SUMMARY
Acinetobacter baumannii is a leading cause of ventilator-associated pneumonia and a critical threat due to multidrug resistance. The A. baumannii outer membrane is an asymmetric lipid bilayer composed of inner leaflet glycerophospholipids and outer leaflet lipooligosaccharides. Deleting mlaF of the maintenance of lipid asymmetry (Mla) system causes A. baumannii to become more susceptible to pulmonary surfactants and antibiotics and decreases bacterial survival in the lungs of mice. Spontaneous suppressor mutants isolated from infected mice contain an ISAba11 insertion upstream of the ispB initiation codon, an essential isoprenoid biosynthesis gene. The insertion restores antimicrobial resistance and virulence to ΔmlaF. The suppressor strain increases lipooligosaccharides, suggesting that the mechanism involves balancing the glycerophospholipids/lipooligosaccharides ratio on the bacterial surface. An identical insertion exists in an extensively drug-resistant A. baumannii isolate, demonstrating its clinical relevance. These data show that the stresses bacteria encounter during infection select for genomic rearrangements that increase resistance to antimicrobials.
Graphical Abstract
In Brief
The outer membrane is a critical barrier to resist host and antibiotic stresses for Gram-negative bacteria. Palmer et al. identify an Acinetobacter baumannii suppressor mutation that restores cell size, resistance to membrane stress and antibiotics, and virulence to a ΔmlaF mutant.
INTRODUCTION
Antibiotic-resistant infections represent a significant burden on the global public health system. Acinetobacter baumannii is a frequent cause of multidrug-resistant hospital-acquired infections and was labeled by the World Health Organization as the top priority pathogen for development of new antibiotics (Peleg et al., 2008; Tacconelli et al., 2018; Vincent et al., 2009; Weiner et al., 2016). A. baumannii and other Gram-negative, opportunistic pathogens are intrinsically resistant to many antibiotics because of their outer membrane (OM) (Blair et al., 2015; Geisinger et al., 2019). The OM is an asymmetric lipid bilayer, with a glycerophospholipid (GPL) comprising the inner leaflet and lipopolysaccharide (LPS) or lipooligosaccharide (LOS) comprising the outer leaflet. Bilayer asymmetry and the negatively charged LPS/LOS molecules within the outer leaflet promote bacterial cell shape and stiffness (Rojas et al., 2018) as well as resistance to permeabilization, detergents, membrane stresses, and antibiotics (Nikaido, 2003).
Gram-negative bacteria generally maintain OM lipid asymmetry using the maintenance of lipid asymmetry (Mla) system and the OM phospholipase PldA (Malinverni and Silhavy, 2009; Powers and Trent, 2019; Shrivastava and Chng, 2019). The Mla system is conserved throughout Gram-negative bacteria and in chloroplasts (Awai et al., 2006; Casali and Riley, 2007) and is also known as the mammalian cell entry (MCE) protein family, toluene tolerance gene (Ttg) system, and the trigalactosyldiacylglycerol protein (Tdg). An ATP binding cassette (ABC) transporter, the Mla system consists of the OM protein MlaA in complex with OmpC/F, the soluble periplasmic binding protein MlaC, the inner membrane (IM) periplasmic binding protein MlaD, the IM protein MlaE, the ATPase MlaF, and the cytoplasmic protein MlaB (Abellón-Ruiz et al., 2017; Chong et al., 2015; Ekiert et al., 2017; Malinverni and Silhavy, 2009; Thong et al., 2016; Yeow et al., 2018). Inactivating any component of the Mla system inactivates the entire system in several species (Chong et al., 2015; Malinverni and Silhavy, 2009).
The Mla system transports lipids between the OM and IM. Genetic evidence in Escherichia coli, A. baumannii, and chloroplasts suggests that the Mla system functions as a retrograde transport system that moves GPL from the outer leaflet of the OM to the IM (Awai et al., 2006; Malinverni and Silhavy, 2009; Nagy et al., 2019; Powers and Trent, 2018; Shrivastava et al., 2017; Sutterlin et al., 2016). Crystallographic structures of the OM protein MlaA from Klebsiella pneumoniae and Serratia marcescens, and a structure of E. coli MlaA in complex with the OM porin OmpC suggest the MlaA binding site opens to outer leaflet GPL, consistent with a retrograde transport mechanism (Abellón-Ruiz et al., 2017; Chong et al., 2015; Yeow et al., 2018). However, pulse-chase labeling studies in A. baumannii suggest the Mla system may execute an anterograde transport mechanism that delivers newly synthesized GPL to the inner leaflet of the OM (Kamischke et al., 2019). Biochemical studies have shown that the periplasmic binding protein MlaC has higher binding affinity for GPL than does MlaD, the IM-associated protein and does not require ATP hydrolysis to accept lipid molecules from MlaD (Ercan et al., 2019; Hughes et al., 2019). These findings suggest that anterograde transport could rely on affinity differences and retrograde transport could rely on ATP hydrolysis. The combined evidence supports that Mla may transport GPL bi-directionally across the periplasm.
Because of its role in OM homeostasis, the Mla system is important for the virulence of many pathogens, including Shigella flexneri (Carpenter et al., 2014; Hong et al., 1998; Suzuki et al., 1994), Haemophilus influenzae (Nakamura et al., 2011), Burkholderia pseudomallei (Cuccui et al., 2007), and Pseudomonas aeruginosa (Munguia et al., 2017). In contrast, lack of MlaA promotes virulence in a murine model of Neisseria gonorrhoeae infection (Baarda et al., 2019). Disrupting the Mla system prompts formation of OM vesicles (OMV) for H. influenzae, Vibrio cholerae, N. gonorrhoeae, and A. baumannii (Baarda et al., 2019; Powers and Trent, 2019; Roier et al., 2016; Zingl et al., 2020). The mla genes are transcriptionally regulated by metals (Baarda et al., 2019; Roier et al., 2016), and A. baumannii mlaF expression is induced by metal chelation (Hood et al., 2012; Mortensen et al., 2014). The Mla system is also important for resisting antibiotics and host stresses, such as serum and bile salts for E. coli (Sutterlin et al., 2016), H. influenzae (Nakamura et al., 2011), Burkholderia cepacia complex species (Bernier et al., 2018), A. baumannii (Kamischke et al., 2019), and P. aeruginosa (Munguia et al., 2017; Shen et al., 2012). Therefore, the Mla system has been proposed as a target for drug development (Huang et al., 2019).
This study sought to determine the role of the Mla system in A. baumannii in a murine model of pneumonia. Disrupting the Mla system renders A. baumannii more susceptible to membrane stress and antibiotics and defective in Toll-like receptor 4 (TLR4) evasion and survival during infection. We identify pulmonary surfactants as a membrane stress against which the Mla system protects A. baumannii. Quantitative mass spectrometry demonstrates that, in the absence of Mla, particular GPLs accumulate throughout the envelope, including in the OM. Suppressor mutants isolated from animals infected with mlaF-deficient A. baumannii have restored virulence in mice and resistance to membrane stress and antibiotics. Characterization of the suppressor strains identified a conserved insertion that modulates expression of an isoprenoid biosynthetic gene to restore resistance to membrane stress and antibiotics. The suppressive insertion does not restore the GPL levels, but increases the LOS levels, presumably to offset the GPL accumulation. This insertion is also present in extensively drug-resistant (XDR) A. baumannii clinical isolates, which underscores the clinical relevance of these findings.
RESULTS
MlaF Promotes A. baumannii Resistance to Membrane Stress and Antibiotics
To determine whether the A. baumannii Mla system contributes to pathogenesis during pneumonia, the strain 17978 ΔmlaF::Kn mutant (ΔmlaF) was generated by replacing mlaF with a non-polar kanamycin resistance marker containing a ribosome binding site for the remaining mla operon. The ΔmlaF strain does not display any overt growth defects when grown in lysogeny broth (LB) but has a transparent colony morphology compared with the wild type (Figures 1A and 1C). To assess the role of MlaF in resisting membrane stress, wild-type and ΔmlaF cultures were diluted and spot plated to LB agar plates containing SDS and EDTA. In the presence of SDS and EDTA, the ΔmlaF mutant has a marked defect in colony formation (Figure 1B). mlaF mutants in the A. baumannii AB5075 strain background demonstrate the same phenotype, suggesting that the role of MlaF in resisting membrane stress is conserved in diverse A. baumannii strains (Figures S1A and S1B). Next, the growth of wild type, ΔmlaF, and strains expressing mlaF in trans were compared in the presence of SDS and EDTA. There was no difference in growth between the strains in LB (Figure 1C). However, in the presence of SDS and EDTA, the ΔmlaF mutant had a severe growth defect that is fully complemented by expression of mlaF in trans (Figure 1D). These results show that the A. baumannii Mla system protects against SDS/EDTA membrane stress.
Figure 1. A. baumannii ΔmlaF Has a Growth Defect When Subjected to Membrane Stress and Increased Surface-Accessible Glycero-phospholipids.
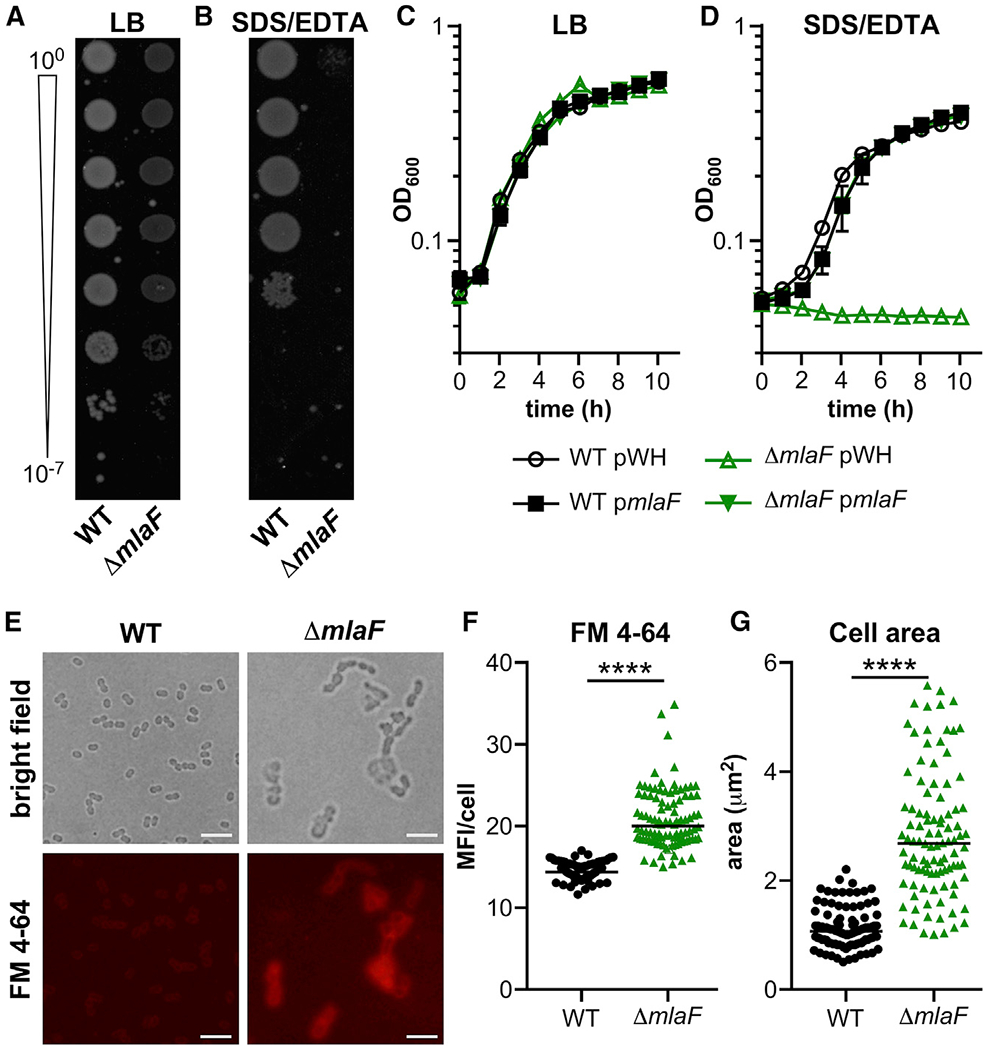
(A and B) Wild-type (WT) and ΔmlaF strains were diluted and spotted to lysogeny broth (LB) plates without (A) and with (B) 0.01% SDS and 0.15 mM EDTA.
(C and D) WT and ΔmlaF strains were grown in LB without (C) and with (D) 0.01% SDS and 0.15 mM EDTA (n = 3; data are means ± SEM).
(E) Live WT and ΔmlaF bacteria were incubated with the lipophilic fluorescent dye FM4-64 and imaged on agar pads. Scale bar is 5 μm.
(F and G) The mean fluorescence intensity (MFI) per cell was measured (F).
(G) The cell area (μm2) was measured.
n = 100 with median; significance by Mann-Whitney test. ****p < 0.0001 (F and G). See also Figure S1.
We hypothesized that MlaF helps promote envelope integrity. To test this, wild-type and ΔmlaF A. baumannii were incubated with the lipophilic dye FM4-64. Live cells were imaged in bright field and red fluorescence to assess FM4-64 exclusion (Figure 1E). Although the wild-type cells largely excluded FM4-64, ΔmlaF cells had significantly elevated FM4-64 fluorescence, demonstrating they have increased surface-accessible GPL (Figure 1F). Additionally, the ΔmlaF cells are larger compared with wild-type cells (Figure 1G). Defects in cell envelope integrity often increase susceptibility to antibiotics, and A. baumannii lacking mlaF has been previously reported to be more susceptible to gentamicin, novobiocin, and rifampicin (Kamischke et al., 2019), which target the ribosome, DNA gyrase, and RNA polymerase, respectively. The antibiotic susceptibility of the wild-type and ΔmlaF strains were compared with a disk diffusion assay (Table 1). The ΔmlaF mutant was significantly more susceptible to multiple antibiotics from different classes, including gentamicin, erythromycin, carbenicillin, meropenem, and bacitracin. The strains were also compared for susceptibility to host-imposed stresses, and the ΔmlaF strain was more susceptible to paraquat, which generates superoxides (Table 1). Together, these data show that the Mla system in A. baumannii is important for maintaining envelope integrity to resist membrane stress, antibiotics, and paraquat.
Table 1.
The ΔmlaF Mutant Has Increased Antibiotic Susceptibility That Is Largely Restored in the Suppressor Strain
| Antibiotic | WTa | ΔmlaFa | ΔmlaF suppa | WT suppa |
|---|---|---|---|---|
| Chloramphenicol | 12 ± 3 | 12 ± 1 | 14 ± 3 | 12 ± 1 |
| Tetracycline | 20 ± 1 | 18 ± 1 | 18 ± 1 | 19 ± 1 |
| Streptomycin | 13 ± 1 | 14 ± 0 | 14 ± 1 | 14 ± 1 |
| Gentamicin | 20 ± 2 | 28 ± 2** | 21 ± 1 | 21 ± 3 |
| Erythromycin | 21 ± 1 | 27 ± 2* | 21 ± 1 | 20 ± 4 |
| Colistin | 12 ± 0 | 13 ± 1 | 11 ± 1 | 11 ± 0 |
| Polymyxin B | 8 ± 0 | 9 ± 0 | 8 ± 0 | 9 ± 1 |
| Daptomycin | 6 ± 1 | 6 ± 0 | 6 ± 1 | 7 ± 1 |
| Carbenicillin | 20 ± 4 | 33 ± 3*** | 19 ± 1 | 23 ± 2 |
| Meropenem | 19 ± 5 | 37 ± 3*** | 18 ± 2 | 24 ± 4 |
| Bacitracin | 10 ± 0 | 16 ± 1**** | 17 ± 1**** | 9 ± 0 |
| Copper chloride | 8 ± 1 | 9 ± 1 | 9 ± 0 | 9 ± 0 |
| Acetic acid | 9 ± 0 | 8 ± 0 | 9 ± 1 | 9 ± 0 |
| Hydrochlorous acid | 8 ± 0 | 8 ± 0 | 8 ± 0 | 8 ± 1 |
| Hydrogen peroxide | 12 ± 1 | 13 ± 1 | 13 ± 1 | 12 ± 1 |
| Paraquat | 10 ± 4 | 18 ± 2* | 13 ± 1 | 12 ± 3 |
Antibiotic susceptibility was determined by a disk-diffusion assay measuring the zone of clearing. Data are two or three replicates and representative of at least two independent experiments. Significance is by one-way ANOVA with Dunnett’s multiple comparisons; significant differences compared to wild-type (WT) are indicated by * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Mean diameter ± standard deviation (mm); limit of detection, 6 mm.
MlaF Contributes to Innate Immune Evasion and Survival in the Lungs
To test whether the Mla system contributes to A. baumannii virulence, the survival of the wild-type and ΔmlaF strains was compared in a murine model of acute pneumonia. A. baumannii was intranasally inoculated into mice, and bacterial burdens in the lungs and spleen were assessed at 8, 12, and 36 h. There was no significant difference in bacterial burdens in the lungs or spleens at 8 or 12 h after infection when comparing the wild-type and ΔmlaFstrains (Figures S2A–S2D). By contrast, by 36 h after infection, the ΔmlaF mutant was recovered at significantly lower burdens than in the wild type in both the lungs (>104-fold) and the spleen (not recovered) (Figures 2A and 2B). Together, these data suggest that MlaF is required for persistence and dissemination during lung infection. To determine the potential mechanisms of clearance by the ΔmlaF mutant, lung inflammation was assessed after hematoxylin and eosin staining at 8 h after infection, when bacterial burdens were similar between the strains (Figure S2E). By qualitative assessment, wild-type bacteria were frequently observed extracellularly in the mouse lung. However, in mice infected with the ΔmlaF mutant, bacteria were more frequently observed intracellularly in macrophages. In addition, there was a trend toward increased inflammation (p = 0.12) in mice infected with the ΔmlaF mutant (Figure S2F). Collectively these findings suggest that bacteria lacking the Mla system may incite increased host immune responses and are more rapidly cleared.
Figure 2. A. baumannii ΔmlaF Is Defective in Infection and Survival in Lung Surfactants and Increases TLR4 Activation.
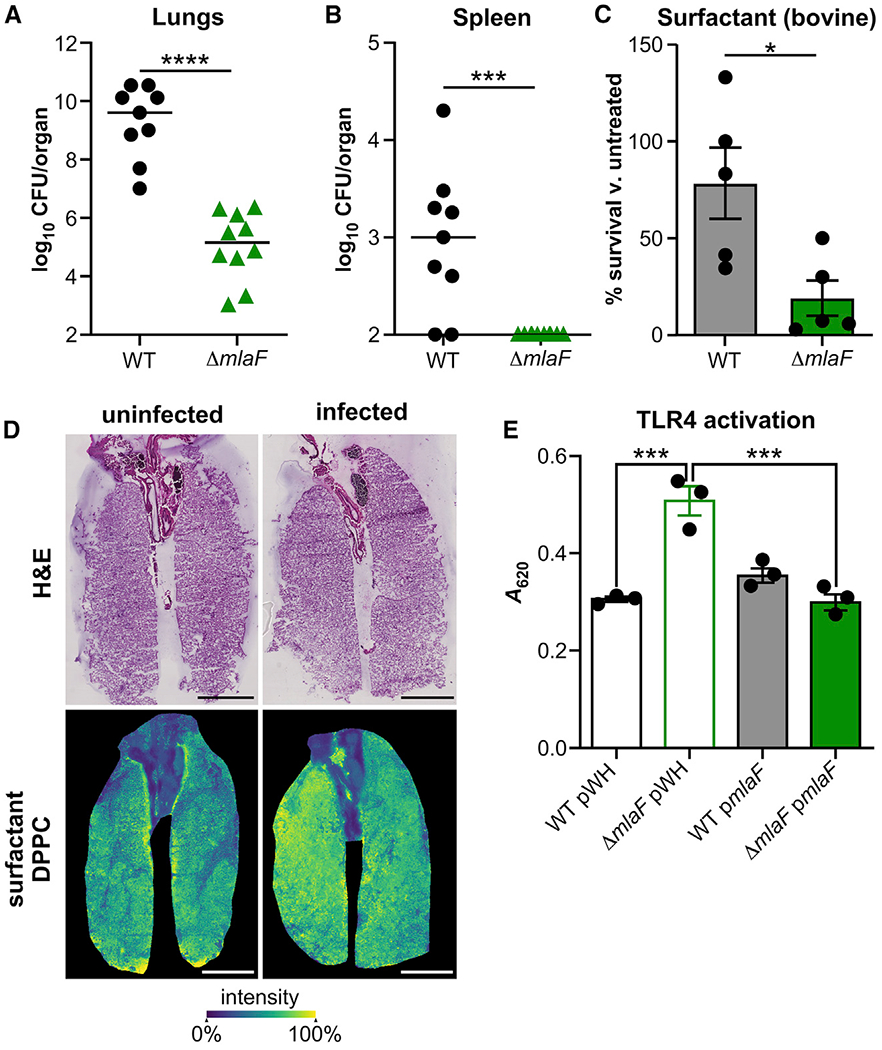
(A and B) Wild-type (WT) and ΔmlaF strains were infected intranasally into mice. At 36 h after infection, lungs (A) and spleens (B) were harvested, and bacterial burdens were enumerated (n = 9–10 with median; significance by Mann-Whitney test).
(C) WT and ΔmlaF strain survival in Infasurf pulmonary surfactant after 90 min (n = 6 from two independent experiments; data are means ± SEM; significance by t test).
(D) Distribution of DPPC (m/z 734.5719) by imaging mass spectrometry. Scale bar is 4 mm.
(E) Activation of HEK-Blue TLR4 reporter cell line at multiplicity of infection 10−2 (n = 3; data are means ± SEM; significance by one-way ANOVA). *p < 0.05, ***p < 0.001, ****p < 0.0001. See also Figure S2.
We hypothesized that the ΔmlaF mutant may be less able to survive lung membrane stresses or may trigger innate immune signaling that leads to effective bacterial clearance. Pulmonary surfactants are essential to lung function and represent a lung-specific membrane stress to pneumonic pathogens (Whitsett et al., 2015). To test whether pulmonary surfactants contribute to decreased persistence of the ΔmlaF mutant in the murine lung, the survival of wild-type and ΔmlaF strains was compared in the presence of the pharmaceutical Infasurf, an ex vivo bovine pulmonary surfactant. The ΔmlaF mutant had significantly reduced survival compared with the wild type after 1.5 h incubation in the pulmonary surfactant (Figure 2C). These data are consistent with a role for the Mla system in protection from lung-specific membrane stresses in the host. To confirm that A. baumannii encounters surfactant stress in the lung, the major surfactant lipid component dipalmitoylphosphatidylcholine (DPPC) was visualized by imaging mass spectrometry (Berry et al., 2011; Spraggins et al., 2019). A phosphatidylcholine mass consistent with DPPC (m/z 734.5719 with 3.4 ppm mass error) was abundant in the lower airways of both uninfected and infected lung tissue (Figure 2D), demonstrating that A. baumannii encounters pulmonary surfactant in the lung.
The fact that the ΔmlaF mutant is more effectively cleared by the immune system and has differences in OM integrity compared to the wild type suggested that the Mla system may affect host detection of the OM component LOS by TLR4. TLR4 binding of the lipid A portion of LOS leads to nuclear factor κB (NF-κB) signaling and inflammatory cytokine production. Therefore, heat-killed wild-type and ΔmlaF bacteria were washed and incubated with HEK-Blue hTLR4 reporter cells that produce extracellular alkaline phosphatase in response to TLR stimulation. The results of this experiment showed that although both wild-type and the ΔmlaF strains activated HEK-Blue hTLR4 reporter expression, the ΔmlaF mutant significantly increased activation (Figure 2E). Furthermore, the increased activation by the ΔmlaF mutant was reversed by expression of mlaF in trans. Interestingly, at 8 h after mouse infection there was no difference in pro-inflammatory cytokines and chemokines such as tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), CXCL1, and IL-12 (p70) in the lung (Figures S2G–S2J), consistent with similar bacterial burdens and suggesting any difference in inflammatory response may occur before 8 h. Together, these data suggest that the Mla system contributes to pneumonia pathogenesis by protecting A. baumannii from membrane stress and may contribute to host immune evasion.
Isolation of an In Vivo ΔmlaF Suppressor Allows Genetic Separation of Membrane Stress Resistance, Innate Immune Evasion, and Virulence
Although the data above demonstrate a role for the Mla system in pathogenesis, it was unclear whether decreased bacterial burdens during infection result from disruption of Mla-dependent membrane stress resistance or immune evasion. Therefore, a suppressor analysis strategy was used to genetically dissect the contributions of Mla-dependent membrane stress resistance and host immune evasion to A. baumannii virulence. We first tested whether previously described suppressors functioned in A. baumannii. In E. coli, the membrane stress phenotype of an mla mutant is suppressed by constitutive expression pldA, which encodes an OM phospholipase with broad substrate specificity, which is necessary to degrade the GPL and lysoGPL that accumulate in the OM outer leaflet (Malinverni and Silhavy, 2009). However, constitutive expression of pldA did not rescue A. baumannii ΔmlaF susceptibility to membrane stress, whereas expression of mlaF from the same promoter complemented that phenotype (Figures S3A and S3B). Because it is possible PldA was not functional in this strain, we chose to take an unbiased approach and characterized suppressors of the ΔmlaF mutant that had lost the transparent colony morphology during lung infection.
We reasoned that the lungs of mice are a selective environment to enrich for mutations that reverse the ΔmlaF mutant phenotypes (i.e., membrane stress or innate immune activation). Thirty suppressors from two independent infections were isolated as opaque colonies against the translucent colonies of the parental genotype, which were not further characterized. Genomes of representative isolates from each infection and the ΔmlaF strain were sequenced. A single genetic difference was observed in each of the suppressor isolates from the two infections: an ISAba11 insertion upstream of the coding sequence of the ispB gene (Figures 3A and S3C). ispB encodes the octaprenyl diphosphate synthase required for elaborating the isoprenoid side chain of ubiquinone and menaquinone (Asai et al., 1994). PCR analysis confirmed the ISAba11 insertion in all 30 suppressor isolates. ISAba11 is a member of an emerging insertion sequence family related to the IS701 family; it includes 13-bp inverted repeats and encodes the transposase TnpISAba11 (A1S_0208) (Moffatt et al., 2011; Rieck et al., 2012). The ISAba11 sequence is inserted with a 6-bp target-site duplication (ATATCA; Figure S3C) and is also maintained in its original site in the genome. In silico analysis identified this same insertion in A. baumannii AB030, an XDR isolate (Figure 3A) (Loewen et al., 2014).
Figure 3. Suppressor Analysis Identifies an Insertion in the 5′ Untranslated Region of ispB That Restores Membrane Stress Resistance and Size, but Not TLR4 Evasion, of the ΔmlaF Strain.
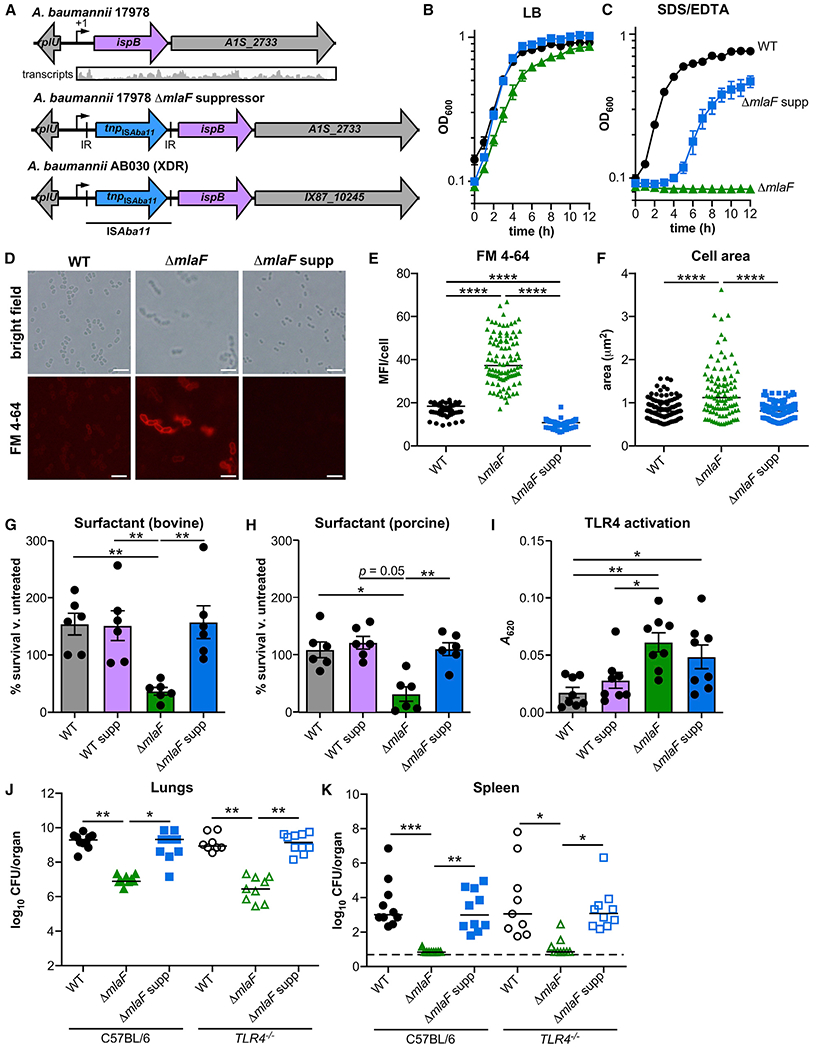
(A) Upstream of ispB and after its predicted transcription start site (+1), insertion ISAba11 includes inverted repeats (IR) and encodes a putative transposase.
(B and C) WT, ΔmlaF, and ΔmlaF suppressor (ΔmlaF supp) strains were grown in LB without (B) and with (C) 0.01% SDS and 0.175 mM EDTA (n = 3, data are means ± SEM).
(D) Live WT and ΔmlaF bacteria were incubated with the lipophilic fluorescent dye FM4-64 and imaged on agar pads. Scale bar is 5 μm.
(E and F) The mean fluorescence intensity (MFI) per cell (E) and the area per cell (F) were measured (n = 100 with median; significance by Kruskal-Wallis with Dunn’s multiple comparisons).
(G and H) WT, ΔmlaF and suppressor strain survival in Infasurf bovine (G) pulmonary surfactant and Curosurf porcine (H) pulmonary surfactant after 90 min (n = 6 from two independent experiments; data are means ± SEM; significance by one-way ANOVA with Tukey’s multiple comparisons).
(I) Activation of HEK-Blue TLR4 reporter cell line at multiplicity of infection 10−2 (n = 8; data are mean ± SEM; significance by one-way ANOVA with Tukey’s multiple comparisons).
(J and K) Wild-type (WT), ΔmlaF, and ΔmlaF suppressor strains were infected intranasally into C57BL/6 and TLR4−/− mice. At 36 h after infection, lungs (J) and spleens (K) were harvested, and bacterial burdens were enumerated (n = 9–10 with median; significance by Kruskal-Wallis with Dunn’s multiple comparisons). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S3.
To ensure that the phenotypes for the suppressor genotype were caused by the insertion in the 5′ region of ispB, the mutation was reconstructed in the wild-type and ΔmlaF strains by allelic exchange using the suppressor genome as a genetic template. The reconstructed insertion mutant strains were used for all subsequent analyses unless otherwise indicated. The reconstructed ΔmlaF suppressor strain was then tested for suppression of the membrane stress, cell envelope integrity, cell size, antibiotic resistance, and TLR4 activation phenotypes of the ΔmlaF mutant compared with the wild type. The ΔmlaF suppressor strain displayed no defect in growth in LB but intermediate resistance to SDS/EDTA membrane stress (Figures 3B and 3C), increased exclusion of FM4-64, and decreased cell size (Figures 3D–3F), largely mimicking the wild-type strain. In addition, the ΔmlaF suppressor strain had restored resistance to gentamicin, erythromycin, meropenem, carbenicillin, and paraquat (Table 1). Resistance to bacitracin was not restored (Table 1). By this assay, the suppressive insertion did not improve resistance to antibiotics in the wild-type background. Restoration of resistance to SDS/EDTA, antibiotics and exclusion of FM4-64 suggests the suppressor insertion restores envelope integrity to the ΔmlaF mutant.
Next, resistance to host surfactant and innate immune activation was assessed. Strains with suppressor insertion in the wild-type and ΔmlaF background were incubated with bovine and porcine pulmonary surfactant, and bacterial survival was compared (Figures 3G and 3H). Consistent with the findings above, the suppressive insertion restored bacterial survival in the ΔmlaF background to wild-type levels and did not affect surfactant resistance in the wild-type background. By contrast, the ΔmlaF suppressor strain maintained higher innate immune activation compared with wild-type A. baumannii using HEK-Blue TLR4 reporter cells (Figure 3I). The insertion did not affect TLR4 activation in the wild-type strain. Because the suppressive insertion appeared to have little effect on fitness in the wild-type background, subsequent assays focused on clarifying the effect of the insertion in the ΔmlaF background. The suppressive insertion restores cell envelope integrity and pulmonary surfactant resistance but not immune evasion to the ΔmlaF mutant. This distinction allows separation of the two phenotypes to identify whether the contribution of MlaF to bacterial envelope integrity or immune evasion is important for A. baumannii lung infection.
To determine whether the suppressive insertion restores virulence to the ΔmlaF mutant and to delineate the contribution of TLR4 activation to pathogenesis, the wild-type strain and the ΔmlaF and ΔmlaF suppressor strains were intranasally inoculated in C57BL/6 or TLR4−/− mice. At 36 h after infection, the ΔmlaF mutant had significantly reduced burdens in the lungs and spleen compared with the wild-type strain (Figures 3J and 3K). The ΔmlaF suppressor strain had significantly greater burdens than the ΔmlaF mutant had in the lungs and spleens and was indistinguishable from the wild-type strain (Figures 3J and 3K). These results demonstrate that the critical role for MlaF in lung infection is due to its function in mediating resistance to membrane stress. In addition, there were no differences in bacterial burdens between the C57BL/6 and TLR4−/− mice (Figures 3J and 3K). Furthermore, this result suggests that, in this infection model with A. baumannii 17978, TLR4 activation does not have a major role in promoting murine resistance to A. baumannii lung infection. Together, these results show that the role of the Mla system in resisting membrane stresses is critical for lung infection.
MlaF Is Necessary for A. baumannii to Balance the Levels of GPL and LOS Molecules within the OM and the Suppressive Insertion Results in Increased LOS Abundance
The Mla system maintains OM-lipid asymmetry and traffics GPL between the OM and IM bilayers (Awai et al., 2006; Kamischke et al., 2019; Malinverni and Silhavy, 2009; Powers and Trent, 2019; Shrivastava and Chng, 2019). We hypothesized that the suppressor insertion had altered OM lipid composition. Therefore, the GPL composition of the wild-type and mlaF-deficient strains was measured. Total membrane and OM fractions were collected from logarithmic phase bacteria (Figure S4A) and GPL was quantified for each fraction by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Cian et al., 2020; Dalebroux et al., 2014, 2015; Osborn et al., 1972). There was no significant difference in the total GPL in the strains (Figure S4B). However, both mutants measured a statistically significant 3-4-fold increase in specific phosphatidyglycerols (PGLs), both in the total and in OM fractions (Figures 4A and 4B; Tables S1 and S2). These PGL molecules are major components of wild-type membranes and carry a m/z of 719, which corresponds to the two isomers of a molecule that carries an sn1/sn2 acyl-chain composition of C16:0/C16:1. The ΔmlaF mutant also had a 1.3-fold increase in total membrane fraction phosphatidylethanolamines (PEs) with an m/z of 716 (C16:0/C18:1) (Table S1). The levels of these and other individual GPLs for the ΔmlaF and ΔmlaF suppressor strains were not statistically different (Tables S1 and 2), suggesting that the suppressor mutation does not restore GPL homeostasis.
Figure 4. The Suppressor Strain Maintains Increased Phosphatidylglycerol Levels but Also Has Increased LOS.
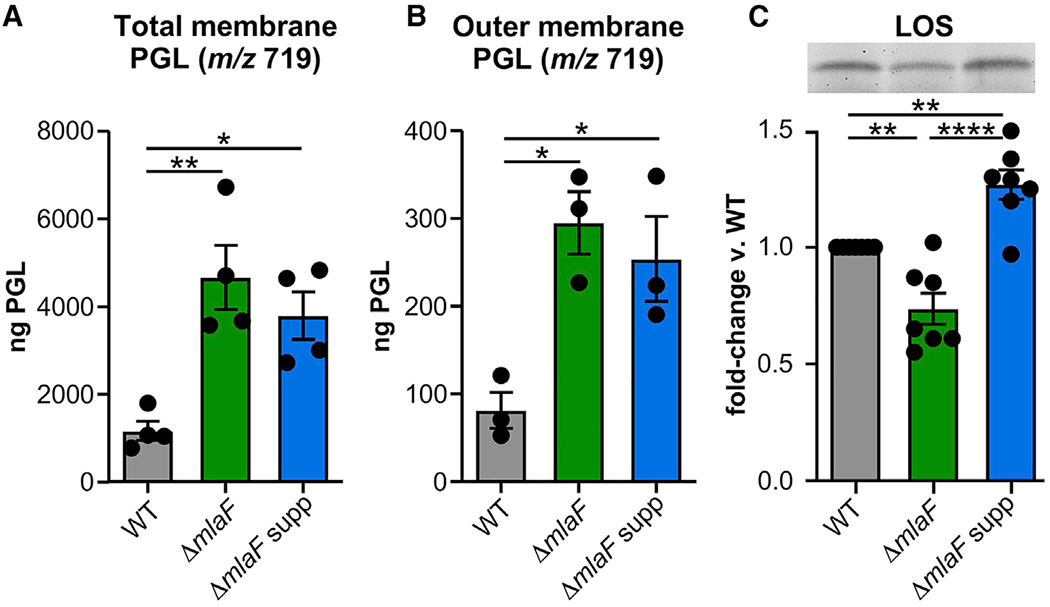
(A and B) Phosphatidylglycerols (PGL) with mass-to-charge ratio (m/z) of 719 in total membrane (n = 4) and outer membrane fractions (n = 3).
(C) Fold-change in lipooligosaccharide (LOS) (n = 7 from 4 independent LOS extractions). Inset shows a representative gel. Data are means ± SEM; significance by one-way ANOVA with Tukey’s multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S4, Table S1, and Table S2.
NADH dehydrogenase activity was assayed to assess the relative cross contamination of the IM and OM bilayers. The OM fractions of the ΔmlaF mutant possessed a slightly higher NADH dehydrogenase activity compared with that of the wild-type and suppressor strain OM fractions, but the difference was not statistically significant (Figures S4C and S4D). Therefore, the statistically significant increase in PGL for the OM fractions of the ΔmlaF mutant and the suppressor strain is not caused by cross contamination with the IM. The IM fractions of the suppressor isolate measured a dramatic decrease in NADH dehydrogenase activity compared with that of the IM fractions of the wild-type and parental mutant genotype (Figures S4C and S4D). Because NADH dehydrogenase is an IM enzyme that requires ubiquinone for activity, this finding suggests that the ΔmlaF suppressor may have lower quinone levels.
In E. coil, a dominant allele of mlaA, which encodes the OM lipoprotein component of the Mla system, causes symmetrical microdomains to form within the OM (Sutterlin et al., 2016). Perturbing asymmetry results in signal transduction across the periplasm and a lipid-mediated regulatory mechanism that controls LPS biosynthesis (Cian et al., 2019; May and Silhavy, 2018). Accordingly, we hypothesized that the Mla system and the suppressive insertion might influence the LOS levels for A. baumannii. To quantify differences in LOS abundance, LOS molecules were extracted from bacterial suspensions that had been normalized to cell number by optical density and colony-forming units (Figure S4E). Using a hot phenol-chloroform extraction method, gradient-gel electrophoresis, and Pro-Q Emerald 300 staining (Figures 4C, S4F, and S4G), LOS levels were quantified for multiple biological and technical replicates from mid-log cultures (Cian et al., 2020). Relative to the wild-type strain, the LOS levels of the ΔmlaF mutant were significantly decreased, whereas the LOS levels of the suppressor mutant were significantly increased (Figure 4C). Compared with the wild-type and ΔmlaF strains, the suppressor mutant measured a significant increase in LOS levels. Therefore, A. baumannii MlaF promotes OM-lipid homeostasis, in part, by balancing PGL and LOS levels.
Reduced ispB Expression Improves A. baumannii Resistance to Membrane Stress
The causative mutation in the ΔmlaF suppressor strain is an insertion upstream of the ispB open reading frame. To identify the relationship of the insertion to the ispB transcript, previously published RNA sequencing (RNA-seq) data from wild-type A. baumannii were mapped to the genome to predict the +1 transcription start site (TSS) (Figures 3A and S3A) (Wang et al., 2019). The putative +1 site determined by transcript read mapping agreed with the start site predicted by the phiSITE Promoter Hunter (Klucar et al., 2010). By this analysis, the suppressive insertion is downstream of the predicted +1 TSS of a transcript that also codes for A1S_2733, a putative membrane protein of unknown function. In E. coli, ispB is essential, consistent with an essential role for either ubiquinone or menaquinone (Okada et al., 1997; Wallace and Young, 1977). Based on pathway analysis, A. baumannii synthesizes ubiquinone-8 and does not synthesize menaquinone. Attempted construction of an ΔispB strain was unsuccessful and insertions in ispB were not recovered in published transposon sequencing datasets, suggesting ispB is also essential in A. baumannii (Gallagher et al., 2015; Wang et al., 2014). Therefore, we hypothesized that the insertion alters ispB expression. Compared with the wild type, ispB transcript abundance was unchanged in the ΔmlaF mutant (Figure 5A). By contrast, ispB transcript abundance was significantly reduced in both independently isolated ΔmlaF suppressor strains (Figure 5A).
Figure 5. A Suppressor Insertion Lowers Expression of ispB and Increases Resistance to Membrane Stress.
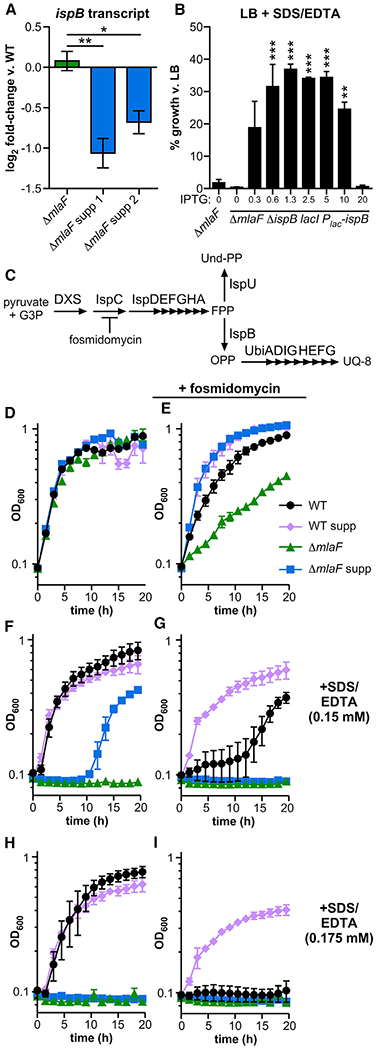
(A) Transcript abundance of ispB in ΔmlaF and independently-isolated ΔmlaF suppressor strains 1 and 2 was measured using RT-qPCR (n = 3; significance by one-way ANOVA with Tukey’s multiple comparisons).
(B) ispB expression was induced by IPTG (mM) in the ΔmlaF lacI Plac-ispB strain and growth in 0.01% SDS and 0.175 mM EDTA was compared to growth in LB at 12 h (n = 2, representative of two independent experiments; significance by one-way ANOVA with Dunnett’s multiple comparisons to ΔmlaF).
(C) Schematic of isoprenoid biosynthesis.
(D–I) Representative growth curves of strains LB without (D, F, and H) and with (E, G, and I) 100 mg/L fosmidomycin, and without (D and E), and with (F and I) 0.01% SDS and EDTA at the concentration indicated (n = 3). Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. G3P, glyceraldehyde 3-phosphate; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; Und-PP, undecaprenyl pyrophosphate; FPP, farnesyl pyrophosphate; UQ-8, ubiquinone-8. See also Figure S5.
To test whether the mechanism of suppression by the insertion upstream of the ispB initiation codon is due to decreased ispB expression, an inducible ispB strain was constructed in the ΔmlaF background. The mini-Tn7 site-specific transposon was engineered to express the E. coli lactose repressor lacI, and the A. baumannii ispB gene, driven by a promoter containing the lactose operator. This mini-Tn7 was introduced into the ΔmlaF strain, and the native A. baumannii ispB was deleted to ensure ispB expression was exclusively driven by isopropyl-β-d-thiogalactoside (IPTG)-dependent induction from the engineered locus. The growth of the ΔmlaF inducible ispB strain was compared with the wild-type and ΔmlaF strains in LB and with SDS/EDTA with varying levels of IPTG to induce expression of ispB. There was no difference in phenotype for any strains or conditions when grown in LB, demonstrating that IPTG induction of ispB is not toxic to A. baumannii (Figure S5A). When subjected to SDS/EDTA membrane stress, the ΔmlaF mutant exhibited no growth (Figures 5B and S5B). By contrast, growth of the ΔmlaF-inducible ispB strain depended on the concentration of IPTG provided, reaching a maximum of 37% of its growth without SDS/EDTA at 1.25 mM IPTG (Figure 5B); the wild type reached 87% ± 2% of its growth without SDS/EDTA. Consistent with the hypothesis that the mechanism of suppression requires lower ispB expression levels, high levels of IPTG decreased growth of the inducible ispB strain in SDS/EDTA but not in LB (Figures 5B, S5A, and S5B). Similarly, constitutive expression of ispB from a multi-copy plasmid suppressed growth of the ΔmlaF mutant generally and the suppressor strain in SDS/EDTA (Figures S5C and S5D). These results demonstrate that modulating expression of the ispB coding sequence is sufficient to restore resistance to membrane stress in the ΔmlaF background.
Furthermore, these results suggest that restricting flux through isoprenoid biosynthesis may restore membrane-stress resistance in the absence of MlaF. Fosmidomycin is an antibiotic that inhibits isoprenoid biosynthesis upstream of ispB and its branchpoint substrate farnesyl-PP (FPP) (Figure 5C). To test whether restricting isoprenoid biosynthesis generally increased fitness in membrane stress, the growth of strains was compared in the presence and absence of fosmidomycin and SDS/EDTA (Figures 5D–5I). Surprisingly, sublethal levels of fosmidomycin inhibited growth of only the ΔmlaF mutant in LB, in direct opposition to the model that restricting isoprenoid biosynthesis generally improves fitness of the ΔmlaF mutant (Figures 5D and 5E). In the presence of SDS/EDTA, fosmidomycin also inhibited growth of the wild-type strain, whereas the suppressive insertion restored growth in the wild-type strain (Figures 5F–5I). These results demonstrate that low ispB expression restores membrane stress resistance to ΔmlaF A. baumannii by a mechanism that requires an intact isoprenoid biosynthetic pathway. Together, these findings suggest that A. baumannii evolves within the lung to offset the ΔmlaF mutant lipid imbalance by decreasing ispB expression and increasing LOS levels.
DISCUSSION
A. baumannii is an important multidrug-resistant pathogen, and the Mla system has been proposed as a novel drug target because of its roles in virulence and resistance to host imposed-stress and antibiotics (Huang et al., 2019; Kamischke et al., 2019). In addition to its previously described role in A. baumannii membrane stress and antibiotic resistance, the data here demonstrate that the Mla system also contributes to A. baumannii resistance to pulmonary surfactants, TLR4 evasion, cell size, and virulence. ΔmlaF suppressor strains selected for during murine infection and isolated based on colony morphology shared a conserved ISAba11 insertion that promoted resistance to membrane stress and antibiotics and restored cell size and virulence. The suppressive insertion was in the 5’ untranslated region of ispB, which encodes an essential isoprenoid biosynthesis protein. The insertion inhibits ispB expression by an uncharacterized mechanism. Potential mechanisms include disruption of elements required for transcript stability or interference by transcriptional elements encoded in the insertion. Quantification of the GPL and LOS molecules produced by A. baumannii revealed that Mla is necessary for maintaining both GPL and LOS levels within the OM. In particular, ΔmlaF mutants accumulated particular PGLs and measured decreased LOS levels relative to the wild type. The ISAba11 insertion increased the LOS levels for the ΔmlaF mutant to offset the PGL increase and rebalanced OM-lipid homeostasis and integrity. The presence of an identical insertion in an XDR clinical isolate of A. baumannii suggests that the mechanism of modulating LOS abundance might have relevance to the evolution of antimicrobial resistance.
In contrast to restoring the cell envelope integrity, the ΔmlaF suppressor insertion did not restore TLR4 evasion to A. baumannii. The lack of significant differences in GPL between the ΔmlaF and ΔmlaF suppressor strains (Figure 5B; Table S1) suggests that the membrane differences that lead to increased TLR4 signaling in the absence of MlaF are maintained in the suppressor strain. Consistent with the suppressor strain maintaining increased TLR4 signaling, TLR4-deficient mice were no more susceptible to A. baumannii than wild-type mice. These results contrast with a previous study that found TLR4-deficient mice had 1-log higher burdens of A. baumannii RUH 2037 in the lungs (Knapp et al., 2006). In bloodstream infections, TLR4-deficient mice survived A. baumannii 17978 infection at a rate similar to that of wild-type mice, but TLR4 was essential for survival of the infection by A. baumannii HUMC1 (Lin et al., 2012). Together, these findings suggest that the role of TLR4 may be strain specific and influenced by GPL.
The ΔmlaF suppressor strain similarly maintained increased susceptibility to bacitracin. Bacitracin acts by binding undecaprenyl pyrophosphate (Und-PP), also known as the C55-isoprenoid, and preventing its recycling in the cell (TouzÉ and Mengin-Lecreulx, 2008). Maintenance of bacitracin susceptibility suggests that Und-PP may be intermediate in the suppressive mechanism. Und-PP is a lipid glycan carrier essential for multiple envelope processes of Gram-negative bacteria, including the elaboration of O-antigen and the biosynthesis of peptidoglycan, glycoproteins, and capsule (TouzÉ and Mengin-Lecreulx, 2008). Although Und-PP is required for capsule formation, there was no striking difference in capsule formation among the strains in this study (Figure S5E). The substrate of IspB, farnesyl-PP, is a shared branchpoint metabolite for quinone and Und-PP biosynthesis (Figure 5C). Therefore, we propose a model in which decreased expression of ispB reduces IspB use of farnesyl-PP, allowing increased flux to Und-PP synthesis, which restores envelope integrity. This model is consistent with data demonstrating that inhibiting isoprenoid biosynthesis upstream of farnesyl-PP with fosmidomycin inhibits growth of the wild-type strain in SDS/EDTA, whereas the suppressive insertion inhibiting ispB expression helps restore growth in SDS/EDTA to the wild-type strain (Figures 5D–5I). Also consistent with this model, A. baumannii and E. coli strains selected for improved envelope integrity have been isolated with mutations in ispB, which presumably lead to decreased IspB activity (Geisinger et al., 2018; Jeong et al., 2012).
However, the data included here do not exclude alternative models. For example, it is possible that the increased LOS levels and high GPL levels for the OM of the suppressor strain does not restore exclusion of bacitracin. Decreased expression of IspB and decreased quinone levels could affect periplasmic disulfide bond formation through DsbA and DsbB or through other metabolic changes. Consistent with reduced quinone levels, the IM fraction of the suppressor mutant has reduced NADH dehydrogenase activity (Figures S4C and S4D), which is quinone dependent. However, the ISAba11 insertion in the ispB 5′ untranslated region does not lead to reduced membrane potential in the wild-type or ΔmlaF background (Figure S5F). These data show that the suppressor strain’s increased resistance to aminoglycosides, such as gentamicin, is not through reduced membrane potential as shown for other organisms (Crabbé et al., 2019; Mates et al., 1982).
The present study confirms a link between maintaining OM lipid asymmetry and regulating the level of LOS and LPS molecules on the surface of Gram-negative bacteria (Malinverni and Silhavy, 2009; May and Silhavy, 2018; Sutterlin et al., 2016). Although our data do not define this regulatory mechanism, the results are consistent with GPL accumulation in the OM causing a decrease in LOS biosynthesis for A. baumannii. Understanding the regulation of LOS biosynthesis in A. baumannii is of particular interest given that this organism genetically removes the LOS biosynthesis pathway as a mechanism to resist the last-resort antibiotic, colistin (Boll et al., 2016; Moffatt et al., 2010). The mutants characterized here did not demonstrate any difference in colistin resistance (Table 1), suggesting that reduced LOS levels in the ΔmlaF strain are not sufficient to increase colistin resistance. However, this work identifies a clear relationship among the Mla system, envelope homeostasis, isoprenoid biosynthesis, and antibiotic resistance in A. baumannii. Therefore, understanding how the Mla system and isoprenoid biosynthesis are regulated to influence LOS biosynthesis and GPL homeostasis will be important to comprehend the mechanisms for the evolution of multidrug resistance.
The mechanisms by which increased Und-PP would promote LOS biosynthesis are unclear, although it is well known that Und-PP influences peptidoglycan assembly and remodeling. The role described for Und-PP in LPS biosynthesis is for elaboration of the O-antigen and the enzymatic addition of amino arabinose to lipid-A-core phosphates (TouzÉ and Mengin-Lecreulx, 2008). A. baumannii lacks O-antigens but decorates lipid A phosphates with a variety of cationic moieties, including galactosamine (Klein et al., 2019). It is conceivable that the galactos-amine-addition machinery requires Und-PP-Gal for decorating lipid A core structures. Future work is necessary to address the mechanism by which decreased ispB expression promotes A. baumannii LOS synthesis and resistance to antibiotics and host stresses.
Together, the results presented here show that the A. baumannii Mla system is critical for resisting antibiotics and membrane stresses, including pulmonary surfactant, and for maintenance of cell size and cellular envelope integrity. In this study, ISAba11 insertions in the 5′ untranslated region of ispB were independently isolated from two infections selecting for survival of mlaF-deficient A. baumannii in the lung. The identical insertion is also present in the A. baumannii XDR strain AB030. ISAba11 has been found in multiple sites in the genome, indicating a lack of insertion site specificity (Moffatt et al., 2011; Rieck et al., 2012). Therefore, independent isolation of this ISAba11 insertion in murine infection and an XDR clinical isolate suggests that A. baumannii experiences similar selective pressures to modulate isoprenoid biosynthesis in both experimental infection models and clinical infections. The fact that this ISAba11 insertion confers increased fitness to the wild-type strain in the presence of membrane stress and the antibiotic fosmidomycin suggests that this insertion could be selected for clinically when A. baumannii is confronted with host-imposed membrane stress and certain antibiotics. In recent years fosmidomycin combination therapies have been used in clinical trials to treat malaria infections (Guggisberg et al., 2016; Mombo-Ngoma et al., 2018), representing one possible mechanism by which this insertion could be selected clinically. Future work may identify additional mechanisms by which modulating isoprenoid biosynthesis could be selected for in a clinical setting.
Importantly, this work suggests bacteria could evolve to resist antibiotics targeting the Mla system and that alleles conferring that resistance are present in the clinical setting. These findings suggest that any antimicrobials targeting the Mla system should be employed as part of a combination-therapy program or other strategy to minimize selection of suppressive genetic rearrangements. Together, these results demonstrate that A. baumannii genetic and metabolic plasticity allows it to rapidly evolve resistance to antibiotics and host-imposed stresses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric Skaar (eric.skaar@vumc.org).
Materials availability
Materials generated in this study will be freely available upon request to Eric Skaar.
Data and code availability
Whole genome sequencing data is available in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProject: PRJNA656143.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and growth
All strains and plasmids used in this study are listed in the Key Resources Table. Strains were grown in LB or on LB plates including 1.5% w/v agar. Antibiotics were included at the following concentrations: carbenicillin, 50 mg/L for E. coli and 75 mg/L for A. baumannii; tetracycline, 10 mg/L; kanamycin, 40 mg/L; chloramphenicol, 15 mg/L; sulfamethoxazole, 100 mg/L. Each overnight culture was initiated from a single colony and incubated at 37°C with shaking for 8-16 h. Growth curves contained 0.1 mL/well in flat bottom 96-well plates and were inoculated with 1 μL overnight culture and incubated at 37°C with shaking. For experiments including SDS/EDTA, multiple EDTA concentrations were included in each experiment due to variability in bacterial growth inhibition. Growth was monitored by optical density at 600 nm (OD600) in a BioTek plate reader (Winooski, VT). Overnight cultures of the inducible ispB strain included 2 mM IPTG.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| A. baumannii 17978 (wild-type; WT) | ATCC | Skaar 13-89 |
| A. baumannii 17978 ΔmlaF(A1S_3103)::Kn | This manuscript | Skaar 47-65 |
| A. baumannii ABUW AB5075 | (Gallagher et al., 2015; Jacobs et al., 2014) | Skaar 56-43 |
| A. baumannii ABUW AB5075 mlaF136::T26(Tc) | (Gallagher et al., 2015) | Skaar 56-44 |
| A. baumannii ABUW AB5075 mlaF188::T26(Tc) | (Gallagher et al., 2015) | Skaar 56-45 |
| A. baumannii ABUW AB5075 mlaF160::T26(Tc) | (Gallagher et al., 2015) | Skaar 56-46 |
| A. baumannii 17978 pWH1266 | This manuscript | Skaar 39-16 |
| A. baumannii 17978 pLDP9 | This manuscript | Skaar 39-17 |
| A. baumannii 17978 ΔmlaF::Kn pWH1266 | This manuscript | Skaar 39-18 |
| A. baumannii 17978 ΔmlaF::Kn pLDP9 | This manuscript | Skaar 39-19 |
| A. baumannii 17978 pLDP29 | This manuscript | Skaar 46-42 |
| A. baumannii 17978 ΔmlaF::Kn pLDP29 | This manuscript | Skaar 46-43 |
| A. baumannii 17978 pLDP35 | This manuscript | Skaar 56-41 |
| A. baumannii 17978 ΔmlaF::Kn pLDP35 | This manuscript | Skaar 46-44 |
| A. baumannii 17978 pLDP28 | This manuscript | Skaar 45-49 |
| A. baumannii 17978 ΔmlaF::Kn pLDP28 | This manuscript | Skaar 45-50 |
| A. baumannii 17978 ΔmlaF::Kn ACX60_03855::ISAba11 (suppressor 1) | This manuscript | Skaar 47-66 |
| A. baumannii 17978 ΔmlaF::Kn ACX60_03855::ISAba11 (suppressor 2) | This manuscript | Skaar 47-67 |
| A. baumannii 17978 ΔmlaF::Kn ACX60_03855::ISAba11 (reconstructed suppressor) | This manuscript | Skaar 56-40 |
| A. baumannii 17978 ΔmlaF::Kn att::Tn7(lacI-Plac-ispB) ΔispB::Tc | This manuscript | Skaar 56-39 |
| A. baumannii 17978 ACX60_03855::ISAba11 (reconstructed suppressor in WT background) | This manuscript | Skaar 56-42 |
| A. baumannii 17978 pLDP53 | This manuscript | Skaar 59-09 |
| A. baumannii 17978 ΔmlaF::Kn ACX60_03855::ISAba11 pLDP29 | This manuscript | Skaar 59-10 |
| A. baumannii 17978 ΔmlaF::Kn ACX60_03855::ISAba11 pLDP53 | This manuscript | Skaar 59-11 |
| A. baumannii 17978 ΔmlaF::Kn pLDP52 | This manuscript | Skaar 59-14 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FM4-64 | ThermoFisher (Waltham, MA) | T13320 |
| Critical Commercial Assays | ||
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel - 32 Plex | EMD Millipore (Burlington, MA) | MCYTMAG70PMX32BK |
| Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit | Molecular Probes/Invitrogen (Carlsbad, CA) | P20495 |
| BacLight Membrane Potential Kit | Invitrogen (Carlsbad, CA) | B34950 |
| Deposited Data | ||
| Whole genome sequencing of Skaar 47-65, Skaar 47-66, and Skaar 47-67 A. baumannii strains | This manuscript | NCBI SRA BioProject: PRJNA656143 |
| Experimental Models: Cell Lines | ||
| HEK-Blue hTLR4 | Invivogen (San Diego, CA) | hkb-htlr4 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | Jackson Laboratory (Bar Harbor, ME) | # 664 |
| B6.B10ScN-Tlr4lps-del/JthJ | Jackson Laboratory (Bar Harbor, ME) | # 7227 |
| Oligonucleotides | ||
| Listed in Table S3 | ||
| Recombinant DNA | ||
| CarbR; allelic exchange vector with sucrose resistance | (Hoang et al., 1998) | pFLP2 |
| CarbR; contains a non-polar kanamycin insertion | (Ménard et al., 1993) | pUC18-K1 |
| KnR; mobilization helper plasmid | (Figurski and Helinski, 1979) | pRK2013 |
| CarbR; helper plasmid encoding the site-specific TnsABCD Tn7 transposition pathway | (Kumar et al., 2010) | pTNS2 |
| mini-Tn7-AmpR on a suicide vector containing the R6Kγ-ori | (Carruthers et al., 2013) | pKNOCK-mTn7-Amp |
| Acinetobacter – E. coli shuttle vector | (Hunger et al., 1990) | pWH1266 |
| Contains an FRT-flanked kanamycin resistance gene | (Datsenko and Wanner, 2000) | pKD4 |
| pMMB67EH with RecAb system | (Tucker et al., 2014) | pAT02 |
| Contains regions upstream and downstream of mlaF to create pLDP8 | This manuscript | pCRBlunt-3103::Km |
| pFLP2-ΔmlaF::Kn | This manuscript | pLDP8 |
| pWH1266-PmlaF-mlaF | This manuscript | pLDP9 |
| pWH1266-PrpsA-pldA | This manuscript | pLDP28 |
| pWH1266-PrpsA | This manuscript | pLDP29 |
| pWH1266-PrpsA-mlaF | This manuscript | pLDP35 |
| pWH1266-PrpsA-ispB | This manuscript | pLDP53 |
| Contains an FRT-flanked tetracycline resistance gene from A. baumannii AYE | This manuscript | pLDP55 |
| pFLP2-ΔispB::Tc | This manuscript | pLDP56 |
| pFLP2-suppIS (upstream of ispB containing ISAba11) | This manuscript | pLDP70 |
| pKNOCK-mTn7-Amp-lacI-Plac-ispB | This manuscript | pLDP77 |
| Software and Algorithms | ||
| Prism 7 | GraphPad (La Jolla, CA) | N/A |
| Canvas X 16 | Canvas GFX | N/A |
| Breseq | (Deatherage and Barrick, 2014) | N/A |
| ISFinder | (Siguier et al., 2006) | https://www-is.biotoul.fr |
| Bowtie2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| Integrated Genomics Viewer (IGV) | (Robinson et al., 2011) | http://software.broadinstitute.org/software/igv/ |
| phiSITE Promoter Hunter | (Klucar et al., 2010) | http://www.phisite.org/ |
| ImageJ Fiji | (Schindelin et al., 2012) | https://imagej.net/Welcome |
| Other | ||
| Curosurf (poractant alfa) | Chiesi (Cary, NC) | NDC 10122-510-01 |
| Infasurf (calfactant) | ONY Inc. (Amherst, NY) | N/A |
Animal models
Mice were purchased from Jackson Laboratory and infected at 6-10 weeks of age. Wild-type mice were C57BL/6J and TLR4−/− mice were B6.B10ScN-Tlr4lps-del/JthJ. Male mice were used for imaging mass spectrometry, all other mice were female. Mice were housed with a 12 h light-dark cycle with food and water provided ad libitum. All animal experiments were approved by the Vanderbilt University Medical Center (VUMC) Institutional Care and Use Committee and conform to policies and guidelines established by VUMC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association.
HEK-Blue hTLR4 cells
HEK-Blue hTLR4 cells were purchased from Invivogen (San Diego, CA) and were maintained according to manufacturer’s instructions.
METHOD DETAILS
Construction of plasmids, allelic exchange mutants, and genetic constructs
Oligonucleotides used in this study are listed in the Table S3. DNA was amplified with 2X Phusion Master Mix (ThermoFisher, Waltham, MA) or GoTaq Green Master Mix (New England Biolabs (NEB), Ipswich, MA). All restriction enzymes were from NEB. To construct ΔmlaF::Kn, 1000-bp regions flanking mlaF were amplified from A. baumannii 17978 genomic DNA, and the kanamycin resistance gene aph was amplified from pUC18K1 (Ménard et al., 1993). The PCR products were joined together using overlap extension PCR and cloned using the Zero Blunt PCR Cloning Kit (Invitrogen, Carlsbad, CA) to generate pCRBlunt-3103::Km. The ΔmlaF::Kn region was then subcloned into pFLP2 using XbaI and SacI. A. baumannii ΔmlaF::Kn was generated by electroporating pLDP8 into A. baumannii 17978 and selecting for KnR, then screening for CarbR and sucroseS. Sucrose sensitive and PCR-confirmed merodiploids were counter selected on LB agar plates containing 10% sucrose and screened for KnR and CarbS. Complementation vectors were constructed in pWH1266 (Hunger et al., 1990) digested with BamHI and SalI. Constitutive complementation vectors were constructed using the rpsA promoter; pLDP29 was generated by digesting pLDP28 with XhoI and KpnI, then re-ligating with the oligonucleotide LP148 by HiFi DNA Assembly Cloning Kit (NEB). Reconstruction of the suppressor mutation was performed by amplifying the ispB region using primers LP270 and LP273 using genomic DNA from strain 47-66 and generating pLDP70 using NEB HiFi. Merodiploids were selected for CarbR and screened for CarbS after sucrose counter selection. To construct the inducible ispB construct, lacI and the promoter including lacO were amplified from pAT02 using primers LP342 and LP356. ispB was amplified from A. baumannii 17978 genomic DNA using primers LP357 and LP345 and the DNA fragments were ligated into pKNOCK-mTn7-Amp digested with BamHI and KpnI by HiFi assembly. The mini-Tn7 was introduced into A. baumannii ΔmlaF::Kn as previously described (Carruthers et al., 2013), using carbenicillin as selection and chloramphenicol to counterselect against E. coli. To construct ΔispB::Tc, the excisable tetracycline resistance cassette was amplified from pLDP55, which was generated from replacing the kanamycin resistance cassette in pKD4 with the tetracycline resistance marker from A. baumannii AYE. pLDP56 was then generated using NEB HiFi. ΔispB::Tc was generated in the ΔmlaF::Kn inducible ispB background in the presence of 5 mM IPTG and using TetR to select for integration of pLDP56 into the genome. All strains were confirmed by PCR and checked for maintenance of the large conjugative plasmid pAB3 by sulfamethoxazole resistance.
HEK-Blue hTLR4 reporter assays
Overnight A. baumannii cultures were diluted 1:1000 in 10 mL LB in a 50-mL conical tube and incubated at 37°C with shaking for 3.5 h. A. baumannii were then washed twice in cold PBS and normalized by optical density, serially diluted and spot plated for CFU enumeration. A. baumannii was heat-inactivated at 75°C for 45 min prior to using in HEK-Blue hTLR4 assays according to manufacturer’s instructions, incubated for 16 h and absorbance at 620 nm was measured.
FM4-64 staining and visualization
Overnight cultures were diluted 1:1000 into 10 mL LB in 50-mL conical tubes and incubated at 37°C with shaking for 3.5 h. Freshly prepared N-(3-triethylammoniumpropyl)-4-(6-(4-(diethyamino)-phenyl)pyridinium dibromide (FM4-64) was added to 0.5 mg/mL and cultures were incubated an additional hour at 37°C with shaking. Live cells were aliquoted to agar pads (1% agarose in PBS) and dried briefly before applying a coverslip and imaging using brightfield and red fluorescence microscopy with a 100X objective on an Olympus BX60 microscope. Images were analyzed using ImageJ Fiji (Schindelin et al., 2012). First, individual cells were identified from brightfield images using background subtraction with a rolling ball radius, separating colors and making binary, and applying the watershed tool to divide cells in close proximity. This matrix was then applied to the corresponding red fluorescent image to quantify mean fluorescence intensity per cell.
Antibiotic and stressor susceptibility
Top agar including 0.8% agar and 0.8% NaCl was seeded with 0.1 mL overnight culture and applied to an LB agar plate. Five μL antibiotic stocks were applied to sterile filter paper disks. After overnight incubation, the diameter of clearing was measured in mm. Antibiotic stocks were at the following concentrations. chloramphenicol, 20 mg/mL; tetracycline, 10 mg/mL; streptomycin, 15 mg/mL; gentamicin, 50 mg/mL; erythromycin, 10 mg/mL; colistin, 2 mg/mL; polymixin B, 2 mg/mL; daptomycin, 100 mg/mL; carbenicillin; 75 mg/mL; meropenem, 20 mg/mL; bacitracin, 50 mg/mL; copper chloride, 1 M; acetic acid, 33%; hydrochlorous acid, 1 N; hydrogen peroxide, 30%; paraquat, 0.1 M.
Murine model of A. baumannii pneumonia
Mice were infected intranasally with approximately 3 X 108 CFU of A. baumannii 17978 and CFU were enumerated as previously described (Palmer et al., 2019). Mice were euthanized at 8 h, 12 h, 24 h or 36 h post infection and lungs, livers, and spleens were removed aseptically. For the Luminex assay, lungs were harvested 8 h post infection, homogenized, and frozen at −80 prior to cytokine/chemokine analysis. Lung homogenates were thawed, and protease inhibitor cocktail (Sigma) was added at approximately 1.100 dilution. The protein concentration was determined by (BCA) assay using bovine serum albumin (BSA) as a standard and then normalized to 10 mg/mL prior to using the Luminex kit (Millipore) as instructed but using 1/3 bead concentration recommended by the manufacturer. Samples were run on the Luminex Flexmap 3D platform (Luminex, Austin TX).
Histological analysis of infected murine lungs
For histology, lungs were harvested at 8 h post infection and inflated with 0.8 mL 10% formalin and fixed in 10% formalin prior to embedding in paraffin blocks. Tissue sections (5 μm) were stained with hematoxylin and eosin stain to assess inflammation using the Leica BOND-MAX autostainer (Wetzlar, Germany). Slides were examined and scored by a pathologist who was blinded to the experimental groups. Inflammation was scored according to the following scale. 0, no inflammatory infiltrates observed in the examined sections; 1, rare perivascular inflammatory infiltration; 2, few perivascular inflammatory infiltrates with rare extension into the interstitium; 3, moderate perivascular inflammatory infiltration and with multifocal interstitial infiltration; 4, marked perivascular inflammatory infiltrates with extensive involvement of the interstitium and alveolar spaces.
Pulmonary surfactant survival assay
Infasurf was from ONY Inc. (Amherst, NY) and Curosurf was from Chiesi (Cary, NC). Overnight cultures of A. baumannii were diluted 1.1000 in 10 mL LB in a 50-mL conical tube and incubated at 37° C with shaking for 3.5 h. A. baumannii were then washed twice in cold PBS and normalized to the same CFU/mL by optical density. A. baumannii was then mixed 1.1 with pulmonary surfactant or PBS and incubated at 37°C for 1.5 h prior to serial dilution and spot plating for CFU enumeration.
Lipid imaging mass spectrometry
Lungs were harvested at 24 h post infection from mock infected or A. baumannii infected mice, inflated with 1.3% carboxymethyl-cellulose (CMC), and flash frozen on dry ice. Frozen lungs were sectioned at −25°C to 10 μm thickness using a Leica CM3050S cryostat (Leica Microsystems, Bannockburn, IL, USA). Tissue sections were thaw-mounted onto indium tin oxide coated glass slides (Delta Technologies, Loveland, CO, USA). Salts were removed by washing with 50 mM ammonium formate for 30 s (Angel et al., 2012). 1,5-Diaminonaphthalene (DAN) matrix was sublimated onto the slide surface (~1.0 mg/cm2) by mounting slides onto a condenser suspended in a sublimation apparatus using approximately 500 mg of DAN at 130°C and 24 mTorr for 3.5 min (Hankin et al., 2007).
Images were acquired using a prototype timsTOF Pro (Bruker Daltonik, Bremen, Germany) equipped with a dual ESI/MALDI source with a SmartBeam 3D 10 kHz frequency tripled Nd:YAG laser (355 nm) (Spraggins et al., 2019). The instrument was operated in qTOF mode where images were collected at 20 μm spatial resolution with a beam scan of 18 μm using 500 laser shots per pixel and 48% laser power. Data were collected in positive ion mode from m/z 400 – 1,500. Lipid identification was determined based on mass accuracy (mass tolerance < 5 ppm) using the LIPIDMAPS lipidomics gateway (lipidmaps.org). Imaging data were visualized using SCiLS Lab Version 2019 (Bruker Daltonics, Bremen, Germany). Post-acquisition, matrix was washed away with ethanol prior to hematoxylin and eosin (H&E) staining on the same tissue sections used for imaging mass spectrometry. H&E images were then acquired using a Leica SCN400 slide scanner (Leica Microsystems, Bannockburn, IL, USA) using 10X magnification.
Genome sequencing and assembly
Genomic DNA was extracted from wild-type, ΔmlaF suppressor 1, and ΔmlaF suppressor 2 strains using the Wizard Genomic DNA Purification Kit (Promega). DNA was quantified using Quant-iT dsDNA Assay Kit (Thermo), and sequenced by Genewiz using Illumina HiSeq, High Output 2×150bp. Adaptor sequences were removed using the FASTX-Toolkit FASTQ/A Clipper with a minimum length of 100 bp (Gordon and Hannon, 2010). Genome sequences of the forward reads were mapped to the ATCC A. baumannii 17978 accession NZ_CP012004 and NZ_CP012005 (Weber et al., 2015) using breseq version 30.0 (Deatherage and Barrick, 2014) with default settings. The ISAba11 insertion was identified using ISfinder (Siguier et al., 2006). The predicted transcription start site was determined by mapping paired-end reads from wild-type replicate 1 in a previously published dataset (Wang et al., 2019) to the ATCC A. baumannii 17978 accession NZ_CP012004 genome using Bowtie2 (Langmead and Salzberg, 2012). The alignment map was converted to a BAM file, sorted, and indexed using Samtools (Li et al., 2009) and visualized with Integrated Genomics Viewer (IGV) (Robinson et al., 2011). The predicted −35 and −10 sites were determined using phiSITE Promoter Hunter (Klucar et al., 2010).
Quantitative RT-PCR
Overnight cultures were diluted 1:1000 into 10 mL LB in 50-mL conical tubes and incubated at 37°C with shaking for 3.5 h. RNA was purified and qRT-PCR was performed as previously described (Lonergan et al., 2019).
Membrane potential
Overnight cultures were diluted 1:1000 into 10 mL LB in 50-mL conical tubes and incubated at 37°C with shaking for 3.5 h. Cultures were then diluted 1:1000 into 10 mL PBS, and 1 mL of diluted sample was used for each condition. Membrane potential was assessed using the BacLight Membrane Potential Kit from ThermoFisher according to manufacturer’s instructions. Fluorescence was measured using an LSRFortessa by Becton Dickinson (Franklin Lakes, NJ).
Membrane isolation and fractionation
Membranes were isolated, fractionated, and characterized similarly to previously described methods (Cian et al. 2020). Overnight cultures were grown in 50 mL LB and diluted 1:100 into 1 L LB in a 2-L flask and incubated at 37°C with shaking until OD600 of 0.6 to 0.8. Pellets were re-suspended in 12.5 mL of 50% sucrose 10 mM Tris solution to which 180 μL of 10 mg/mL of lysozyme was added and stirred for 2 minutes. Next, 12.5 mL of 1.5 mM EDTA was then added and stirred for 7 minutes. The cells were collected in a 50-mL centrifugation tube and centrifuged for 10 min at 9,000-11,000 x g to pellet. The pellets were resuspended in 25 mL of a 20% sucrose 10 mM Tris solution. Fifty-five μL of MgCl2, 1 μL of benzonase, and 1 μL of protease inhibitor were added in preparation for complete lysis by a pressurized cell homogenizer. After several passes the lysed cells were centrifuged at 6,159 x g, and the supernatant collected to be ultracentrifuged. The membranes were centrifuged overnight at 184,500 x g. The following morning the membrane pellets were homogenized in 1 mL of 20% sucrose. One hundred fifty μL was saved for total membrane analysis while 850 μL was used for fractionation. An adjusted sucrose gradient was prepared as follows: 2 mL of 73% sucrose, 4 mL of 45% sucrose, the sample, and the void filled in by 20% sucrose. Forty-five percent sucrose was used in place of the 53% described in previous protocols to allow for the separation of OM; this strain of Acinetobacter the OM was not sufficiently dense to flow through the 53% sucrose (Cian et al., 2020). The sucrose gradients were then centrifuged overnight at 288,000 x g using a SW-41 swinging bucket rotor in a Beckman ultracentrifuge. The lower density IM fractions were collected from the upper portion of the gradient at the interface of the 20% and the 45% sucrose. The higher density OM was collected at the lower portion of the gradient at the interface of the 45% and the 73% sucrose. These membranes were collected and washed in 10 mM Tris and centrifuged at 184,500 x g for 1 hour. The pellets were then collected, and homogenized in 300 μL of 10 mM Tris, and stored at −20°C until analysis by LC-MS/MS.
GPL extraction and LC-MS/MS
The GPLs were extracted from the total and OM fractions using Bligh-Dyer method (Bligh and Dyer, 1959). For total membranes, the equivalent of 1.5 mg of protein was extracted into chloroform and dried under N2. The dried total membrane GPLs were resuspended in 150 μL of mobile phase A [CHCl3/CH3OH/NH4OH (800:195:5, vol/vol/vol)] for the LC-MS/MS analysis. Substantially less material was recovered for the OM fractions, so 0.5 mg of protein was extracted, dried, and resuspended in 50 μL of mobile phase A. The data for the total membranes and OM were acquired independently and represent 3-4 independent biological samples per A. baumannii genotype. Samples were delivered to an Applied Biosystems Sciex API 4000 Triple Quad Mass Spectrometer using a Waters Acquity H-class UPLC system interfaced with an Agilent Zorbax Rx-SIL column (2.1 × 100 mm, 1.8 μm). Analytes were ionized by electrospray in [M-H]-1 mode with a voltage of −4.5 kV. The source temperature was 450°C. Nitrogen was used as the curtain gas (setting 10), nebulizer gas (setting 20) and turbo gas (setting 20). MS/MS was performed using nitrogen as the collision gas (setting 4.0). The declustering, entrance, and collision cell exit potential was −120, −10, and −15, respectively. Retention of acyl-PGL, PGL, CL, and PE was achieved at a flow rate of 0.35 ml/min using mobile phase A and mobile phase B [CHCl3/CH3OH/H2O/NH4OH (600:340:50:5, vol/vol/vol/vol). A three-step gradient used started at 0% B for 1 min, continued at 0%–50% B over the next 3 min, was held at 50% B for 4 min, re-turned to starting conditions in 0.1 min, and was allowed to equilibrate for an additional 3 min, giving a total run time of ~11.1 min. The following precursor > product ion transitions were monitored [multiple reaction monitoring (MRM)]: 955 > 253 a-PGL, 958 > 255 a-PGL, 986 > 255 a-PGL, 1012 > 255 a-PGL, 719 > 253 PGL, 733 > 267 PGL, 747 > 281 PGL, 759 > 267 PGL, 773 > 281 PGL, 1322 > 253 CL, 1348 > 253 CL, 1376 > 281 CL, 1402 > 281 CL, 688 > 253 PE, 702 > 267 PE, 714 > 253 PE, 716 > 281 PE, 742 > 281 PE. In the case of acyl-PGL, PGL, and PE molecules, singly charged [M-H]– precursor > product ion transitions were monitored. For CL, doubly charged [M-2H]2- precursor ions were monitored because they were more abundant than singly charged ions. Collision energies ranged from −35 to −40V for the individual analytes. The data were acquired with the Sciex Analyst software version 1.6.2 (Applied Biosystems, Foster City, CA, USA).
The flow through needle delivered 1 μL and 10 μL injections for the total and OM fractions, respectively. The retention times for the major phospholipid families on the silica column were confirmed and standard curves were generated using commercial standard from Avanti Polar lipids. These included 1-palmitoyl-2- oleoyl-sn-glycero-3-phosphoethanolamine, (C16:0, C18:1) PE, m/z 716.7, 1-palmitoyl-2- oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol), (C16:0, C18:1) PGL, m/z 747.6, and 1’,3′-bis[1-palmitoyl-2-oleoyl-sn-glycero-3-phospho]-glycerol, (C16:0, C18:1, C16:0, C18:1) [M-H]– CL m/z 1404 or [M-2H]-2 m/z 702. Integrated peak areas were plotted against standard concentration to generate a linear equation. Peak area values for the targeted precursor > product ion transitions were applied to the curve to determine the ng of the PE, PGL, and acyl-PGL per μl of sample (ng/μl). The starting volume of the resuspended dried extract (150 μL for the total membranes and 50 μL for the OMs) was accounted for in our calculation to determine the mass of the individual GPL in each extract (Figures 4A, 4B, and S4A).
NADH dehydrogenase activity
A total of 50 μg of protein from the IM and OM fractions of A. baumannii was incubated with 140 nmol NADH to assay NADH dehydrogenase activity. NADH oxidation was measured by monitoring the absorbance of the sample at 340 nm (A340) for 5 min. To graph the change overtime, the absorbance for each time point was normalized to the 0 min time point for each sample. The slope was determined from raw data.
LOS extraction and analysis
A. baumannii strains were inoculated into 8 mL LB and incubated with shaking at 37°C overnight. The following morning, cell cultures were diluted 1:100 into 8 mL of LB and incubated at 37°C on a culture rotator until reaching mid-log phase (OD600 of 0.6-0.8). Each culture was then normalized to an OD600 of 2.5, which was confirmed to produce an equal number of CFU, and the required volume was aliquoted into a 2-mL microcentrifuge tube. The bacterial cultures were pelleted by centrifugation at 18,400 x g for 5 minutes. The supernatant was carefully discarded and each cell pellet was resuspended in 200 μL 0.2% SDS solution by vigorous pipetting without vortexing. Samples were denatured via a boiling lysis method and allowed to re-equilibrate to room temperature. Once cooled, samples were treated with 5 μL of Proteinase K solution (20 mg/mL) and incubated in a 59°C water bath overnight. Utilizing a hot-phenol technique, 180 μL of pre-warmed (68°C) Tris-saturated phenol (pH 8.0) was added to each sample and vortexed for approximately 5-10 s. Samples were then incubated in a 68°C water bath for 15 minutes and then immediately transferred to an ice-cold water bath for 10 min. To achieve proper phase separation, samples were centrifuged at 1,900 x g for 10 minutes at 4°C. The upper aqueous phase containing the lipids (~150 μL) was carefully extracted using a P200 pipette and placed in a clean microcentrifuge tube while the remaining solution was discarded. For SDS-PAGE, 30 μL of lipid samples was diluted into 10 μL of 4X Laemmli buffer (Bio-Rad). Lipid samples (20 μL sample per well) were then separated on a 4%–20% Tris-glycine gradient gel (Bio-Rad) and electrophoresed at 100V for 45 minutes. Gels were stained according to Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (Molecular Probes) and the relative fold change in band intensity was quantified using BioRad Image Lab 6.0.1 software.
Capsule staining
Colonies of A. baumannii were suspended in 10 μL 1% Congo red and spread in a thin film on a glass slide. After drying, the slide was saturated with Maneval’s stain, incubated for 5 min, then gently washed with dH2O and blotted dry (Maneval, 1941). Capsule was then visualized by brightfield microscopy with a 100X objective on an Olympus BX60 microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data represent the mean of biological duplicate or triplicate ± SEM, unless otherwise indicated. Graphs were generated using GraphPad Prism 7 or Prism 8 (La Jolla, CA). Significance was determined by the tests described in the figure legend and is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. All figures were generated in Canvas X16.
Supplementary Material
Highlights.
mlaF helps A. baumannii resist pulmonary surfactants and antibiotics
Infection selects for genetic rearrangements that alter isoprenoid gene expression
A suppressive insertion restores virulence and antibiotic resistance to ΔmlaF
The insertion is conserved in an extensively drug-resistant (XDR) clinical isolate
ACKNOWLEDGMENTS
We thank members of the Skaar Laboratory for critical reading of the manuscript. We thank Brittany L. Nairn for constructing pCRBlunt-3103::Km, Erin R. Green for capsule staining, Brittany Matlock of the VUMC Flow Cytometry Shared Resource for assistance with the membrane potential assays, Margaret Allaman for assistance with the Luminex assay, and Sebastian Joyce for art in the graphical abstract. This work was funded by National Institutes of Health grants R01 AI101171 to E.P.S., T32 HL094296 to Dr. Timothy Blackwell (L.D.P.), F32 AI122516 and K99 HL143441 to L.D.P., P20 GM10344 and R01 AI139248 to Z.D.D., and R01 AI138581 to J.M.S. and E.P.S, and Vanderbilt Digestive Disease Research Center (VDDRC) P30 DK058404. Additional funding was provided by the Ernest W. Goodpasture professorship to E.P.S. and the Parker B. Francis fellowship program to L.D.P.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108129.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, and van den Berg B (2017). Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol 2, 1616–1623. [DOI] [PubMed] [Google Scholar]
- Angel PM, Spraggins JM, Baldwin HS, and Caprioli R (2012). Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem 84, 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K, Fujisaki S, Nishimura Y, Nishino T, Okada K, Nakagawa T, Kawamukai M, and Matsuda H (1994). The identification of Escherichia coli ispB (cel) gene encoding the octaprenyl diphosphate synthase. Biochem. Biophys. Res. Commun 202, 340–345. [DOI] [PubMed] [Google Scholar]
- Awai K, Xu C, Tamot B, and Benning C (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. USA 103, 10817–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarda BI, Zielke RA, LeVan A, Jerse AE, and Sikora AE (2019). Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog. 15, e1007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier SP, Son S, and Surette MG (2018). The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J. Bacteriol 200, e00156–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, and Murphy RC (2011). MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev 111, 6491–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, and Piddock LJ (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol 13,42–51. [DOI] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Boll JM, Crofts AA, Peters K, Cattoir V, Vollmer W, Davies BW, and Trent MS (2016). A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 113, E6228–E6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, and Payne SM (2014). The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect. Immun 82, 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers MD, Nicholson PA, Tracy EN, and Munson RS Jr. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS ONE 8, e59388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N, and Riley LW (2007). A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong ZS, Woo WF, and Chng SS (2015). Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol 98, 1133–1146. [DOI] [PubMed] [Google Scholar]
- Cian MB, Giordano NP, Masilamani R, Minor KE, and Dalebroux ZD (2019). Salmonella enterica serovar Typhimurium uses PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect. Immun 88, e00758–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian MB, Giordano NP, Mettlach JA, Minor KE, and Dalebroux ZD (2020). Separation of the cell envelope for gram-negative bacteria into inner and outer membrane fractions with technical adjustments for Acinetobacter baumannii. J. Vis. Exp (158) 10.3791/60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé A, Ostyn L, Staelens S, Rigauts C, Risseeuw M, Dhaenens M, Daled S, Van Acker H, Deforce D, Van Calenbergh S, and Coenye T (2019). Host metabolites stimulate the bacterial proton motive force to enhance the activity of aminoglycoside antibiotics. PLoS Pathog. 15, e1007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, and Wren BW (2007). Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun 75, 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, and Miller SI (2014). PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. USA 111, 1963–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, and Miller SI (2015). Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, and Barrick JE (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol 1151, 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, and Vale RD (2017). Architectures of lipid transport systems for the bacterial outer membrane. Cell 769, 273–285.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan B, Low WY, Liu X, and Chng SS (2019). Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-Mla system. Biochemistry 58, 114–119. [DOI] [PubMed] [Google Scholar]
- Figurski DH, and Helinski DR (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, and Manoil C (2015). Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol 197, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Mortman NJ, Vargas-Cuebas G, Tai AK, and Isberg RR (2018). A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 14, e1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Huo W, Hernandez-Bird J, and Isberg RR (2019). Acinetobacter baumannii: Envelope determinants that control drug resistance, virulence, and surface variability. Annu. Rev. Microbiol 73, 481–506. [DOI] [PubMed] [Google Scholar]
- Gordon A, and Hannon GJ (2010). FASTX-toolkit: FASTQ/A short-reads pre-processing tools.
- Guggisberg AM, Sundararaman SA, Lanaspa M, Moraleda C, González R, Mayor A, Cisteró P, Hutchinson D, Kremsner PG, Hahn BH, et al. (2016). Whole-genome sequencing to evaluate the resistance landscape following antimalarial treatment failure with fosmidomycin-clindamycin. J. Infect. Dis 214, 1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JA, Barkley RM, and Murphy RC (2007). Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom 18, 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, and Schweizer HP (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Hong M, Gleason Y, Wyckoff EE, and Payne SM (1998). Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun 66, 4700–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, and Skaar EP (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8, e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YM, Munguia J, Miao Y, Nizet V, and McCammon JA (2019). Docking simulation and antibiotic discovery targeting the MlaC protein in Gram-negative bacteria. Chem. Biol. Drug Des 93, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, et al. (2019). Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol 4, 1692–1705. [DOI] [PubMed] [Google Scholar]
- Hunger M, Schmucker R, Kishan V, and Hillen W (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87, 45–51. [DOI] [PubMed] [Google Scholar]
- Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, et al. (2014). AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio 5, e01076–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Kim SH, Han SS, Kim MH, and Lee KC (2012). Changes in membrane fatty acid composition through proton-induced fabF mutation enhancing 1-butanol tolerance in E. coli. J. Korean Phys. Soc 61, 227–233. [Google Scholar]
- Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, and Miller SI (2019). The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. eLife 8, e40171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DR, Powers MJ, Trent MS, and Brodbelt JS (2019). Top-down characterization of lipooligosaccharides from antibiotic-resistant bacteria. Anal. Chem 91, 9608–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucar L, Stano M, and Hajduk M (2010). phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res. 38, D366–D370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Wieland CW, Florquin S, Pantophlet R, Dijkshoorn L, Tshimbalanga N, Akira S, and van der Poll T (2006). Differential roles of CD14 and toll-like receptors 4 and 2 in murine Acinetobacter pneumonia. Am. J. Respir. Crit. Care Med 173, 122–129. [DOI] [PubMed] [Google Scholar]
- Kumar A, Dalton C, Cortez-Cordova J, and Schweizer HP (2010). Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J. Microbiol. Methods 82, 296–300. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Sub-group (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, et al. (2012). Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. MBio 3, e00312–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen PC, Alsaadi Y, Fernando D, and Kumar A (2014). Genome sequence of an extremely drug-resistant clinical isolate of Acinetobacter baumannii strain AB030. Genome Announc. 2, e01035–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, Chazin WJ, Trinidad JC, Van Nieuwenhze MS, Giedroc DP, et al. (2019). An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep. 26, 2009–2018.e2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni JC, and Silhavy TJ (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 106, 8009–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneval WE (1941). Staining bacteria and yeasts with acid dyes. Stain Technol. 16, 13–19. [Google Scholar]
- Mates SM, Eisenberg ES, Mandel LJ, Patel L, Kaback HR, and Miller MH (1982). Membrane potential and gentamicin uptake in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 79, 6693–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KL, and Silhavy TJ (2018). The Escherichia coli phospholipase PldA regulates outer membrane homeostasisvia lipid signaling. MBio 9, e00379–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R, Sansonetti PJ, and Parsot C (1993). Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol 175, 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother 54, 4971–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Adler B, Nation RL, Li J, and Boyce JD (2011). Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother 55, 3022–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombo-Ngoma G, Remppis J, Sievers M, Zoleko Manego R, Endamne L, Kabwende L, Veletzky L, Nguyen TT, Groger M, Lötsch F, et al. (2018). Efficacy and safety of fosmidomycin-piperaquine as nonartemisinin-based combination therapy for uncomplicated falciparum malaria: a single-arm, age de-escalation proof-of-concept study in Gabon. Clin. Infect. Dis 66, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen BL, Rathi S, Chazin WJ, and Skaar EP (2014). Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J. Bacteriol 196, 2616–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, and Nizet V (2017). The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J. Mol. Med. (Berl.) 95, 1127–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Losick R, and Kahne D (2019). Robust suppression of lipopolysaccharide deficiency in Acinetobacter baumannii by growth in minimal medium. J. Bacteriol 201, e00420–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, and Weiser JN (2011). Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 7, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Minehira M, Zhu X, Suzuki K, Nakagawa T, Matsuda H, and Kawamukai M (1997). The ispB gene encoding octaprenyl diphosphate synthase is essential for growth of Escherichia coli. J. Bacteriol 179, 3058–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, and Carson J (1972). Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem 247, 3962–3972. [PubMed] [Google Scholar]
- Palmer LD, Green ER, Sheldon JR, and Skaar EP (2019). Assessing Acinetobacter baumannii virulence and persistence in a murine model of lung infection. Methods Mol. Biol 1946, 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Seifert H, and Paterson DL (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev 21, 538–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, and Trent MS (2018). Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl. Acad. Sci. USA 115, E8518–E8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, and Trent MS (2019). Inter membrane transport: Glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc. Natl. Acad. Sci. USA 116, 17147–17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieck B, Tourigny DS, Crosatti M, Schmid R, Kochar M, Harrison EM, Ou HY, Turton JF, and Rajakumar K (2012). Acinetobacter insertion sequence ISAba11 belongs to a novel family that encodes transposases with a signature HHEK motif. Appl. Environ. Microbiol 78, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, et al. (2016).A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun 7, 10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, and Huang KC (2018). The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J,Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Gao X, Wei J, Chen L, Zhao X, Li B, and Duan K (2012). PA2800 plays an important role in both antibiotic susceptibility and virulence in Pseudomonas aeruginosa. Curr. Microbiol 65, 601–609. [DOI] [PubMed] [Google Scholar]
- Shrivastava R, and Chng SS (2019). Lipid trafficking across the Gram-negative cell envelope. J. Biol. Chem 294, 14175–14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava R, Jiang X, and Chng SS (2017). Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol 106, 395–408. [DOI] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, and Chandler M (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggins JM, Djambazova KV, Rivera ES, Migas LG, Neumann EK, Fuetterer A, Suetering J, Goedecke N, Ly A, Van de Plas R, and Caprioli RM (2019). High performance molecular imaging with MALDI trapped ion mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem 91, 14552–14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, and Silhavy TJ (2016). Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. USA 113, E1565–E1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, and Sasakawa C (1994). Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol 11, 31–41. [DOI] [PubMed] [Google Scholar]
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, et al. ; WHO Pathogens Priority List Working Group (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis 18, 318–327. [DOI] [PubMed] [Google Scholar]
- Thong S, Ercan B, Torta F, Fong ZY, Wong HY, Wenk MR, and Chng SS (2016). Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5, e19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TouzÉ T, and Mengin-Lecreulx D (2008). Undecaprenyl phosphate synthesis. EcoSal Plus 3, 4.7.1.7. [DOI] [PubMed] [Google Scholar]
- Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, and Davies BW (2014). Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio 5, e01313–e01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, and Reinhart K; EPIC II Group of Investigators (2009). International study of the prevalence and outcomes of infection in intensive care units. JAMA 302, 2323–2329. [DOI] [PubMed] [Google Scholar]
- Wallace BJ, and Young IG (1977). Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA− menA− double quinone mutant. Biochim. Biophys. Acta 461, 84–100. [DOI] [PubMed] [Google Scholar]
- Wang N, Ozer EA, Mandel MJ, and Hauser AR (2014). Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5, e01163–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lonergan ZR, Gonzalez-Gutierrez G, Nairn BL, Maxwell CN, Zhang Y, Andreini C, Karty JA, Chazin WJ, Trinidad JC, et al. (2019). Multi-metal restriction by calprotectin impacts de novo flavin biosynthesis in Acinetobacter baumannii. Cell Chem. Biol 26, 745–755.e747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BS, Ly PM, Irwin JN, Pukatzki S, and Feldman MF (2015). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 112, 9442–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, and Sievert DM (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA, Wert SE, and Weaver TE (2015). Diseases of pulmonary surfactant homeostasis. Annu. Rev. Pathol. 10, 371–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow J, Tan KW, Holdbrook DA, Chong ZS, Marzinek JK, Bond PJ, and Chng SS (2018). The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J. Biol. Chem 293, 11325–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingl FG, Kohl P, Cakar F, Leitner DR, Mitterer F, Bonnington KE, Rechberger GN, Kuehn MJ, Guan Z, Reidl J, and Schild S (2020). Outer membrane vesiculation facilitates surface exchange and in vivo adaptation of Vibrio cholerae. Cell Host Microbe 27, 225–237.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequencing data is available in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProject: PRJNA656143.


