Abstract
Background
LncRNAs play important roles in papillary thyroid carcinoma (PTC). LINC02471 has been reported to be related to PTC prognosis. The current study aimed to investigate the effects of LINC02471 on human PTC cells.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to examine LINC02471 expression in PTC tissues and cells and miR-375 expression in PTC cells. SiLINC02471, miR-375 mimic and miR-375 inhibitor were used for cell transfection. Cell proliferation, apoptosis, migration, and invasion were detected by performing Cell Counting Kit-8 (CCK-8), clone formation assay, flow cytometry, scratch assay, and transwell assay. Western blot was carried out to detect protein levels of E-cadherin, N-cadherin and Snail. The target gene for LINC02471 was verified by dual-luciferase reporter assay.
Results
LINC02471 was highly expressed in PTC tissues and cells. After silencing LINC02471, cell proliferation, migration and invasion were reduced, but cell apoptosis was increased. SiLINC02471 increased the expressions of E-cadherin and miR-375, and inhibited the expressions of N-Cadherin and Snail. LINC02471 directly targeted miR-375 in PTC cells. Overexpression of miR-375 inhibited the proliferation, migration, invasion of PTC cells and reduced the expressions of N-Cadherin and Snail but promoted the cell apoptosis and increased E-cadherin expression, while miR-375 inhibitor produced opposite effects to overexpressed miR-375. After inhibiting miR-375 expression, siLINC02471 reversed the effect of miR-375 inhibitor.
Conclusion
LINC02471 could promote the development of PTC. Knocking down LINC02471 could inhibit invasion and metastasis and promote PTC cell apoptosis through directly targeting miR-375.
Keywords: papillary thyroid carcinoma, long non-coding RNAs, miR-375, epithelial-mesenchymal transition, EMT
Introduction
Thyroid cancer is one of the most common endocrine malignant tumors, and its incidence is increasing annually.1,2 According to histological characteristics, thyroid cancer can be subdivided into papillary thyroid carcinoma (PTC), thyroid follicular cancer, anaplastic thyroid carcinoma, and medullary thyroid carcinoma.3 The majority of thyroid cancer patients are PTC patients.4 Currently, 131I radiotherapy, surgical and TSH inhibition treatments are the main strategy for the management of PTC.5 Although these treatments have been constantly improved, PTC patients still suffer from cancer recurrence, invasion and/or metastasis.6 Therefore, developing more effective methods of diagnosis and treatment for managing PTC is highly necessary.
Long non-coding RNAs (lncRNAs) are noncoding RNAs with a length of more than 200 nucleotides, and they have gene-specific regulatory functions.7 The roles of lncRNAs in the occurrence and development of tumors have been increasingly discovered.8,9 LncRNAs are also involved in the proliferation, invasion and metastasis of thyroid cancer,10,11 for example, Li et al12 found that lncRNA n340790 is increased in thyroid cancer, and it directly adsorbs miR-1254 to promote thyroid cancer cell proliferation through a sponging effect. LINC02471 has a length of 705 nt, and is located in p7 region of chromosome 12. However, the role of LINC02471 in thyroid cancer is less studied.13 Cai et al reported that the combination of two DEGs (METTL7B and KCTD16) and two DElncRNAs (LINC02454 and LINC02471) could more accurately predict the prognosis of PTC.14 Therefore, the present study explored the internal correlation between LINC02471 and PTC, aiming at understanding its function and finding its target gene. Our findings provide scientific basis for gene diagnosis and targeted therapy of PTC.
Materials and Methods
Clinical Specimens
The surgical cancer tissues and paired adjacent tissues 2 cm away from the cancer tissues were collected from PTC patients who received surgery in Xiasha Branch of Sir Run Run Shaw Hospital from 2016 to 2018. All the patients were diagnosed as PTC by postoperative pathological examination, and did not have previous antitumor treatment history such as radiotherapy, chemotherapy or cell biological therapy. Surgical specimens were immediately frozen in liquid nitrogen until subsequent experiments. The current experiment was approved by the Ethics Committee of Xiasha Branch of Sir Run Run Shaw Hospital (approved number: ZJ2016090122), and informed consent was signed by all the participants.
Cell Culture
Human thyroid follicular epithelial cell line Nthy-ori3-1, human PTC cell line TPC-1, IHH4 and BCPAP were purchased from the Cell Bank of the Typical Culture Preservation Committee of the Chinese Academy of Sciences (http://www.cellbank.org.cn/). The cells were cultured in RPMI-1640 medium (Gibco, USA) containing 10% FBS (Gibco, USA) in a humidified incubator with 5% CO2 at 37°C.
Cell Transfection
Transfections of TPC-1 and IHH4 were realized using Lipofectamine 2000 Transfection Reagent (Invitrogen, USA). MiR-375 mimic (M), miR-375 inhibitor (I), mimic control (MC) and inhibitor control (IC) were mixed with OPTIM-MEM medium, respectively. Lipofectamine 2000 reagent was then mixed with OPTIM-MEM medium, and the diluted miR-375 was further mixed with the diluted Lipofectamine 2000 reagent. The cells at a density of 5⨰104 cells/well were inoculated into a 96-well plate, added with the mixture, and incubated at 37°C with 5% CO2 for 48 h. The cells were harvested for further study after transfection for 48 h.
The siRNAs against LINC02471 (siLINC02471) and the negative control (siNC) were designed and synthesized by Shanghai GenePharma Co., Ltd (China). The siRNAs were transfected into TPC-1 and IHH4 cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s instruction. The cells were harvested for further study after transfection for 48 h. The sequences used for transfection were listed in Table 1.
Table 1.
The Sequences Used for Transfection
| Name | Sequences: 5ʹ-3’ |
|---|---|
| siRNAs negative control (siNC) | AATTCACTCCAAGTCTCTTCC |
| miR-375 mimic | UUUGUUCGUUCGGCUCGCGUGA |
| miR-375 mimic control | UUUGUACUACACAAAAGUACUG |
| miR-375 inhibitor | UAACGAGCCGAACGAACAAA |
| miR-375 inhibitor control | CAGUACUUUUGUGUAGUACAA |
Cell Counting Kit-8 (CCK-8)
Cell viability was determined by CCK-8 according to manufacturer’s protocol. TPC-1 and IHH4 cells with or without transfection were seeded into 96-well plates (5⨰103 cells/well) and cultured with 5% CO2 at 37°C for 24, 48, and 72 h. Then, CCK-8 reagent was added to the wells and cultured at 37°C for two hours. The absorbance (OD: 450 nm) was measured by microplate (Model 680, Bio-Rad, USA) to calculate cell viability.
Clone Formation Assay
Approximately 500 TPC-1 and IHH4 cells with or without transfection were seeded into 24-well plates, and cultured in RPMI-1640 containing 10% FBS and 1% agarose for 14 days with 5% CO2 at 37°C. Subsequently, the cells were fixed by 4% methanol for 20 min, and then stained by 0.1% crystal violet for 15 min. The images were observed under an inverted phase contrast microscope (CK40, Olympus, Japan) and the colonies were counted.
Cell Apoptosis
TPC-1 and IHH4 cells with or without transfection were cultured for 48 h and the harvested. The cells were stained by annexin V-FITC and propidium iodide and incubated in the dark for 10 min. Following the instructions of Annexin V-FITC Apoptosis Detection kit, cell apoptosis was detected using an Annexin V-FITC Apoptosis Detection kit (Key GEN, Shanghai, China) by flow cytometry (BD FACSCalibur, BD Biosciences, USA) equipped with Cell Quest software (BD Biosciences).
Scratch Assay
The cells at a density of 5⨰105 (cells/mL) were seeded into a 24-well plate and scratched by a pipette tip in a predetermined straight position. The images of the cells were captured under an inverted phase-contrast microscope (Eclipse TS100, Nikon, Japan) at 0 and 48 h. NIS-Element Basic Research v3.2 software (Nikon, Japan) was used to determine the scratch area for examining cell migration capacity.
Transwell Assay
The cells (5⨰103 cells/mL) in the 24-well plate were seeded into the upper Transwell chamber (8-μm pores, Corning Company, USA) coated with matrigel, while the lower chamber contained RPMI-1640 medium with 10% FBS. After incubation for 48 h at 37°C with 5% CO2, noninvading cells on the upper chamber were removed by cotton swabs, whereas invaded cells were fixed by paraformaldehyde and then stained by 0.1% crystal violet for 10 min. The images were captured under an inverted microscope (TS100, Nikon, Japan).
Dual-luciferase Reporter Assay
Direct target gene of LINC02471 was predicted by The DIANA tools LncBase Predicted v.2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted). The wild-type (LINC02471-WT) or mutant (LINC02471-MUT) LINC02471 vectors were synthesized and purchased from Shanghai GenePharma Co., Ltd (China). For dual-luciferase reporter assay, LINC02471-MUT or LINC02471-WT vectors were cotransfected with miR-375 mimic or with transfection reagent into TPC-1 and IHH4 cells by using Lipofectamine 2000 Transfection Reagent (Invitrogen, USA). After transfection for 48 h, the cells were harvested and subjected to dual-luciferase reporter assay kit (Promega, USA). Relative luciferase of the cells was normalized to that of Renilla luciferase.
RNA Pull-down Assay
The TPC-1 and IHH4 cells were treated by 50 nM biotin-labeled WT-bio-LINC02471 RNA and MUT-bio-LINC02471 RNA for 48 h. The cells were collected to incubate with specific lysate buffer (Ambion, USA) for 10 min. The lysates were incubated with the M-280 streptavidin beads (S3762, Sigma-Aldrich, USA), which were precoated with RNase free BSA and yeast tRNA (TRNABAK-RO, Sigma-Aldrich, USA), at 4°C for three hours. Next, the beads were first washed twice with precooled lysate buffer solution and low-salt buffer solution three times, and then with high-salt buffer solution once. The combined RNAs were purified by Trizol, and miR-375 enrichment was examined by quantitative real-time polymerase chain reaction (qRT-PCR).
RNA Immunoprecipitation (RIP) Assay
The binding of LINC02471 RNA to argonaute-2 (AGO2) protein was detected using Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Temecula, USA). The cells were then washed by precooled PBS, and the supernatant was discarded. Next, the cells were lysed by an equal amount of radioimmunoprecipitation assay (RIPA) lysis (P0013B, Beyotime Biotechnology Co., Ltd, Beijing, China) in an ice bath for five minutes, and the supernatant was collected by centrifugation (at 14000 rpm, at 4°C for 10 min). A section of the cell extracts were used as the input, while the rest was incubated with antibody AGO2 (ab32381, 1:50, Abcam, UK) at room temperature for 30 min for coprecipitation. IgG (ab109489, 1:100, Abcam, UK) served as a negative control (NC).
RT-qPCR
Total RNAs were isolated from the cancer tissues, adjacent tissues, and cell lines using Trizol reagent (Invitrogen, USA). Purity and concentration of RNA were determined by Nano Drop 2000 (Thermo Fisher Scientific, USA). RNAs (2 μg) were reverse-transcribed into cDNAs using a PrimeScript RT Master Mix kit (Takara, China). The PCR was performed using a SYBR miRNA detection assays (Takara, China) in Opticon real-time PCR Detection System (ABI 7500, Life technology, USA). The reaction conditions were set as follows: at 95°C for five minutes, at 95°C for 30 seconds, at 60°C for 30 seconds, at 72°C for 15 seconds, for a total of 40 cycles. The relative gene expression levels were calculated by 2-ΔΔCT method.15 U6 and GAPDH served as internal controls. The primers used were listed in Table 2.
Table 2.
The Primers Used for qPCR
| Name | Forward: 5ʹ-3’ | Reverse: 5ʹ-3’ |
|---|---|---|
| LncRNA LINC02471 | ATGCTAAACACTGCCTCATCTCT | GATTAGCTGCTTCCCAGTGT |
| GAPDH | CATGGAATCCTGTGGCATCC | TGATCTTCATGGTGCTGGG |
| U6 | CTCAGAGCGTGTTCTCCGTCAC | TATAAATCTTTACCCTGTTGGCAGT |
| miR-375 | TCGCACAAACGTCGTATCCA | GTATCCAGTGCGTGTCGTGG |
Western Blot
Total proteins were extracted from cells using RIPA buffer (Beyotime, China), the protein concentration was determined by BCA protein assay kit (Pierce, Germany). The protein (20 μg) were separated by 10% SDS-PAGE (Invitrogen, USA) and transferred onto PVDF membranes (Merck, Germany). The membranes were blocked by 5% nonfat milk for one hour and then incubated with the following primary antibodies overnight at 4°C: E-cadherin (E-Cad, Cat#14472, 1:1000, CST, USA), N-Cadherin (N-Cad, Cat #14215, 1:1000, CST, USA), Snail (ab53519, 1:1000, Abcam, USA), GAPDH (ab8245, 1:2000, Abcam, USA). After that, the membranes were further incubated with secondary antibodies, donkey anti-goat IgG H&L (HRP) (ab205723, 1:2000, Abcam, USA), goat anti-mouse IgG H&L (HRP) (ab205719, 1:2000, Abcam, USA) for two hours, and visualized by ECL chemiluminescence (Thermo Scientific, USA) using a Band Scan 5.0 system (Bio-Rad, Hercules, USA). GAPDH served as a control.
Statistical Analysis
The experiments were performed in triplicate. The statistical analysis was performed by Student’s tests or one-way ANOVA using SPSS 20.0 system (IBM Corporation, Armonk, NY, USA). The data were shown as the mean ±SD. P<0.05 was considered to be statistically significant.
Results
LINC02471 Expression was Increased in PTC Tissues and Cells
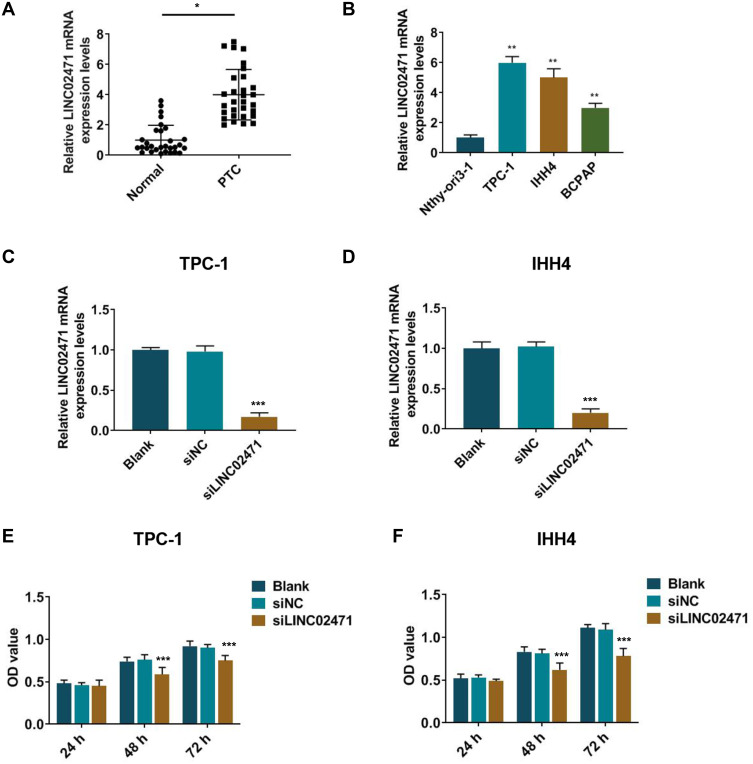
LINC02471 expression of PTC tissues was detected by qPCR, and the results showed that LINC02471 level of PTC cancer tissues was observably increased (Figure 1A, P<0.05). Moreover, compared with Nthy-ori3-1, LINC02471 was greatly increased in TPC-1, IHH4 and BCPAP cells (Figure 1B, P<0.05), especially in the first two cells. Thus, TPC-1 and IHH4 cells were used in the following study. These results showed that upregulated expression of LINC02471 may be involved in PTC progression.
Figure 1.
Long non-coding RNAs (lncRNA) LINC02471 in human papillary thyroid carcinoma (PTC) tissues and cell lines. (A) The expression of LINC02471 in human PTC tissues and paired adjacent tissues (Normal). (B) The expression of LINC02471 in human thyroid follicular epithelial cell line (Nthy-ori3-1) and human PTC cell lines (TPC-1, IHH4 and BCPAP). (C) In TPC-1, siRNAs against LINC02471 (siLINC02471) and the negative control (siNC) were used for cell transfection, and quantitative real-time polymerase chain reaction (qPCR) was performed to detect the knockdown efficiency. (D) In IHH4, siLINC02471 and siNC were used for cell transfection, and qPCR was performed to detect the knockdown efficiency. (E) In TPC-1, Cell Counting Kit-8 (CCK-8) was performed to determine the cell viability. (F) In IHH4, CCK-8 was performed to determine the cell viability. (A) *P<0.01 vs Normal; (B) **P<0.01 vs Nthy-ori3-1; (C–F) ***P<0.01 vs siNC.
Knocking Down LINC02471 Inhibited Cell Proliferation and Promoted Apoptosis of PTC Cells
The siLINC02471 was transfected into the PTC cell lines to explore the biological effects of LINC02471 on PTC cells, and the knockdown efficiency of LINC02471 was detected by qPCR. In TPC-1 (Figure 1C) and IHH4 (Figure 1D) cells, LINC02471 was low-expressed in the siLINC02471 group compared with siNC group (P<0.05), indicating that LINC02471 expression was successfully knocked down in the PCT cells.
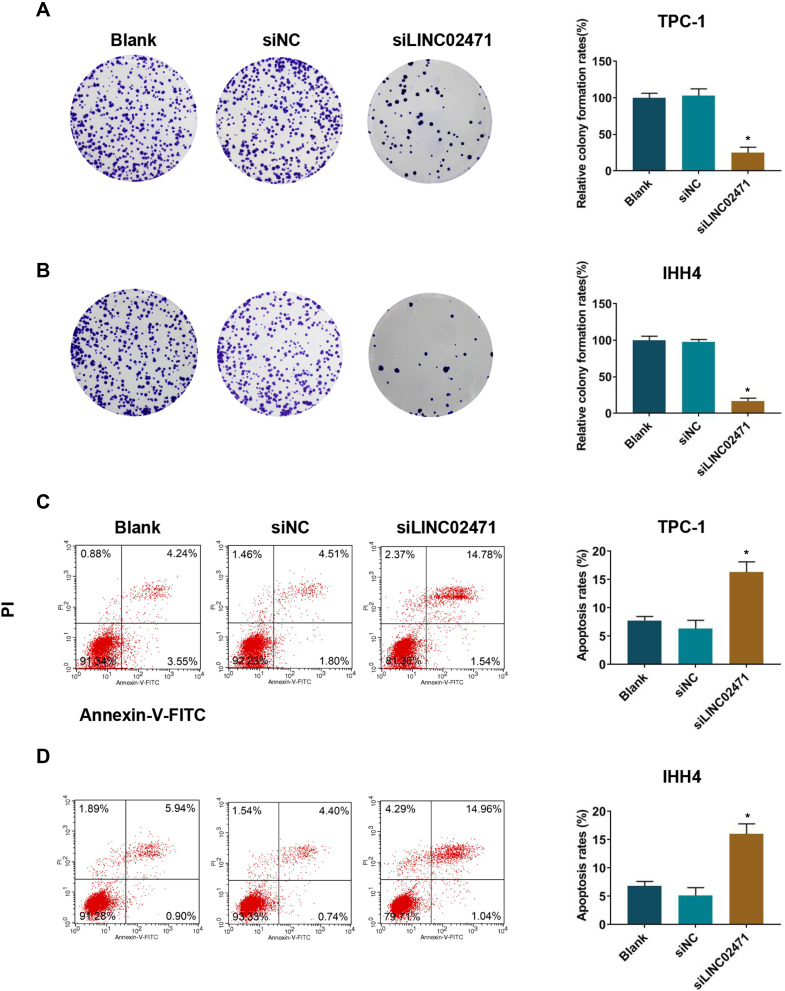
Following the transfection of siLINC02471, cell proliferation of PCT cell was examined by CCK-8 and clone formation assay. CCK-8 results demonstrated that siLINC02471 greatly reduced the cell viabilities of TPC-1 (Figure 1E) and IHH4 (Figure 1F) at 48 h and 72 h compared with siNC group (P<0.05). Meanwhile, the clones of TPC-1 (Figure 2A) and IHH4 (Figure 2B) cells treated by siLINC02471 were markedly fewer than that of the siNC group (P<0.05).
Figure 2.
Effect of siLINC02471 on PTC cell proliferation and apoptosis. (A) In TPC-1, clone formation assay was performed to determine the cell clones. (B) In IHH4, clone formation assay was performed to determine the cell clones. (C) In TPC-1, flow cytometer was performed to determine the apoptosis. (D) In IHH4, flow cytometer was performed to determine the apoptosis. *P<0.01 vs siNC.
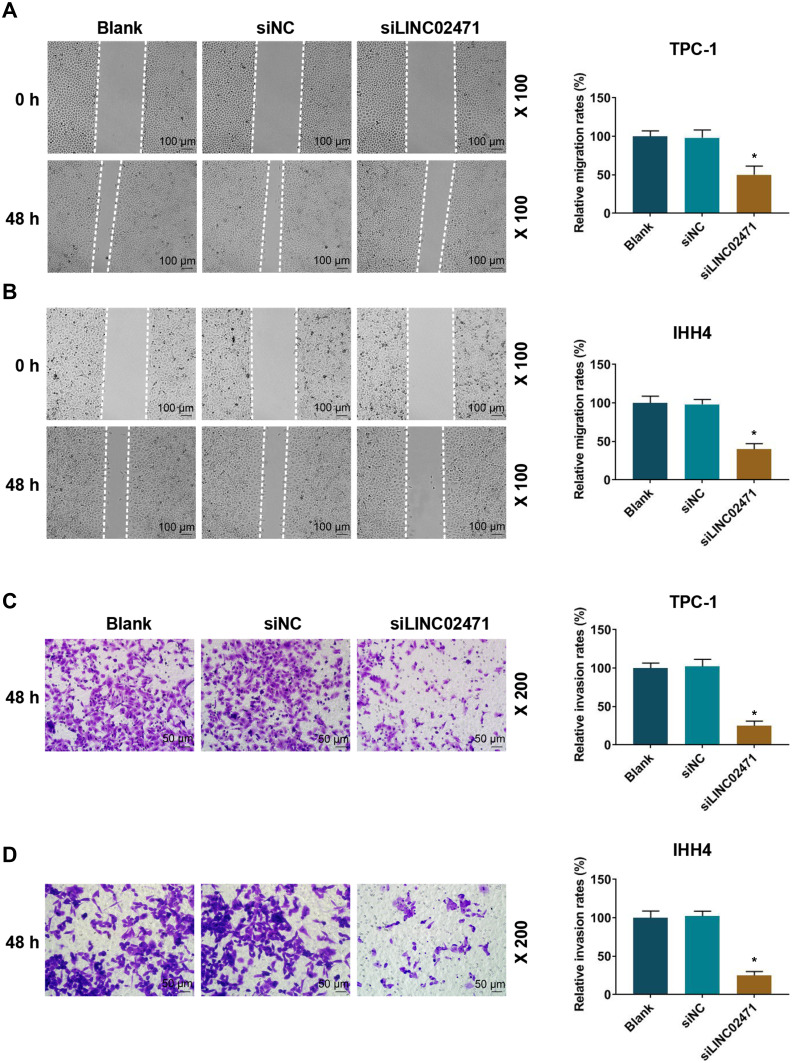
The effect of LINC02471 on apoptotic of transfected PCT cell lines was examined by flow cytometry, and the results revealed that the apoptosis of TPC-1 (Figure 2C) and IHH4 (Figure 2D) cells treated by siLINC02471 were higher than that of the siNC group (P<0.05). At the same time, the invasion and migration of PCT cell lines were determined by transwell and scratch assay. As shown in Figure 3, the migration (Figure 3A and B) and invasion (Figure 3C and D) of TPC-1 and IHH4 cells were reduced in siLINC02471 group compared with siNC group (P<0.05), indicating that knocking down LINC02471 obviously reduced the proliferation, migration and invasion of the PCT cells but induced the apoptosis.
Figure 3.
Effect of siLINC02471 on PTC cell migration and invasion. (A) In TPC-1, scratch assay was performed to determine the cell migration. (B) In IHH4, scratch assay was performed to determine the cell migration. (C) In TPC-1, transwell assay was performed to determine the cell invasion. (D) In IHH4, transwell assay was performed to determine the cell invasion. *P<0.01 vs siNC.
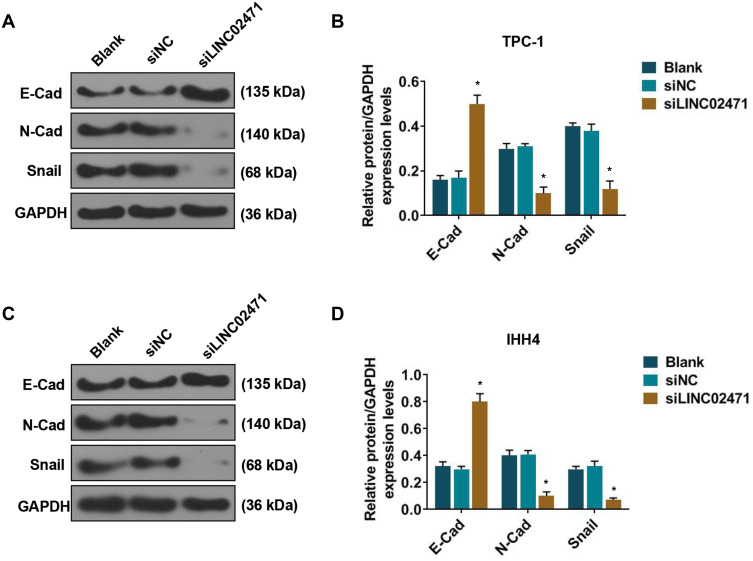
LINC02471 Knockdown Increased E-Cad Level, While Decreased N-Cad and Snail Levels in PTC Cells
Western blot was performed to analyze the effect of siLINC02471 treatment on epithelial-mesenchymal transition (EMT) of PCT cells. The results showed that for TPC-1 (Figure 4A and B) and IHH4 (Figure 4C and D) cells, the expressions of N-Cad and Snail was reduced and E-Cad protein was increased in siLINC02471 group, compared with the siNC group (P<0.05).
Figure 4.
Effect of siLINC02471 on PTC cell epithelial-mesenchymal transition (EMT). (A) In TPC-1, Western blot was performed to determine the EMT-relative protein expression. (B) The expressions of E- cadherin (E-Cad), N-Cadherin (N-Cad) and Snail in each group. (C) In IHH4, Western blot was performed to determine the EMT-relative protein expression. (D) The expressions of E-Cad, N-Cad and Snail in each group. *P<0.01 vs siNC.
LINC02471 Directly Targeted miR-375 in PTC Cell Lines
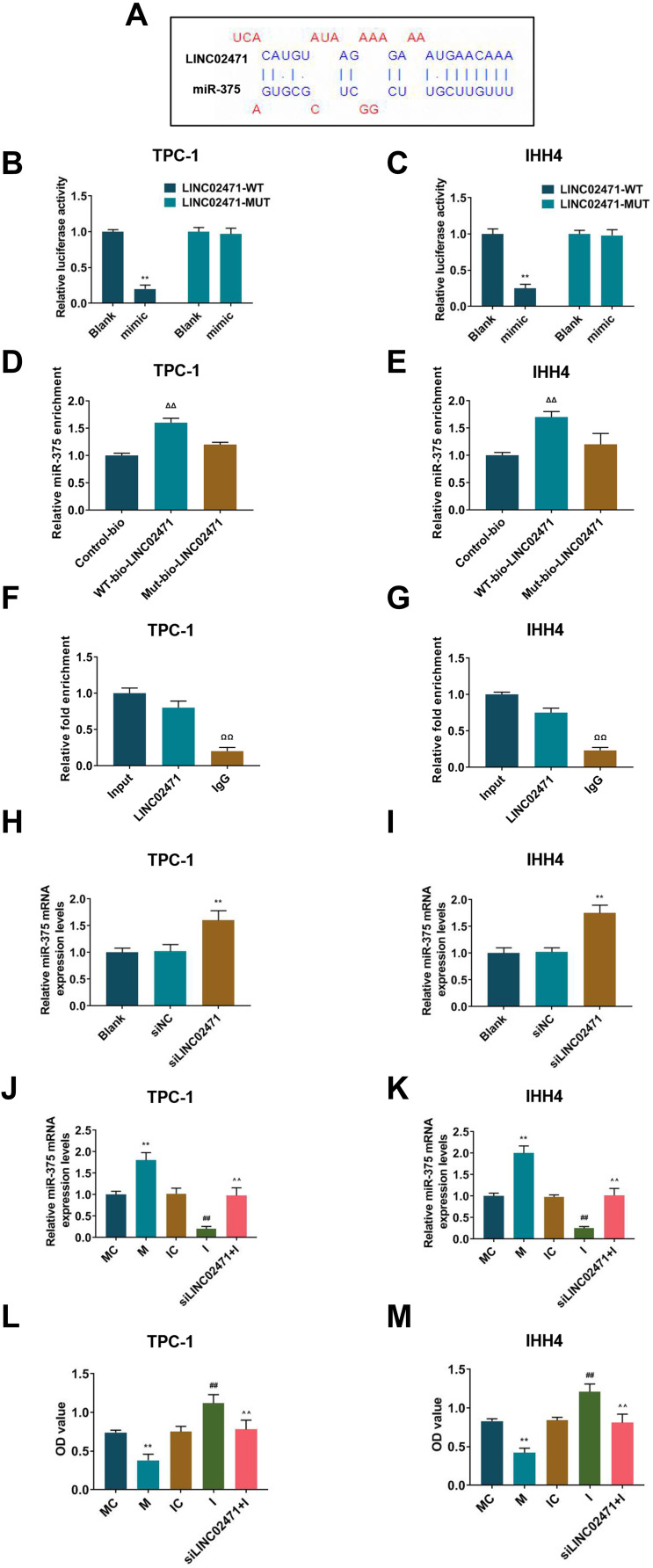
To understand the mechanism of LINC02471 on PTC cell lines, LncBase Predicted v.2, that miR-375 was a potential target of LINC02471 (Figure 5A). Then, dual-luciferase assay was performed to validate whether miR-375 bound to LINC02471. The results demonstrated that luciferase activities of TPC-1 (Figure 5B) and IHH4 (Figure 5C) cells cotransfected with miR-375 mimic and LINC02471-WT were reduced (P<0.05), while the luciferase activities of TPC-1 and IHH4 cells cotransfected with LINC02471-MUT did not change (P>0.05). RNA-pull down and RIP assays were conducted to further verify the interaction between LINC02471 and miR-375. The results of RNA pull-down assay indicated when compared with LINC02471-Mut, relative miR-375 enrichment was significantly increased in LINC02471-WT (P<0.05, Figure 5D and E), suggesting that miR-375 and LINC02471 could directly bind to each other. The RIP assay demonstrated when compared with IgG, LINC02471 enrichment in AGO2 was notably increased (P<0.05, Figure 5F and G), indicating that LINC02471 could bind to AGO2 protein. To confirm that LINC02471 targeted and suppressed miR-375 expression level of PCT cells, qPCR was performed to detect miR-375 level in PCT cell lines after knocking down LINC02471. For TPC-1 (Figure 5H) and IHH4 (Figure 5I) cells, the downregulation of LINC02471 resulted in a significant increase of miR-375 level compared with the siNC group (P<0.05). Taken together, miR-375 was the direct target of LINC02471 in human PCT cell lines.
Figure 5.
LINC02471 directly targeted miR-375 in PTC cell lines. (A) Prediction of binding sites of LINC02471 and miR-375. (B) In TPC-1, dual-luciferase reporter assay was performed to detect whether miR-375 binds to LINC02471. (C) In IHH4, dual-luciferase reporter assay was performed to detect whether miR-375 binds to LINC02471. (D) In TPC-1, relative enrichment of miR-375 to LINC02471-WT and LINC02471-MUT analyzed by RNA pull-down assay. (E) In IHH4, ralative enrichment of miR-375 to LINC02471-WT and LINC02471-MUT analyzed by RNA pull-down assay. (F) In TPC-1, relative enrichment of AGO2 to miR-375 and LINC02471 detected by RIP assay. (G) In IHH4, relative enrichment of AGO2 to miR-375 and LINC02471 detected by RIP assay. (H) In TPC-1, qPCR was performed to detect the miR-375 level in siLINC02471 cells. (I) In IHH4, qPCR was performed to detect the miR-375 level in siLINC02471 cells. (J) In TPC-1, miR-375 mimic, miR-375 inhibitor and siLINC02471 were used for cell transfection, and qPCR was performed to detect the transfection efficiency. (K) In IHH4, miR-375 mimic, miR-375 inhibitor and siLINC02471 were used for cell transfection, and qPCR was performed to detect the transfection efficiency. (L) In TPC-1, CCK-8 was performed to determine the cell viability. (M) In IHH4, CCK-8 was performed to determine the cell viability. (B and C) **P<0.01 vs blank; (H and I) **P<0.01 vs siNC; (J–M) **P<0.01 vs MC; ##P<0.01 vs IC; ^^P<0.01 vs I.
LINC02471 Knockdown Regulated PTC Cells Progression by Mediating miR-375
Whether LINC02471 regulated PCT cell proliferation and migration through targeting miR-375 expression was examined by transfecting miR-375 inhibitor, miR-375 mimic into the TPC-1 (Figure 5J) and IHH4 (Figure 5K) cells in the absence or presence of siLINC02471. For TPC-1 and IHH4 cells, RT-qPCR showed that miR-375 mRNA level was markedly upregulation in M group compared with MC group, and it was lower in the I group than that in the IC group (P<0.05). After treating the two cells with miR-375 inhibitor and siLINC02471, the miR-375 mRNA level was upregulated and higher than that in the I group (P<0.05). MiR-375 mimic increased the miR-375 expression, while miR-375 inhibitor reduced the miR-375 expression, moreover, the expression of miR-375 was restored by miR-375 inhibitor and siLINC02471.
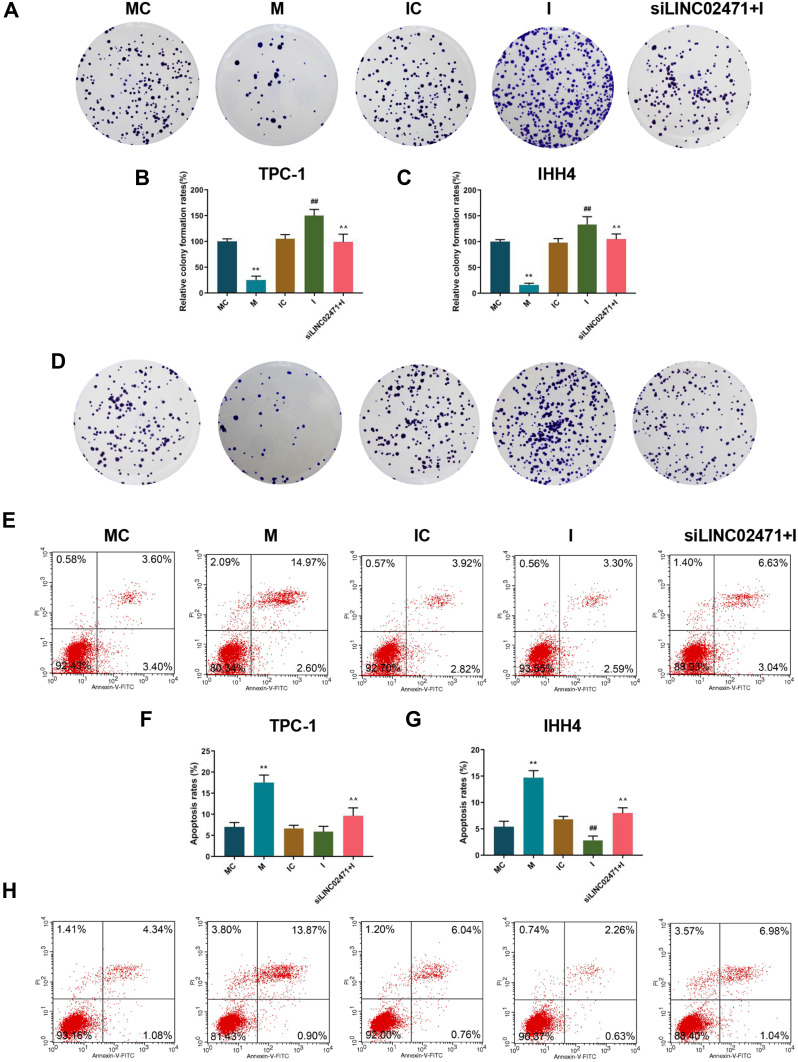
Cell proliferation was determined by CCK-8 and clone formation assay. The data revealed that the cell viability (Figure 5L and M) and clones (Figure 6A–D) of TPC-1 and IHH4 cells in the M group were reduced compared with the MC group, while those in the I group were increased compared with the IC group (P<0.05). Moreover, the cell viability (Figure 5L and M) and clones (Figure 6A–D) of the cells treated by miR-375 inhibitor and siLINC02471 were lower and fewer than those of cells treated by miR-375 inhibitor alone (P<0.05).
Figure 6.
Effect of siLINC02471 and miR-375 on PTC cell proliferation and apoptosis. (A) In TPC-1, clone formation assay was performed to determine the cell clones. (B) The clone formation rates of TPC-1 in each group. (C) The clone formation rates of IHH4 in each group. (D) In IHH4, clone formation assay was performed to determine the cell clones. (E) In TPC-1, flow cytometer was performed to determine the apoptosis. (F) The apoptosis rate of TPC-1 in each group. (G) The apoptosis rate of IHH4 in each group. (H) In IHH4, flow cytometer was performed to determine the apoptosis. (B–G) **P<0.01 vs MC; ##P<0.01 vs IC; ^^P<0.01 vs I.
Subsequently, cell apoptosis was detected by flow cytometer, and the data showed that the apoptosis of TPC-1 (Figure 6E and F) and IHH4 (Figure 6G and H) cells treated by miR-375 mimic were higher than those of cells in the MC group (P<0.05), while the apoptosis of cells in siLINC02471+I group were higher than that in the I group (P<0.05). In addition, no significant difference was observed in the apoptosis of TPC-1 cells between the IC and I groups (P>0.05), while the apoptosis of IHH4 cells treated by miR-375 inhibitor was lower than those of cells in the IC group (P<0.05).
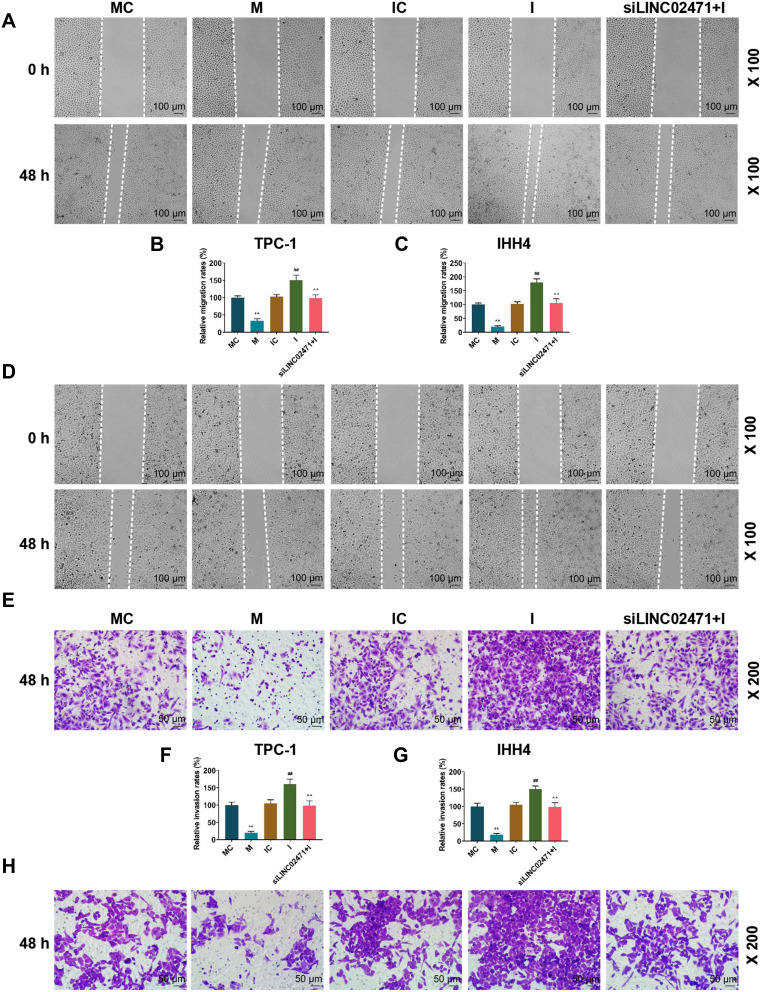
The detection of cell invasion and migration by transwell and scratch assays showed that cell migration (Figure 7A–D) and invasion (Figure 7E–H) of the cells treated by miR-375 mimic were reduced, while those of the cells treated by miR-375 inhibitor were increased (P<0.05). Still, the cell migration and invasion of the cells in siLINC02471+I group were lower than those in I group (P<0.05).
Figure 7.
Effect of siLINC02471 and miR-375 on PTC cell migration and invasion. (A) In TPC-1, scratch assay was performed to determine the cell migration. (B) The migration rate of TPC-1 in each group. (C) The migration rate of IHH4 in each group. (D) In IHH4, scratch assay was performed to determine the cell migration. (E) In TPC-1, transwell assay was performed to determine the cell invasion. (F) The invasion rate of TPC-1 in each group. (G) The invasion rate of IHH4 in each group. (H) In IHH4, transwell assay was performed to determine the cell invasion. (B–G) **P<0.01 vs MC; ##P<0.01 vs IC; ^^P<0.01 vs I.
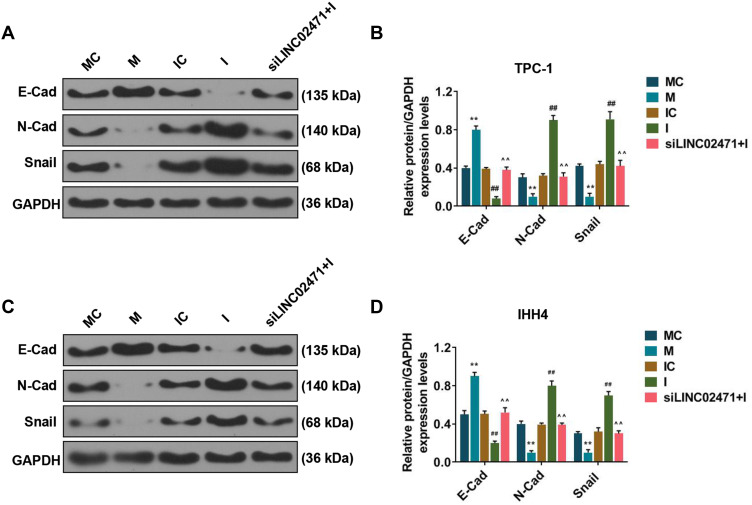
As shown in Figure 8, Western blot results demonstrated that miR-375 mimic increased the E-Cad, and decreased N-Cad and Snail levels of the TPC-1 (Figure 8A and B) and IHH4 (Figure 8C and D) cells, while miR-375 inhibitor produced completely opposite effects (P<0.05). Moreover, siLINC02471 increased the E-Cad level of TPC-1 and IHH4 cells previously reduced by miR-375 inhibitor, and reduced the N-Cad and Snail levels of the cells previously increased by miR-375 inhibitor (P<0.05).
Figure 8.
Effect of siLINC02471 and miR-375 on the EMT of PTC cell. (A) In TPC-1, Western blot was performed to determine the EMT-relative protein expression. (B) The expressions of E-Cad, N-Cad and Snail in each group. (C) In IHH4, Western blot was performed to determine the EMT-relative protein expression. (D) The expressions of E-Cad, N-Cad and Snail in each group. (B and D) **P<0.01 vs MC; ##P<0.01 vs IC; ^^P<0.01 vs I.
The above data indicated that miR-375 mimic inhibited the cell proliferation, invasion, migration and EMT, promoted apoptosis, while miR-375 inhibitor exerted opposite effects to miR-375 mimic. Moreover, after knocking down the expressions of miR-375 and LINC02471, the effects of miR-375 inhibitor were reversed by siLINC02471, suggesting that LINC02471 regulated human PTC progression through mediating miR-375.
Discussion
Recently, studies have confirmed that many lncRNAs are differentially expressed in the thyroid gland and can affect the occurrence of thyroid cancer.16,17 In this study, we observed that LINC02471 expression in PTC tissues was increased, and at the same time also increased in the three PTC cell lines, especially in TPC-1 and IHH4 cells, however, it was low-expressed in normal thyroid cell lines, suggesting that LINC02471 might be an oncogenic gene for PTC. Subsequently, in the present study, to verify the biological role of LINC02471 expression changes in PTC development, TPC-1 and IHH4 cell lines were selected as research models, and the LINC02471 expression was successfully knocked down in TPC-1 and IHH4 cells by siRNA technology.
Further studies were conducted to determine the effect of LINC02471 knockdown on PTC cells, and the results that knocking down LINC02471 inhibited proliferation, invasion and migration of the PTC cell lines, while downregulating LINC02471 expression could promote the apoptosis of PTC cell lines. The occurrence and development of malignant tumors are closely related to the rapid proliferation of cells, inhibition of apoptosis, enhanced invasion and metastasis abilities.18–20 Consistent with our findings, we speculate that LINC02471 may affect the development of PTC by regulating biological behaviors of PTC cells. In addition, LINC02471 is possibly an oncogene for PTC, and might be a new direction of targeted gene therapy for PTC.
Invasion and metastasis are detected in many PTC patients.21,22 EMT refers to the process during which cells transform from the epithelium to the interstitial under the action of certain factors.23,24 EMT is also associated with the invasion of PTC.25,26 The reduction or loss of E-Cad expression marks EMT change. Reduced expression of E-Cad can lead to decreased adhesion between cells and enhanced invasion and metastasis of tumors.27,28 In other words, low expression of E-Cad is involved in the malignant transformation of tumors. Snail is a transcription factor regulating the EMT process, and it could induce EMT through inhibiting the transcription expression of E-Cad.29,30 N-Cad, a mesenchymal cell marker, does not express in normal epithelial tissues, but its ectopic expression in epithelial tissues can induce EMT, causes tumor cells to develop invasive phenotype and enables distant metastasis.31,32 The current study showed that knockdown of LINC02471 increased the E-Cad expression level and reduced the N-Cad and Snail expression level in PTC cell lines, suggesting that LINC02471 could enhance the metastasis of PTC by inducing EMT.
LncRNAs can serve as ceRNAs binding to miRNAs through “sponge” adsorption to reduce miRNA activity, affect miRNA levels of downstream target genes, and participate in the regulation of cell biological behaviors.33,34 MiRNAs are noncoding endogenous small molecular RNAs with tissue specificity, and can directly act on the relevant genes, affect tumor migration and invasion.35,36 Other scholars have found that lncRNAs bind miRNAs through molecular sponge adsorption and participates in the PTC process, for instance, as found by Zhang et al,37 LncRNA BISPR can competitively bind with miR-21-5p to promote the progression of PTC; Sun et al38 showed that LncRNA CRNDE can promote the development of PTC cells by regulating miR-384. In this study, whether LINC02471 regulated cell biological behaviors by binding with miRNAs should be further studied. The results showed that LINC02471 negatively regulated the miR-375 expression in PTC cell lines.
Wang et al39 pointed out that miR-375 expression was reduced in human PTC tissues and cell lines, and overexpressed miR-375 could inhibit PTC progression. Consistently, we also found that increased miR-375 expression inhibited the proliferation and invasion of PTC cell lines, and promoted the cell apoptosis, while the inhibition of miR-375 showed the opposite results. Moreover, overexpression of miR-375 inhibited EMT of PTC cell lines, while inhibition of miR-375 produced opposite effects. After miR-375 expression was inhibited, silencing LINC02471 expression in PTC cells reversed the proliferation, invasion and EMT promoted by miR-375 inhibitor and its inhibition of the cell apoptosis. Thus, siLINC02471 could negatively regulate the miR-375 expression to inhibit the proliferation, invasion and EMT of PTC cells in vitro.
Our study indicated that LINC02471 expression was upregulated in PTC tissues and cell lines. Further cytological experiments preliminarily confirmed that LINC02471 knockdown inhibited the proliferation, invasion and metastasis of PTC cells, reduced expressions of EMT-related genes (N-Cad and Snail), and promoted cell apoptosis and increased E-Cad expression. LINC02471 may be a potential molecule in promoting the development of PTC. In addition, we demonstrated that LINC02471 could regulate PTC cell behaviors by directly targeting miR-375. This study provides a theoretical basis for further study on the molecular regulatory mechanism of LINC02471 in PTC progression, but the specific molecular mechanism should be further confirmed.
Funding Statement
There is no funding to report.
Abbreviations
PTC, papillary thyroid carcinoma; lncRNAs, long non-coding RNAs; CCK-8, Cell Counting Kit-8; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation; EMT, epithelial-mesenchymal transition.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Experimental procedure was approved by the Ethics Committee of Xiasha Branch of Sir Run Run Shaw Hospital, and obtained informed consent form for all patients. No animals were involved in this research.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.La Vecchia C, Malvezzi M, Bosetti C, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer. 2015;136(9):2187–2195. doi: 10.1002/ijc.29251 [DOI] [PubMed] [Google Scholar]
- 2.Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12(11):646–653. doi: 10.1038/nrendo.2016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagin JA, Wells SA. Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375(11):1054–1067. doi: 10.1056/NEJMra1501993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zivancevic-Simonovic S, Mihaljevic O, Majstorovic I, et al. Cytokine production in patients with papillary thyroid cancer and associated autoimmune Hashimoto thyroiditis. Cancer Immunol Immunother. 2015;64(8):1011–1019. doi: 10.1007/s00262-015-1705-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen QT, Lee EJ, Huang MG, Park YI, Khullar A, Plodkowski RA. Diagnosis and treatment of patients with thyroid cancer. Am Health Drug Benefits. 2015;8(1):30–40. [PMC free article] [PubMed] [Google Scholar]
- 6.Grau JJ, Caballero M, Langdon C, Bernal-Sprekelsen M, Blanch JL. Electrochemotherapy as palliative treatment in patients with thyroid papillary carcinoma. Braz J Otorhinolaryngol. 2016;82(3):285–288. doi: 10.1016/j.bjorl.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 8.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14(3):4655–4669. doi: 10.3390/ijms14034655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhiqiang W, Qian L, Tieqiang L, et al. Abnormal expressed long non-coding RNA IRAIN inhibits tumor progression in human renal cell carcinoma cells. Open Life Sci. 2016;11(1):200–205. doi: 10.1515/biol-2016-0026 [DOI] [Google Scholar]
- 10.Sui F, Ji M, Hou P. Long non-coding RNAs in thyroid cancer: biological functions and clinical significance. Mol Cell Endocrinol. 2018;469:11–22. doi: 10.1016/j.mce.2017.07.020 [DOI] [PubMed] [Google Scholar]
- 11.Murugan AK, Munirajan AK, Alzahrani AS. Long noncoding RNAs: emerging players in thyroid cancer pathogenesis. Endocr Relat Cancer. 2018;25(2):R59–r82. doi: 10.1530/ERC-17-0188 [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Shen W, Li X, Zhang L, Jin X. The lncRNA n340790 accelerates carcinogenesis of thyroid cancer by regulating miR-1254. Am J Transl Res. 2017;9(5):2181–2194. [PMC free article] [PubMed] [Google Scholar]
- 13.Luo YH, Liang L, He RQ, et al. RNA-sequencing investigation identifies an effective risk score generated by three novel lncRNAs for the survival of papillary thyroid cancer patients. Oncotarget. 2017;8(43):74139–74158. doi: 10.18632/oncotarget.18274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai WY, Chen X, Chen LP, Li Q, Du XJ, Zhou YY. Role of differentially expressed genes and long non-coding RNAs in papillary thyroid carcinoma diagnosis, progression, and prognosis. J Cell Biochem. 2018;119(10):8249–8259. doi: 10.1002/jcb.26836 [DOI] [PubMed] [Google Scholar]
- 15.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostatistics Bioinformatics Biomathematics. 2013;3(3):71–85. [PMC free article] [PubMed] [Google Scholar]
- 16.Fu XM, Guo W, Li N, et al. The expression and function of long noncoding RNA lncRNA-ATB in papillary thyroid cancer. Eur Rev Med Pharmacol Sci. 2017;21(14):3239–3246. [PubMed] [Google Scholar]
- 17.Liyanarachchi S, Li W, Yan P, et al. Genome-wide expression screening discloses long noncoding RNAs involved in thyroid carcinogenesis. J Clin Endocrinol Metab. 2016;101(11):4005–4013. doi: 10.1210/jc.2016-1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, Zhu P, Liao Y, Li J, Liao W, He S. Elevated preoperative aspartate aminotransferase to lymphocyte ratio index as an independent prognostic factor for patients with hepatocellular carcinoma after hepatic resection. Oncotarget. 2015;6(22):19217–19227. doi: 10.18632/oncotarget.4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner VP, Martins MD, Guimaraes DM, et al. Reduced chromatin acetylation of malignant salivary gland tumors correlates with enhanced proliferation. J Oral Pathol Med. 2017;46(9):792–797. doi: 10.1111/jop.12557 [DOI] [PubMed] [Google Scholar]
- 20.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15(2):2494–2516. doi: 10.3390/ijms15022494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Y, Liu X, Yu Y, et al. Association of ATM gene polymorphism with PTC metastasis in female patients. Int J Endocrinol. 2014;2014:370825. doi: 10.1155/2014/370825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Wang Y, Meng X, et al. Research on influence factors and clinical significance of delphian lymph node metastasis in papillary thyroid carcinoma. 2016.
- 23.Heerboth S, Housman G, Leary M, et al. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith BN, Bhowmick NA. Role of EMT in metastasis and therapy resistance. J Clin Med. 2016;5(2):17. doi: 10.3390/jcm5020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv N, Gao Y, Guan H, et al. Inflammatory mediators, tumor necrosis factor-α and interferon-γ, induce EMT in human PTC cell lines. Oncol Lett. 2015;10(4):2591–2597. doi: 10.3892/ol.2015.3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui D, Zhao Y, Xu J. Activated CXCL5-CXCR2 axis promotes the migration, invasion and EMT of papillary thyroid carcinoma cells via modulation of β-catenin pathway. Biochimie. 2018;148:1–11. doi: 10.1016/j.biochi.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 27.Matos ML, Lapyckyj L, Rosso M, et al. Identification of a novel human E-cadherin splice variant and assessment of its effects upon EMT-related events. J Cell Physiol. 2017;232(6):1368–1386. doi: 10.1002/jcp.25622 [DOI] [PubMed] [Google Scholar]
- 28.Meng X, Kong DH, Li N, et al. Knockdown of BAG3 induces epithelial-mesenchymal transition in thyroid cancer cells through ZEB1 activation. Cell Death Dis. 2014;5:e1092. doi: 10.1038/cddis.2014.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papiewska-Pajak I, Kowalska MA, Boncela J. Expression and activity of SNAIL transcription factor during Epithelial to Mesenchymal Transition (EMT) in cancer progression. Postepy Higieny I Medycyny Doswiadczalnej. 2016;70:968–980. doi: 10.5604/17322693.1219401 [DOI] [PubMed] [Google Scholar]
- 30.Kyung SY, Kim DY, Yoon JY, et al. Sulforaphane attenuates pulmonary fibrosis by inhibiting the epithelial-mesenchymal transition. BMC Pharmacol Toxicol. 2018;19(1):13. doi: 10.1186/s40360-018-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han B, Cui H, Kang L, et al. Metformin inhibits thyroid cancer cell growth, migration, and EMT through the mTOR pathway. Tumour Biol. 2015;36(8):6295–6304. doi: 10.1007/s13277-015-3315-4 [DOI] [PubMed] [Google Scholar]
- 32.Jung CW, Han KH, Seol H, et al. Expression of cancer stem cell markers and epithelial-mesenchymal transition-related factors in anaplastic thyroid carcinoma. Int J Clin Exp Pathol. 2015;8(1):560–568. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, Du X. Noncoding RNAs in gastric cancer: research progress and prospects. World J Gastroenterol. 2016;22(29):6610–6618. doi: 10.3748/wjg.v22.i29.6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Wang H, Wu C, et al. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39(3):1197–1206. doi: 10.3892/or.2018.6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oom AL, Humphries BA, Yang C. MicroRNAs: novel players in cancer diagnosis and therapies. Biomed Res Int. 2014;2014:959461. doi: 10.1155/2014/959461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Cai Y, Zheng L, Zhang Z, Lin X, Jiang N. LncRNA BISPR promotes the progression of thyroid papillary carcinoma by regulating miR-21-5p. Int J Immunopathol Pharmacol. 2018;32:2058738418772652. doi: 10.1177/2058738418772652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H, He L, Ma L, et al. LncRNA CRNDE promotes cell proliferation, invasion and migration by competitively binding miR-384 in papillary thyroid cancer. Oncotarget. 2017;8(66):110552–110565. doi: 10.18632/oncotarget.22819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XZ, Hang YK, Liu JB, Hou YQ, Wang N, Wang MJ. Over-expression of microRNA-375 inhibits papillary thyroid carcinoma cell proliferation and induces cell apoptosis by targeting ERBB2. J Pharmacol Sci. 2016;130(2):78–84. doi: 10.1016/j.jphs.2015.12.001 [DOI] [PubMed] [Google Scholar]