To the editor,
Currently, we are facing SARS-CoV-2; the gold standard for the COVID-19 diagnosis is through nucleic acid analysis, that is, the demonstration of SARS-CoV-2 RNA in respiratory samples. Within the contribution of laboratory tests are serological tests, which are still an integral part of the general response and can complement the diagnosis based on the real-time reverse transcription polymerase chain reaction (RT-PCR) test, by confirming the antibody response during the early stage of infection [1]. The traditional scheme of specific IgM and IgG detection for SARS-CoV-2 allows classifying the temporary state of the infection; although the dynamics of the immune response in COVID-19 is not fully understood, typically IgM antibodies are produced by immune cells of the host during the early stages of a viral infection [2]. However, the mucosal and systemic IgA responses that may play a critical role in the pathogenesis of the disease have received much less attention.
The detection of these antibodies can be performed by a wide variety of methods; there are four main types of methods for serological diagnosis: lateral flow immunochromatographic assay (LFIA), enzyme-linked immunosorbent assay (ELISA), chemiluminescent immunoassay (CLIA), and the neutralization assay [3]. Among the advantages of LFIA, also called “rapid test,” we have that is easy to use, with a time to obtain results between 10 and 30 min; in addition to the shortage of molecular tests, it has been the technique chosen within the workflow for countries with limited resources [4]. Generally, rapid tests has a poor diagnostic performance compared with ELISA tests, which can be explain not only by the known technical differences between the two methodologies but also for the possible low concentrations of antibodies that can contribute even more to false negatives or false positives because of a non-specific binding observed with LFIA method. A lot of researches demonstrate the great variability in the performance of LFIA; given this, it seems that other methodologies such ELISA and CLIA have shown a better performance [5].
Yu et al. detected the IgA seroconversion on day 2 and IgM/IgG on day 5 after the symptoms onset. Furthermore, the study reported that 100% of the cases had detectable levels of IgA, IgG, and IgM on day 32 after the symptoms onset [6].
It seems that the IgM detection despite being performed by other methodologies different to LFIA maintains a great variability in the results. Therefore, there is an urgent need for an alternative that can help to assess early seroconversion; this could be in the detection of IgA, which seems to be detected earlier than IgM and IgG antibodies [7].
Padoan et al. described the characteristics of the kinetics of IgA antibodies compared with IgM antibodies. IgA response appeared and grew earlier, reached its peak in the third week, and maintained a stronger and more persistent response than IgM [8].
We performed the detection of specific IgA for SARS-CoV-2 in samples from the seroteca of the Hospital Nacional Dos de Mayo laboratory; 65 sera were randomly analyzed that had a request for detection of IgM/IgG by LFIA due to suspicion of COVID-19 and also RT-PCR result. All samples were stored at − 20 °C before use. Statistical analysis was performed with GraphPad Prism (version 8.4.2).
Of 65 samples, 39 were PCR positive and 26 PCR negative. Samples were processed with the EUROIMMUN Analyzer I and the EUROIMMUN (Euroimmun Medizinische Laboradiagnostika, Luebeck, Germany) anti-SARS-CoV-2 IgA ELISA kit, according to manufacturer’s instructions. Microplate’s wells are covered with recombinant structural s1 protein. Results are examined semi quantitatively by calculating a relationship between the absorbance of samples and the absorbance of calibrator. The interpretation of the relationship was as follows: < 0.8 = negative, ≥ 0.8 to < 1.1 = limit (o borderline), and ≥ 1.1 = positive. Limit (o borderline) data were considered positive for the statistical analyzes. In total, 46 positive and 19 negatives were obtained. Of RT-PCR-positive samples, 36 were positive and 3 were negative for IgA (92.3% of positive agreement, 95% CI: 79.7–97.3), whereas 24 samples had IgM positive and 15 had IgM negative (61.5% of positive agreement, 95% CI: 45.9–75.1) (Fig. 1). The results of the ROC curve analysis is shown in Fig. 2a. The area under the curve (AUC) of anti-SARS-CoV-2 IgA was 0.98 (95% CI, 0.97–1.00; p > 0.0001).
Fig. 1.
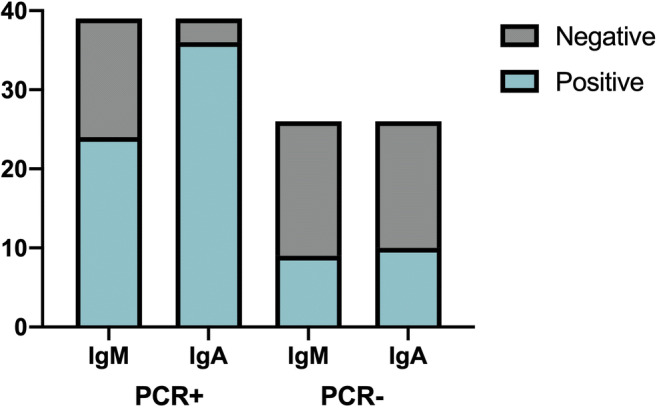
Frequency of results of serological tests for detection of SARS-CoV-2. IgM antibody detection using LFIA and IgA antibody detection using ELISA in serums that had tested positive by PCR (39 cases) or negative for the virus by PCR (26 cases). Abbreviations: ELISA, enzyme-linked immunosorbent assay; Ig, immunoglobulin; LFIA, lateral flow immunochromatographic assay; PCR, polymerase chain reaction; +, positive; −, negative
Fig. 2.
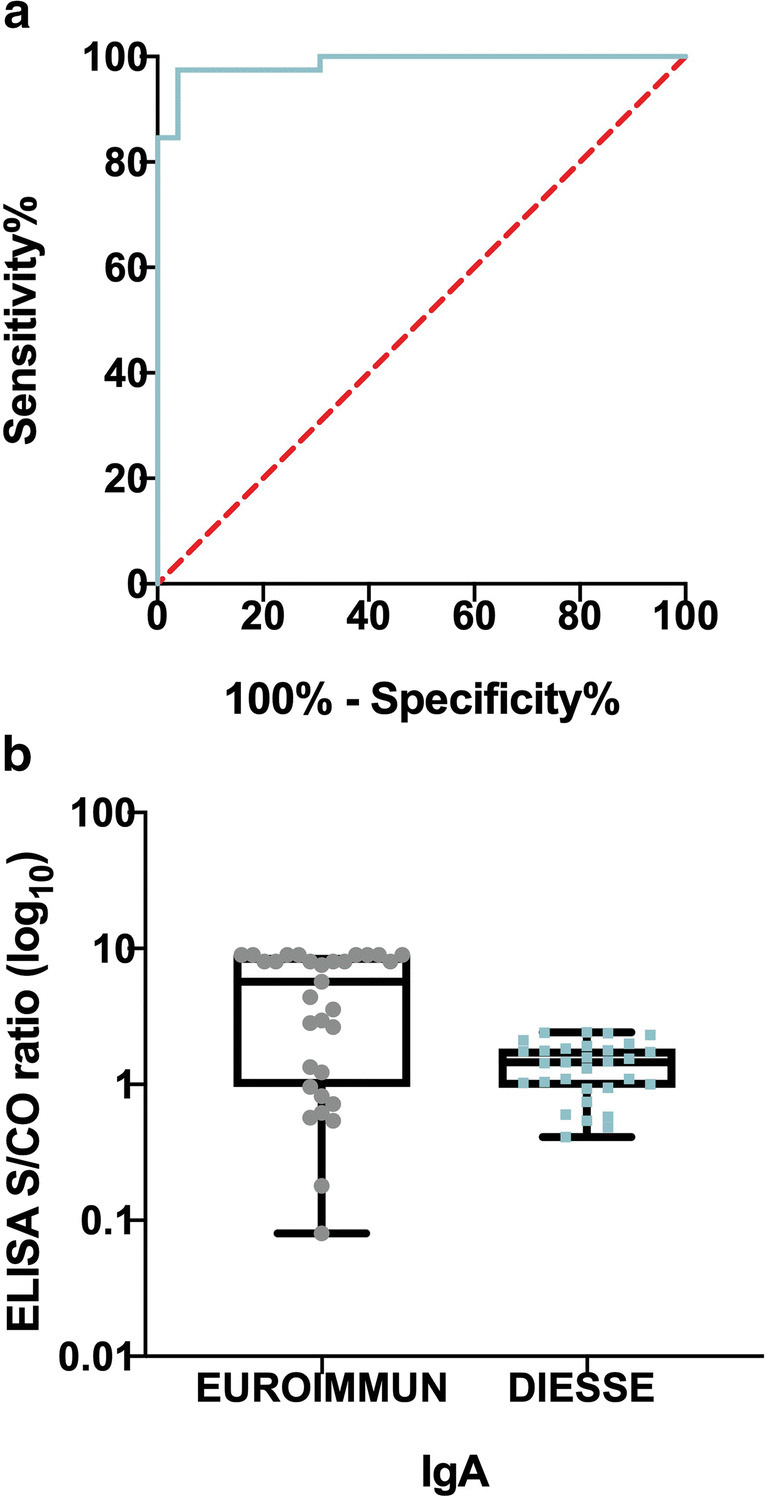
Analysis of SARS-CoV-2-specific IgA. a Receiver operating characteristic (ROC) curve analysis of EUROIMMUN anti-SARS-CoV-2 IgA ELISA kit compared with PCR. The overall concordance of test positive is 98%. b Distribution of signal/cut-off ratios obtained for the ELISA between commercial serological assays for detection of SARS-CoV-2-IgA (Euroimmun and DIESSE). The boxplots show medians (middle line) and third and first quartiles (boxes), while the whiskers show the interquartile range (IQR) above and below the box
Despite being different immunoglobulin classes, we performed a comparison between IgM positive/negative results reported by LFIA versus ELISA anti-SARS-Cov-2 IgA positive/negative results. We found a concordance of 74% (kappa statistic = 0.474; 95% CI, 0.25–0.70; p = 0.001).
We extend our observation by comparing the results of anti-SARS-CoV-2 IgA EUROIMMUN with ENZY-WELL SARS-CoV-2 IgA commercial kit developed by DIESSE Diagnostica Senese S.p.A. which is based on native antigen obtained from Vero E6 cells infected with SARS-CoV-2 strain “2019-nCoV/Italia-INMI1” (EVAg Ref-SKU: 008V-03893). The interpretation of the result (absorbance of sample/absorbance of calibrator) < 0.9 = negative, ≥ 0.9 to < 1.1 = borderline, ≥ 1.1 = positive, the borderline data were also considered positive for statistical analyzes. This comparison was made in 45 of 65 samples (Fig. 2b). The results obtained show a concordance of 89% (kappa statistic = 0.71; 95% CI, 0.41–0.99; p = 0.001).
Our results, although based on a small number of samples, are consistent with the study previously published by Pauline H Herroelen et al. [9], who reported a positivity for anti-SARS-CoV-2 IgA of 91.1% (51/56), using the same immunoassay.
The results show a greater positivity for IgA than IgM in samples RT-PCR positive (92% vs 61.5%), with a moderate kappa statistic. It seems that IgM remains limited for use, as has been reported for other kits that test IgM, suggesting research into technical improvements [10].
Regarding antigenic target, it is essential to compare tests that target the detection of the same antibodies. In this study, we compared for the first time two ELISA tests that detect specific IgA antibodies, finding a concordance of 89% with a good kappa statistic, although antigenic target was different (S1 protein vs inactivated virus antigen).
The authors are conscious of the limitations of the study by not considering the time of symptoms onset, but we believe that presenting our results provides information to future research.
The difficult access to molecular tests in low-income countries, as the majority in Latin America, proposes the resource of antibody detection by methodologies with short response time and easy to perform; however, the performance of the called rapid tests seems not to be adequate. Therefore, the use of different methodologies with better performance is necessary.
In our opinion, we suggest that IgA detection could be added to the classically used IgM and IgG antibodies as a complementary aid in the diagnosis of COVID-19.
Acknowledgments
We wish to thank the personnel of laboratory Delfina Soto y Carmen Ramos for its technical assistance.
Author’s Contributions
All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no conflict of interest.
Ethical Approval
The Local Institutional Review Board deemed the study exempt from review.
Footnotes
This article is part of the Topical Collection on Covid-19
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Infantino M, Damiani A, Gobbi FL, et al. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J. 2020;22(203):210. [PubMed] [Google Scholar]
- 2.Sethuraman N, Jeremiah S, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 3.Özçürümez MK, Ambrosch A, Frey O. SARS-CoV-2 antibody testing – questions to be asked. J Allergy Clin Immunol. 2020;146:35–43. doi: 10.1016/j.jaci.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai WR, Rodriguez W, Bassett IV. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect Dis. 2014;14(3):239–249. doi: 10.1016/S1473-3099(13)70250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui LP, Johnston JC, Lan Z, Law S, MacLean E, Trajman A, Menzies D, Benedetti A, Ahmad Khan F. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H-Q, Sun B-Q, Fang Z-F, Zhao JC, Liu XY, Li YM, Sun XZ, Liang HF, Zhong B, Huang ZF, Zheng PY, Tian LF, Qu HQ, Liu DC, Wang EY, Xiao XJ, Li SY, Ye F, Guan L, Hu DS, Hakonarson H, Liu ZG, Zhong NS. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020;2001526:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padoan A, Sciacovelli L, Basso D, et al. Respuesta de IgA - Ab a la glucoproteína de pico de SARS - CoV - 2 en pacientes con COVID - 19: un estudio longitudinal. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herroelen PH, Martens GA, De Smet D, Swaerts K, Decavele AS. Humoral immune response to SARS-CoV-2 [published online ahead of print, 2020 Aug 18]. Am J Clin Pathol. 2020:aqaa140. 10.1093/ajcp/aqaa140. [DOI] [PMC free article] [PubMed]
- 10.Long Q-X, Deng H-J, Chen J, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv preprint.


