Abstract
Purpose
Glioblastoma (GBM) is the most aggressive type of brain tumor and has poor survival outcomes, even after a combination of surgery, radiotherapy, and chemotherapy. Temozolomide is the only agent that has been shown to be effective against GBM, suggesting that combination of temozolomide with other agents may be more effective. Niclosamide, an FDA approved anthelmintic agent, has shown anti-cancer effects against human colon, breast, prostate cancers as well as GBM. However, the efficacy of the combination of niclosamide with temozolomide against GBM tumorspheres (TSs) has not been determined. We hypothesized that the combined treatment could effectively suppress GBM TSs.
Methods
GBM TSs (TS15-88, GSC11) were treated with niclosamide and/or temozolomide. Combined effects of two drugs were evaluated by measuring viability, neurosphere formation, and 3D-invasion in collagen matrix. Transcriptional profiles of GBM TS were analyzed using RNA sequencing. In vivo anticancer efficacy of combined drugs was tested in a mouse orthotopic xenograft model.
Results
Combination treatment of niclosamide and temozolomide significantly inhibited the cell viability, stemness, and invasive properties of GBM TSs. This combined treatment significantly down-regulated the expression of epithelial mesenchymal transition-related markers, Zeb1, N-cadherin, and β-catenin. The combined treatment also significantly decreased tumor growth in orthotopic xenograft models.
Conclusion
The combination of niclosamide and temozolomide effectively decreased the stemness and invasive properties of GBM TSs, suggesting that this regimen may be therapeutically effective in treating patients with GBM.
Electronic supplementary material
The online version of this article (10.1007/s00432-020-03330-7) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma, Invasion, Niclosamide, Temozolomide, Tumorsphere
Introduction
Patients with glioblastoma (GBM), the most common type of primary brain tumor, have a poor prognosis, with a median overall survival of about 17.5 months in Korea (Kim et al. 2017a). Standard treatments in patients with GBM include chemotherapy with the alkylating agent temozolomide, along with surgical resection and radiotherapy (Roh et al. 2017, 2020; Stupp et al. 2005). Outcomes may be improved by combining temozolomide with other therapeutic agents, especially old or failed drugs (Chong and Sullivan 2007). Because the pharmacokinetics of existing drugs have been determined, and because many of these agents have been approved for human use, many clinical trials have evaluated new uses of old drugs (Chong and Sullivan 2007).
Niclosamide is an anthelmintic agent approved by the U.S. Food and Drug Administration (FDA) that has been used to treat tapeworm infection for approximately 50 years (Garin et al. 1964). Recent drug screening has identified niclosamide as a potential anticancer agent (Li et al. 2014). This agent has been found to inhibit cancer-associated signaling cascades, including Wnt (Sack et al. 2011), mTORC1 (Balgi et al. 2009), STAT3 (Ren et al. 2010), NF-κB (Wieland et al. 2013) and Notch (Wang et al. 2009). In addition, the anti-cancer effects of niclosamide have been demonstrated in human colon (Sack et al. 2011), breast (Londono-Joshi et al. 2014), prostate (Lu et al. 2011), lung (Stewart et al. 2016) cancers as well as in GBM (Wieland et al. (2013); Cheng et al. 2017).
GBM tumorspheres (TSs) are cells refractory to treatment (Kang et al. 2015; Kong et al. 2013a, b), making them a good testing platform to evaluate the efficacy of drugs (Kim et al. 2017b; Choi et al. 2016; Park et al. 2018). Niclosamide was found to have anticancer effects against GBM TSs in vitro (Wieland et al. 2013; Cheng et al. 2017) and in vivo (Wieland et al. 2013). However, the effects of the combination of niclosamide and temozolomide on GBM TSs in vivo have not yet been determined. In this study, we examined combined effects of niclosamide and temozolomide using patient-derived TSs and a mouse orthotopic xenograft model. Our findings suggest that this combined treatment could be a new treatment option for GBM.
Materials and methods
Cell culture and reagents
Two primary tumor cells derived from GBM patients, TS15-88 and GSC11, were used to create TS models in this study. TS15-88 was established from fresh GBM tissue specimens, as approved by the institutional review board of Yonsei University College of Medicine (4-2014-0649). Patient-derived GSC11 cells were provided by Dr. Frederick F. Lang (Department of Neurosurgery, The University of Texas, M. D. Anderson Cancer Center, Houston, Texas, USA). For TS culture, cells were cultured in TS complete media composed of DMEM/F-12 (Mediatech, Manassas, VA, USA), 1 × B27 (Invitrogen, San Diego, CA, USA), 20 ng/ml of bFGF, and 20 ng/ml of EGF (Sigma-Aldrich, St. Louis, MO, USA) (Kwak et al. 2013). All in vitro experiments were performed under TS culture conditions. For in vitro treatments, niclosamide and TMZ were dissolved in DMSO and added to cell cultures to the desired concentration.
Characterization of GBM TSs
TS formation from human GBM specimens followed previous methods (Kong et al. 2013b), and their expression of stemness markers, CD133, nestin, musashi, and podoplanin (Abcam, Cambridge, UK), was tested by immunocytochemistry. Neuroglial differentiation in GBM TSs was evaluated by monitoring the expression of GFAP (Dako, Carpinteria, CA, USA), MBP, NeuN, and TUBB3 (Chemicon, Temecula, CA, USA).
Cell viability assay
The effects of niclosamide, TMZ, and the combination of niclosamide and TMZ on cell survival were determined using MTS viability assays (Promega, Madison, WI, USA) (Mosmann 1983). GBM TS cells seeded in 96-well plates (1 × 104 cells/well) were incubated at 37 °C for 24 h and treated with niclosamide and/or temozolomide for 3 days. MTS reagent (20 μl/well) was added, the cells were incubated at 37 °C for 2 h, and the absorbance of each well was measured at 595 nm. Each experiment was repeated three times in triplicate, with the results expressed as the percentage of viable cells relative to controls. Synergy score for combination treatment of niclosamide and TMZ was calculated using Bliss method. Combination indices (CIs) of combined treatment with niclosamide and temozolomide were calculated by CompuSyn software.
Sphere formation assay
Dissociated 10 single GBM TSs were cultured in 96-well plates in medium containing DMEM/F-12 (Mediatech, Manassas, VA, USA), supplemented with 1 × B27 (Invitrogen, San Diego, CA, USA), 20 ng/ml of bFGF, 20 ng/ml of EGF (Sigma-Aldrich, St. Louis, MO, USA), and 50 U/ml penicillin/50 mg/ml streptomycin. After 3 weeks of incubation under different conditions, the number of sphere-positive wells was counted, and the proportion of sphere-positive wells in the treatment group relative to that in controls was calculated and presented as a percentage. Images of sphere positive wells were captured and analyzed using ToupView software (ToupTek Photonics, Zhejiang, China).
Invasion assay
GSC-11 and TS15-88 cells grown as single spheroids were seeded and cultured in individual wells of a 96-well plate. Each well was filled with mixed matrix composed of Matrigel, collagen type I (Corning), and TS complete media. Single spheroids were seeded inside the matrix prior to gelation. Then, TS complete media was added over the gelled matrix to prevent drying. The invaded area was quantified as occupied area at (72–0 h)/0 h.
Western blot analyses
Cell lysates were separated by SDS-PAGE on 10% Tris–glycine gels. Proteins were transferred to nitrocellulose membranes and probed with antibodies against Sox2 (Merck Millipore, Billerica, MA, USA); Nestin (Novus Biologicals, Littleton, CO, USA); PDPN and β-catenin (Cell Signaling Technology, Beverly, MA, USA); N-cadherin (R&D Systems); Zeb1 (Sigma-Aldrich); STAT3 (Cell Signaling Technology); MGMT (MT3.1); Phosphorylated STAT3 (Cell Signaling Technology); and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Proteins were detected using horseradish peroxidase-conjugated IgG (Santa Cruz Biotechnology) in conjunction with Western Lightning Plus-enhanced chemiluminescence reagent (PerkinElmer, Waltham, MA, USA). Images were captured using an ImageQuant LAS 4000 mini (GE Healthcare Life Sciences, Little Chalfont, UK).
RNA QC, library construction, and sequencing
The quality and quantity of total RNA were assessed by Agilent 2100 Bioanalyzer with a Eukaryotic Total RNA Pico chip (Agilent Technologies). Libraries were quantified using the Agilent TapeStation 4200 HSD1000 screen tapes (Agilent Technologies) and KAPA Library Quantification Kit (KK4824, Kapa Biosystems). The individual samples were pooled and sequenced on the Illumina NovaSeq6000 with 150 bp paired-end by following the manufacturer's protocols. Image analysis were performed using the NovaSeq6000 control Software version 1.3.1 and the output data was demultiplexed with bcl2fastq v2.2 generating fastqc files. Detailed description of experimental materials and methods is given in the supplementary experimental procedures.
Preprocessing of transcriptome data
The quality of the reads was checked using fastQC (v.0.10.1) and the sequencing adapters were removed using trimmomatic (v. 0.38). Low quality reads were filtered according to the following criteria; reads contain more than 10% of skipped bases (marked as ‘N’s), reads contain more than 40% of bases whose quality scores are less than 20, and reads whose average quality scores of each read are less than 20. Filtered reads were mapped to the human reference genome (Ensembl release 72 (Flicek et al. 2013)) using the aligner Tophat (Trapnell et al. 2009). Gene expression level was measured with Cufflinks v2.1.1 (Trapnell et al. 2012) using the gene annotation database of Ensembl release 72. Non-coding region was removed with—mask option.
Functional annotation to DEGs
A total of 1391 DEGs (One-way ANOVA with Tukey’s post hoc test; P < 0.001) was identified between control and combination groups. Functional annotation to these DEGs was performed by over-representation analysis (ORA) using GO gene sets, and then visualized as an enrichment map using Cytoscape (Shannon et al. 2003) and ClueGO (Bindea et al. 2009) plug-in. Enriched GO terms were functionally categorized based on their kappa scores (> 0.4). Statistical significance was determined using two-sided hypergeometric test, and only nodes with Bonferroni-adjusted P value < 0.05 were displayed.
Mouse orthotopic xenograft model
Male athymic nude mice (4–8 weeks old; Central Lab. Animal Inc., Seoul, Korea) were used in this study. Mice were housed in micro-isolator cages under sterile conditions and monitored for at least 1 week before study initiation to ensure proper health. Lighting, temperature, and humidity were controlled centrally. Mice were anesthetized with a solution of Zoletil (30 mg/kg; Virbac Korea, Seoul, Korea) and xylazine (10 mg/kg; Bayer Korea, Seoul, Korea), which was administered intraperitoneally. GBM TSs (GSC11) were pretreated by niclosamide (500 nM), TMZ (250 µM), and combination of niclosamide and TMZ for 3 days with reference to the pretreatment method from several studies (Liu et al. 2016; Natale et al. 2018; Wang et al. 2019; Xia et al. 2017). DMSO-control (n = 5), niclosamide (n = 5), TMZ (n = 5), and combination of niclosamide and TMZ (n = 5)—pretreated GBM TSs were implanted into the right frontal lobe of nude mice using a guide-screw system (Lal et al. 2000). total of 5 × 105 cells was injected to a depth of 4.5 mm using a Hamilton syringe (Dongwoo Science Co., Seoul, Korea). Mice were euthanized according to the approved protocol if daily monitored body weight had decreased by more than 15% compared to the original body weight.
Bioluminescence imaging
Bioluminescence acquisition and analyses were performed using an IVIS imaging system and Living Image v4.2 software (Caliper Life Sciences, Hopkinton, MA, USA). Mice were injected intraperitoneally with 100 μl d-luciferin (30 mg/ml; Promega) 15 min prior to signal acquisition (5 s), which took place under 2.5% isoflurane anesthesia. Grayscale photographic images and bioluminescence color images were superimposed.
Statistical analysis
Levels of significance for comparisons among treatment groups were determined using one-way ANOVA with Tukey’s post hoc test for multiple comparisons. Bliss expectation was calculated as (A + B) – A × B, where A and B are the fractional growth inhibitions of drug A and B at a given dose. Survival analysis was performed using Kaplan–Meier curves with log-rank test. Results were considered statistically significant at a P value < 0.05. GraphPad Prism 6 (GraphPad Software Inc.) was used for quantitative analysis.
Results
Characterization of GBM TSs
Morphologically, GSC11 and TS15-88 cells cultured in TS complete media were spheroid shaped. Immunocytochemistry showed that both GSC11 and TS15-88 TSs were positive for the stem cell markers, CD133 and Nestin, whereas only GSC11 TSs were further positive for Podoplanin (PDPN) and Musashi. Neuroglial differentiation was successfully induced in all TSs, which was confirmed by positive GFAP, MBP, NeuN and TUBB3 stains (Supplementary Fig. 1).
Combined treatment with niclosamide and TMZ synergistically reduces cellular viability of GBM TSs
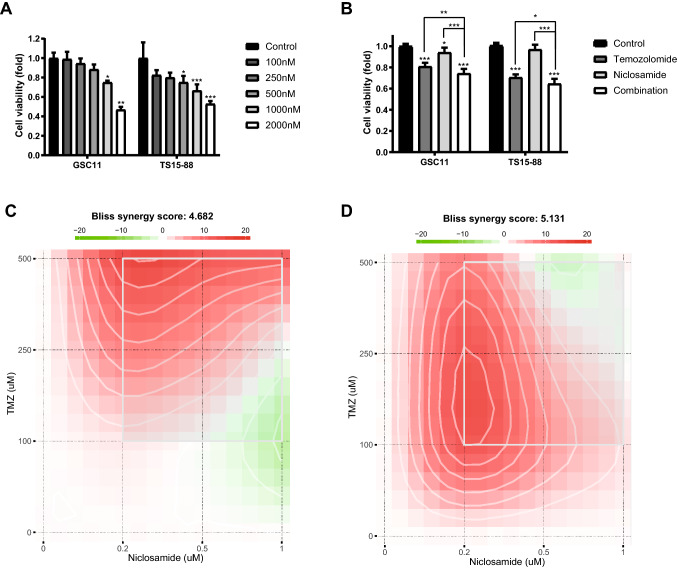
To determine an optimal working concentration of niclosamide, its cytotoxicity against GBM TSs was tested. Treatment of both GSC11 and TS15-88 TSs with niclosamide reduced cell viability in a concentration dependent manner (Fig. 1a). MTS assays testing the viability of GBM TS cells treated with niclosamide (500 nM), and/or temozolomide (250 μM) for 3 days showed that the combination treatment of niclosamide and TMZ more effectively inhibited the proliferation of GBM TSs than either agent alone or untreated controls (Fig. 1b). The synergy scores are calculated across all the tested concentration combinations by bliss method, showing that combined treatment of niclosamide and TMZ synergistically inhibited both GBM TSs (GSC11 and TS15-88) (Fig. 1c, d). The combination index of combined treatment of niclosamide (500 nM) and TMZ (250 μM) is shown in supplementary Fig. 2, suggesting an existence of synergism between the drugs.
Fig. 1.
Effects of combined treatment with niclosamide and TMZ on the viability of GBM TSs. a The viability of GBM TSs was measured 72 h after treatment with niclosamide alone at different concentration. b Cell viability was measured 72 h after treatment with niclosamide and/or TMZ. The drug interaction landscapes based on the bliss model. The bliss synergy score of drug combination with niclosamide and TMZ against c GSC11 d TS15-88 is shown. Differences among groups were compared by one-way ANOVA with Tukey’s post hoc test; means ± SD; *P < 0.05, ***P < 0.001, where asterisks denote significant differences between the indicated groups or compared with controls (asterisks over the bar)
Fig. 2.
Evaluation of stemness of GBM TSs after treatment with niclosamide and TMZ. a spheroid shaped GBM TSs. Stemness was determined by neurosphere formation assays, quantified as the percentage of b sphere-positive wells and c sphere radius. d Expression of genes related to stemness was measured by western blot analysis. Differences among groups were compared by one-way ANOVA with Tukey’s post hoc test; means ± SD; *P < 0.05, ***P < 0.001, where asterisks denote significant differences between the indicated groups or compared with controls (asterisks over the bar)
Combined treatment with niclosamide and TMZ suppresses stemness of GBM TSs
Sphere-formation assays showed that temozolomide, niclosamide, and their combination differed in their ability to inhibit the stemness of GBM TSs. GBM TS morphology was more affected by combination treatment than by either agent alone (Fig. 2a). Although temozolomide alone somewhat reduced the proportion of sphere-positive wells and niclosamide alone was ineffective, niclosamide enhanced the anti-stemness activity of temozolomide (Fig. 2b), reducing the sphere radii (Fig. 2c). Western blot analyses showed that the combination of niclosamide and temozolomide reduced the expression of stemness-related proteins, including nestin and Oct3/4 (Fig. 2d).
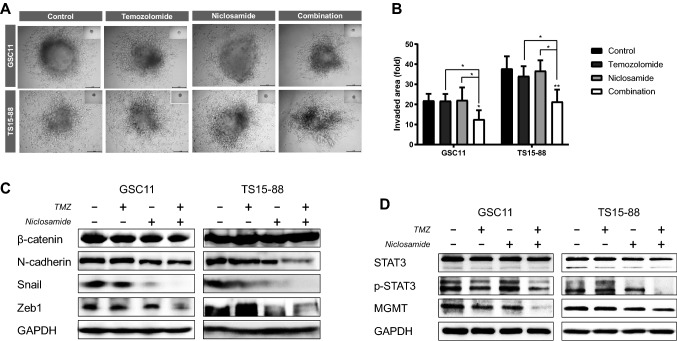
Combined treatment with niclosamide and TMZ suppresses invasiveness of GBM TSs
For 3D invasion assays, we implanted GFP-GBM TSs in a collagen type 1 matrix and evaluated the anti-invasion effects of niclosamide and/or temozolomide after 72 h. Invasion morphology was more influenced by combination treatment than by either temozolomide or niclosamide alone (Fig. 3a). Compared with untreated GBM TSs, the combination of niclosamide (500 nM) and temozolomide (250 μM) significantly inhibited invasion by the GBM TSs, GSC11 and TS15-88 cells (Fig. 3b). Western blot analyses showed that combined treatment with niclosamide and temozolomide of both TSs reduced the expression of the EMT-related markers, N-cadherin, Snail, and Zeb1 (Fig. 3c). The combination treatment of niclosamide and temozolomide also suppressed expressions of MGMT and phosphorylated STAT3 (Fig. 3d).
Fig. 3.
Effects of treatment with niclosamide and/or TMZ on the invasive properties of GBM TSs and expression of mesenchymal transition markers. Invasiveness were measured after 72 h of treatment with niclosamide and TMZ. a Decreased invasiveness of implanted GBM TSs was demonstrated using 3D collagen matrix invasion assays, under treatment with niclosamide and TMZ, alone and in combination (scale bar = 50 μM). b Inhibitory effects of drug treatment on invasiveness were quantified by measuring the area of invasion. c Expression of proteins related to mesenchymal transition and invasion was measured by western blot analysis. d Immunoblot analysis of MGMT, STAT3, and p-STAT3 in TSs treated with combination of niclosamide and temozolomide. Differences among groups were compared by one-way ANOVA with Tukey’s post hoc test; means ± SD; *P < 0.05, ***P < 0.001, where asterisks denote significant differences between the indicated groups or compared with controls (asterisks over the bar)
Transcriptional profiles following niclosamide and/or TMZ treatment
We next used RNA sequencing to examine the effect of niclosamide and TMZ on transcriptional profiles of GBM TS (GCS11). Hierarchical clustering of differentially expressed genes (DEGs) with expression levels of top 30% showed strong intragroup clustering and distinct expression patterns compared with controls (Fig. 4a). A subset of genes, which encode proteins that regulate cell stemness and EMT such as NES (nestin), SNAI2 (TWIST), and ZEB1 were significantly downregulated, validating western-blotting data treated with combination of niclosamide and TMZ (Fig. 4b). Among 1391 DEGs from control and combination groups, 892 genes from combination group were upregulated. Functional annotation of these DEGs using Gene Ontology (GO) database revealed that protein catabolism and autophagy-related gene sets were exclusively enriched by combined therapy (Fig. 4c).
Fig. 4.
GSC11 cells were treated with niclosamide and TMZ alone or in combination for 72 h, and gene expression profile was obtained using RNA-sequencing. a For genes with average expression levels of top 30%, average linkage hierarchical clustering was performed with Euclidean distance as a distance metric, and expression levels were depicted as a heat map using GENE-E software. b Expression levels of stemness- and invasiveness-associated genes were displayed as a heat map. c Among 1391 DEGs between control and combination groups, 892 genes whose expression levels were upregulated in combination group were functionally annotated, clustered, and visualized as an enrichment map. Each node represents a GO term, with the node size reflecting statistical significance for over-representation. An edge between two nodes denotes kappa score relationship. Node colors reflect clustered modules; the most significant GO terms for each module have highlighted labels
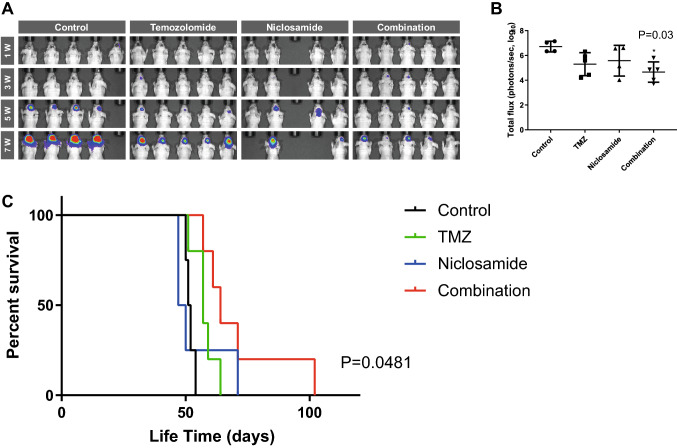
Therapeutic responses in a mouse orthotopic xenograft model
The in vivo therapeutic effects of niclosamide and/or temozolomide on tumor growth were analyzed in a mouse orthotopic xenograft model using GBM TSs (GSC11). Bioluminescence imaging showed that, compared with either agent alone, the combination of niclosamide and temozolomide significantly reduced the tumor burden compared to the control in this orthotopic xenograft model (Fig. 5a, b). In the Kaplan–Meier survival analysis, different anti-cancer effects were shown accordingly to treatment groups (Fig. 5c). Combined treatment with niclosamide and TMZ confers significant survival benefits compared to the control.
Fig. 5.
Therapeutic responses in a mouse orthotopic xenograft model. a Tumor volume was measured by bioluminescence imaging. b Signal intensity was quantified as total photon flux from tissues on the 5th week (*P < 0.05; one-way ANOVA with Tukey’s post hoc test). c Kaplan–Meier survival curve showed increased survival of mice treated with the combination of niclosamide and temozolomide compared to the control (P = 0.0481)
Discussion
Despite recent progress in understanding the biology of GBM, many tested chemotherapy strategies have been found that they do not confer significant survival advantages over treatment with temozolomide alone. Accordingly, more effective therapeutic agents are required for clinical management of GBM. Because TSs derived from GBM are highly resistant to radiation and chemotherapy (D'Alessandris et al. 2017), we screened drug regimens against GBM TSs (Kim et al. 2017b; Choi et al. 2016; Park et al. 2018).
Although niclosamide has anti-cancer effects against various types of cancer including GBM (Sack et al. 2011; Wieland et al. 2013; Londono-Joshi et al. 2014; Lu et al. 2011; Stewart et al. 2016), the in vivo combination effects of niclosamide and TMZ against GBM TSs are still unknown. The present study evaluated the effects of treatment with niclosamide and temozolomide, alone and in combination, on the stemness and invasiveness of GBM TSs, demonstrating that combination treatment effectively reduced the stemness and invasive properties (Lim et al. 2020) of GBM TSs. Furthermore, we confirmed for the first time that the combination treatment of niclosamide and TMZ significantly prolong overall survival compared to the control group in a mouse orthotopic xenograft model.
Firstly, we found that niclosamide significantly reduced proliferation of GBM TSs in a dose dependent manner. Chien et al. (2018) reported that niclosamide inhibited invasiveness but could not alter cell viability in hepatocellular carcinoma. However, several studies have revealed that niclosamide inhibits the proliferation of cancer cells in colon (Sack et al. 2011), breast (Londono-Joshi et al. 2014), prostate (Lu et al. 2011), and lung cancers (Stewart et al. 2016), as well as GBM (Wieland et al. 2013). Moreover, niclosamide enhanced the anti-proliferative activities of oxaliplatin in human colorectal cancer (Osada et al. 2011) and of cisplatin in lung cancer (Zuo et al. 2018). Wieland et al. also reported that niclosamide acts as a natural inducer of NFKBIA and combined treatment of niclosamide and TMZ synergistically inhibited cellular viability in NFKBIA ± glioblastoma genotype (Wieland et al. 2013). Our results are consistent with those of previous studies, showing the anti-proliferative activities of niclosamide as well as synergistic effects of combined treatments.
In the case of GBM, the cell of origin is in the subventricular zone (Yoon et al. 2020; Lee et al. 2018), but the cells in which the tumor is present are a realistic therapeutic target. Cancer stem cells (CSCs) have been tested as targets of therapeutic agents because such cells are resistant to chemotherapy (Dean et al. 2005). In large tumors, however, cancer cells may acquire or lose stemness, resulting in tumors containing large numbers of CSCs (Kang et al. 2015). We therefore tested the effects of niclosamide and/or temozolomide on heterogenous GBM TSs. Some studies reported that niclosamide reduces the stemness of CSCs in breast cancer (Wang et al. 2013) and chronic myelogenous leukemia (Jin et al. 2017) by inhibiting the Wnt/β-catenin pathway. Similarly, it was confirmed that GBM cells pre-exposed to niclosamide had decreased stemness in neurosphere assays (Wieland et al. 2013). Consistent with previous results, we found that combined treatment with niclosamide and temozolomide significantly reduced the stemness of GBM TSs, with western blotting showing that this combination reduced the expression of stemness markers.
The EMT has been implicated in cancer progression, and the canonical Wnt signaling cascade has been found to control transcription factor Snail, which triggers EMT in human cancer by suppressing the expression of epithelial cell genes (Barrallo-Gimeno and Nieto 2005). Niclosamide is known for a potent inhibitor of Wnt/β-catenin signaling in various types of cancer (Sack et al. 2011; Wieland et al. 2013). Ahn et al. (2017) revealed that niclosamide directly targets Axin-GSK3 interactions, suppressing Wnt/Snail-mediated EMT in human colon cancer. Some studies reported that niclosamide significantly reduced invasiveness of breast (Ye et al. 2014) and lung (Stewart et al. 2016) cancer cells. However, the ability of the combination of niclosamide and temozolomide to inhibit invasiveness of GBM TSs had not yet been reported. We confirmed for the first time that the combination treatment of niclosamide and TMZ effectively reduced the invasiveness of GBM TSs, suppressing Snail mediated EMT.
Temozolomide, known as alkylating agent, makes DNA methylation of guanine at O6 position, leading to double strand break and induction of cancer cell apoptosis (Trivedi et al. 2005). Because DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) removes this methylation (Kokkinakis et al. 2001), the methylation extent of the MGMT promoter has been regarded as a prognostic factor for GBM (Hegi et al. 2005). Kohsaka et al. (2012) found that STAT3 inhibition downregulated MGMT expression and overcame temozolomide resistance in GBM cell lines. Because it is well known that niclosamide acts as a STAT3 pathway inhibitor (Ren et al. 2010; Gyamfi et al. 2019), we conducted immunoblotting and found that the combination treatment of niclosamide and temozolomide suppresses the expression of phosphorylated STAT3 as well as MGMT. This result suggests that STAT3 inhibition could be a potential synergistic mechanism of the combination treatment.
Transcriptome analyses comparing expression profiles before and after combined treatment of niclosamide and TMZ revealed that a subset of genes related to stemness and EMT were significantly downregulated. Gene Set Enrichment Analysis (GSEA) also showed that autophagy related gene set was upregulated after combined treatment, suggesting that the combination treatment has anti-proliferation effect as well as anti-stemness and anti-invasiveness effects.
Several orthotopic xenograft models have shown that niclosamide inhibits tumor growth and metastases in various types of cancers (Osada et al. 2011; Li et al. 2013). One glioblastoma study also reported that niclosamide-pretreated group had survival benefit compared with untreated group in a mouse orthotopic xenograft model (Wieland et al. 2013). However, the therapeutic potential of combination treatment of niclosamide and TMZ against GBM TSs in vivo is still unknown. Here, we found that the combination treatment of niclosamide and TMZ significantly prolonged survival period in our experiments. These results could be explained by RNA sequencing and immunoblotting data that niclosamide treatment might have impact on the cancer signaling cascades which are associated with stemness, invasiveness, and proliferation.
In summary, the combination of niclosamide and temozolomide effectively reduced the viability, stemness and invasion capacity of GBM TSs as well as prolonged survival period in a mouse orthotopic xenograft models, suggesting that this combination could be treatment option for GBM patients in clinical setting.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The GBM TS cell line GSC11, derived from primary GBM specimens was provided by Dr. Frederick F. Lang, M. D. Anderson Cancer Center, Houston, Texas, USA. This work was supported by grants from the Korean Health Technology R&D Project, the Ministry of Health & Welfare, Republic of Korea (HI17C2586 and HI14C1324), and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2019R1A2C3004155), and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIP&MOHW) (NRF-2016M3A9B6945832)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving animals were in accordance with the ethical standards of the Yonsei University College of Medicine Institutional Animal Care and Use Committee. This article does not contain any studies involving human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jong In Yook, Email: jiyook@yuhs.ac.
Seok-Gu Kang, Email: seokgu9@gmail.com.
References
- Ahn SY, Kim NH, Lee K, Cha YH, Yang JH, Cha SY, Cho ES, Lee Y, Cha JS, Cho HS, Jeon Y, Yuk YS, Cho S, No KT, Kim HS, Lee H, Choi J, Yook JI (2017) Niclosamide is a potential therapeutic for familial adenomatosis polyposis by disrupting Axin-GSK3 interaction. Oncotarget 8(19):31842–31855. 10.18632/oncotarget.16252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M (2009) Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS ONE 4(9):e7124. 10.1371/journal.pone.0007124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132(14):3151–3161. 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8):1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Morales LD, Zhang Y, Mito S, Tsin A (2017) Niclosamide induces protein ubiquitination and inhibits multiple pro-survival signaling pathways in the human glioblastoma U-87 MG cell line. PLoS ONE 12(9):e0184324. 10.1371/journal.pone.0184324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien MH, Ho YC, Yang SF, Yang YC, Lai SY, Chen WS, Chen MJ, Yeh CB (2018) Niclosamide, an oral antihelmintic drug, exhibits antimetastatic activity in hepatocellular carcinoma cells through downregulating twist-mediated CD10 expression. Environ Toxicol 33(6):659–669. 10.1002/tox.22551 [DOI] [PubMed] [Google Scholar]
- Choi J, Lee JH, Koh I, Shim JK, Park J, Jeon JY, Yun M, Kim SH, Yook JI, Kim EH, Chang JH, Kim SH, Huh YM, Lee SJ, Pollak M, Kim P, Kang SG, Cheong JH (2016) Inhibiting stemness and invasive properties of glioblastoma tumorsphere by combined treatment with temozolomide and a newly designed biguanide (HL156A). Oncotarget 7(40):65643–65659. 10.18632/oncotarget.11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong CR, Sullivan DJ Jr (2007) New uses for old drugs. Nature 448(7154):645–646. 10.1038/448645a [DOI] [PubMed] [Google Scholar]
- D'Alessandris QG, Biffoni M, Martini M, Runci D, Buccarelli M, Cenci T, Signore M, Stancato L, Olivi A, De Maria R, Larocca LM, Ricci-Vitiani L, Pallini R (2017) The clinical value of patient-derived glioblastoma tumorspheres in predicting treatment response. Neuro Oncol 19(8):1097–1108. 10.1093/neuonc/now304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S (2005) Tumour stem cells and drug resistance. Nat Rev Cancer 5(4):275–284. 10.1038/nrc1590 [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM (2013) Ensembl 2013. Nucleic Acids Res 41(Database issue):D48–55. 10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin JP, Despeignes J, Billerau M (1964) Present treatment of taeniasis with niclosamide. Lyon Med 212:1581–1588 [PubMed] [Google Scholar]
- Gyamfi J, Lee YH, Min BS, Choi J (2019) Niclosamide reverses adipocyte induced epithelial-mesenchymal transition in breast cancer cells via suppression of the interleukin-6/STAT3 signalling axis. Sci Rep 9(1):11336. 10.1038/s41598-019-47707-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352(10):997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- Jin B, Wang C, Li J, Du X, Ding K, Pan J (2017) Anthelmintic niclosamide disrupts the interplay of p65 and FOXM1/beta-catenin and eradicates leukemia stem cells in chronic myelogenous leukemia. Clin Cancer Res 23(3):789–803. 10.1158/1078-0432.CCR-16-0226 [DOI] [PubMed] [Google Scholar]
- Kang SG, Cheong JH, Huh YM, Kim EH, Kim SH, Chang JH (2015) Potential use of glioblastoma tumorsphere: clinical credentialing. Arch Pharm Res 38(3):402–407. 10.1007/s12272-015-0564-0 [DOI] [PubMed] [Google Scholar]
- Kim BS, Seol HJ, Nam DH, Park CK, Kim IH, Kim TM, Kim JH, Cho YH, Yoon SM, Chang JH, Kang SG, Kim EH, Suh CO, Jung TY, Lee KH, Kim CY, Kim IA, Hong CK, Yoo H, Kim JH, Kang SH, Kang MK, Kim EY, Kim SH, Chung DS, Hwang SC, Song JH, Cho SJ, Lee SI, Lee YS, Ahn KJ, Kim SH, Lim DH, Gwak HS, Lee SH, Hong YK (2017a) Concurrent chemoradiotherapy with temozolomide followed by adjuvant temozolomide for newly diagnosed glioblastoma patients: a retrospective multicenter observation study in Korea. Cancer Res Treat 49(1):193–203. 10.4143/crt.2015.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Lee JH, Oh Y, Koh I, Shim JK, Park J, Choi J, Yun M, Jeon JY, Huh YM, Chang JH, Kim SH, Kim KS, Cheong JH, Kim P, Kang SG (2017b) Inhibition of glioblastoma tumorspheres by combined treatment with 2-deoxyglucose and metformin. Neuro Oncol 19(2):197–207. 10.1093/neuonc/now174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka S, Wang L, Yachi K, Mahabir R, Narita T, Itoh T, Tanino M, Kimura T, Nishihara H, Tanaka S (2012) STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol Cancer Ther 11(6):1289–1299. 10.1158/1535-7163.MCT-11-0801 [DOI] [PubMed] [Google Scholar]
- Kokkinakis DM, Bocangel DB, Schold SC, Moschel RC, Pegg AE (2001) Thresholds of O6-alkylguanine-DNA alkyltransferase which confer significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin Cancer Res 7(2):421–428 [PubMed] [Google Scholar]
- Kong BH, Shin HD, Kim SH, Mok HS, Shim JK, Lee JH, Shin HJ, Huh YM, Kim EH, Park EK, Chang JH, Kim DS, Hong YK, Kim SH, Lee SJ, Kang SG (2013a) Increased in vivo angiogenic effect of glioma stromal mesenchymal stem-like cells on glioma cancer stem cells from patients with glioblastoma. Int J Oncol 42(5):1754–1762. 10.3892/ijo.2013.1856 [DOI] [PubMed] [Google Scholar]
- Kong BH, Park NR, Shim JK, Kim BK, Shin HJ, Lee JH, Huh YM, Lee SJ, Kim SH, Kim EH, Park EK, Chang JH, Kim DS, Kim SH, Hong YK, Kang SG, Lang FF (2013b) Isolation of glioma cancer stem cells in relation to histological grades in glioma specimens. Childs Nerv Syst 29(2):217–229. 10.1007/s00381-012-1964-9 [DOI] [PubMed] [Google Scholar]
- Kwak J, Shin HJ, Kim SH, Shim JK, Lee JH, Huh YM, Kim EH, Park EK, Chang JH, Kim SH, Hong YK, Kim DS, Lee SJ, Kang SG (2013) Isolation of tumor spheres and mesenchymal stem-like cells from a single primitive neuroectodermal tumor specimen. Childs Nerv Syst 29(12):2229–2239. 10.1007/s00381-013-2201-x [DOI] [PubMed] [Google Scholar]
- Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF (2000) An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg 92(2):326–333. 10.3171/jns.2000.92.2.0326 [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um JY, Kim WK, Lee JK, Park J, Kim EH, Lee JH, Lee JH, Chung WS, Ju YS, Park SH, Chang JH, Kang SG, Lee JH (2018) Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560(7717):243–247. 10.1038/s41586-018-0389-3 [DOI] [PubMed] [Google Scholar]
- Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, Ramalingam SS, Curran WJ, Khuri FR, Deng X (2013) Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Mol Cancer Ther 12(10):2200–2212. 10.1158/1535-7163.MCT-13-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li PK, Roberts MJ, Arend RC, Samant RS, Buchsbaum DJ (2014) Multi-targeted therapy of cancer by niclosamide: a new application for an old drug. Cancer Lett 349(1):8–14. 10.1016/j.canlet.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-J, Kim S, Oh Y, Suh Y, Kaushik N, Lee J-H, Lee H-J, Kim M-J, Park M-J, Kim R-K, Cha J, Kim SH, Shim J-K, Choi J, Chang JH, Hong YK, Huh YM, Kim P, Kang S-G, Lee S-J (2020) Crosstalk between GBM cells and mesenchymal stemlike cells promotes the invasiveness of GBM through the C5a/p38/ZEB1 axis. Neuro Oncol. 10.1093/neuonc/noaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Cui W, Yuan X, Lin L, Cao Q, Wang N, Li Y, Guo W, Zhang X, Wu C, Yang J (2016) Targeting ALDH1A1 by disulfiram/copper complex inhibits non-small cell lung cancer recurrence driven by ALDH-positive cancer stem cells. Oncotarget 7(36):58516–58530. 10.18632/oncotarget.11305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Joshi AI, Arend RC, Aristizabal L, Lu W, Samant RS, Metge BJ, Hidalgo B, Grizzle WE, Conner M, Forero-Torres A, Lobuglio AF, Li Y, Buchsbaum DJ (2014) Effect of niclosamide on basal-like breast cancers. Mol Cancer Ther 13(4):800–811. 10.1158/1535-7163.MCT-13-0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lin C, Roberts MJ, Waud WR, Piazza GA, Li Y (2011) Niclosamide suppresses cancer cell growth by inducing Wnt co-receptor LRP6 degradation and inhibiting the Wnt/beta-catenin pathway. PLoS ONE 6(12):e29290. 10.1371/journal.pone.0029290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65(1–2):55–63. 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- Natale CA, Li J, Zhang J, Dahal A, Dentchev T, Stanger BZ, Ridky TW (2018) Activation of G protein-coupled estrogen receptor signaling inhibits melanoma and improves response to immune checkpoint blockade. Elife. 10.7554/eLife.31770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada T, Chen M, Yang XY, Spasojevic I, Vandeusen JB, Hsu D, Clary BM, Clay TM, Chen W, Morse MA, Lyerly HK (2011) Antihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutations. Cancer Res 71(12):4172–4182. 10.1158/0008-5472.CAN-10-3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shim JK, Kang JH, Choi J, Chang JH, Kim SY, Kang SG (2018) Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex I suppresses glioblastoma tumorspheres. Neuro Oncol 20(7):954–965. 10.1093/neuonc/nox243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, Pan J, Pei D, Ding K (2010) Identification of niclosamide as a new small-molecule inhibitor of the STAT3 signaling pathway. ACS Med Chem Lett 1(9):454–459. 10.1021/ml100146z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TH, Park HH, Kang SG, Moon JH, Kim EH, Hong CK, Ahn SS, Choi HJ, Cho J, Kim SH, Lee SK, Kim DS, Kim SH, Suh CO, Lee KS, Chang JH (2017) Long-term outcomes of concomitant chemoradiotherapy with temozolomide for newly diagnosed glioblastoma patients: a single-center analysis. Medicine (Baltimore) 96(27):e7422. 10.1097/MD.0000000000007422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TH, Kang SG, Moon JH, Sung KS, Park HH, Kim SH, Kim EH, Hong CK, Suh CO, Chang JH (2020) Survival benefit of lobectomy over gross-total resection without lobectomy in cases of primary glioblastoma in non-eloquent area: a retrospective study. J Neurosurg 132(3):895–901. 10.3171/2018.12.JNS182558 [DOI] [PubMed] [Google Scholar]
- Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, Fichtner I, Schlag PM, Shoemaker RH, Stein U (2011) Novel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancer. J Natl Cancer Inst 103(13):1018–1036. 10.1093/jnci/djr190 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RL, Carpenter BL, West DS, Knifley T, Liu L, Wang C, Weiss HL, Gal TS, Durbin EB, Arnold SM, O'Connor KL, Chen M (2016) S100A4 drives non-small cell lung cancer invasion, associates with poor prognosis, and is effectively targeted by the FDA-approved anti-helminthic agent niclosamide. Oncotarget 7(23):34630–34642. 10.18632/oncotarget.8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for R, Treatment of Cancer Brain T, Radiotherapy G, National Cancer Institute of Canada Clinical Trials G (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW (2005) The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res 65(14):6394–6400. 10.1158/0008-5472.CAN-05-0715 [DOI] [PubMed] [Google Scholar]
- Wang AM, Ku HH, Liang YC, Chen YC, Hwu YM, Yeh TS (2009) The autonomous notch signal pathway is activated by baicalin and baicalein but is suppressed by niclosamide in K562 cells. J Cell Biochem 106(4):682–692. 10.1002/jcb.22065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Chao TK, Chang CC, Yo YT, Yu MH, Lai HC (2013) Drug screening identifies niclosamide as an inhibitor of breast cancer stem-like cells. PLoS ONE 8(9):e74538. 10.1371/journal.pone.0074538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu H, Fu W, Lu Z, Guo M, Wu X, Sun M, Liu Y, Yu X, Sui D (2019) 20(S)-Protopanaxadiol inhibits angiotensin II-induced epithelial- mesenchymal transition by downregulating SIRT1. Front Pharmacol 10:475. 10.3389/fphar.2019.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland A, Trageser D, Gogolok S, Reinartz R, Hofer H, Keller M, Leinhaas A, Schelle R, Normann S, Klaas L, Waha A, Koch P, Fimmers R, Pietsch T, Yachnis AT, Pincus DW, Steindler DA, Brustle O, Simon M, Glas M, Scheffler B (2013) Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res 19(15):4124–4136. 10.1158/1078-0432.CCR-12-2895 [DOI] [PubMed] [Google Scholar]
- Xia W, Lo CM, Poon RYC, Cheung TT, Chan ACY, Chen L, Yang S, Tsao GSW, Wang XQ (2017) Smad inhibitor induces CSC differentiation for effective chemosensitization in cyclin D1- and TGF-beta/Smad-regulated liver cancer stem cell-like cells. Oncotarget 8(24):38811–38824. 10.18632/oncotarget.16402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Xiong Y, Yan Y, Xia Y, Song X, Liu L, Li D, Wang N, Zhang L, Zhu Y, Zeng J, Wei Y, Yu L (2014) The anthelmintic drug niclosamide induces apoptosis, impairs metastasis and reduces immunosuppressive cells in breast cancer model. PLoS ONE 9(1):e85887. 10.1371/journal.pone.0085887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SJ, Park J, Jang DS, Kim HJ, Lee JH, Jo E, Choi RJ, Shim JK, Moon JH, Kim EH, Chang JH, Lee JH, Kang SG (2020) Glioblastoma cellular origin and the firework pattern of cancer genesis from the subventricular zone. J Korean Neurosurg Soc 63(1):26–33. 10.3340/jkns.2019.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Yang D, Yu Y, Xiang M, Li H, Yang J, Li J, Jiang D, Zhou H, Xu Z, Yu Z (2018) Niclosamide enhances the cytotoxic effect of cisplatin in cisplatin-resistant human lung cancer cells via suppression of lung resistance-related protein and c-myc. Mol Med Rep 17(3):3497–3502. 10.3892/mmr.2017.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.