Abstract
Autophagy is a highly conserved lysosome-dependent degradation system in eukaryotic cells. This process removes long-lived intracellular proteins, damaged organelles, and recycles biological material to maintain cellular homeostasis. Dysfunction of autophagy triggers a wide spectrum of human diseases, including cancer and neurodegenerative diseases. In the present study, we show that RNF115, an E3 ubiquitin ligase, regulates autophagosome–lysosome fusion and autophagic degradation under both nutrient-enriched and stress conditions. Depletion of the RNF115 gene caused the accumulation of autophagosomes by impairing fusion with lysosomes, which results in an accumulation of autophagic substrates. Further investigation suggests that RNF115 interacts with STX17 and enhances its stability, which is essential for autophagosome maturation. Importantly, we provide in vitro and in vivo evidence that RNF115 inactivation inhibits the tumorigenesis and metastasis of BGC823 gastric cancer cells. We additionally show that high expression levels of RNF115 mRNA correlate with poor prognosis in gastric cancer patients. These findings indicate that RNF115 may play an evolutionarily conserved role in the autophagy pathway, and may act to maintain protein homeostasis under physiological conditions. These data demonstrate the need to further evaluate the potential therapeutic implications of RNF115 in gastric cancer.
Subject terms: Macroautophagy, Gastric cancer
Introduction
Autophagy is a complex biological process that is crucial for maintaining cellular homeostasis1. Autophagy is a multistep process, initiated by autophagosome formation, followed by docking and fusion of the autophagosome with the endosome/lysosome to form the autolysosome, resulting in the elimination of engulfed substrates and eventual reformation of the autophagic lysosome2. The mechanisms of autophagy initiation and autophagosome formation have been clearly described, but the mechanisms of autophagosome–lysosome fusion remain to be elucidated.
In eukaryotic cells, protein degradation is primarily mediated through the ubiquitin proteasome system (UPS) and autophagy, and there is extensive cross talk and interplay between the UPS and autophagy3. In addition, many E3 ubiquitin ligases participate in regulation of autophagy, including NEDD44, BRUCE5, Parkin6, and RNF1857. RNF115 (Rabring7 in mice) is a highly conserved protein, and is a RAB7 target protein. RNF115 directly binds the GTP-bound form of Rab7, and can be recruited efficiently to the late endosome/lysosome8. Moreover, RNF115 regulates the endosomal sorting of the EGFR protein, and knockdown of RNF115 decreases the number of multivesicular bodies (MVBs)9. Functional MVBs are required for autophagic clearance of protein aggregates10. In addition, RNF115 participates in innate immunity and can target the HIV-I virus for lysosomal degradation11–13.
In the present study, we demonstrate that RNF115 regulates autophagy and promotes autophagosome–lysosome fusion by interacting with the STX17 protein. We also show that blocking autophagy by inactivating RNF115 inhibits the growth of gastric cancer cells in vitro and in vivo, which maybe a potential therapeutic target for cancers.
Results
Knockdown of RNF115 impairs autophagic flux
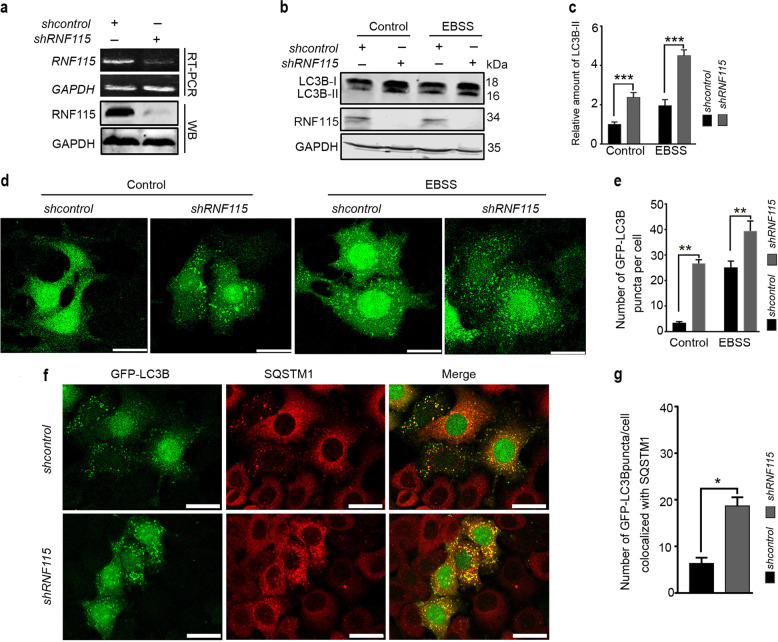
To investigate the physiological effects of RNF115 knockdown on the regulation of autophagy, experiments were conducted in RNF115-depleted cells. Using RT-PCR and western blot to evaluate the expression of gene and protein products, an effective shRNA against RNF115 (shRNF115) was identified (Fig. 1a). Data from experiments proved that depletion of RNF115 elevated the levels of endogenous LC3B-II protein (Fig. 1b, c) under both nutrient-rich and starvation conditions, compared with shcontrol-transfected cells. In the presence of rapamycin, the knockdown of RNF115 also increased the accumulation of LC3B-II (Supplementary Fig. S1a). Similar results were observed in HEK293T, BGC823, and MCF7 cell lines (Supplementary Fig. S1b, d, f). In line with these results, RNF115 knockdown increased the number of GFP-LC3B puncta per cell compared with the shcontrol group in HeLa cells. Autophagosome/autolysosome accumulation is also identified by colocalization of GFP-LC3B and P62/SQSTM1. Knockdown of RNF115 led to a significant increase in the number of GFP-LC3 puncta colocalized with SQSTM1 per cell (Fig. 1f, g). These data show that RNF115 knockdown increased the accumulation of autophagosome/autolysosome.
Fig. 1. Depletion of RNF115 casued autophagosome accumulation.
a RT-PCR and western blotting detected the levels of RNF115 in Hela cells. b, c HeLa cells were transfected with shcontrol or shRNF115 for 48 h, with or without EBSS for another 2 h, then LC3B-II levels were analyzed by western blotting. The relative amount of LC3B-II levels relative to GAPDH was analyzed. Average value in shcontrol-transfected cells without EBSS was normalized as 1. Data are means ± s.d. of results from at least three independent experiments. d, e Representative confocal microscopy images of GFP-LC3B distribution in stable GFP-LC3B Hela cells transfected with shcontrol or shRNF115 for 48 h, and treated with or without EBSS for another 2 h. The number of GFP-LC3B puncta/cell was calculated. Data are means ± s.d. of at least 50 cells scored. f, g Representative confocal microscopy images were shown in stable GFP-LC3B HeLa cells transfected with shcontrol or shRNF115 for 48 h, stained with anti-SQSTM1 antibody, and then observed by confocal microscopy. The number of GFP-LC3 puncta/cell colocalized with SQSTM1 aggregates was calculated. Data are means ± s.d. of at least 50 cells scored. Scale bar, 25 μm. *p < 0.05; **p < 0.01; ***p < 0.001.
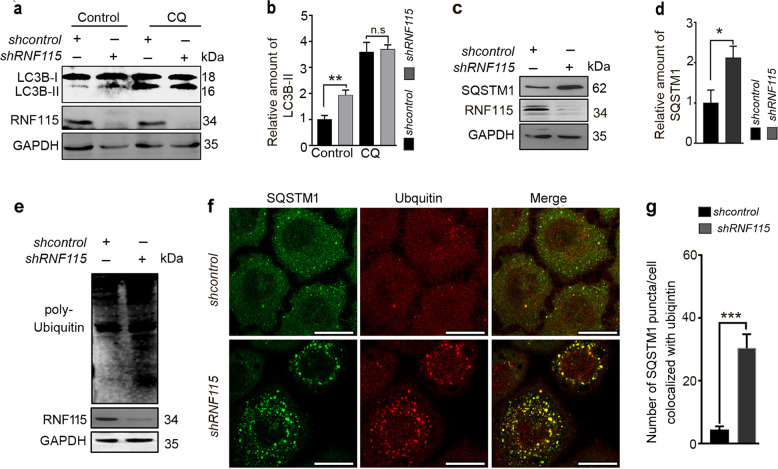
The increased levels of LC3B-II could be caused by either elevated autophagosome formation or blockade of autophagic flux14. To distinguish between these two possibilities, RNF115 knockdown cells were treated with chloroquine (CQ), which blocks autophagosome–lysosome fusion, thereby causing an accumulation of autophagosomes15. As shown in Fig. 2a, b, the levels of LC3B-II were not further increased in RNF115-depleted cells treated with CQ compared with control cells, indicating that the loss of RNF115 may block autophagic flux. We further examined the levels of autophagic substrates in RNF115 knockdown cells. SQSTM1 is a well-characterized autophagic substrate that mediates the formation and autophagic degradation of ubiquitin-positive protein aggregates16. In RNF115-depleted HeLa cells, levels of SQSTM1 and ubiquitinated proteins were increased (Fig. 2c–e). The accumulation of SQSTM1 was also presented in RNF115-silenced HEK293T, BGC823, and MCF7 cell lines (Supplementary Fig. S1c–e). Confocal microscopy revealed that SQSTM1 and ubiquitin-positive aggregates were increased and colocalized (Fig. 2f, g). In addition, in RNF115-overexpressing HeLa cells, the levels of endogenous LC3B-II protein and SQSTM1 protein were reduced (Supplementary Fig. S2a), indicating that RNF115 may promote the rate of autophagic flux and/or increase the degradation of autophagic substrates. At the same time, we performed a recovery experiment in RNF115-depleted HeLa cells. Accumulation of SQSTM1 protein was significantly decreased after transfection with the RNF115 expression plasmid in RNF115-silencing cells (Supplementary Fig. S2b), implying that RNF115 ablation modulated autophagy directly. Collectively, these findings suggest that the loss of RNF115 impairs autophagic flux in mammalian cells.
Fig. 2. Depletion of RNF115 blocks autophagic flux.
a, b HeLa cells were transfected with shcontrol or shRNF115 for 48 h, with or without CQ (50 μM) for 4 h, then the levels of LC3B-II were analyzed by western blotting. The quantification of LC3B-II levels relative to GAPDH was analyzed. Average value in shcontrol-transfected cells without CQ was normalized as 1. Data are means ± s.d. of results from at least three independent experiments. c, d HeLa cells were transfected with shcontrol or shRNF115 for 48 h, the SQSTM1 levels were analyzed by western blotting. The quantification of SQSTM1 levels relative to GAPDH was analyzed. Average value in shcontrol-transfected cells was normalized as 1. Data are means ± s.d. of results from at least three independent experiments. e Cells were treated as in c, the levels of ubiquitinated protein were detected by western blotting. f, g Hela cells were transfected with shcontrol or shRNF115 for 48 h, stained with anti-SQSTM1 and anti-ubiquitin antibodies, and observed by confocal microscope. The number of SQSTM1 puncta/cell colocalized with ubiquitin aggregates was calculated. Data are means ± s.d. of at least 50 cells scored. Scale bar, 25 μm. *p < 0.05; **p < 0.01; ***p < 0.001; n.s not significance.
RNF115 inactivation impairs autophagosome–lysosome fusion
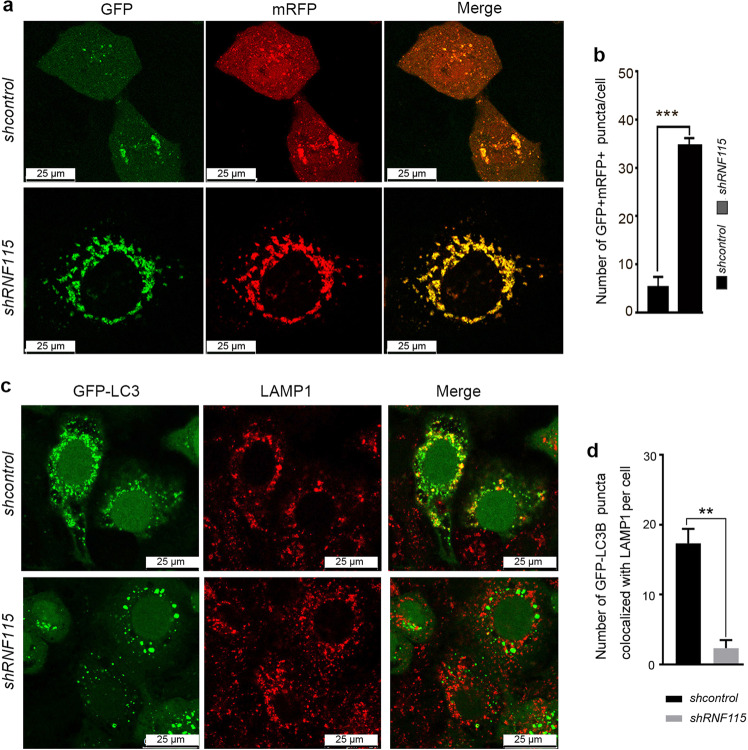
We next determined at which step in autophagic flux is impaired by RNF115 knockdown. ZFYVE1/DFCP1-labeled omegasomes, which act as cradles for autophagosome formation, exhibited no difference in control vs. RNF115 knockdown cells (Supplementary Fig. S3a, b). Next, the expression of the tandem fluorescent-tagged LC3 (mTagRFP-GFP-LC3) was examined to monitor autophagosome–lysosome fusion (autophagosome maturation). In this assay, the fluorescent signal of GFP is quenched in the lysosome, while RFP shows more stable fluorescence in acidic compartments17. Therefore, autophagosomes and amphisomes are labeled with yellow, while red fluorescence signals are detected in autolysosome. As shown in Fig. 3a, b, we observed more yellow (RFP+GFP+) puncta in RNF115-depleted HeLa cells after EBSS treatment, compared with control cells. Simultaneously, RNF115 deletion significantly decreased colocalization of GFP-LC3 with LAMP1 (Fig. 3c, d). These data suggested that RNF115 knockdown suppressed the autolysosome–lysosome fusion in the autophagy process.
Fig. 3. Knockdown of RNF115 impairs the fusion of autophagosome with lysosome.
a, b HeLa cells were transfected with shRNA or shRNF115 for 24 h, then with mTagRFP-GFP-LC3 for another 24 h and EBSS treatment for 2 h before confocal microscopy analysis. The quantification of GFP+RFP+ puncta/cell was calculated. Data are means ± s.d. of at least 50 cells scored. c, d GFP-LC3 stably expressing HeLa cells were cultured on confocal dishes and transfected with shcontrol or shRNF115 for 48 h and EBSS treatment for 2 h. Then cells were stained with anti-LAMP1, and observed by confocal microscopy. The number of GFP-LC3 puncta/cell colocalized with LAMP1 was calculated. Data are means ± s.d. of at least 50 cells scored. Scale bar, 25 μm. **p < 0.01; ***p < 0.001.
RNF115 interacts with STX17 and enhances its stability
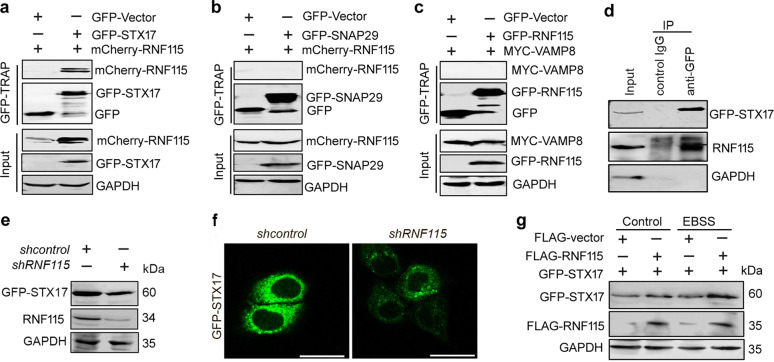
The fusion of autophagosomes with lysosomes requires the concerted actions of SNARE proteins, Rab GTPases, and tethering factors18. RNF115 binds the GTP-bound form of RAB7 at the N-terminal portion, and acts as a RAB7 effector8. Thus, we explored whether RNF115-regulated autophagosome maturation involved SNARE complexes. Using GFP-TRAP assays, we found that GFP-STX17 precipitated mcherry-RNF115 and endogenous RNF115 (Fig. 4a, d). However, RNF115 failed to interact with SNAP29 and VAMP8 protein (Fig. 4b, c). Further evaluation indicated that depletion of RNF115 downregulated the expression of STX17 protein (Fig. 4e, f). By contrast, RNF115 overexpression increased the accumulation of STX17 protein under both nutrient-rich and starvation conditions (Fig. 4g). We next measured the half-life of STX17 using the protein translation inhibitor cycloheximide (50 μg/ml) in RNF115-silenced cells. As shown in Supplementary Fig. S4, knockdown of RNF115 promoted the decay of STX17 protein comparing with that control group. These data suggest that RNF115 might regulate the homeostasis of the STX17 protein.
Fig. 4. RNF115 interacts with STX17 and enhances its stability.
a–c HEK293T cells were cotransfected with indicated plasmids for 24 h, then the cell lysates were subjected to GFP-TRAP beads. The interactions were analyzed by western blotting with anti-RNF115, anti-GFP, or anti-MYC antibodies, respevtively. GAPDH was used as the loading control. d HEK293T cells were transfected with GFP-STX17 for 24 h, the cell lysates were subjected to IP using either an anti-GFP antibody or an IgG isotype control. The interaction was analyzed by western blotting with anti-RNF115 antibodies. GAPDH was used as the loading control. e Hela cells were cotransfected with GFP-STX17 and shcontrol or shRNF115 for 24 h, the levels of GFP-STX17 were analyzed by western blotting. f Cells were treated as in e, the fluorescence of GFP-STX17 was analyzed by confocal microscope. Scale bar, 25 μm. g HEK293T cells were cotransfected with indicated plasmids for 24 h, with or without EBSS for another 2 h, then the levels of GFP-STX17 were analyzed by western blotting.
Protein ubiquitination is associated with the destabilization and degradation of proteins. Therefore, we examined whether STX17 instability caused by RNF115 knockdown was associated with ubiquitination degradation. Immunoprecipitation and western blot analysis showed that RNF115 did not significantly affect either ubk48-linked or ubk63-linked STX17 polyubiquitination levels (Supplementary Fig. S5), implying that another mechanism may be involved in the downregulation of STX17 in the RNF115-silenced cells.
RNF115 deletion inhibits the tumorigenesis and metastasis of BGC823 cells
RNF115 is highly expressed in ERα-positive breast cancer cell lines and tumors, and positively regulates the growth of breast cancer cells19,20. However, the function of RNF115 in other cancers is unknown. The Gene Expression profiling Interactive Analysis database21 showed that the expression levels of RNF115 mRNA in patients with gastric carcinoma is higher than in the normal population (Supplementary Fig. S6a). Using the Kaplan–Meier Plotter online database (http://kmplot.com/analysis/index.php?p=service&cancer=gastric), we analyzed the correlation between RNF115 mRNA levels and survival time in 876 patients, with gastric cancer. Low levels of RNF115 mRNA correlated with better overall survival in gastric cancer patients (Supplementary Fig. S6b), indicating that decreased expression of RNF115 may be a favorable prognostic indicator in patients with gastric cancer. The expression trend of STX17 is similar to that of RNF115 (Supplementary Fig. S6c, d). Correlation analysis (http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=boxplot) suggested a positive correlation between RNF115 and STX17 expression in gastric cancer (Supplementary Fig. S6e). Data from tissue microarray and immunohistochemistry demonstrate that RNF115 protein displayed moderate or high expression levels in most gastric adenocarcinoma cells (Supplementary Fig. S6f). RNF115 protein was mainly located in the cell cytoplasm of the mucosa gland, and exhibited a diffuse expression pattern.
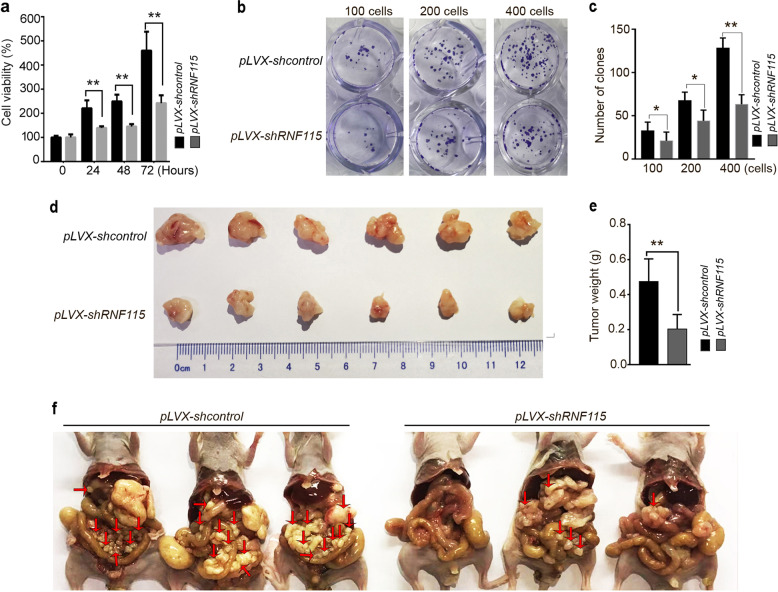
To further explore the role of RNF115 in the development and progression of gastric cancer, we constructed lentiviral knockdown vector, pLVX-shRNF115, and screened BGC823 cell lines for stable knockdown of RNF115. qRT-PCR and western blot analysis confirmed that the RNF115 mRNA and protein were significantly decreased in BGC823 cells, respectively (Supplementary Fig. S7a, b). In addition, the accumulation of both SQSTM1 and LC3B-II was observed by western blot analysis, and CQ treatment failed to affect the levels of these proteins (Supplementary Fig. S7c), indicating the blockade of autophagic flux in RNF115-depleted BGC823 cells. Furthermore, cell viability and colony formation assays suggested that knockdown of RNF115 arrests the growth of BGC823 cells (Fig. 5a–c).
Fig. 5. RNF115 knockdown in BGC823 cells inhibits cell growth, reduces tumorigenicity and metastasis.
a BGC823 cells stable infected with pLVX-shcontrol or pLVX-shRNF115 were seeded in 96-well plates (3 × 103 cells/well; five replicates), serum starved for 18 h and then pulsed with 10% FBS for indicated time. Cell viability was detected. Data are means ± s.d. of results from three independent experiments. b Representative images of colony formation by indicated cells. c Number of clones counted in three independent experiments. d, e BGC823 cells stable infected with pLVX-shcontrol or pLVX-shRNF115 (4 × 106 cells/mouse) were injected subcutaneously in BALB/c nude mice. At day 19, excised xenograft tumors were imaged and weighed (n = 6). f BGC823 cells stable infected with pLVX-shcontrol or pLVX-shRNF115 (6 × 106 cells/mouse) were injected into the abdominal cavity of mouse. At day 28, the mice were sacrificed and the metastatic nodules was observed and photographed. Arrows indicate the disseminated tumor nodules. *p < 0.05; **p < 0.01.
The in vivo effects of RNF115 were evaluated using a gastric cancer xenograft model established in BALB/C nude mice. These mice were subcutaneously injected with pLVX-shcontrol/BGC823 cells or pLVX-shRNF115/BGC823 cells. The control group developed grossly visible tumors at the site of injection (Fig. 5d). By comparison, the pLVX-shRNF115 group displayed smaller tumors. The tumor weight in the pLVX-shRNF115 group was significantly lower than that in the control group (Fig. 5e), suggesting that knockdown of RNF115 inhibits the growth of BGC823 cells in vivo.
We further established an animal model of peritoneal metastasis by injection of pLVX-shcontrol/BGC823 cells or pLVX-shRNF115/BGC823 cells into mice to explore the effects of RNF115 on tumor metastasis. All mice were euthanized 28 days after injection. A large number of metastatic nodules were found in the peritoneal cavity in control mice (Fig. 5f). By contrast, the pLVX-shRNF115 group had few or no tumor nodules. These data suggested that the inactivation of RNF115 might prevent peritoneal homing of gastric cancer cells and growth of metastatic nodules.
Discussion
In this study, we demonstrate that RNF115 is a positive regulator that participates in the autophagosome–lysosome fusion. Inactivation of RNF115 impaired autophagy, and caused autophagic substrate accumulation under basal or starvation conditions. We report that RNF115 interacts with STX17 and enhances its stability, which can promote autophagosome maturation. In addition, RNF115 silencing can inhibit the tumorigenicity and metastasis of gastric cancer cells.
Autophagosome–lysosome fusion is an important autophagic process for cargo degradation. ATG8 family members, tethering factors, Rab GTPases (such as RAB7), and SNARE proteins act coordinately to mediate this process. Dysfunction of autophagosome maturation is associated with various human diseases, including neurodegenerative diseases, cancer, and lysosomal storage disorders22. Understanding the molecular mechanisms underlying autophagosome maturation will provide new insights into the pathogenesis and treatment of these diseases. STX17 is a well-studied SNARE protein that mediates autophagosome–lysosome fusion. Upon starvation stimulus, STX17 translocates to autophagosomes, where it forms a complex with SNAP29, VAMP8, or VAMP723. STX17 inactivation prevents autophagosome–lysosome fusion. Furthermore, ATG14 and UVRAG can also promote the fusion of autophagosomes and lysosomes. ATG14 can bind to STX17, and stabilizes the STX17–SNAP29 binary t-SNARE complex on autophagosomes, further priming the autophagosome for VAMP8 interaction and promoting autophagosome–lysosome fusion. Our results show that RNF115 can interact with STX17 and enhance its stability, but not SNAP29 or VAMP8. We suspect that RNF115 may not bind the individual VAMP8 and SNAP29 proteins, but may associate with the STX17–SNAP29–VAMP8 complex through interacting with STX17, thereby promoting the maturation of the autophagosome or increasing the fusion rate. In addition, our results also show that RNF115 can mediate the stability of STX17. As an E3 ubiquitin ligase, RNF115 does not influence the ubiquitination of STX17, implying that another mechanism may be involved in the homeostatic regulation of STX17 protein by RNF115, which merits further investigation.
RNF115 has been reported to be overexpressed in >50% of invasive breast tumors, and is important for regulating breast cancer cell proliferation, migration, and invasion20. Moreover, high expression of RNF115 is associated with regional recurrence, lymph node metastasis, and unfavorable prognosis in breast cancer. Mechanistic investigations reveal that RNF115 promotes breast cancer cell proliferation through targeting the cyclin-dependent kinase inhibitor p21Waf/Cip1 for ubiquitin-dependent degradation. In the present study, it was found that the expression of RNF115 is increased in gastric adenocarcinoma samples compared to normal gastric tissue. Furthermore, the low levels of RNF115 mRNA in gastric tissues correlated to better prognosis in patients with gastric cancer, indicating that the expression of RNF115 may be a potential independent prognostic factor for patients with gastric cancer. In vitro and in vivo investigation demonstrated that RNF115 knockdown in gastric cancer cells can decrease cell growth and reduce tumorigenicity in a xenograft model. This phenomenon may be related to the blockade of autophagy, suggesting that the inactivation of RNF115 exerted antitumor activity in gastric cancer cells. The relationship between autophagy and tumor growth and metastasis regulated by RNF115 requires further elucidation.
Materials and methods
Antibodies, plasmids, and reagents
Primary antibodies used in this study include monoclonal antibodies against RNF115 (Sigma Aldrich, St. Louis, MO, USA, 019130), SQSTM1 (MBL, Woburn, MA, USA, PM045), LC3 (Cell Signaling Technology, Danvers, MA, USA, 2775 s), ubiquitin (Cell Signaling Technology, 3936), GAPDH (Sungene, Tianjin, China, KM9002), MYC (Sungene, KM8003), and GFP (Sungene, KM8009). Secondary antibodies included DyLight 800/DyLight 680-conjugated IgG against mouse (Rockland, Philadelphia, PA, USA, 610-145-002/610-144-002) or rabbit (Rockland, 611-145-002/611-144-002).
GFP-STX17, MYC-VAMP8, and GFP-SNAP29 plasmids were kindly provided by Dr. Hong Zhang (Institute of Biophysics, Chinese Academy of Sciences). RNF115 was constructed in the pEGFP-C1 vector and mCherry-C1 vector as GFP-RNF115 or mCherry-RNF115, respectively. GFP-LC3 and mTagRFP-GFP-LC3 plasmids were produced in our lab. CQ (C6628) and MG132 (C2211) were purchased from Sigma Aldrich.
Cells culture and transfection
HeLa cells, BGC823 cells, and HEK293T cell lines from American Type Culture Collection were cultured in Dubelcco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA, 12800-017) supplemented with 10% fetal bovine serum and maintained at 37 °C in a humidified incubator with 5% CO2. HeLa cells stably expressing GFP-LC3 were kindly provided by Dr. Li Yu (Tsinghua University).
The shRNA targeting the RNF115 gene was synthesized by GenePharma (Suzhou, China) and the targeting sequence was CGTCTGAATAGAATTAATT. The negative control shRNA sequence consisted of an oligonucleotide sequence with no sequence homology to any known human gene. NEOFECTTM DNA transfection reagent was used for transient transfection of plasmids, according to manufacturer’s instructions. For shRNA transfection, cells were transfected with Lipofectamine3000 according to manufacturer’s instructions (Invitrogen, L3000015).
Western blot analysis
Treated cells were washed twice with cold phosphate-buffered saline (PBS) solution and lysed in RIPA buffer containing a cocktail of protease inhibitors. The cells were then placed on ice for 30 min, and the lysates were centrifuged at 15,000 r.p.m. for 15 min at 4 °C. The total protein concentration was measured with a Protein Quantification Kit (BCA Assay). Equal amounts of proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels and transferred onto nitrocellulose membranes. After blocking the membrane with 5% nonfat milk for 1 h, blots were visualized using an IRDye 800CW-conjugated or Alexa Fluo 680 secondary antibody, and imaged using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA).
GFP-TRAP assays
HEK293T cells were transiently cotransfected with the indicated plasmids for 24 h, then treated with MG132 (20 μM) for 4–6 h. Cell lysates were immunoprecipitated by GFP-TRAP agarose beads for 4 h at 4 °C. Then the GFP-TRAP agarose beads were washed with PBS five times and boiled for 10 min in 2 × SDS–PAGE loading buffer. The samples were analyzed by western blotting.
Immunofluorescence, fluorescence, and confocal microscopy assays
HeLa cells were cultured in confocal dishes and transfected with the indicated plasmids. Cells were then washed with PBS and fixed in 4% PFA (dissolved in PBS) for 30 min at 37 °C. The fixed cells were permeabilized with 0.2% Triton X-100 for 15 min at 37 °C and blocked with BSA for 1 h. The cells were then incubated with primary antibodies overnight at 4 °C. After washing three times with PBS-T, cells were stained with FITC/TRITC-conjugated secondary antibodies and imaged by a Leica TCSSP8 Confocal Microscope.
For the tandem mRFP-GFP-LC3 assay, HeLa cells were transfected with mRFP-GFP-LC3 or cotransfected with the indicated plasmids for 24 h. Next, cells were fixed with 4% PFA at 37 °C for 15 min, coverslipped, sealed with nail polish, and observed under a confocal fluorescent microscope. Representative cells were randomly selected for analysis, and the numbers of GFP+ mRFP+ puncta per cell were calculated.
Cell viability and colony formation assays
BGC823 cells stably transduced with pLVX-shcontrol or pLVX-shRNF115 were treated with serum deprivation for 18 h. Cells were then cultured in complete culture medium for indicated time. Cell viability assays were performed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, G1111) according to the manufacturer’s instructions. Absorbance at 490 nm was measured on an EL-311SX ELISA Reader (Bio-Tec Instruments, USA). Cell viability was calculated as follows: cell viability = absorbance of test group/absorbance of control cell group × 100%. Each experiment was performed in biological triplicate and independently repeated three times.
For the colony formation assay, cells were plated in triplicate at 100, 200, and 400 cells/well, and cultured for 2 weeks in 24-cell plates. Then cells were fixed with methanol for 15 min and stained with crystal violet for 30 min. Colonies were counted and photographed.
Tumorigenicity in nude mice
A nude mouse xenograft model was established using 8-week-old male BALB/c nude mice (Experimental Animal Center, Peking University Health Sciences Center, Beijing, China). Mice were housed and maintained in a pathogen-free facility, and all experimental procedures were approved by the Institutional Authority for Laboratory Animal Care of Peking University (LA2019203). BGC823 cells stably transduced with pLVX-shcontrol or pLVX-shRNF115 were subcutaneously injected in the right axilla of randomized BALB/c nude mice (3 × 106 cells/mouse, n = 6). Mice were euthanized at 19 days after cell inoculation. Tumors were excised and photographed.
Peritoneal metastasis assay in nude mice
BGC823 cells stably transduced with pLVX-shcontrol or pLVX-shRNF115 (6 × 106 cells/mouse, n = 6) were injected into the peritoneum of randomized BALB/c nude mice (8-week-old male). The body weight of the mice was monitored every 4 days after injection. At 28 days after inoculation, the mice were euthanized and metastatic nodules were observed and photographed.
Statistics analysis
Data are presented as the mean ± s.d. Differences between groups were analyzed using the Student’s t test for continuous variables. Statistical significance in this study was set at p < 0.05. All reported p values are two sided.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (91954116 and 31872827), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT320006).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by B. Zhivotovsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Riyong Li, Zhaohui Gu
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03011-w).
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 2017;130:1209–1216. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 3.Ji CH, Kwon YT. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells. 2017;40:441–449. doi: 10.14348/molcells.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun A, et al. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy. 2017;13:522–537. doi: 10.1080/15548627.2016.1268301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebner P, et al. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat. Commun. 2018;9:599. doi: 10.1038/s41467-018-02823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang F, et al. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS ONE. 2011;6:e24367. doi: 10.1371/journal.pone.0024367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol. Biol. Cell. 2003;14:3741–3752. doi: 10.1091/mbc.e02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CJ, Berry DM, McGlade CJ. The E3 ubiquitin ligases RNF126 and Rabring7 regulate endosomal sorting of the epidermal growth factor receptor. J. Cell Sci. 2013;126:1366–1380. doi: 10.1242/jcs.116129. [DOI] [PubMed] [Google Scholar]
- 10.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyakawa K, et al. BCA2/Rabring7 promotes tetherin-dependent HIV-1 restriction. PLoS Pathog. 2009;5:e1000700. doi: 10.1371/journal.ppat.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nityanandam R, Serra-Moreno R. BCA2/Rabring7 targets HIV-1 Gag for lysosomal degradation in a tetherin-independent manner. PLoS Pathog. 2014;10:e1004151. doi: 10.1371/journal.ppat.1004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colomer-Lluch M, Serra-Moreno R. BCA2/Rabring7 interferes with HIV-1 proviral transcription by enhancing the SUMOylation of IκBα. J. Virol. 2017;91:e02098–16. doi: 10.1128/JVI.02098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 15.Mauthe M, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 17.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2014;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 18.Zhao YG, Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J. Cell Biol. 2019;218:757–770. doi: 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, et al. RNF115/BCA2 E3 ubiquitin ligase promotes breast cancer cell proliferation through targeting p21Waf1/Cip1 for ubiquitin-mediated degradation. Neoplasia. 2013;15:1028–1035. doi: 10.1593/neo.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger AM, et al. A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65:10401–10412. doi: 10.1158/0008-5472.CAN-05-2103. [DOI] [PubMed] [Google Scholar]
- 21.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun, C. W. & Lee, S. H. The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466 (2018). [DOI] [PMC free article] [PubMed]
- 23.Itakura E, Mizushima N. Syntaxin 17: the autophagosomal SNARE. Autophagy. 2013;9:917–919. doi: 10.4161/auto.24109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.