 9A). In fungi, Ilyonectria and Hypocreales were negatively correlated with various environmental variables. Mortierellaceae Mortierellomycota, Mortierellomycetes Mortierellales and Mortierella were positively related to the AN and OM. However, there is no strong correlation between Solicoccozyma, Tremellomycetes and Filobasidiales and environmental variables.
Figure 9.
Redundancy analysis (RDA) of soil samples; (A) RDA plots of bacteria in soil samples; (B) RDA plots of fungi in soil samples. HG: healthy ginseng soil group; GRR: ginseng rusty root symptom soil group.
Discussion
In this study, GRR rhizosphere soil and HG rhizosphere soil were compared comprehensively and carefully. Most of the previous studies were based on soil samples from different ginseng farms in different regions10. To reduce the influence of unknown differences, the soil samples collected are from the same ginseng farm with a high rusty root index. To study the relationship between ginseng rusty root and soil, this study compared the soil physical and chemical properties and nutrient content, enzyme activity, and microbial community. This study examined the soil physical and chemical properties, nutrient content, enzyme activity, and microbial community to study the relationship between ginseng rusty root and soil.
Soil pH seriously affects crop growth and microbial community structure in the soil11,12. Compared with healthy ginseng rhizosphere soil, the pH of GRR rhizosphere soil decreased significantly (Fig. 1A). Ginseng in northern China usually grows in acid soil, but the lower pH value may affect the healthy growth of ginseng. The pH has a particular influence on microbial community structure, diversity, and richness13,14. Therefore, fungi or bacteria related to GRR may appear in the soil with pH changes. There are many redox systems in soil, such as oxygen systems, iron system, manganese system. Under certain conditions, each soil has its Eh value15. In this study, Eh value in GRR soil decreased significantly compared with that in HG soil (Fig. 1B). Its higher water content may cause the change of the Eh value of GRR soil, or some chemical reactions may occur in the soil. For a long time, high soil moisture content has been considered one of the factors causing GRR, and the part of this rust root is deemed to be the product of some chemical reactions2.
The growth of ginseng has high requirements for various substances in the soil. Phosphorus is an essential component of nuclear and membrane structure in plants. Normal phosphorus levels can complete the healthy metabolism of protein in plants, stimulate the growth of plant roots, increase the absorption of mineral nutrients by rhizomes, to alleviate the damage of plant diseases16,17. In our results, GRR rhizosphere soil phosphorus significantly decreased compared with HG rhizosphere soil (Fig. 2C, E). Besides, potassium can affect the metabolism of plants. Furthermore, potassium can promote the development of the thick outer wall of epidermal cells, thus preventing the occurrence of diseases16,19. As shown in Fig. 2F, the GRR rhizosphere soil had significantly less potassium than the HG rhizosphere soil. This result seems to imply that GRR is closely related to the potassium content of the soil. Overall, the decrease of some nutrients in GRR soil may affect the healthy growth of ginseng. The GRR may be related to reducing plant disease resistance caused by the reduction of phosphorus and potassium in the soil.
Soil enzymes are the products of residual decomposition of plants and animals, exudation of plant roots, and metabolism of soil microorganisms, which involve many critical biochemical processes in soil20. Soil enzyme activity is an essential index of soil fertility, quality, and health21. The catalase, invertase, urease and phosphatase activities in rhizosphere soil of ginseng were measured in this experiment. The results showed that except urease, the activities of the other three enzymes in ginseng rhizosphere soil decreased significantly (Fig. 3). Urease catalyzes the hydrolysis of urea to carbon dioxide and ammonia, which plays an essential role in the nitrogen cycle in soil22. In our results, it did not show a significant correlation with GRR. Invertase is a ubiquitous enzyme in soil, which plays an essential role in the release of fructose and glucose from sucrose and provides a carbon source for the growth of soil microorganisms23. The organic phosphorus in the soil can be absorbed and utilized by crops under the action of phosphatase. Therefore, soil phosphatase activity can be an essential indicator of the hydrolysis of soil phosphorus compounds24. The decrease of phosphorus in GRR soil may be related to the reduction of the enzyme activity. Catalase can drive the decomposition and transformation of peroxides in soil, eliminating the adverse effects of peroxides on soil quality25. In other words, the decrease of catalase activity in GRR soil may make ginseng suffer more oxidative stress26. In brief, the reduction of these three enzymes in GRR soil indicated that the production of ginseng rusty root might be related to the decrease of carbon source and available phosphorus, and the content of peroxides in the soil.
In this study, high-throughput sequencing technology was used to study the microbial diversity of GRR rhizosphere soil. The alpha diversity analysis of GRR and HG rhizosphere soil, the Chao1 estimator27, Shannon28, and Simpson29 diversity of ginseng rhizosphere soil with rusty root showed a significant decline (Fig. 4). Therefore, in this study, the richness and diversity of fungi or bacteria in the rhizosphere soil of GRR significantly decreased. Then, we conducted the beta diversity analysis based on PCoA, Jaccard, and Bray–Curtis distance was estimated in this study (Fig. 5). Jaccard distance and Bray–Curtis distance are measures used to measure the difference in species composition of different soil samples. They can calculate the characteristics of different species composition of samples. The focus of the two analyses is different: Jaccard only considers the presence or absence of species in the sample, while Bray–Curtis considers the presence or absence of species in the sample and considers the relative richness of different species. Whether fungi or bacteria, the GRR group, and HG group had visible separation, which indicated that GRR rhizosphere soil microorganism had significant changes compared with HG soil microorganism. It should be noted that six samples of the HG group showed high aggregation. In comparison, six samples of the GRR group were very dispersed, which indicated that the microbial structure of HG rhizosphere soil was very stable. In contrast, that of GRR rhizosphere soil might have microbial community disorder. Finally, we analyzed the species that caused these differences. Using the ASVs abundance table to make Venn diagrams for community analysis shows the ASV level variation between the two groups (Fig. 6). To further compare the differences in species composition between samples, we use the abundance data of the top 50 genera of average abundance to draw a heatmap to display the genera abundance distribution trend of each sample (Figs. 7A, 8A). In this study, LEfSe analysis was used to find the biomarkers between groups, and LDA effect histograms and cladograms were drawn for bacteria and fungi, respectively (Figs. 7B, 8B and Fig. S4). We found possible pathogens in the GRR rhizosphere soil. However, the other results of LEfSe analyse were different from previous studies, which may be related to the sample collection site and the cultivation conditions.
In terms of fungi, the biomarker’s analysis found Ilyonectria, which is thought to be the pathogen of GRR30. Hypocreales is often reported to be related to pests' pathogenicity or some bacteria31–33, and it can survive in harsher soil conditions34. Nectriaceae is also listed as a biomarker. It includes numerous important plant and human pathogens, and several species used extensively in industrial and commercial applications as biodegraders and biocontrol agents31. We found a possible pathogen Plectosphaerella, it has been reported to be associated with plant root diseases35, but it has not yet been reported on ginseng disease. In HG rhizosphere soil, Mortierellales, Tremellomycetes orders predominated. Mortierella is a biomarker in HG rhizosphere soil, but rarely in GRR soil. Mortierella is the producer of polyunsaturated fatty acids36, whether it is related to ginseng growth has not been reported. There are reports that Tremellomycetes could efficiently act as opportunists that utilize the decomposition products provided by other microbes37. RDA reveals the correlation between fungal taxa and environmental variables (Fig. 9B). Ilyonectria was negatively correlated with several environmental variables, especially pH, catalase, and invertase.
On the other hand, in the analysis of bacteria, more proteobacteria appeared in GRR rhizosphere soil. Although proteobacteria have been reported to dominate pesticide and herbicide contaminated soils38,39, it is generally one of the most abundant phyla in many soils and rhizosphere soils. The reasons for the increase of proteobacteria in GRR rhizosphere soil need to be further explored. A significant increase in Proteobacteria often indicates the dysbiosis in the microbiota. Further, a higher abundance of Alphaproteobacterial (including Sphingomonadales) and Gammaproteobacterial (including Xanthomonadales) indicates that GRR rhizosphere soil may have stronger nitrification and the turnover of OM40. Interestingly, anaerobic Deltaproteobacteria (including Myxococcales) is more abundant in HG rhizosphere soil41. Myxococcus is considered to be an essential and active predator that regulates bacterial communities in agricultural land42. Cyanobacteria are the main constituent of biological bacteria and can survive in more extreme environments43. Some cyanobacteria have increasing soil stability and moisture-holding capacity44,45. Their abundance increased in GRR rhizosphere soil, confirming the deterioration of GRR rhizosphere soil conditions compared to HG. Chloroflexi (including TK10) is abundant in HG rhizosphere soil. It seems to play a useful role in the soil in providing the filamentous scaffolding around which flocs are formed feed on the debris from lysed bacterial cells. It can ferment carbohydrates and degrade other complex polymeric organic compounds to low molecular weight substrates to support their growth and that of different bacterial populations46. Also, Acidobacteria subgroup 6 and Gemmatimonadetes emerged as the keystone taxa in HG rhizosphere soils, and they may play an important ecological role by degrading polysaccharides of plant and fungal origin47,48. These differences could be partially due to the different physicochemical characteristics of the two soils49. Fungal plant pathogens live in a specific environment, and the GRR rhizosphere soil environment may be more suitable for the survival of pathogenic fungi such as Ilyonectria.
Many studies have pointed out that ginseng, American ginseng (Panax quiquefolium L), (Panax notoginseng (Burkill) F. H. Chen ex C. H.), and other medicinal plants have rusty root symptoms30,50. Furthermore, they all seem to be closely related to Ilyonectria. The same results were found in our study. However, in Wang et al.51, Ilyonectria does not have a large abundance in GRR rhizosphere soil. Also, in Liu et al. research results, Ilyonectria is positively correlated with N and OM10. In our analysis, there is a negative correlation. Plectosphaerella, as A potential pathogen, also showed greater abundance in our results, which had not been found in the study of Liu et al. In agreement with previous studies, we also found large abundances of Proteobacteria, Gammaproteobacterial and Rhizobiales in GRR rhizosphere soil. What is different is that we also found a large abundance of Cyanobacteria. Interestingly, our analysis of HG rhizosphere soil bacterial and fungal abundance showed significant differences from previous studies. Thus, the microbial composition of the rhizosphere soil of ginseng is likely to be closely related to the planting site, cultivation method, fertilizer and pesticide application.
In conclusion, pH and Eh in GRR soil decreased significantly, which may be related to the metabolism of the ginseng root system and soil aeration. GRR also showed significant changes in soil nutrient content, reducing TP, TK, and AK, making ginseng absorb fewer nutrients and may affect the disease resistance of ginseng. The decrease in the activities of many enzymes in GRR soil indicates that the metabolism of soil substances is greatly affected. Besides, compared with HG rhizosphere soil, microbial abundance and diversity of GRR decreased significantly, community structure also changed dramatically, and community disorder occurred. Finally, through Lefse analysis, the number of microorganisms in GRR rhizosphere soil that may be beneficial to the soil decreased, and the pathogenic bacteria (Ilyonectria) mentioned in previous studies were also found. Soil is a complex environment. Physical and chemical properties, nutrients, and microbial communities affect the growth of ginseng. RDA analysis of the correlation between different microorganisms and environmental factors will help improve ginseng planting soil in the future. This study mainly studies the change of ginseng rusty root symptom soil, and ginseng rusty root symptom may be caused by many factors that need further investigation.
Materials and methods
Soil information and sample collection
All soil samples were collected from the same ginseng farm with ginseng (5-year-old) in Hunchun city, Jilin province, China (42.86′ N and 130.37′ E). In detail, this is a ginseng farm with an area of about 21,000 square meters, with an altitude of 40 m, belongs to the temperate marine climate, with an annual average temperature of 5.65 °C, average precipitation of 617.9 mm, a frost-free period of 140–160 days, and an average temperature of 21.2 °C in August. The farm was the first time to cultivate ginseng and was cultivating transplanted 2-year-old ginseng seedlings. The pesticides applied in ginseng cultivation are “carbendazim (systemic broad-spectrum fungicide)” bought from Shandong Xinnongji Biotechnology Co. LTD (China), “tiandashenbao (plant cell membrane stationary agent)” bought from Shandong Tianda Bio-pharmaceutical Co. LTD (China) and “metalaxyl (Phenylamide fungicides)” bought from Alta Technologies LTD (China). All pesticides are used strictly following the “Ginseng safe production technical specification of pesticide application (DB22/T 1233-2019)”.
We randomly collected 200 ginseng and calculated the rusty root index9,52 to evaluate the severity of the disease.
In this formula: n = The quantity of ginseng in different rusty root grades, N = Total number of ginsengs used for statistics. Rusty root grade was divided into 0–4, which were healthy ginseng root, rust area < 10%, 10–25%, 25–50%, and > 50% in turn.
Then, six plots with ginseng grade 0 and six plots with ginseng grade 4 were selected to collect soil samples, about 1 square meter per plot. The soils were sampled from each plot at depths of 5–15 cm, representing soils from the root zone and below the root zone. The 12 soil samples are divided into the HG group (healthy ginseng group) and GRR group (ginseng rusty root symptom group). Part of each soil sample is kept wet for the determination of soil Eh.
Finally, six HG rhizosphere soils and six GRR (grade 4) rhizosphere soils were collected from the 12 plots above (one rhizosphere soil sample was collected at each polt). Part of the soil microbial DNA was extracted, and the samples were kept in the sterile centrifuge tube at—80 °C until use. The other part of the soil was air-dried at ambient temperature, crushed, and sieved through a 2 mm plastic mesh to determine soil pH, soil nutrition, and soil enzyme activity53. In this part, only the soil attached to the ginseng root is considered as rhizosphere soil54.
Analysis of basic physical and chemical properties
The pH value of soil was measured with a pH meter/potentiometer under the soil: water ratio of 1:2.5. The Eh was determined by potentiometry (China HJ 746-2015)55. The soil OM content was measured by the potassium dichromate external heating method. The TN was determined by Kjeldahl method55, and the TP was determined by alkali fusion molybdenum antimony colorimetry (China HJ 632-2011). The AP was determined by NaHCO3 extraction molybdenum-antimony colorimetry; the AN was determined by the alkali diffusion method; AK was determined with the fame photometric method57.
Analysis of soil enzyme activity
The activities of catalase, invertase, urease, and phosphatase in the soil of ginseng were measured. The activity of catalase was determined by KMnO4 titration; soil invertase activity was determined by 3,5-dinitrosalicylic acid colorimetry; urease activity was determined by indophenol blue colorimetry; acid phosphatase activity was determined by disodium phenyl phosphate colorimetry method38.
DNA extraction and Illumina sequencing
DNA was extracted from 0.5 g of soil (wet weight) using the Mag-Bind soil DNA kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s protocols. Then, DNA was quantified by Nanodrop (Thermo Scientific, Waltham, MA, U.S.) and the quality of DNA was detected by 1.2% agarose gel electrophoresis, the OD260/280 values of DNA extraction were between 1.8 and 2.0.
The V3-V4 region of bacterial 16s rRNA gene was amplified by bacterial specific primers 338F and 806R, and the internal transcribed spacer (ITS1) was amplified by eukaryotic specific primers ITS5F and ITS1R. In this step, the sample is uniformly diluted to 20 ng/ul. Amplification system (25 μL): 5 × reaction buffer 5 μL, 5 × GC buffer 5 μL, Dntp (2.5 Mm) 2 μL, Forwardprimer (10 uM) 1 μL, Reverseprimer (10 uM) 1 μL, DNA Template 2 μL, ddH2O 8.75 μL, Q5 DNA Polymerase 0.25 μL (NEB). Amplification parameters: Initial denaturation 98 °C 2 min, Denaturation 98 °C 15 s, Annealing 55 °C 30 s, Extension 72 °C 30 s, Final extension 72 °C 5 min, 10 °C Hold. 25–30 Cycles. The sequencing library was prepared by using Illumina’s TruSeq Nano DNA LT Library Prep Kit, the final fragment was selected and purified by 2% agarose gel electrophoresis.
The library was quantified by Quant-iT PicoGreen dsDNA Assay Kit in Promega QuantiFluor fluorescence quantitative system, and then libraries were sequenced on the Illumina HiseqPE250 platform at Personal Biotechnology Co., Ltd. (Shanghai, China).
Bioinformatics and data analysis
Microbiome bioinformatics was performed with QIIME 2 2019.458 with slight modification according to the official tutorials (https://docs.qiime2.org/2019.4/tutorials/). Briefly, raw sequence data were demultiplexed using the demux plugin following by primers cutting with cutadapt plugin59. Sequences were then quality filtered, denoised, merged, and chimera removed using the DADA2 plugin60. Non-singleton ASVs were aligned with mafft61 and used to construct a phylogeny with fasttree262. To analyze different samples using the same sequencing depth, all samples were re-sampled into the same sequence number: 119938 sequences from fungi and 91,828 sequences from bacteria. Alpha diversity metrics: The Chao1 estimator, Simpson and Shannon diversity index. Beta diversity metrics: Jaccard distance and Bray–Curtis dissimilarity. Taxonomy was assigned to ASVs using the classify-sklearn naïve Bayes taxonomy classifier in feature-classifier plugin63. LEfSe was used to elucidate the biomarkers in each group64. In addition, RDA was used to reveal the relationships between microbiota and soil properties. This analysis was carried out with the “vegan” package in R.
Statistical analyses
All data were analyzed using the SPSS software (IBM Corporation, Armonk, NY, USA), and the results were expressed as the arithmetic mean value ± standard deviation. The differences in the means were compared by the Student’s t test at P < 0.05.
Supplementary information
Acknowledgements
The authors are grateful for the National key R&D program (Grant No. 2017YFC1702100).
Author contributions
L.Z. and X.B. designed the study. X.B. and H.Y. conducted the experiments. S.X., Y.Z., Y.X. and X.B. analysed the data. L.Z., S.X. and X.B. prepared the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71024-8.
References
- 1.Zhou Y, et al. Changes in element accumulation, phenolic metabolism, and antioxidative enzyme activities in the red-skin roots of Panax ginseng. J. Ginseng Res. 2016;41:307–315. doi: 10.1016/j.jgr.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman M, Punja ZK. Biochemistry of ginseng root tissues affected by rusty root symptoms. Plant Physiol. Biochem. 2005;43:1103–1114. doi: 10.1016/j.plaphy.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Reeleder RD, Hoke SMT, Zhang Y. Rusted root of ginseng (Panax quinquefolius) is caused by a species of rhexocercosporidium. Phytopathology. 2006;96:1243–1254. doi: 10.1094/PHYTO-96-1243. [DOI] [PubMed] [Google Scholar]
- 4.Lu XH, et al. Taxonomy of fungal complex causing red-skin root of Panax ginseng in China. J. Ginseng Res. 2020;44:506–518. doi: 10.1016/j.jgr.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Kim KY, Lee JE, Kim S, An G. Enzymes hydrolyzing structural components and ferrous ion cause rusty-root symptom on ginseng (Panax ginseng) J. Microbiol. Biotechnol. 2011;21:192–196. doi: 10.4014/jmb.1008.08010. [DOI] [PubMed] [Google Scholar]
- 6.Yuan X, Song TJ, Yang JS, Huang XG, Shi JY. Changes of microbial community in the rhizosphere soil of Atractylodes macrocephala when encountering replant disease. S. Afr. J. Bot. 2019;127:129–135. doi: 10.1016/j.sajb.2019.08.046. [DOI] [Google Scholar]
- 7.Mazzola M, Manici LM. Apple replant disease: role of microbial ecology in cause and control. Annu. Rev. Phytopathol. 2012;50:45–65. doi: 10.1146/annurev-phyto-081211-173005. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. Comparison of the characteristics of artificial ginseng bed soils in relation to the incidence of ginseng red skin disease. Exp. Agric. 2014;50:59–71. doi: 10.1017/S0014479713000367. [DOI] [Google Scholar]
- 9.Wang, Q. X. et al. Analysis of the relationship between rusty root incidences and soil properties in Panax ginseng. 41, 012001 (2016)
- 10.Liu D, Sun H, Ma H. Deciphering microbiome related to rusty roots of Panax ginseng and evaluation of antagonists against pathogenic ilyonectria. Front. Microbiol. 2019;10:1350. doi: 10.3389/fmicb.2019.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JH, et al. Significant acidification in major Chinese croplands. Science. 2010;327:1008–1010. doi: 10.1126/science.1182570. [DOI] [PubMed] [Google Scholar]
- 12.Wan W, et al. Responses of the rhizosphere bacterial community in acidic crop soil to pH: changes in diversity, composition, interaction, and function. Sci. Total Environ. 2020;700:134418. doi: 10.1016/j.scitotenv.2019.134418. [DOI] [PubMed] [Google Scholar]
- 13.Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 14.Carrino-Kyker SR, Coyle KP, Kluber LA, Burke DJ. Fungal and bacterial communities exhibit consistent responses to reversal of soil acidification and phosphorus limitation over time. Microorganisms. 2019;8:1. doi: 10.3390/microorganisms8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Chen S, Chao L, Sun T. Effects of flooding on changes in Eh, pH and speciation of cadmium and lead in contaminated soil. Bull. Environ. Contam. Toxicol. 2007;79:514–518. doi: 10.1007/s00128-007-9274-8. [DOI] [PubMed] [Google Scholar]
- 16.Dordas C. Role of nutrients in controlling plant diseases in sustainable agriculture: a review. Agron. Sustain. Dev. 2008;28:33–46. doi: 10.1051/agro:2007051. [DOI] [Google Scholar]
- 17.Warren SLJH. Mineral nutrition of crops: fundamental mechanisms and implications. HortScience. 2004;39:462–462. doi: 10.21273/HORTSCI.39.2.462B. [DOI] [Google Scholar]
- 18.Sun, H., Zhang, Y. Y. & Song, X. X. Study on the soil nutrients and enzyme activity of cultivate ginseng soil in the farmland and wild ginseng soil under forest by canonical correlation analysis. Acta Agriculturae Boreali-Sinica S2 (2010).
- 19.Sharma S, Duveiller E, Basnet R, Karki CB, Sharma R. Effect of potash fertilization on Helminthosporium leaf blight severity in wheat, and associated increases in grain yield and kernel weight. Field Crops Res. 2005;93:142–150. doi: 10.1016/j.fcr.2004.09.016. [DOI] [Google Scholar]
- 20.Floch C, Capowiez Y, Biology S. Enzyme activities in apple orchard agroecosystems: how are they affected by management strategy and soil properties. Soil Biol. Biochem. 2009;41:61–68. doi: 10.1016/j.soilbio.2008.09.018. [DOI] [Google Scholar]
- 21.Aon MA, Coloneri AC., II Temporal and spatial evolution of enzymatic activities and physico-chemical properties in an agricultural soil. Appl. Soil Ecol. 2001;18:255–270. doi: 10.1016/S0929-1393(01)00161-5. [DOI] [Google Scholar]
- 22.Cai Z, et al. Effects of the novel pyrimidynyloxybenzoic herbicide ZJ0273 on enzyme activities, microorganisms and its degradation in Chinese soils. Environ. Sci. Pollut. Res. 2015;22:4425–4433. doi: 10.1007/s11356-014-3674-1. [DOI] [PubMed] [Google Scholar]
- 23.Antonious GF. Impact of soil management and two botanical insecticides on urease and invertase activity. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes. 2003;38:479–488. doi: 10.1081/PFC-120021667. [DOI] [PubMed] [Google Scholar]
- 24.Makoi JHJR, Ndakidemi PA. Selected soil enzymes: examples of their potential roles in the ecosystem. Afr. J. Biotechnol. 2008;7:181–191. [Google Scholar]
- 25.Jian S, Li J, Chen J, Wang G, Mayes MA, Dzantor KE, Hui D, Luo Y. Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: a meta-analysis. Soil Biol. Biochem. 2016;101:32–43. doi: 10.1016/j.soilbio.2016.07.003. [DOI] [Google Scholar]
- 26.Schutzendubel A, Polle A. Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002;53:1351–1365. [PubMed] [Google Scholar]
- 27.Chao A. Non-parametric estimation of the classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- 28.Shannon CEJBSTJ. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 29.Simpson EH. Measurement of diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 30.Farh ME, Kim YJ, Kim YJ, Yang DC. Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 2018;42:9–15. doi: 10.1016/j.jgr.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombard L, Der Merwe NAV, Groenewald JZ, Crous PW. Generic concepts in Nectriaceae. Stud. Mycol. 2015;80:189–245. doi: 10.1016/j.simyco.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinezbarrera OY, et al. Does Beauveria bassiana (Hypocreales: Cordycipitaceae) affect the survival and fecundity of the parasitoid Coptera haywardi (Hymenoptera: Diapriidae)? Environ. Entomol. 2019;48:156–162. doi: 10.1093/ee/nvy182. [DOI] [PubMed] [Google Scholar]
- 33.Migiro LN, Maniania NK, Chabi-Olaye A, Wanjoya A, Control JVJB. Effect of infection by Metarhizium anisopliae (Hypocreales: Clavicipitaceae) on the feeding and oviposition of the pea leafminer Liriomyza huidobrensis (Diptera: Agromyzidae) on different host plants. Biol. Control. 2011;56:179–183. doi: 10.1016/j.biocontrol.2010.09.013. [DOI] [Google Scholar]
- 34.Tardy V, et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 2015;90:204–213. doi: 10.1016/j.soilbio.2015.08.010. [DOI] [Google Scholar]
- 35.Yang L, Lu X, Li S, Wu B. First report of common bean (Phaseolus vulgaris) root rot caused by Plectosphaerella cucumerina in China. Plant Dis. 2018;102:1849–1849. doi: 10.1094/PDIS-10-17-1659-PDN. [DOI] [Google Scholar]
- 36.Baldeweg F, Warncke P, Fischer D, Gressler MJOL. Fungal biosurfactants from Mortierella alpina. Org. Lett. 2019;21:1444–1448. doi: 10.1021/acs.orglett.9b00193. [DOI] [PubMed] [Google Scholar]
- 37.Masinova T, Yurkov A, Baldrian PJFE. Forest soil yeasts: decomposition potential and the utilization of carbon sources. Fungal Ecol. 2018;34:10–19. doi: 10.1016/j.funeco.2018.03.005. [DOI] [Google Scholar]
- 38.Hu H, et al. Fomesafen impacts bacterial communities and enzyme activities in the rhizosphere. Environ. Pollut. 2019;253:302–311. doi: 10.1016/j.envpol.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 39.Barriuso J, Marín S, Mellado RP. Effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities: a comparison with pre-emergency applied herbicide consisting of a combination of acetochlor and terbuthylazine. Environ. Microbiol. 2010;12:1021–1030. doi: 10.1111/j.1462-2920.2009.02146.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, et al. Effects of organic–inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice–wheat cropping system. Appl. Soil Ecol. 2016;99:1–12. doi: 10.1016/j.apsoil.2015.11.006. [DOI] [Google Scholar]
- 41.Davidova, I. A., Marks, C. R., & Suflita, J. M. Anaerobic hydrocarbon-degrading Deltaproteobacteria. Handbook of Hydrocarbon and Lipid Microbiology 207–243 (2019).
- 42.Wang W, et al. Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land. Appl. Soil Ecol. 2020;146:103365. doi: 10.1016/j.apsoil.2019.103365. [DOI] [Google Scholar]
- 43.Yadav, S. et al. Cyanobacteria: role in agriculture, environmental sustainability, biotechnological potential and agroecological impact. In Plant-Microbe Interactions in Agro-Ecological Perspectives 257–277 (2017).
- 44.Rossi F, Li H, Liu Y, De Philippis R. Cyanobacterial inoculation (cyanobacterisation): perspectives for the development of a standardized multifunctional technology for soil fertilization and desertification reversal. Earth Sci. Rev. 2017;171:28–43. doi: 10.1016/j.earscirev.2017.05.006. [DOI] [Google Scholar]
- 45.Muñoz-Rojas M, et al. Effects of indigenous soil cyanobacteria on seed germination and seedling growth of arid species used in restoration. Plant Soil. 2018;429:91–100. doi: 10.1007/s11104-018-3607-8. [DOI] [Google Scholar]
- 46.Speirs LBM, Rice DTF, Petrovski S, Seviour RJ. The phylogeny, biodiversity, and ecology of the chloroflexi in activated sludge. Front. Microbiol. 2019;10:2015. doi: 10.3389/fmicb.2019.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitman T, et al. Dynamics of microbial community composition and soil organic carbon mineralization in soil following addition of pyrogenic and fresh organic matter. ISME J. 2016;10:2918–2930. doi: 10.1038/ismej.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lladó S, et al. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol. Fertil. Soils. 2016;52:251–260. doi: 10.1007/s00374-015-1072-6. [DOI] [Google Scholar]
- 49.Bousset, L., Ermel, M., Soglonou, B. & Husson, O. Fungal growth is affected by and affects pH and redox potential (Eh) of the growth medium. bioRxiv 401182 (2018). [DOI] [PubMed]
- 50.Mi C, et al. Unveiling of dominant fungal pathogens associated with rusty root rot of Panax notoginseng based on multiple methods. Plant Dis. 2017;101:2046–2052. doi: 10.1094/PDIS-01-17-0135-RE. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, et al. Analysis of rhizosphere bacterial and fungal communities associated with rusty root disease of Panax ginseng. Appl. Soil Ecol. 2019;138:245–252. doi: 10.1016/j.apsoil.2019.03.012. [DOI] [Google Scholar]
- 52.Li Z, Guo S, Tian S, Liu Z, Long B. Study on the causes for ginseng red skin sickness occurred in albic bed soil. Acta Pedol. Sin. 1997;34:328–335. [Google Scholar]
- 53.Shi J, Yuan X, Lin H, Yang Y, Li ZY. Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int. J. Int. J. Mol. Sci. 2011;12:3770–3785. doi: 10.3390/ijms12063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol. Biochem. 2019;41:919–925. doi: 10.1016/j.soilbio.2008.10.027. [DOI] [Google Scholar]
- 55.Pei G, Zhu Y, Wen J, Pei Y, Li H. Vinegar residue supported nanoscale zero-valent iron: remediation of hexavalent chromium in soil. Environ. Pollut. 2019;256:113407. doi: 10.1016/j.envpol.2019.113407. [DOI] [PubMed] [Google Scholar]
- 56.Fawcett JK. The semi-micro Kjeldahl method for the determination of nitrogen. J. Med. Lab Technol. 1954;12:1–22. [PubMed] [Google Scholar]
- 57.Li Y, et al. Humic acid fertilizer improved soil properties and soil microbial diversity of continuous cropping peanut: a three-year experiment. Sci. Rep. 2019;9:12014. doi: 10.1038/s41598-019-48620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 60.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin APJPO. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:1–18. doi: 10.1186/1465-6906-12-S1-P1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.