Abstract
MicroRNAs-199a-5p (miR-199a-5p) plays critical regulatory roles in various types of human cancers. However, the biological function and regulatory mechanisms of miR-199a-5p in colorectal cancer (CRC) remain unclear. The aim of this study was to investigate the role of miR-199a-5p in CRC and possible mechanisms of its action. The expression of miR-199a-5p in CRC tumor tissues was validated using quantitative real-time PCR (qRT-PCR). The effects of miR-199a-5p on cell proliferation and apoptosis were evaluated in vitro. Then, the association of miR-199a-5p and its downstream target was investigated in both cell line and clinical specimens. Furthermore, gain- and loss-of-function studies of cytoplasmic activation/proliferation-associated protein-1 (Caprin1) were performed to assess whether the suppressive effect of on CRC cells were via targeting Caprin1. Using a microarray platform, we focused on miR-199a-5p for further research, which was one of the most markedly downregulated miRNAs in CRC tumor tissues. Functionally, the overexpression of miR-199a-5p inhibited proliferation and induced apoptosis in both HTC116 and SW480 cells. Furthermore, cytoplasmic activation/proliferation-associated protein-1 (Caprin1), a well-known oncogene, was directly targeted by miR-199a-5p. It was also observed that Caprin1 was upregulated, and inversely correlated with miR-199a-5p levels in CRC tissues. Further investigations revealed that knockdown of Caprin1 by siRNA has similar role with miR-199a-5p overexpression in CRC cells, suggesting the oncogenic role of Caprin1 in CRC. In the contrast, we found that overexpression of Caprin1 reversed the suppressive effects of miR-199a-5p on CRC cells. Collectively, our study suggests that miR-199a-5p/Caprin1 axis may serve as potential therapeutic targets for the treatment of CRC.
Keywords: MicroRNA-199a-5p, Tumor suppressor, Colorectal cancer, Proliferation, Apoptosis, Caprin1
Introduction
Colorectal cancer (CRC) is one of the most prevalent malignancies and the leading causes of cancer mortality worldwide (Gaedcke et al. 2012; Siegel et al. 2016). Recently, the morbidity of CRC has been increasing in Asian countries, including China, Singapore, Korea and Japan, due to demographic trends and adaption to westernized lifestyle in developing countries (Walker et al. 2014). Despite considerable work on the treatment options, such as radical surgery and adjuvant chemotherapies, CRC patients still have a poor prognosis mainly because of tumor metastasis (Study 2013). Therefore, the identification of novel promising applicable therapeutic candidates and approaches for clinical management of CRC is urgent.
MicroRNAs (miRNAs) belong to a broad class of small non-coding RNAs usually with 21–25 nucleotides, which act as a unique regulator of gene expression at the posttranscriptional level through suppressing translation or inducing RNA degradation (Ambros 2003). Growing evidence has suggested that miRNAs are involved in a variety of biological and pathological processes, including carcinogenesis, cellular apoptosis, proliferation and differentiation (Bartel; Sage and Agami 2006; Croce 2009). The miRNAs have been confirmed to function either as oncomiRs or tumor suppressors in CRC development (Mohammadi et al. 2016; Yamamoto and Mori 2016). For example, miR-21 is highly expressed in tumor tissues of advanced CRC patients, which function as a potential biomarker for diagnosis of CRC (Valeri et al. 2010). Ren et al. showed that miR-382 inhibited cell growth and migration in colorectal cancer by targeting SP1 (Ren et al. 2018). Zhang et al. found that miR-301a was elevated in CRC tissues and overexpression of miR-301a promoted tumor metastasis and cell invasion in vivo (Zhang et al. 2014). Thus, identification of other miRNAs may be the important step in construction of a new treatment strategy for CRC.
In the present study, we performed a miRNA microarray analysis to detect the miRNA expression profiles in CRC tissues and adjacent normal tissues. Furthermore, we further investigated the functions and molecular mechanism of miR-199a-5p in the development of CRC.
Materials and methods
Clinical samples
35 pairs of tumor samples and the paired adjacent non-tumor mucosa (about 5 cm away from the tumor margin) were collected from CRC patients undergoing laparoscopic surgery from November 2017 to January 2019 at the Tianjin Medical University Cancer Institute and Hospital. The patients enrolled in this study did not received radiotherapy or chemotherapy before surgical resection. Samples were snap-frozen in liquid nitrogen and stored at − 80 °C until use. All tissue samples were confirmed by pathological examinations. Written informed consent was obtained from each patient before the surgery. The study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. The experimental procedures were performed in accordance with the Declaration of Helsinki.
Cell culture
CRC cell lines including HT29, SW620, SW480, and HCT116 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to the manufacturer’s instructions. SW620 and SW480 cells were grown in RPMI 1640 (Gibco, Grand Island, NY, USA), HT29 and HCT116 cells were grown DMEM medium (Gibco, Grand Island, NY, USA). The medias were supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO), and 1% penicillin/streptomycin (Sigma, St. Louis, MO). The normal human colon mucosal epithelial cell line NCM460 was obtained from Incell Corporation (San Antonio, TX, USA) and cultured in F-12/Ham medium (Gibco, Grand Island, NY, USA) supplemented with 20% FBS, 0.29 g/L glutamine and 1% penicillin/streptomycin. All cells were grown at 37 °C in a humidified atmosphere containing 5% CO2.
Cell transfection
The miR-199a-5p mimics/inhibitor or corresponding negative control (NC) (mimics/inhibitor NC), small interfering (si) RNA sequences, si-Caprin1, and NC si-RNA were designed and purchased from GenePharma Co., Ltd (Shanghai, China). For plasmid construction (pcDNA-Caprin1), the Caprin1 3ʹ-UTR was cloned into the XhoI and KpnI sites of the pcDNA3.1 expression vector (Invitrogen, Carlifornia, America), an empty pcDNA3.1 vector (pcDNA-vector) used as a control. For transfection, SW480 or HCT116 cells were plated in 6-well plates (2 × 106 cells/well) and were transfected with miR-199a-5p mimics/inhibitor, mimics/inhibitor NC, si-Caprin1/si-RNA or pcDNA-Caprin1/pcDNA-vector at a final concentration of 100 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer’s protocol. Then cells were cultured at 37 °C in an incubator containing 5% CO2 for 48 h. Transfection efficiency was assessed using RT-qPCR.
miRNA microarray analysis
Total RNAs were extracted from CRC and the adjacent tissues by TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the miRNA fraction was further purified by a mirVana miRNA isolation kit (Ambion, Austin, TX) according to manufacturer’s instructions. The isolated miRNAs were labeled with Hy3 using the miRCURY array labeling kit (Exiqon, Vedbaek, Denmark) and hybridized with miRCURY locked nucleic acid (LNA) microRNA arrays (v8.0; Exiqon). Microarray images were taken with a Genepix 4000B scanner (Axon Instruments, Foster City, CA, USA) and analyzed with Genepix Pro 6.0 software (Axon Instruments).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocols. TaqMan Gene Expression Assay and TaqMan MicroRNA Reverse Transcription kits (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for reverse transcription prior to Caprin1 and miRNA detection according to manufacturer’s instructions. PCR was performed using the TaqMan™ MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The amplifications were conducted at 95 °C for 10 s and at 60 °C for 60 s using the StepOnePlus™ Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 served as an endogenous control for miR-199a-5p and GAPDH served as a reference control for Caprin1. Fold changes were determined using relative quantification (2−ΔΔCt) method.
Cell viability analysis
The Cell counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Gaithersburg, MD, USA) assay was performed to detect cell proliferation according to the manufacturer’s instructions (He et al. 2014b). Briefly, SW480 or HCT116 cells were seeded into 96-well plates (2 × 104 cells/well) and then transfected with matching oligonucleotides or plasmids. At the indicated time point (1, 2, 3 days), 10 μl of CCK-8 solution was added into each well and the mixture was incubated for 1 h at 37 °C with 5% CO2. The absorbance rate at 450 nm was determined by Microplate Reader (Bio-Rad, Hercules, CA, USA). All experiments were performed in quintuplicate on three separate occasions.
Caspase 3 activation assay
Forty-eight hours after transfection, Caspase 3 activity was measured using Caspase 3 Activity Detection Kit, BioAssay(TM) (CPP-32, Apoptain, Yama, SCA-1) according to the manufacturer’s instruction.
Immunofluorescence staining
SW480 or HCT116 cells were transfected with oligonucleotides, and fixed with 4% paraformaldehyde for 30 min, and then were permeabilized with 0.1% Triton X-100 in PBS for 15 min. Cells were blocked with 5% BSA for 1 h at room temperature, and then incubated with indicated primary antibody (cleaved-caspase-3, 1:200) at 4 °C overnight. After three washes with PBS, the cells were incubated with secondary antibodies (anti-rabbit IgG antibodies). Images were photographed using a fluorescence microscope (DX51; Olympus, Tokyo, Japan).
Luciferase reporter assay
miRNA target prediction tools, including PicTar version 2007 (https://pictar.mdc-berlin.de/) and TargetScan Release 7.0 (https://targetscan.org/) were used to search for the putative targets of miR-199a-5p. The fragment of the 3ʹ-UTR of Caprin1 was amplified and cloned into the luciferase reporter vector pGL3cM (Promega) (wild type, pGL3cM-Caprin1-3ʹ-UTR). Site-directed mutagenesis of the Caprin1 3ʹ-UTR at the putative miR-199a-5p binding site was performed by a QuikChange Kit (Qiagen) (mutation, pGL3cM-Caprin1-mut-3ʹ-UTR). Subsequently, SW480 cells (2 × 105 per well) were seeded into 24-well plates and co-transfected with 0.8 μg of pGL3cM-Caprin1-3ʹ-UTR or pGL3cM-Caprin1-mut-3ʹ-UTR, 50 nM miR-199a-5p mimic/inhibitor or corresponding NC using Lipofectamine 2000 reagent (Invitrogen). The ratio of firefly to Renilla luciferase was used to normalize relative firefly luciferase activity 48 h post-transfection, and luciferase activity was determined using the Dual-Light luminescent reporter gene assay (Applied Biosystems), according to manufacturer’s protocol.
Western blot analysis
SW480 or HCT116 cells were transfected with miR-199a-5p mimics for 48 h, after which total protein was extracted using RIPA lysis buffer (Solarbio, China). The protein concentration was measured using a BCA protein assay kit (Pierce, Rockford, IL). Protein samples (50 μg/sample) were separated by 10% SDS-PAGE gel (Sigma-Aldrich, St. Louis, MO) and then transferred to polyvinylidene difluoride (PVDF, Millipore, Bedford, MA, USA). After blocking with 5% non-fat milk at room temperature for 1 h, the PVDF membranes were incubated primary antibodies against Caprin1 (Cat no. ab244360, 1:1000) at 4 °C overnight. β-actin (Cat no. ab179467, 1:1000) was used as an internal control for protein loading. The goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) was used as the secondary antibody. All antibodies were obtained from Abcam, United States. The relative intensity of each band was semi-quantified using Alpha Imager software version 2000 (ProteinSimple, San Jose, CA, USA).
Statistical analysis
All statistical analyses were performed using SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are presented as the means ± standard deviation. Student’s t test or one-way ANOVA were used to analyze the difference among/between sample groups. The relationship between Caprin1 and miR-199a-5p expressions was tested with two-tailed Pearson’s correlation. P < 0.05 was considered to indicate a statistically significant difference.
Results
miR-199a-5p expression is downregulated in CRC tissues and cell lines
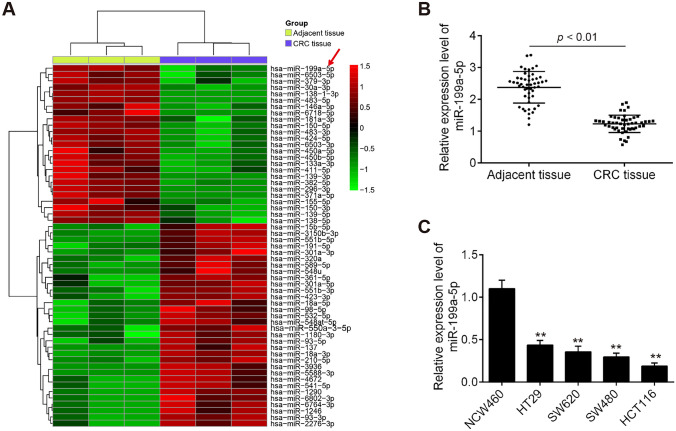
To explore the role of miRNAs in CRC, we first analyzed the differential expressed miRNAs using miRNA microarray analysis. Cluster analysis based on the miRNA expression pattern indicated a significant difference between CRC tissue and the adjacent normal tissues (Fig. 1a). Among the downregulated miRNAs, miR-199a-5p is the most significantly downregulated. In addition, miR-199a-5p has also been found to be downregulated in CRC tissues (Tan et al. 2018; Zhu et al. 2018). Interestingly, several studies have shown that miR-199a-5p functions as a tumor suppressor in various types of human cancers (Ren et al. 2016; Zhang et al. 2019); however, the actions of miR-199a-5p in CRC remain largely unclear. Thus, miR-199a-5p was selected for subsequent studies.
Fig. 1.
The miR-199a-5p expression levels in colorectal cancer (CRC) tissues and adjacent normal tissues. a Heat map of the differentially expressed miRNAs. The miRNA expression profiles in CRC tissues and adjacent normal tissues (n = 3) were identified by miRNA microarray analysis. Red or green color separately represents high or low expression. b miR-199a-5p level was significantly decreased in CRC tissues compared with that in adjacent normal tissues (n = 50) (**P < 0.01 vs. adjacent tissue group). c miR-199a-5p level was significantly decreased in CRC cell lines HT29, SW620, SW480, HCT116, compared with that in normal human colon mucosal epithelial cell line NCM460. (**P < 0.01 vs. NCM460 group)
To validate the expression of miR-199a-5p obtained from miRNA microarray assay, miR-199a-5p expression in 50 paired CRC tissues and adjacent normal tissues were further determined using RT-qPCR assay. We found that the expression level of miR-199a-5p in CRC tissues was significantly lower than that in adjacent normal tissues (Fig. 1b; P < 0.01). To validate whether downregulation of miR-199a-5p was also present in CRC cell lines, miR-199a-5p expression was determined in four human CRC cell lines (HT29, SW620, SW480, and HCT116) and normal colon mucosal epithelial cell line (NCM460) by qRT-PCR. The results showed that miR-199a-5p was obviously downregulated CRC cell lines, as compared to NCM460 cells (Fig. 1c; P < 0.01), especially in HTC116 and SW480 cells. These results indicated that miR-199a-5p may act as an important role in the development of CRC.
Overexpression of miR-199a-5p inhibits cell growth
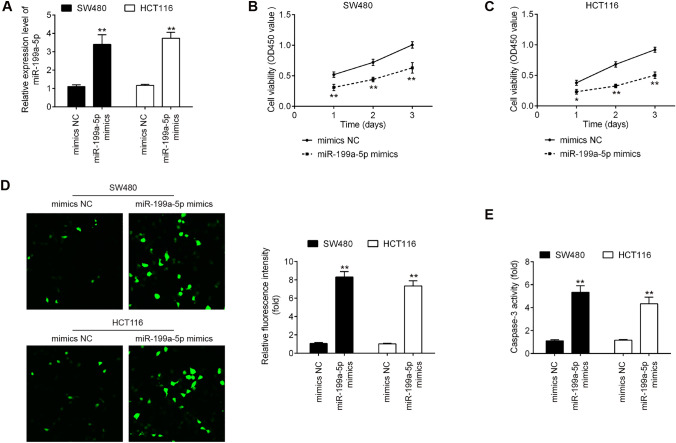
Given the downregulation of miR-199a-5p in CRC tissues and cell lines, we predicted that miR-199a-5p may function as a tumor suppressor. To verify our hypothesis, miR-199a-5p mimics was transfected into HTC116 and SW480 cells, because HTC116 and SW480 cells exhibited the lowest expression levels of miR-199a-5p in these CRC cell lines, respectively. qRT-PCR analysis showed that miR-199a-5p expression was effectively enhanced by miR-199a-5p mimic (Fig. 2a; P < 0.01). CCK-8 assay demonstrated that overexpression of miR-199a-5p significantly suppresses cell proliferation in HTC116 or SW480 cells compared with mimics NC (Fig. 2b, c; P < 0.01). To verify whether miR-199a-5p modulates cell apoptosis, we performed the immunofluorescence staining assay to detect the marker of intrinsic apoptosis pathway, cleaved-caspase-3 in HTC116 or SW480 cells. The results showed that upregulation of miR-199a-5p dramatically elevated the cleaved-caspase-3 level in CRC cells compared with mimics NC (Fig. 2d; P < 0.01). It was also observed that the activity of caspase 3 was obviously increased by miR-199a-5p (Fig. 2e; P < 0.01). Overall, these results suggested that overexpression of miR-199a-5p inhibits cell proliferation and induces cell apoptosis in CRC cells.
Fig. 2.
Effects of miR-199a-5p on the cell proliferation and cleaved-caspase-3 level in CRC cells. The HTC116 and SW480 cells were transfected with miR-199a-5p mimics or mimics NC for 48 h, and then the cells were harvested for further experiments. a miR-199a-5p expression level in miR-199a-5p mimics group was significantly increased in HTC116 and SW480 cells compared with mimic NC group. b and c Cell proliferation of HTC116 and SW480 cells in miR-199a-5p mimics group was markedly reduced compared with mimic NC group. d The cleaved-caspase-3 protein expression in miR-199a-5p mimics group was significantly increased compared with mimic NC group. e The activity of caspase 3 in miR-199a-5p mimics group was significantly increased compared with mimic NC group. (*P < 0.05, **P < 0.01 vs. mimics NC)
miR-199a-5p suppresses Caprin1 expression by directly targeting its 3ʹ-UTR
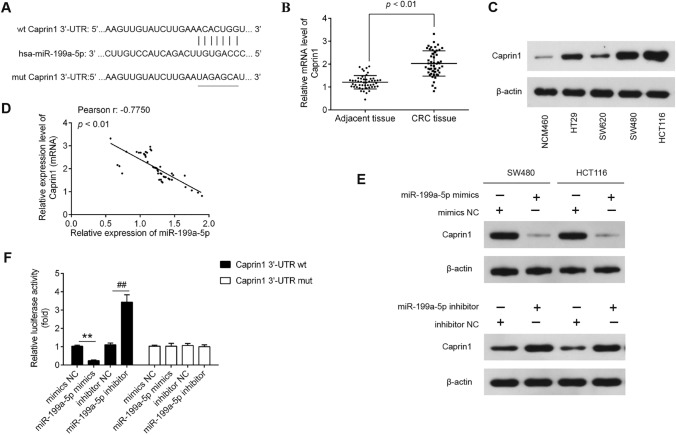
To better understand the mechanisms involved in the suppressive role of miR-199a-5p in CRC, we searched for potential target genes of miR-199a-5p using PicTar version 2007 (https://pictar.mdc-berlin.de/) and TargetScan Release 7.0 (https://targetscan.org/). Caprin1 was chosen as a potential target gene of miR-199a-5p because Caprin1 acts as oncogenic gene in various types of cancers, including osteosarcoma (Sabile et al. 2013), human breast cancer (Gong et al. 2013a), and hepatocellular carcinoma (Tan et al. 2017). The predicted binding sites for miR-199a-5p in the Caprin1 sequence are illustrated in Fig. 3a. To verify the Caprin1 expression in CRC, we then performed the qRT-PCR to detect the Caprin1 mRNA level in CRC tissues and adjacent normal tissues. The results showed that the Caprin1 mRNA level was dramatically increased in CRC tissues (n = 50) compared with adjacent tissues (n = 50) (Fig. 3b; P < 0.01). We also measured the expression of Caprin1 protein in four human CRC cell lines (HT29, SW620, SW480, and HCT116) and NCM460 by western blot. The results showed that Caprin1 was obviously upregulated CRC cell lines compared with those in NCM460 cells (Fig. 3c). In addition, the correlation analysis revealed that miR-199a-5p expression level was negatively correlated with mRNA expression of Caprin1 level in CRC tissues (r = − 0.7750, P < 0.01; Fig. 3d). Moreover, Caprin1 protein expression was identified in SW480 or HCT cells after transfection with miR-199a-5p mimic/inhibitor using western blot analysis. The results indicated that upregulation or downregulation of miR-199a-5p dramatically decreased or increased the Caprin1expression in HTC116 or SW480 cells compared with NC (P < 0.01; Fig. 3e). To further investigate whether the Caprin1 is the direct target of miR-199a-5p, we conducted a luciferase reporter assay to measure luciferase activity in HTC116 cells co-transfected with pGL3cM-Caprin1-3ʹ-UTR or pGL3cM-Caprin1-mut-3ʹ-UTR, miR-199a-5p mimic/inhibitor or corresponding NC. The results showed that overexpression of miR-199a-5p significantly reduced the luciferase activity of the luciferase reporter containing wild-type 3ʹ-UTR of Caprin1 vector, whereas miR-199a-5p inhibition caused an increased luciferase activity; however, no changes were observed in the cells co-transfected with pGL3cM-Caprin1-mut-3ʹ-UTR with miR-199a-5p (Fig. 3f). Collectively, these data suggested that miR-199a-5p suppress Caprin1 expression by targeting its 3ʹ-UTR in CRC cells.
Fig. 3.
Caprin1 is a direct target of miR-199a-5p in CRC cells. a Predicted miR-199a-5p binding sites within the 3′-UTR of Caprin1 mRNA. b The mRNA level of Caprin1 was significantly increased in CRC tissues and adjacent normal tissues (n = 50). c Caprin1 protein levels were was significantly increased in CRC cell lines HT29, SW620, SW480, HCT116, compared with that in NCM460 cells. d The negative correlation between Caprin1 and miR-199a-5p levels in CRC tissues (r = − 0.7750, P < 0.01). e The SW480 and HTC116 cells were transfected with miR-199a-5p mimic/inhibitor or corresponding NC, and the Caprin1 protein level was measured using western blot analysis. f The HTC116 cells co-transfected with pGL3cM-Caprin1-3ʹ-UTR or pGL3cM-Caprin1-mut-3ʹ-UTR, miR-199a-5p mimic/inhibitor or corresponding NC for 48 h, and the relative luciferase activity were measured. β-actin was used as an internal control
Caprin1 silencing inhibits CRC cell growth
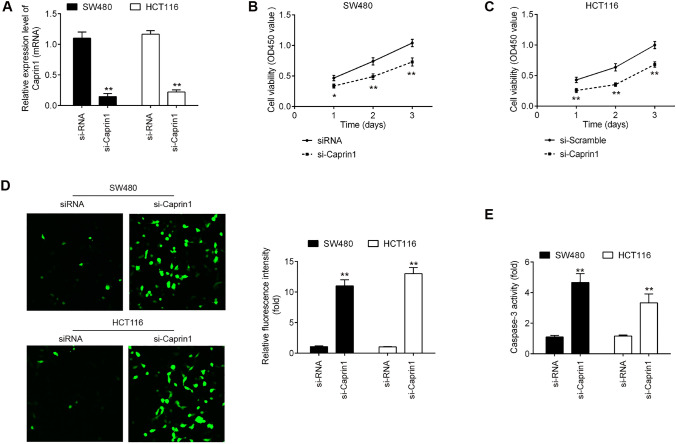
To further confirm the role of Caprin1 in CRC, we next preformed CCK-8 assay and immunofluorescence staining assay to investigate whether Caprin1 inhibition could suppress proliferative abilities of CRC cells using Caprin1-specific small interfering RNA (siRNA). The transfection efficiency of si-Caprin1 was first evaluated using RT-qPCR assay. As shown in Fig. 4a, si-Caprin1 significantly inhibited Caprin1 mRNA in both HTC116 and SW480 cells compared with NC si-RNA (P < 0.01). CCK-8 assay demonstrated that inhibition of Caprin1 by si-Caprin1 markedly repressed the cell proliferation in both HTC116 and SW480 cells compared with si-RNA (P < 0.01; Fig. 4b, c). Moreover, we found that knockdown of Caprin1 dramatically increased the cleaved-caspase-3 expression level and activity of caspase 3 compared with those in si-RNA transfected cells (P < 0.01; Fig. 4d, e). These data suggested that Caprin1 acts as an oncogenic gene in the development of CRC.
Fig. 4.
Effects of silencing Caprin1 on the cell proliferation and apotosis in CRC cells. The HTC116 or SW480 cells were transfected with si-Caprin1 and NC si-RNA for 48 h. a Caprin1 mRNA level in si-Caprin1 group was significantly decreased compared with NC si-RNA group. b and c Cell proliferation of HTC116 and SW480 cells in si-Caprin1 group was markedly reduced compared with NC si-RNA group. d The cleaved-caspase-3 protein expression in si-Caprin1 group was significantly increased compared with NC si-RNA group. e The activity of caspase 3 in si-Caprin1 group was significantly increased compared with NC si-RNA group (*P < 0.05, **P < 0.01 vs. si-RNA)
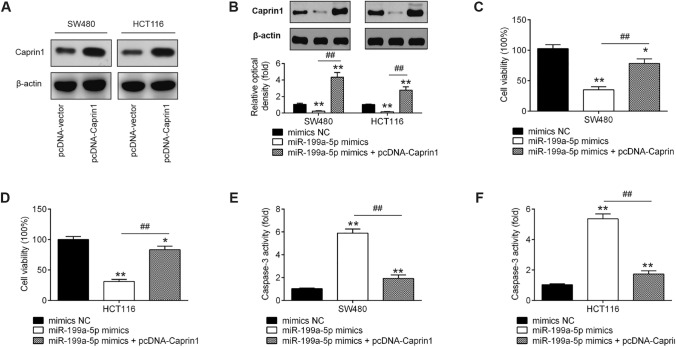
Overexpression of Caprin1 rescues the suppressive effects of miR-199a-5p on CRC cells
To evaluate if Caprin1 is responsible for the tumor-suppressive potential of miR-199a-5p in CRC cells, gain-of-function studies of Caprin1 were performed. First, the transfection efficiency of pcDNA-Caprin1 was identified by western blot. As shown in Fig. 5a, Caprin1 was notably upregulated in both HTC116 and SW480 cells after pcDNA-Caprin1 transfection. Moreover, decreased Caprin1 levels caused by miR-199a-5p mimics were also reversed by pcDNA-Caprin1 in CRC cells (P < 0.01; Fig. 5b). Subsequently, cell proliferation and the activity of caspase 3 were assessed by CCK-8 assay and a commercial Caspase 3 Activity Detection Kit, respectively. As shown in Fig. 5c and d, the reduction of cell proliferation caused by miR-199a-5p mimics was reversed by upregulation of Caprin1. Meanwhile, the increased activity of caspase 3 induced by miR-199a-5p mimics was also attenuated by upregulation of Caprin1 in HTC116 and SW480 cells (P < 0.01; Fig. 5e, f). These results indicated that miR-199a-5p inhibited CRC cell growth via suppressing Caprin1.
Fig. 5.
Restoration of Caprin1 reverses the suppressive effects of miR-199a-5p on cell growth. The HTC116 or SW480 cells were transfected with miR-199a-5p mimics or mimics NC, or were co-transfected with miR-199a-5p mimics and pcDNA-Caprin1 for 48 h. a The transfection efficiency of pcDNA-Caprin1 was identified in HTC116 or SW480 cells transfected with pcDNA-Caprin1 or pcDNA-vector by western blot. b Caprin1 protein expression was detected using western blot. c and d The reduction of cell proliferation caused by miR-199a-5p mimics was reversed by upregulation of Caprin1. e and f The increased activity of caspase 3 induced by miR-199a-5p mimics was also attenuated by upregulation of Caprin1 in HTC116 and SW480 cells (**P < 0.01 vs. mimics NC; ##P < 0.01 vs. miR-199a-5p mimics)
Discussion
In the present study, our results showed that miR-199a-5p was downregulated in CRC tissues and cell lines. It is also the first study to show that Caprin1 acts as an oncogene in CRC. More importantly, we demonstrated that miR-199a-5p suppressed the CRC cell growth by targeting oncogene, Caprin1.
Recently, miR-199a-5p was identified to serve as a tumor suppressor and be downregulated in various types of cancers, including gastric and breast cancer (He et al. 2014a; Yi et al. 2013). For example, Wei et al. (2019) found that a low expression of miR-199a-5p was associated with tumor differentiation, lymph node metastasis and TNM stage in oral squamous cell carcinoma (OSCC), suggesting that miR-199a-5p may serves as a substantial novel biomarker for tumor diagnosis and therapy. Moreover, Li et al. (2019) found that miR-199a-5p suppressed cell proliferation and arrested cell cycle in G1 phase in non-small cell lung cancer (NSCLC) cells via targeting MAP3K11. Kimberly A Byrnes et al. also found that overexpression of miR-199a-5p decreased esophageal cancer cell proliferation through MAP3K11 (Byrnes et al. 2016). Shen et al. showed that miR-199a-5p was capable to modulate tumor cell invasion at least in part by targeting discoidin domain receptor 1 (DDR1) in HCC cells (Shen et al. 2010). Zhou et al. found that miR-199a-5p overexpression inhibited cell migration and invasion through decreasing the expression of CCR7 in bladder cancer cells (Zhou et al. 2016). A previous study demonstrated that miR-199a levels were significantly lower in serum of CRC patients, and have been suggested as potential biomarkers for CRC (Tan et al. 2018). However, the functional role and mechanistic action of miR-199a-5p in CRC remain largely unclear. In the present study, our results showed that miR-199a-5p is downregulated in CRC tissues and cell lines. In addition, upregulation of miR-199a-5p suppresses proliferation and induces apoptosis in CRC cells. Thus, miR-199a-5p may be novel prognostic biomarkers and common therapeutic target for treating CRC.
Cytoplasmic activation/proliferation-associated protein-1 (Caprin1), a protein of 709 amino acids, belongs to a highly conserved protein family observed throughout vertebrate evolution (Zhang et al. 2018). It has been widely accepted that Caprin1 acts as oncogenic gene in various types of cancers, with marked effects on many cellular processes, including cell proliferation, apoptosis, migration, and invasion (Sabile et al. 2013; Gong et al. 2013a). It was also shown that upregulation of Caprin1 is involved in poor prognosis in hepatocellular carcinoma (HCC) (Tan et al. 2017). Sabile et al. (2013) have reported that increased expression of Caprin1 can promote osteosarcoma tumor growth and lung metastasis from primary tumors, and shorten survival time in mice. However, the expression patterns and biological functions of Caprin1 in CRC have not been established. In this study, Caprin1 was significantly upregulated in CRC tissues compared with adjacent tissues. It was also demonstrated that knockdown of Caprin1 by si-Caprin1 suppressed the cell proliferation and reduces cleaved-caspase-3 expression in CRC cells. To the best of our knowledge, the present results revealed for the first time that Caprin1 functions as an oncogene in CRC.
It is well known that miRNAs regulate gene expression at the posttranscriptional level. In this study, Caprin1 was identified as a target of miR-199a-5p in CRC cells. We also observed a highly significant negative correlation between miR-199a-5p and Caprin1 mRNA in tumor tissues. Thus, we postulated that miR-199a-5p may suppress CRC cell growth by targeting Caprin1. This postulation was verified by further experimental studies on CRC cell lines. Our results showed that Caprin1 upregulation reversed the tumor-suppressive effect of miR-199a-5p on CRC cell proliferation and apoptosis. These results indicated that miR-199a-5p functions as a tumor suppressor in CRC at least in part by targeting oncogenic gene, Caprin1. Indeed, other mechanisms and targets of miR-199a-5p besides Caprin1 may play roles in mediating CRC cell growth and apoptosis. Nevertheless, these other mechanisms still need to be elucidated.
Although recent genomic studies have provided an emerging understanding of the role of Caprin1 in the development of cancers, the detailed mechanisms underlying Caprin1 function are still complex in distinct cancer types. Given its role as a transcriptional regulator, many efforts have been dedicated to the identification of downstream targets or pathways that driven by Caprin1. Caprin1 can affect cell survival and growth through selectively binding a variety of mRNAs that are involved in cell growth, differentiation and migration, including c-Myc and cyclin-D2 (Xiao et al. 2015; Qiu et al. 2015). Gong et al. found that Caprin1 regulated the proliferation and invasion of human breast cancer cells (Gong et al. 2013b). Researches revealed that Caprin1 contributes to carcinogenesis by acting as oncogene through activation of the Akt and ERK1/2 signaling pathways (Sabile et al. 2013). In this study, we proved that miR-199-5p functions as a tumor suppressor by depression of Caprin1. However, the detailed mechanism of why knockdown of Caprin1 inhibited cell viability and cell invasion and migration ability needs to be further studied in future.
However, there are still some limitations to the present study. Due to the limitation in experimental conditions and funds, further research is required in the future to investigate the expression levels of miR-199a-5p in more clinical samples. Furthermore, other targets of miR-199a-5p or other miRNAs that could regulate Caprin1 needs to be identified in the following studies.
In summary, our results, for the first time, illustrated the miR-199a-5p-Caprin1 axis in CRC, in which miR-199a-5p targets Caprin1 to inhibit the tumorigenesis of CRC. The data from present study suggested that miR-199a-5p may serve as a biomarker for diagnosis and a potential therapeutic target in CRC.
Author contributions
XY, YH, YW, YT, YG, JY, JB, and TY performed the experiments, contributed to data analysis, and wrote the paper. XY, YH, and YW analyzed the data. XW conceptualized the study design, contributed to data analysis and experimental materials. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81473441) and the Program for New Century Excellent Talents in University (NO.NCET-11-1068).
Data availability
All data generated or analyzed during this study are included in this published article.
Compliance with ethical standards
Conflict of interest
The authors declare that no competing interests exist.
Ethics approval and consent to participate
All individuals provided informed consent for the use of human specimens for clinical research. The present study was approved by Tianjin Medical University Cancer Institute and Hospital Ethics Committees.
Consent for publication
Written informed consent for publication was obtained from all participants.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s13205-022-03345-6"
Change history
9/29/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s13205-022-03345-6
References
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Byrnes KA, Phatak P, Mansour D, Xiao L, Zou T, Rao JN, Turner DJ, Wang JY, Donahue JM. Overexpression of miR-199a-5p decreases esophageal cancer cell proliferation through repression of mitogen-activated protein kinase kinase kinase-11 (MAP3K11) Oncotarget. 2016;7(8):8756–8770. doi: 10.18632/oncotarget.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedcke J, Grade M, Camps J, Søkilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Møller S, Beissbarth T, Ried T, Litman T. The rectal cancer microRNAome – microrna expression in rectal cancer and matched normal mucosa. Clin Cancer Res. 2012;18(18):4919. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Hu H, Chen J, Cao S, Yu J, Xue J, Chen F, Cai Y, He H, Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67(7):629–636. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Gong B, Hu H, Chen J, Cao S, Yu J, Xue J, Chen F, Cai Y, He H, Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67(7):629–636. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- He X-J, Ma Y-Y, Yu S, Jiang X-T, Lu Y-D, Tao L, Wang H-P, Hu Z-M, Tao H-Q. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14(1):218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- He Y, Meng X-M, Huang C, Wu B-M, Zhang L, Lv X-W, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang D, Li X, Shao Y, He Y, Yu H, Ma Z. MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J Cancer. 2019;10(11):2472–2479. doi: 10.7150/jca.29426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- Qiu YQ, Yang CW, Lee YZ, Yang RB, Lee CH, Hsu HY, Chang CC, Lee SJ. Targeting a ribonucleoprotein complex containing the caprin-1 protein and the c-Myc mRNA suppresses tumor growth in mice: an identification of a novel oncotarget. Oncotarget. 2015;6(4):2148–2163. doi: 10.18632/oncotarget.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Li T, Zhang W, Ren J, Li Z, Wu G. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J Biomed Sci. 2016;23(1):79. doi: 10.1186/s12929-016-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Zhang H, Jiang P. MicroRNA-382 inhibits cell growth and migration in colorectal cancer by targeting SP1. Biol Res. 2018;51(1):51. doi: 10.1186/s40659-018-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabile AA, Arlt MJ, Muff R, Husmann K, Hess D, Bertz J, Langsam B, Aemisegger C, Ziegler U, Born W. Fuchs B (2013a) Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochem Biophys Acta. 1832;8:1173–1182. doi: 10.1016/j.bbadis.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Sabile AA, Arlt MJE, Muff R, Husmann K, Hess D, Bertz J, Langsam B, Aemisegger C, Ziegler U, Born W, Fuchs B. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochimica et Biophysica Acta (BBA) 2013;1832(8):1173–1182. doi: 10.1016/j.bbadis.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Sage CL, Agami R. Immense promises for tiny molecules: uncovering miRNA functions. Cell Cycle. 2006;5(13):1415–1421. doi: 10.4161/cc.5.13.2890. [DOI] [PubMed] [Google Scholar]
- Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, Beckebaum S. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mole Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Study APC Chemotherapy use and patient treatment preferences in advanced colorectal cancer. Cancer. 2013 doi: 10.1002/cncr.27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N, Dai L, Liu X, Pan G, Chen H, Huang J, Xu Q. Upregulation of caprin1 expression is associated with poor prognosis in hepatocellular carcinoma. Pathology. 2017;213(12):1563–1567. doi: 10.1016/j.prp.2017.07.014. [DOI] [PubMed] [Google Scholar]
- Tan HY, Zheng YB, Liu J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8657–8663. doi: 10.26355/eurrev_201812_16630. [DOI] [PubMed] [Google Scholar]
- Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, Nuovo GJ, Fishel R, Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107(49):21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AS, Johnson EK, Maykel JA, Stojadinovic A, Nissan A, Brucher B, Champagne BJ, Steele SR. Future directions for the early detection of colorectal cancer recurrence. J Cancer. 2014;5(4):272–280. doi: 10.7150/jca.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Shen B, Wang W, Zhou Y, Yang X, Lu G, Yang J, Shao Y. MicroRNA199a5p functions as a tumor suppressor in oral squamous cell carcinoma via targeting the IKKβ/NFκB signaling pathway. Int J Mol Med. 2019;43(4):1585–1596. doi: 10.3892/ijmm.2019.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J, Tang K, Zhou H, Huang Q, Li A, Li Y, Ye Z, Wang J, Xu H. MiR-1 downregulation correlates with poor survival in clear cell renal cell carcinoma where it interferes with cell cycle regulation and metastasis. Oncotarget. 2015;6(15):13201–13215. doi: 10.18632/oncotarget.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Mori M. MicroRNAs as therapeutic targets and colorectal cancer therapeutics. In: Slaby O, Calin GA, editors. Non-coding RNAs in colorectal cancer. Cham: Springer International Publishing; 2016. pp. 239–247. [DOI] [PubMed] [Google Scholar]
- Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013;587(5):436–443. doi: 10.1016/j.febslet.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang T, Jin R, Zhao H, Hu J, Feng B, Zang L, Zheng M, Wang M. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J Exp Clin Cancer Res. 2014;33:113. doi: 10.1186/s13046-014-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, You W, Zhou H, Chen Z, Han G, Zuo X, Zhang L. Downregulated miR-621 promotes cell proliferation via targeting CAPRIN1 in hepatocellular carcinoma. Am J Cancer Res. 2018;8(10):2116–2129. [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen Q, Zhu JW, Liu ZF. MicroRNA-199a-5p regulates glioma progression via targeting MARCH8. Eur Rev Med Pharmacol Sci. 2019;23(17):7482–7487. doi: 10.26355/eurrev_201909_18858. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang S, Hu L, Liu F, Zhang Q, Zhang D. miR-199a-5p suppresses human bladder cancer cell metastasis by targeting CCR7. BMC Urol. 2016;16(1):64. doi: 10.1186/s12894-016-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QD, Zhou QQ, Dong L, Huang Z, Wu F, Deng X. MiR-199a-5p inhibits the growth and metastasis of colorectal cancer cells by targeting ROCK1. Technol Cancer Res Treat. 2018;17:1533034618775509. doi: 10.1177/1533034618775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.