Abstract
This study is aimed at evaluating the regulatory mechanism of quercetin on lipid metabolism in the ileum of broilers to better understand these pathways decreasing abdominal fat. 480 chickens were randomly divided into 4 groups (control, 0.02% quercetin, 0.04% quercetin, and 0.06% quercetin). Breast muscle, thigh muscle, and abdominal fat pad were removed and weighed at 42 d of age. Serum was obtained by centrifuging blood samples from the jugular vein (10 ml) to determine high-density lipoprotein (HDL), total cholesterol (TC), low-density lipoprotein (LDL), triglyceride (TG), leptin, and adiponectin using ELISA. About 5 g of the ileum was harvested and immediately frozen in liquid nitrogen for RNA-seq. Then, the confirmation of RNA-seq results by the Real-Time Quantitative PCR (RT-qPCR) method was evaluated using Pearson's correlation. Compared with control, abdominal fat percentage was significantly decreased with increasing quercetin supplementation, and the best result was obtained at 0.06% dietary quercetin supplementation (P < 0.01). Breast muscle percentage was significantly decreased at 0.02% quercetin (P < 0.01), and thigh muscle percentage tended to increase (P = 0.078). Meanwhile, 0.04% and 0.06% quercetin significantly decreased TG (P < 0.01), TC (P < 0.01), and LDL content (P < 0.05) in serum. Serum leptin and adiponectin contents were significantly increased by 0.04% and 0.06% dietary quercetin supplementation, compared with the control (P < 0.01). Analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used to identify differently expressed genes and lipid metabolism pathways. Quercetin decreased abdominal fat percentage through regulating fat digestion and absorption, glycerophospholipid metabolism, AMPK signaling pathway, fatty acid degradation, and cholesterol metabolism.
1. Introduction
Meat is an important protein source, and chicken is one of the most popular food commodities in the world and the second most preferred meat for Chinese consumers. The fat of meat is an important component in meat quality and impacts animal productivity. The breeding of poultry focused on increasing growth performance and improving breast and thigh meat yields over the past decades. Growth performance made great progress; however, there has been excessive deposition of abdominal fat. In order to reduce fat deposition and improve feed efficiency, different feed additives have been adopted in broiler production. As a kind of safe feed additives, quercetin is impressive [1].
Quercetin is a flavonoid abundant in onions, apples, tea, and red wine, which exhibits antiviral, antitumorigenic, and anti-inflammatory effects [2–5]. Moreover, chronic administration of quercetin markedly improved dyslipidemia, hypertension, and hyperinsulinemia; reduced body weight gain; and increased blood adiponectin levels in obese Zucker rats [6]. Quercetin reduced hepatic fat accumulation in mice fed a high-fat diet and high-sucrose diet [5, 7]. It also decreased TG accumulation in cultured adipocytes in vitro [8]. Significant decreases in serum TG, TC, and epididymal adipose tissue were observed in mice fed a diet supplemented with 0.025% quercetin for 9 weeks [5]. Another study found a significant reduction of visceral fat (-15.5%) in mice fed a diet supplemented with 0.05% quercetin for 20 weeks [7]. Quercetin attenuated abdominal obesity (-37%) in rats fed a high-fat diet supplemented with 0.08% quercetin for 8 weeks [9]. It also exerted protective effects against the development of nonalcoholic fatty liver disease (NAFLD), partly by overexpression of adiponectin and reduction of inflammatory cytokine levels in ob/ob mice [10]. Additionally, it significantly decreased TC contents of the liver, heart, kidney, and small intestine in rats [11].
The intestine plays a vital role in fat digestion and absorption; both the jejunum and ileum were involved in the absorption of fatty acid (FA) in laying hens and broilers [12, 13]. Estimations for endogenous losses of protein and amino acids in the ileum have been published [14]; however, studies on endogenous fat losses in poultry are scant [15]. No examination was done in the specific intestinal segments where the mechanism of quercetin regulated lipid metabolism, and no systematic studies have been reported on quercetin regulating lipid metabolism in the ileum of broilers at the molecular level.
The transcriptome is a necessary link between genomic/genetic information and the biological functions of the proteome. Of these, RNA-seq has been widely utilized to detect differentially expressed genes (DEGs) between two gene expression patterns and causative variants. Many studies of RNA-seq have been conducted in the intestinal mucosa [16], heart [17], uterine [18], and ovarian tissues in broilers [19].
The objective of this study was to evaluate the regulatory mechanism of quercetin on carcass characteristics in broilers. The results of this experiment will provide scientific basis for quercetin application in animal production.
2. Materials and Methods
2.1. Animal Feeding and Diets
Arbor Acre broilers (480 chickens, 1 day old, healthy, similar body weight) from a commercial company (Yinong Poultry Limited Company, Harbin, China) were purchased and randomly divided into 4 groups with 6 replicates per group and 20 broilers per replicate. The broilers were housed in wire cages (four-stacked cages; width: 52.6 cm; depth: 42.3 cm; height: 38.1 cm). Lighting (16 h light : 8 h darkness) was provided during the 6-week experimental period, and temperature was maintained at 32 to 34°C in the starting 3 days and decreased by 2 to 3°C per week to a final temperature of 24°C. Humidity of the experimental room varied from 60% to 65%. Experimental diets and water were available ad libitum. The composition of the basal diets is shown in Table 1 (NY/T33-2004), with 4 levels of quercetin: 0.00%, 0.02%, 0.04%, and 0.06% of quercetin in the diet. Quercetin dihydrate powder with 97% purity was purchased from Sigma-Aldrich Company and mixed with basal diets and was offered in mash form (5 mm) after grinding according to the methods of Yang et al. in 2020 [20].
Table 1.
Analysis composition of basal diets and nutrient level (air-dry basis, %).
| Item | Content (1 to 3 week) | Content (4 to 6 week) |
|---|---|---|
| Ingredient | ||
| Corn | 57.50 | 62.30 |
| Soybean meal | 34.50 | 30.00 |
| Vegetable oil | 3.00 | 3.00 |
| Fish meal | 1.00 | 1.00 |
| Methionine | 0.20 | 0.20 |
| Dicalcium phosphate | 1.62 | 1.67 |
| Limestone | 1.55 | 1.20 |
| Sodium chloride | 0.30 | 0.30 |
| Multivitamin premix1 | 0.03 | 0.03 |
| Mineral premix1 | 0.20 | 0.20 |
| Choline | 0.10 | 0.10 |
| Total | 100.00 | 100.00 |
| Nutrient2 | ||
| Metabolizable energy (ME) (MJ/kg) | 12.33 | 12.50 |
| CP | 21.75 | 19.72 |
| Total lysine (%) | 1.18 | 1.04 |
| Methionine (%) | 0.91 | 0.86 |
| Ca | 1.07 | 0.96 |
| Total P | 0.70 | 0.68 |
| Available P | 0.46 | 0.45 |
1Amount provided per kilogram of diet: vitamin A = 1,500 IU; vitamin D3 = 3,200 IU; vitamin E = 10 IU; vitamin K = 0.5 mg; vitamin B1 = 1.8 mg; vitamin B2 = 3.6 mg; vitamin B6 = 3.5 mg; vitamin B12 = 0.01 mg; biotin = 0.15 mg; folic acid = 0.55 mg; niacin = 30 mg; pantothenic acid = 10 mg; Cu (CuSO4·5H2O) = 8 mg; I (KI) = 0.35 mg; Fe (FeSO4·7H2O) = 80 mg; Mn (MnSO4·H2O) = 60 mg; Se (NaSeO3) = 0.15 mg; Zn (ZnO) = 40 mg. 2Based on composition of ingredients provided by NY/T33-2004.
2.2. Sample Preparation
At the end of experiment (42 d), two broilers per replicate were selected (n = 12 per group). After 12 h fasting, blood samples from the jugular vein (10 ml) were obtained from each group and kept in ice. Serum was obtained after centrifugation at 4°C, 3000 × g for 15 min, and stored at -20°C until analysis. Breast muscle, thigh muscle, and abdominal fat pad (including fat surrounding the gizzard, bursa of Fabricius, cloaca, and adjacent muscles) from one bird of average BW per replicate were removed and weighed at 42 d of age. To compensate for the differences in carcass weight, these values were expressed as a percentage of carcass weight. About 5 g of ileal mucosa was harvested and immediately frozen in liquid nitrogen, and kept at -80°C for further RNA isolation. All procedures used in this study were approved by the Animal Care and Use Committee of the University. Housing, management, and care of the birds confirmed to the guidelines of the Agricultural Animal in Agricultural Research and Teaching of Heilongjiang Province (HEI Animal Management Certificate No. 11928).
2.3. Enzyme-Linked Immunosorbent Assay
Content of high-density lipoprotein (HDL), LDL, TG, TC, leptin, and adiponectin were determined using an enzyme-labeled instrument according to ELISA kit instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China).
2.4. RNA Sequencing
In this endeavor, twelve samples of ileal mucosa were sequenced using the HiSeq 2000 System (Illumina, Inc., USA) by Genesis (Beijing) Co. Ltd. The data analysis included sequencing data filtering, read mapping, transcript and gene identification, analysis of differential gene expression, and functional annotation.
2.5. Real-Time Quantitative PCR
To confirm differential expression of genes, real-time quantitative polymerase chain reaction (RT-qPCR) assays for 12 randomly selected DEGs in the same RNA samples were determined using RNA-seq. All gene expression levels were measured using RT-qPCR (7500 Real-Time PCR System, Singapore). The primer sequences for broilers (Table 2) were designed to span an intron to avoid genomic DNA contamination using Primer 5.0. Total RNA was isolated from ileal mucosa using TRIzol (Invitrogen, San Diego, CA, USA), and total cDNA was synthesized following the manufacturer's instructions. Briefly, a total 20 μl of reaction mixture containing 5 μl of total RNA, 4 μl of 5 × Prime Script Buffer, 1 μl of PrimeScript RT Enzyme Mix I, 1 μl of Oligo(dT) Primer (50 μmol/l), 1 μl of Random Primers 6 mers (10 μM moI/l), and 8 μl of RNase Free dH2O (TaKaRa Biotechnology, Co., Ltd. Dalian, P. R. China) was incubated in the following conditions: reverse transcription at 37°C for 15 min and inactivation of reverse transcriptase at 85°C for 5 min, until temperature was decreased to 4°C. Successful cDNA synthesis was confirmed by amplifying the β-actin amplicon using PCR. cDNA was amplified using a 20 μl PCR reaction system containing 2 μl of cDNA, 10 μl of 2 × SYBR Green PCR Mix, 0.4 μl of 50 × ROX Reference Dye II (TaKaRa Biotechnology, Co., Ltd. Dalian, P. R. China), 0.8 μl of PCR Forward Primer, 0.8 μl of PCR Reverse Primer (Sangon Biological Engineering Technology & Service Co., Ltd. Shanghai, P. R. China), and 6 μl of ddH2O. The following PCR conditions were used: initial denaturation at 95°C for 30 s, followed by PCR reaction at 40 cycles of 95°C for 5 s and 60°C for 34 s, and melting curve analysis at 95°C for 15 s, 60°C for 2 s, and 95°C for 15 s. The PCR products were verified by electrophoresis on 1% agarose gel and DNA sequencing. Standard curves were generated using pooled cDNA from the assayed samples, and the comparative cycle threshold method (2−ΔΔCT) was used for quantifying mRNA levels.
Table 2.
Parameters of primer pairs for the genes.
| Gene | Primer sequence | GenBank accession |
|---|---|---|
| MTTP | F: 5′-GCTGGTAATGCTGAGGTGGATTCC-3′ | NM_001109784.2 |
| R: 5′-AGGAGAGCCAACTCTGTCCATCTG-3′ | ||
|
| ||
| APOA1 | F: 5′-CCTTCTGGCAGCACGATGAGC-3′ | XM_015297971.2 |
| R: 5′-CAGCGTGTCCAGGTTGTCAGC-3′ | ||
|
| ||
| CD36 | F: 5′-ACCAGACCAGTAAGACCGTGAAGG-3′ | NM_001030731.1 |
| R: 5′-ATGTCTAGGACTCCAGCCAGTGTG-3′ | ||
|
| ||
| STRADA | F: 5′-ACACCACCACAATTCCTGCTGATG-3′ | XM_025143971.1 |
| R: 5′-TGACTCTCCGTTGGCTGCTCTC-3′ | ||
|
| ||
| APOA4 | F: 5′-GACAACGCCGACAGCATCCAG-3′ | NM_204938.2 |
| R: 5′-TCCACGCTCTGTGCCACCTG-3′ | ||
| APOB | F: 5′-AGGTGGTGGTGAAGAGGTGGAGAG-3′ | NM_001044633.1 |
| R: 5′-GAGCAGCAAGAGCCGCACAG-3′ | ||
|
| ||
| CYP3A5 | F: 5′-AGCCTGCGGTTGTTGTCATGG-3′ | NM_001001751.2 |
| R: 5′-CTGCGGTTGGTGAAGGTGGAG-3′ | ||
|
| ||
| LOC107080643 | F: 5′-ATCCTCTGCGTCGCTCTCCATC-3′ | NM_001318851.1 |
| R: 5′-TACACTCGCGTCCACCGTCAG-3′ | ||
|
| ||
| ACSL5 | F: 5′-GGTTCACAAGGAGAGTGCAGGAAG-3′ | NM_001031237.1 |
| R: 5′-TCTGAGGCTAGGAGCAGGAAGTTC-3′ | ||
|
| ||
| ADH1C | F: 5′-TTGCCACAACTACGGAGTCAG-3′ | NM_001305183.1 |
| R: 5′-CCAGGTGCGACCACTGAAGATAAG-3′ | ||
|
| ||
| SELENOI | F: 5′-GCCTCTGAACTGGATGCTGCTG-3′ | NM_001031528.3 |
| R: 5′-TGGCTCACCACGACCACTCC-3′ | ||
|
| ||
| PPARα | F: 5′-TGCTGTGGAGATCGTCCTGGTC-3′ | XM_015289959.2 |
| R: 5′-CTGTGACAAGTTGCCGGAGGTC-3′ | ||
|
| ||
| β-Actin | F: 5′-TGCGTGACATCAAGGAGAAG-3′ | L08165 |
| R: 5′-TGCCAGGGTACATTGTGGTA-3′ | ||
|
| ||
| 18sRNA | F: 5′-TAGATAACCTCGAGCCGATCGCA-3′ | AF 173612 |
| R: 5′-GACTTGCCCTCCAATGGATCC TC-3′ | ||
2.6. Statistical Analysis
The data from this experiment were subjected to one-way ANOVA as a completely randomized design with 4 treatments and 6 replicates in each treatment. The data were submitted to ANOVA, using SPSS 20.0 software (2011, IBM). Calculated△Ct (corrected sample) = the mean value of the target gene − the mean value of the internal reference gene; △△Ct = △Ct − the mean value of the control group. Differences among treatment means with a probability level of P<0.05 were accepted as statistically significant, and all the results were expressed as the “mean values ± standard deviation.”
3. Results
3.1. Carcass Characteristics
Thigh muscle percentage tended to increase (P = 0.078); however, abdominal fat percentage was significantly decreased with increasing quercetin (P < 0.01) (Table 3), and breast muscle percentage was significantly decreased by 0.02% quercetin (P < 0.01) compared with the control.
Table 3.
Effect of quercetin on carcass characteristic in broilers.
| Items | Control | 0.02% quercetin | 0.04% quercetin | 0.06% quercetin | P |
|---|---|---|---|---|---|
| Breast muscle (%) | 29.65 ± 0.84A | 27.11 ± 0.51B | 29.28 ± 0.57AB | 30.58 ± 0.4A | 0.001 |
| Thigh muscle (%) | 19.56 ± 0.5 | 21.58 ± 0.43 | 20.57 ± 0.6 | 21.00 ± 0.63 | 0.078 |
| Abdominal fat (%) | 1.68 ± 0.08A | 1.65 ± 0.07A | 1.49 ± 0.04AB | 1.36 ± 0.06B | 0.004 |
Note: in the same row, values with different small letter superscripts mean significant difference (P < 0.05); values with different capital letter superscripts mean significant difference (P < 0.01); Values with no letter or the same letter superscripts mean no significant difference (P < 0.05). Values are expressed as mean ± SEM, and n = 6 for all groups.
3.2. Serum Biochemical Parameters
Comparing with the control, quercetin did not affect HDL content (P > 0.05); however, 0.04% and 0.06% quercetin significantly decreased the content of TG (P < 0.01), TC (P < 0.01), and LDL (P < 0.05) in serum. Contents of serum leptin and adiponectin were significantly increased by 0.04% and 0.06% dietary quercetin supplementation (P < 0.01) (Table 4).
Table 4.
Effect of quercetin on serum biochemical parameters in broilers.
| Items | Control | 0.02% quercetin | 0.04% quercetin | 0.06% quercetin | P |
|---|---|---|---|---|---|
| TG (mg/dl) | 0.68 ± 0.03A | 0.60 ± 0.03AB | 0.54 ± 0.03BC | 0.50 ± 0.03C | 0.003 |
| LDL (mg/dl) | 14.5 ± 1.03a | 12.4 ± 1.39ab | 9.65 ± 1.05b | 9.56 ± 0.99b | 0.014 |
| HDL (mg/dl) | 1.76 ± 0.16 | 1.69 ± 0.18 | 2.08 ± 0.16 | 1.99 ± 0.15 | 0.298 |
| TC (mg/dl) | 4.51 ± 0.19A | 3.76 ± 0.31AB | 3.33 ± 0.25B | 3.36 ± 0.73B | 0.008 |
| Leptin (ng/ml) | 12.47 ± 0.20A | 10.47 ± 0.19B | 14.23 ± 0.26aC | 15.01 ± 0.20bC | 0.000 |
| Adiponectin (mg/ml) | 95.44 ± 4.37A | 94.55 ± 1.68A | 109.48 ± 4.47B | 111.02 ± 3.99B | 0.004 |
Note: in the same row, values with different small letter superscripts mean significant difference (P < 0.05); Values with different capital letter superscripts mean significant difference (P < 0.01); Values with no letter or the same letter superscripts mean no significant difference (P > 0.05). Values are expressed as mean ± SEM, and n = 6 for all groups.
3.3. Summary of the Raw Sequence Reads
Sequence data from the 12 samples were mapped to the reference genome (Gallus gallus, 5.0). Table 5 presents a summary of the RNA-seq analysis including the number of mapped reads and detection of corresponding broilers. The RNA-seq libraries of the 12 samples were sequenced on the Illumina HiSeq 2500 platform, generating 6.22 Gb raw paired-end reads. Comparison of clean reads with the reference genome sequence was done using HISAT. The average ratio of each sample reached 79.09%, and the uniform ratio between samples indicates that the data among the samples were comparable. These 436 million (total) clean reads were subjected to further analysis; 29,186,694 (79.96%), 28,367,936 (78.96%), 29,334,130 (79.55%), and 29,185,088 (80.26%) reads for the control, 0.02%, 0.04%, and 0.06% quercetin groups, respectively, were mapped to the chicken reference genome. The average mapping frequency was 79.68% alignment to the chicken reference genome. On average, 62.27% of the reads were uniquely mapped to the galGal5 (Gallus_gallus-5.0) assembly of the chicken genome, and the mapping frequencies were 62.19%, 61.35%, 62.49%, and 63.05% for the control, 0.02%, 0.04%, and 0.06% quercetin groups, respectively (Table 5). Variant/reference quality ≥ 30 and quality ≥ 20 means that the alternate allele was supported by minimum Phred mass fractions of 30 and 20, and had averages of 98.01% and 93.93%. The above results showed that the next analysis was performed.
Table 5.
Summary statistics for sequence quality and alignment information of 12 samples from ileal mucosa.
| Groups | Control | 0.02% quercetin | 0.04% quercetin | 0.06% quercetin |
|---|---|---|---|---|
| Clean reads | 36,504,566 | 35,926,566 | 36,864,268 | 36,359,240 |
| Q20 (%) | 97.94 | 97.97 | 98.12 | 98.00 |
| Q30 (%) | 93.85 | 93.89 | 94.03 | 93.95 |
| Total mapped reads | 29,186,694 | 28,367,936 | 29,334,130 | 29,185,088 |
| Uniquely mapped reads | 22,698,908 | 22,040,620 | 23,043,483 | 22,925,773 |
| Multiple mapped reads | 6,487,786 | 6,327,316 | 6,290,646 | 6,259,315 |
| Total mapping ratio (%) | 79.96 | 78.96 | 79.55 | 80.26 |
| Uniquely mapping ratio (%) | 62.19 | 61.35 | 62.49 | 63.05 |
1Uniquely mapped reads = reads that matched only one position in the genome. 2Mapping ratio = mapped reads/clean reads. 3Unique mapping ratio = mapped unique reads/clean reads. 4n = 3 for all groups.
3.4. Identification of Differentially Expressed Genes
The detection and analysis of DEGs between control and 0.02%, 0.04%, and 0.06% quercetin help elucidate the regulation of genes. Compared with control, 10,095 significant DEGs were found in this study, including 4276 downregulated and 5819 upregulated genes in 0.02% quercetin; 13,033 significant DEGs were found in this study, including 4117 downregulated and 8916 upregulated genes in 0.04% quercetin; and 11,056 significant DEGs were found in this study, including 4417 downregulated and 6639 upregulated genes in 0.06% quercetin (Figure 1).
Figure 1.
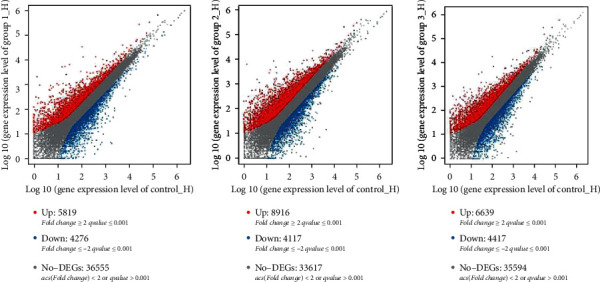
Comparison of differentially expressed genes between control and quercetin (0.02%, 0.04%, and 0.06%). Scatter plot shows the correlation of gene abundance. Red points represent genes upregulated by at least twofold at FDR < 0.05, blue points represent genes downregulated at the same thresholds, and grey dots indicate transcripts that did not change significantly. Group1_H, Group2_H, and Group3_H mean 0.02% quercetin, 0.04% quercetin, and 0.06% quercetin, respectively.
The top 10 genes were significantly upregulated and downregulated among the samples according to the log2FC (Table 6). H3 histone family 3C (H3F3C), histone-lysine N-methyltransferase 2D (KMT2D), PNN-interacting serine- and arginine-rich protein (PNISR), TNF receptor superfamily member 11a (TNFRSF11A), and LOC107052719 were significantly downregulated in 0.02% quercetin/control, 0.04% quercetin/control, and 0.06% quercetin/control. Isocitrate dehydrogenase (NADP (+)) 1 (IDH1), CD36, SON DNA-binding protein (SON), prosaposin (PSAP), and nuclear receptor corepressor 1 (NCOR1) were significantly upregulated in 0.02% quercetin/control, 0.04% quercetin/control, and 0.06% quercetin/control.
Table 6.
The upregulated and downregulated genes compared with control.
| Gene ID | Gene name | Log2FC | q value | P value | Type |
|---|---|---|---|---|---|
| Downregulated genes | |||||
| 0.02% quercetin/control | |||||
| NM_001031482.2 | H3F3C | -10.898 | 3.40E − 138 | 2.41E − 139 | H3F3C: H3 histone, family 3C |
| XM_025145484.1 | KMT2D | -10.7913 | 1.16E − 130 | 8.94E − 132 | KMT2D: histone-lysine methyltransferase 2D |
| XM_025148752.1 | PNISR | -10.5709 | 2.55E − 116 | 2.28E − 117 | PNISR: PNN-interacting serine- and arginine-rich protein |
| XM_004939689.3 | TNFRSF11A | -10.5566 | 1.92E − 115 | 1.73E − 116 | TNFRSF11A: TNF receptor superfamily member 11a |
| 0.04% quercetin/control | |||||
| XM_015282677.2 | LOC107052719 | -13.92476 | 0 | 0 | Uncharacterized LOC107052719 |
| NM_001031482.2 | H3F3C | -10.9586 | 5.51E − 140 | 7.24E − 141 | H3 histone, family 3C |
| XM_025145484.1 | KMT2D | -10.8519 | 2.22E − 132 | 3.15E − 133 | Lysine methyltransferase 2D |
| XM_025148752.1 | PNISR | -10.6903 | 7.98E − 117 | 1.34E − 117 | PNN-interacting serine- and arginine-rich protein |
| XM_004939689.3 | TNFRSF11A | -10.6172 | 5.08E − 117 | 8.47E − 118 | TNF receptor superfamily member 11a |
| 0.06% quercetin/control | |||||
| XM_015282677.2 | LOC107052719 | -13.9034 | 0 | 0 | Uncharacterized LOC107052719 |
| NM_001031482.2 | H3F3C | -10.9373 | 3.19E − 139 | 2.53E − 140 | H3 histone, family 3C |
| Upregulated genes | |||||
| 0.02% quercetin/control | |||||
| XM_015289550.2 | IDH1 | 13.64523 | 5.78E − 153 | 3.61E − 154 | Isocitrate dehydrogenase (NADP (+)) 1, cytosolic |
| XM_025147445.1 | CD36 | 12.3756 | 2.82E − 288 | 8.11E − 290 | CD36 molecule |
| XM_003640519.4 | SON | 13.03347 | 0 | 0 | SON DNA-binding protein |
| XM_015288195.2 | PSAP | 11.55249 | 2.69E − 189 | 1.34E − 190 | Prosaposin |
| XM_004946661.2 | NCOR1 | 11.06195 | 1.02E − 146 | 6.73E − 148 | Nuclear receptor corepressor 1 |
| 0.04% quercetin/control | |||||
| XM_015288195.2 | IDH1 | 12.66957 | 7.90E − 97 | 1.61E − 97 | Isocitrate dehydrogenase (NADP (+)) 1, cytosolic |
| XM_015288195.2 | PSAP | 13.97177 | 0 | 0 | Prosaposin |
| XM_003640519.4 | SON | 12.38731 | 2.36E − 296 | 1.18E − 297 | SON DNA-binding protein |
| 0.06% quercetin/control | |||||
| XM_025147445.1 | CD36 | 10.91717 | 6.73E − 138 | 5.43E − 139 | CD36 molecule |
| XM_004946661.2 | NCOR1 | 10.70961 | 1.18E − 123 | 1.11E − 124 | Nuclear receptor corepressor 1 |
| XM_004946661.2 | NCOR1 | 11.06195 | 1.02E − 146 | 6.73E − 148 | Nuclear receptor corepressor 1 |
1 q value: the corrected P value. The smaller the q value, the more significant the difference in gene expression. 2P value: significant statistical value. 3Log2fold change(sample 2/sample 1): differential expression multiple between samples (groups) after log2 conversion.
3.5. Functional Analysis of Differentially Expressed Genes
The functional distribution of the DEGs was analyzed in the ileum of broilers supplemented with dietary quercetin (0.02%, 0.04%, and 0.06%) compared with the control using GO enrichment and KEGG pathway analyses to understand the regulatory network of lipid metabolism. GO is a type of biological ontology language, which is divided into three parts, including biological process, cellular component, and molecular function. Unlike the functional annotation of a single gene, a gene functional enrichment analysis is based on GO entries, and the results may directly reveal the overall functional characteristics of an entire list of genes. Metabolic processes accounted for the second in the most significantly enriched GO term (Figure 2).
Figure 2.
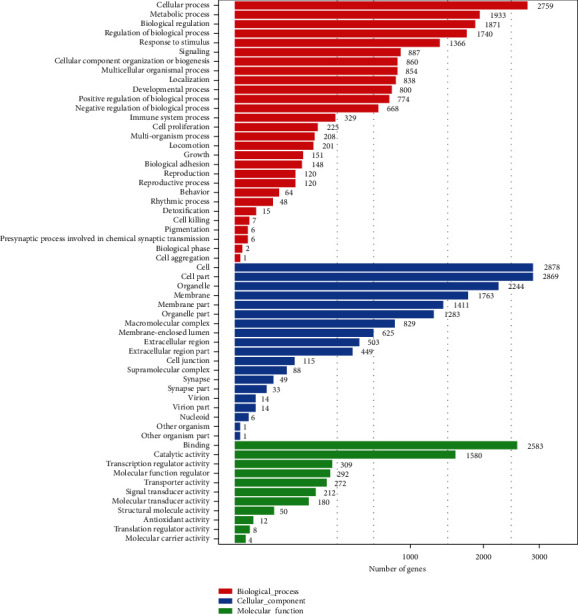
GO analyses of differentially expressed genes in control and quercetin (0.02%, 0.04%, and 0.06%).
The results showed that 9039 DEGs of control/0.02% quercetin were annotated into 336 pathways, 11842 DEGs of control/0.04% quercetin were annotated into 337 pathways, and 9970 DEGs of control/0.06% quercetin were annotated into 337 pathways, including three different classifications (Table 6). The KEGG database revealed that genes were enriched in the transcriptome participating in 33 pathways related to lipid metabolism.
Compared with control, there were differential gene enrichment pathways including glycerophospholipid metabolism (P < 0.01), AMPK signaling pathway (P < 0.01), cholesterol metabolism (P < 0.01), steroid hormone biosynthesis (P < 0.01), insulin resistance (P < 0.05), bile secretion (P < 0.01), and metabolic pathway (P < 0.05) in 0.02% quercetin. There were differential gene enrichment pathways including cholesterol metabolism (P < 0.01), insulin resistance (P < 0.05), and fatty acid metabolism (P < 0.05) in 0.04% quercetin. And there were differential gene enrichment pathways including fat digestion and absorption (P < 0.01), glycerophospholipid metabolism (P < 0.01), fatty acid degradation (P < 0.05), metabolic pathways (P < 0.05), and ether lipid metabolism (P < 0.05) in 0.06% quercetin (Table 7). The main DEGs in significant signaling pathways were as follows: fat digestion and absorption which included microsomal triglyceride transfer protein (MTTP), apolipoprotein A1 (APO A1), secreted phospholipase A2 (SPLA2) G12B (PLA2G12B), and ATP-binding cassette (ABC) transporter (ABCG5/8); glycerophospholipid metabolism which included selenoprotein I (SELEDI), phospholipase D1 (PLD1), and lysophosphatidylcholine acyltransferase 3 (LPCAT3); AMPK signaling pathway which included CD36, STE20-related kinase adaptor alpha (STRADA), protein kinase AMP-activated noncatalytic subunit beta 2 (PRKAB2), TSC2, and phosphatidylinositol 3-kinase 1 (PI3KR1); cholesterol metabolism which included apolipoprotein A4 (APO A4), apolipoprotein C3 (APO C3), and scavenger receptor class B member 1 (SCARB1); fatty acid metabolism which included acyl-CoA synthetase long-chain family member 4 (ACSL4), acyl-CoA synthetase long-chain family member 3 (ACSL3), acyl-CoA synthetase long-chain family member 5 (ACSL5), alcohol dehydrogenase 1C (ADH1C), and carnitine palmitoyltransferase 1A (CPT1A); and steroid hormone biosynthesis which included cytochrome P450 family 3 subfamily A member 5 (CYP3A5), LOC107080643, and steroid sulfatase (STS) (Table 8).
Table 7.
Important lipid metabolic pathways.
| Pathway ID | Pathway definition | P value | All genes | ||
|---|---|---|---|---|---|
| Control/0.02% quercetin | Control/0.04% quercetin | Control/0.06% quercetin | |||
| Ko04975 | Fat digestion and absorption | 8.137809E − 05 | 0.05434665 | 0.004701606 | 115 |
| Ko00564 | Glycerophospholipid metabolism | 0.01281478 | 0.0535793 | 0.009868102 | 105 |
| Ko04152 | AMPK signaling pathway | 0.001574863 | 1.896442E − 07 | 0.09219744 | 505 |
| Ko04979 | Cholesterol metabolism | 0.001932857 | 0.008406933 | 0.08742154 | 217 |
| Ko00071 | Fatty acid degradation | 0.06928025 | 0.06733572 | 0.03222657 | 117 |
| Ko00140 | Steroid hormone biosynthesis | 0.003068663 | 0.09103691 | 0.1157436 | 188 |
| Ko04931 | Insulin resistance | 0.0307667 | 0.03297987 | 0.5826344 | 477 |
| Ko00120 | Primary bile acid biosynthesis | 0.3000608 | 0.9761336 | 0.1666659 | 42 |
| Ko00561 | Glycerolipid metabolism | 0.22205 | 0.4289608 | 0.3153083 | 457 |
| Ko04910 | Insulin signaling pathway | 0.2856451 | 0.541523 | 0.6822437 | 702 |
| Ko04923 | Regulation of lipolysis in adipocytes | 0.3564451 | 0.1735033 | 0.7989723 | 177 |
| Ko00591 | Linoleic acid metabolism | 0.3872519 | 0.746023 | 0.4859596 | 112 |
| Ko00592 | Alpha-linolenic acid metabolism | 0.4011347 | 0.620726 | 0.28578 | 96 |
| Ko00590 | Arachidonic acid metabolism | 0.4455902 | 0.2346687 | 0.1989776 | 178 |
| Ko03320 | PPAR signaling pathway | 0.5193936 | 0.719966 | 0.9634836 | 501 |
| Ko00061 | Fatty acid biosynthesis | 0.6108374 | 0.04648987 | 0.5227091 | 72 |
| Ko00073 | Cutin, suberine, and wax biosynthesis | 0.6533068 | 0.1904358 | 0.7331227 | 25 |
| Ko00785 | Lipoic acid metabolism | 0.6816958 | 0.4859596 | 6 | |
| Ko01040 | Biosynthesis of unsaturated fatty acids | 0.7195848 | 0.3515127 | 0.759634 | 84 |
| Ko00072 | Synthesis and degradation of ketone bodies | 0.7962817 | 0.2089213 | 0.2889255 | 57 |
| Ko00062 | Fatty acid elongation | 0.798622 | 0.1710817 | 0.9398205 | 83 |
| Ko04151 | PI3K/AKT signaling pathway | 0.9998879 | 0.6963632 | 0.8758061 | 1862 |
| Ko04024 | cAMP signaling pathway | 0.9999875 | 0.9999477 | 0.9999958 | 993 |
| Ko04920 | Adipocytokine signaling pathway | 0.70824614 | 0.07274647 | 0.2555452 | 278 |
| Ko00565 | Ether lipid metabolism | 0.09687652 | 0.08454478 | 0.04087001 | 183 |
| Ko01100 | Metabolic pathways | 0.02313553 | 0.1280044 | 0.044613536 | 5281 |
| Ko00062 | Fatty acid elongation | 0.798622 | 0.1710817 | 0.9398205 | 83 |
| Ko00603 | Glycosphingolipid biosynthesis—globo and isoglobo series | 0.1741715 | 0.1247949 | 0.4047945 | 506 |
| Ko04911 | Insulin secretion | 0.999995 | 1 | 0.9999993 | 549 |
| Ko00600 | Sphingolipid metabolism | 0.4227905 | 0.1776699 | 0.5005832 | 165 |
| Ko00604 | Glycosphingolipid biosynthesis—ganglio series | 0.4087944 | 0.1547651 | 0.449979 | 74 |
| Ko04976 | Bile secretion | 0.001000504 | 0.3618376 | 0.4639476 | 230 |
| Ko01212 | Fatty acid metabolism | 0.45523 | 0.0157721 | 0.4646036 | 173 |
1 P value: significant statistical value.
Table 8.
Summary of DEGs involved in lipid accumulation.
| Gene ID | Gene name | Log2FC | Type | ||
|---|---|---|---|---|---|
| 0.02% quercetin/control | 0.04% quercetin/control | 0.06% quercetin/control | |||
| NM_001109784.2 | MTTP | 2.01 | 2.16 | 1.61 | Fat digestion and absorption |
| XM_015297971.2 | APOA1 | 2.1 | 1.37 | 1.95 | Fat digestion and absorption |
| XM_421584.6 | PLA2G12B | 1.63 | 1.02 | 1.2 | Fat digestion and absorption |
| XM_419457.6 | ABCG5 | 1.12 | 0.82 | 0.8 | Fat digestion and absorption |
| XM_419458.6 | ABCG8 | 1.56 | 0.53 | 1.34 | Fat digestion and absorption |
| NM_001031528.3 | SELENOI | 1.21 | 0.92 | 0.73 | Glycerophospholipid metabolism |
| XM_015291712.1 | PLD1 | 6.86 | 5.66 | 7.22 | Glycerophospholipid metabolism |
| XM_416516.5 | LPCAT3 | 1.38 | 1.15 | 1.34 | Glycerophospholipid metabolism |
| NM_001030731.1 | CD36 | 12.38 | 9.72 | 10.92 | AMPK signaling pathway |
| XM_025143971.1 | STRADA | -2.41 | -2.29 | -1.55 | AMPK signaling pathway |
| XM_015290312.2 | PRKAB2 | 8.1 | 6.67 | 6.91 | AMPK signaling pathway |
| XM_015294450.2 | TSC2 | -8.23 | -8.29 | -8.27 | AMPK signaling pathway |
| XM_004937312.3 | PIK3R1 | -5.93 | -5.99 | -5.97 | AMPK signaling pathway |
| NM_204938.2 | APOA4 | 3.42 | 2.94 | 3.48 | Cholesterol metabolism |
| NM_001044633.1 | APOB | 1.53 | 1.34 | 0.93 | Cholesterol metabolism |
| NM_001302127.1 | APOC3 | 3.29 | 3.15 | 3.55 | Cholesterol metabolism |
| XM_015275626.1 | SCARB1 | 8.06 | 6.02 | 6.62 | Cholesterol metabolism |
| NM_001001751.2 | CYP3A5 | 1.02 | 0.7 | 0.88 | Steroid hormone biosynthesis |
| NM_001318851.1 | LOC107080643 | 1.71 | 1.46 | 1.71 | Steroid hormone biosynthesis |
| XM_025146627.1 | STS | 3.64 | 3.64 | 3.8 | Steroid hormone biosynthesis |
| XM_015278614.2 | ACSL4 | 8.67 | 9.35 | 6.45 | Fatty acid degradation |
| XM_0152777033.2 | ACSL3 | 3.59 | 3.99 | 2.67 | Fatty acid degradation |
| NM_001031237.1 | ACSL5 | 1.17 | 0.68 | 0.96 | Fatty acid degradation |
| NM_001305183.1 | ADH1C | 2.1 | 1.2 | 3.94 | Fatty acid degradation |
| XM_015286797.2 | CPT1A | 7.63 | 7.63 | 8.6 | Fatty acid degradation |
1Log2fold change(sample 2/sample 1): differential expression multiple between samples (groups) after log2 conversion.
To verify the accuracy of the RNA-seq results in the transcriptome, the 12 genes from the main lipid metabolic pathways were randomly selected, including 11 significantly upregulated genes and 1 significantly downregulated gene. The expression levels of these 12 genes were quantified using RT-qPCR, and β-actin was used as the internal reference gene. The results showed that the vast majority of genes selected by RT-qPCR validation were similar to sequencing data in the expression pattern, and it suggested that the detection and expression abundance of genes in the present transcriptome sequencing was highly accurate (Figures 3 and 4).
Figure 3.
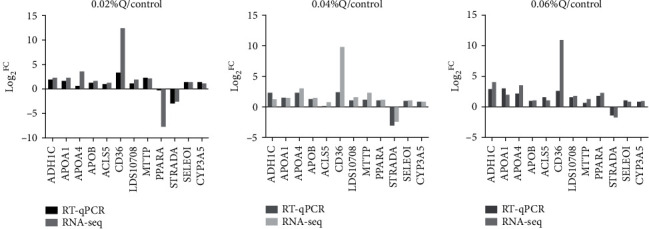
Correlations of mRNA expression level of 12 random DEGs between high and low polyunsaturated fatty acid percentages using RNA-seq and RT-qPCR. Note. The x- and y-axes correspond to the log2(ratio of quercetin/control) measured by RNA-seq and RT-qPCR, respectively. Values are mean ± SEM (n = 6). 0.02%Q, 0.04%Q, and 0.06%Q mean 0.02% quercetin, 0.04% quercetin, and 0.06% quercetin, respectively.
Figure 4.
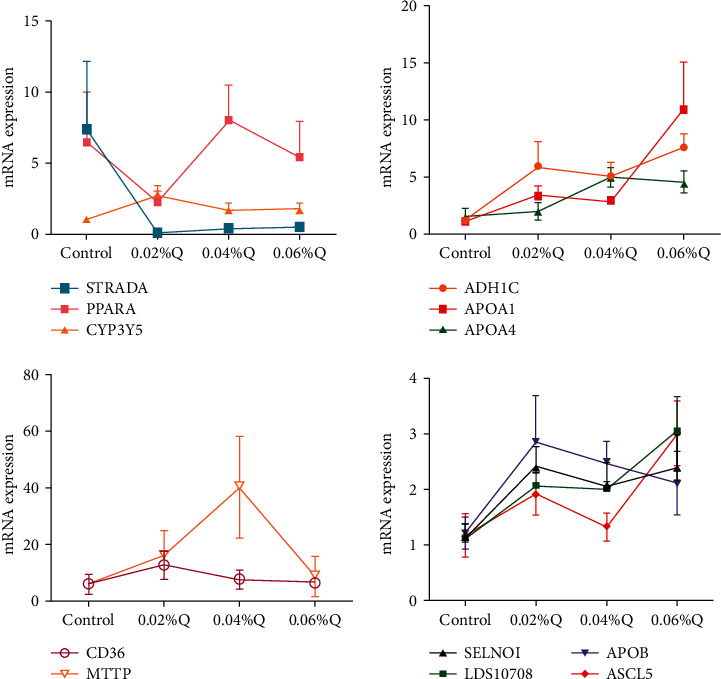
Effects of quercetin on content and mRNA expression of enzymes associated with lipid metabolism in ileal mucosa. Note. (1) The results of relative quantification are expressed as 2−ΔΔCT. The quantification of control is 1, namely 2−ΔΔCT = 1. The value 2−ΔΔCT of the treatment group is a multiple of control. n = 6. (2) Mean values without a common letter are significantly different, P < 0.05. Values are mean ± SEM (n = 6). 0.02%Q, 0.04%Q, and 0.06%Q mean 0.02% quercetin, 0.04% quercetin, and 0.06% quercetin, respectively.
4. Discussion
4.1. The Effect of Quercetin on Carcass Characteristics in Broilers
Flavonoids belong to a group of natural substances, which have a variable phenolic structure and are abundant in fruit, vegetables, tea, and wine. Quercetin is one of the most abundant flavonoids [21]. Different from mammals, the chickens synthesize fatty acids predominantly in the liver and then export fatty acids to other tissues including muscle and adipose tissue by the peripheral vascular system. Therefore, the blood lipid index is related to the carcass characteristics. However, reports concerning the effects of flavonoids on carcass characteristics of animals were also not consistent. 0.20% flavones of sea buckthorn significantly increased dressing percentage [22]. However, the results of the present study showed that no significant responses were found in breast and thigh muscle percentage in quercetin treatments. There was a trend to increase thigh muscle percentage by dietary quercetin supplementation (P = 0.078), affirming the findings of Ma et al. [23]. Too much abdominal fat may adversely affect feed conversion rate and the commercial value of the carcass [24]. Diet supplemented with fermented Ginkgo biloba leaves (including abundant flavonoids) in broilers decreased abdominal fat deposition [25]. Moreover, abdominal fat deposition was reduced in chickens that consumed Hawthorn extract in drinking water [26]. Our results showed that abdominal fat percentage was lowered by 0.06% quercetin, compared with the other groups (P < 0.05) in broilers. The current results are supported by previous studies on the effect of flavonoids on chicken quality in broilers [27, 28].
4.2. Effect of Quercetin on Serum Biochemical Parameters in Broilers
Lipids mainly include triglyceride (TG), phospholipids, and cholesterol (CHO), and the content of TG and CHO are key indicators of lipid metabolism. Hesperidin and naringin mainly regulated lipid metabolism by reducing the content of TG and CHO in the human liver [29]. Furthermore, Hawthorn leaf flavonoid (HLF) extract significantly reduced serum TG and TC levels in mice [30]. In addition, naringenin supplementation significantly decreased concentrations of LDL in ethanol-fed rats [31]. Chrysanthemum indicum ethyl acetate (CIEA) significantly decreased serum lipid profiles, including TG and LDL [29]. The current results showed that 0.04% and 0.06% quercetin significantly decreased the content of serum TG, TC (P < 0.01), and LDL (P < 0.05) in broilers. This further confirmed the hypolipidemic effects of quercetin, just like the other flavonoids.
Leptin is a hormone synthesized by white adipose tissue (WAT) and plays an important role in weight control by suppressing food intake and increasing energy expenditure. Leptin is also a regulator of cellular TG content [32]. Adiponectin is involved in fatty acid oxidation and glucose regulation in liver [33, 34]. And, adiponectin decreased TG content in the muscles and livers of obese mice [35]. CIEA increased adiponectin levels in HFD-induced obese mice [29]. Moreover, the level of leptin (556.7 pg/ml) was significantly increased in mice that received 150 mg/kg M. citrifolia leaf extracts (MLE60) compared to control (535.3 pg/ml) [36]. The result of adiponectin was consistent with the study on Poncritus trifoliate leaf extracts which increased adiponectin levels in serum of HFD-fed mice [37]. The present results showed that 0.04% and 0.06% quercetin significantly increased serum adiponectin and leptin levels (P < 0.01). Meanwhile, together with the results for TG in our experiment, our findings were supported by Nepali et al. and Ma et al. [23, 29], who found that leptin and adiponectin directly interacted with TG, thus decreasing abdominal fat percentage.
4.3. The Effect of Quercetin on Lipid Metabolism Mechanism in Ileum of Broilers
Recently, the RNA-seq technique has been a powerful and revolutionary approach to quantify gene expression levels and survey detailed transcriptome profiles at unprecedented resolution and sensitivity [38, 39]. In chickens, the intestinal mucosa [17], heart [18], and uterine [19] and ovarian tissues [20] have been determined using RNA-seq. Some studies found that quercetin regulated lipid metabolism [30, 31]. Nonetheless, the precise regulating mechanism is not clear in the ileum of broilers. In the present study, RNA-seq was used for determining gene expression profiles and metabolic pathways in broilers fed by diet supplemented with dietary quercetin. Meanwhile, multiple signaling pathways were detected, including the metabolism process in the ileum of broilers. The current RNA-seq data provided greater sequence depth and obtained 79.09% proportions of mapped reads. The high-quality sequences and superior mapping rates enabled the accuracy and reliability of further differential gene expression (Table 4).
In the present study, among the top 10 of the significantly upregulated and downregulated genes between quercetin (0.02%, 0.04%, and 0.06%) and the control group according to the log2FC, the upregulated genes are mainly related to amino acid metabolism (H3F3C, KMT2D, and PNISR), while the downregulated genes are mainly related to lipid metabolism (IDH1, PSAP, and CD36). IDH1 is involved in the tricarboxylate cycle and plays a role in energy metabolism and biosynthesis in organisms. PSAP is a lysosomal gene which delivers bound sphingolipids to cell plasma membranes and enters an endocytotic pathway, responsible for digesting the lipoproteins [39–41]. CD36 is a membrane protein associated with fatty acid transportation [42].
After functional enrichment analyses, GO terms and KEGG pathways were mainly involved in cellular and metabolism processes (Figure 3). The results were also in accordance with the previous studies in chickens [43]. These results suggested that lipid metabolism mainly proceeded in the intestine. In the meantime, to confirm the putative results from RNA-seq, several genes of the lipid metabolic pathway for RT-qPCR assays were randomly selected. Overall, there was excellent agreement and high concordance between the computational and experimental results, which were similar to some previous results in cows [44], broilers [45], and chickens [46]. In the present study, thirty-three of the significant lipid metabolic pathways were detected compared to the control, and most of them were involved in fat digestion and absorption, glycerophospholipid metabolism, AMPK signaling pathway, fatty acid degradation, and cholesterol metabolism (Table 6).
4.3.1. Fat Digestion and Absorption
There were 115 DEGs in this pathway, and 0.06% quercetin was significantly different from the control (P < 0.01). APO A1, MTTP, ABCG5/8, and PLA2G12B are important genes involved in fat digestion and absorption, which are highly expressed in 0.02%, 0.04%, and 0.06% quercetin treatments. Secreted PLA2G12B is a novel mediator of TG metabolism [47]. Intriguingly, the plasma TG, TC, and fatty acid levels were apparently decreased in PLA2G12B-null mice, which are attributed to the compromised hepatic VLDL-TG secretion. ABCG5/ABCG8 belongs to the ABC transporter family members and mediates the biliary secretion of cholesterol. Polydatin increased the secretion of cholesterol into bile by ABCG5/ABCG8, thus improving cholesterol metabolism [48]. The protein expression of MTTP was significantly increased by fisetin [49]. MTTP mainly acts on the synthesis and secretion of chylomicrons and VLDL in the lumen of the endoplasmic reticulum of the liver and the intestine [50, 51]. Numerous studies proved that MTTP plays a distinct role in lipid transport [52, 53]. MTTP is involved in delivering TG to nascent APO B molecules during the assembly of lipoprotein particles [54]. Our results showed that 0.02%, 0.04%, and 0.06% quercetin upregulated MTTP with a log2FC of 2.01, 2.16, and 1.61, respectively. Furthermore, another significantly differential gene, APO A1, belongs to an APO family that encodes important regulators of lipid biosynthesis and metabolism [55]. APO A1 is involved in cholesterol transport [54] and the major constituent of the protein fraction of HDL. Quercetin increased APO A1 mRNA and gene promoter activity in HepG2 cells [56]. According to our log2FC records, the expression of APO A1 was increased by 2.1-, 1.37-, and 1.95-fold as much as the control for 0.02%, 0.04%, and 0.06% quercetin. Meanwhile, together with the results for HDL in this experiment, our findings were supported by Zhou et al. [57], who found that APO A1 directly interacted with HDL. The above results indicate that quercetin regulated lipid metabolism through fat digestion and the absorption pathway of upregulating APO A1, MTTP, ABCG5/8, and PLA2G12B.
4.3.2. Glycerophospholipid Metabolism
PLD1 and LPCAT3 are the two main genes that are associated with the glycerophospholipid metabolism. There were 105 DEGs in this pathway, and 0.02% quercetin and 0.06% quercetin were significantly different from control (P < 0.01). There are four isoforms for LPCAT (LPCAT1, LPCAT2, LPCAT3, and LPCAT4) [58]. Plasma VLDL-TG levels were reduced in the liver of mice lacking LPCAT3 [59]. LPCAT3 deficiency significantly reduced polyunsaturated phosphatidylcholines (PCs) on the hepatocytes and enterocytes, and impacted plasma lipid metabolism [60]. Again, LPCAT3 activity was an important determinant of SREBP-1c activation and lipogenesis [61]. PLD hydrolyzes phosphatidylcholine producing phosphatidic acid, and there are two main isoforms: PLD1 and PLD2. PLD1, the key enzyme involved in lipid metabolism, is associated with metastasis [62]. PLD1 knockout mice resulted in mitochondrial abnormalities and subsequent lipid accumulation [63]. PLD1 knockout mice consume more food due to defects in the hypothalamus, which results in obesity [64]. Furthermore, hepatic TG and TC were increased by 26.5% and 60.4%, respectively, in 4-week high-fat-diet- (HFD-) fed PLD1 knockout mice [63]. The above results indicate that quercetin regulated lipid metabolism through the glycerophospholipid metabolism pathway of upregulating LPCAT3 and PLD1.
4.3.3. AMP-Activated Protein Kinase (AMPK) Signaling Pathway
There were 505 DEGs in this pathway, and 0.02% quercetin was significantly different from control (P < 0.01). The CD36, PRKAB2, and PI3KR1 genes involved in the AMPK signaling pathway were differentially expressed compared with the control. CD36 is a membrane receptor that facilitates long-chain fatty acid uptake and plays an important role in mitochondrial oxidation [65, 66]. A series of evidence supports that CD36 may promote the clearance of chylomicrons from plasma [67, 68] as well as the lipid metabolism and fatty acid transport [65, 66, 69, 70]. CD36 enhanced cellular fatty acid (FA) uptake using the overexpressing CHO model [71]. Our results showed that 0.02%, 0.04%, and 0.06% quercetin increased CD36 expression, which was consistent with Cabanillas et al.'s findings [72]. PI3KR encodes p58 [73]; meanwhile, p58 plays a negative role in PI3K/AKT signaling axis [74, 75]. The PI3K/AKT signaling pathway regulates lipid metabolism. Therefore, quercetin downregulated the PI3KR1 gene that activated the PI3K/AKT signaling axis, thus regulating lipid metabolism. However, PRKAB2 encodes AMPK, which is an energy sensing/signaling intracellular protein activated by an increase in the cellular AMP : ATP ratio after ATP depletion. Once activated, AMPK inhibits fatty acid synthesis [76]. Quercetin regulated lipid metabolism by the AMPK signaling pathway in the liver (data not published). In addition, apigenin suppresses adipogenesis in 3T3-L1 cells via activation of the AMPK pathway [77]. Our results are in line with previous studies, in which quercetin upregulated the AMPK signaling pathway [78].
4.3.4. Fatty Acid Degradation
In the present study, compared with the control, 0.06% quercetin was significantly different from the control in the fatty acid degradation pathway (P < 0.05). ACSLs (ACSL3, ACSL4, and ACSL5), ADH1C, and CPT1A were upregulated by dietary quercetin supplementation. ACSLs are essential for de novo lipid synthesis, fatty acid catabolism, and remodeling of membranes [79]. A previous study identified the sequences of the genes of the ACSL family (ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6) in the sable [80] via transcriptome sequencing. ACSL3 mediated hepatic lipogenesis through transcriptional regulation of lipogenic gene expression [81]. It was transcriptionally upregulated by the cytokine oncostatin M (OSM) in HepG2 cells, accompanied by reduced cellular TG content and enhanced β-oxidation [82]. Polymorphisms of the ACSL4 gene were significantly correlated with liver and intramuscular fat content [83, 84]. Recently, the first evidence in vivo showed that ACSL4 plays a role in plasma TG and hepatic phospholipid synthesis of hyperlipidemic mice [85]. Meanwhile, ACSL5 is an important regulator of whole-body energy metabolism and is implicated in TG synthesis and fat deposition [86–88]. Knockdown of ACSL5 in isolated rat hepatocytes reduces TG accumulation and increases fat oxidation [89]. Our results further confirmed that 0.06% quercetin significantly decreased the plasma's TG content via increasing expression of ACSL3, ACSL4, and ACSL5 (P < 0.05). ADH1C (also known as ADH3) is the predominant isozyme expressed in duodenal, jejunal, and ileum mucosa [90], which has two alleles, namely gamma 1 and gamma 2 (γ1 and γ2). It also directly interacted with the high-density lipoprotein (HDL) [91]. The present study showed that 0.06% quercetin significantly increased mRNA expression of ADH1C in broilers (P < 0.01), while ACSLs and ADH1C directly interacted with TG and HDL, respectively, together with the results on TG and HDL in this experiment, thereby decreasing abdominal fat percentage.
4.3.5. Cholesterol Metabolism
APO A4, APO B, APO C3, and SCARB1 are key transcription factors in the regulation of cholesterol metabolism. There were 217 DEGs in this pathway, and 0.02% quercetin and 0.04% quercetin were significantly different from the control (P < 0.01). APO A4, APO B, and APO C3 also belong to an APO family that encodes important regulators of lipid biosynthesis and metabolism [54]. APO A4 is involved in TG metabolism [59]. Additionally, APO A4 knockout mice as a regulator of TG metabolism increased plasma TG [92]. The results of the current study showed that 0.04% quercetin significantly increased the expression of APO A4 in the ileum of broilers (P < 0.05). APO B located on the surface of the lipoprotein particles is the main lipoprotein of low-density lipoprotein taking charge of transferring cholesterol into tissues. Total flavonoids extracted from Polygonum perfoliatum L. (TFP) decreased the level of APO B and increased the level of APO A, thus adjusting the ratio of APO A/APO B and the metabolic disturbance of lipoprotein by TFP treatment in hyperlipidemia rats [93]. However, our results showed that 0.02%, 0.04%, and 0.06% quercetin increased expression of APO B in the ileum of broilers. Since ileum mucosa has several characteristics different from the liver, the regulatory mechanisms of the respective genes possibly differ [94]. APO C3 is an exchangeable lipoprotein produced by both the liver and the intestine, and found on both chylomicrons and VLDLs. An increase in intestinal APO C3 expression may be of clinical importance for the control of hypertriglyceridemia and the metabolic syndrome [95]. Meanwhile, studies suggested that HDL stimulates scavenger receptor class B type 1 (SR-B1) to promote hepatic uptake of cholesterol. SR-B1 is encoded by the SCARB1 gene in humans, which has been linked to cholesterol metabolism in humans [96]. Our results showed that quercetin decreased TG, TC, and LDL via upregulating APO A4, APO B, APO C3, and SCARB1 expression.
In summary, our results showed that leptin and adiponectin directly interacted with TG, thus decreasing abdominal fat percentage. Meanwhile, quercetin decreased TG, TC, and LDL via regulating APO A4, APO B, APO C3, APO A1, MTTP, ABCG5/8, PLA2G12B, CD36, PRKAB2, PIK3R1, ACSLs, ADH1C, LPCAT3, PLD1, and SCARB1 gene expression in ileal mucosa of broilers.
5. Conclusions
Quercetin decreased abdominal fat percentage through regulating the signaling pathway, including fat digestion and absorption, glycerophospholipid metabolism, the AMPK signaling pathway, fatty acid degradation, and cholesterol metabolism.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (31872377). We thank our members for their assistance in sample collection and laboratory work.
Abbreviations
- HDL:
High-density lipoprotein
- TC:
Total cholesterol
- TG:
Triglyceride
- LDL:
Low-density lipoprotein
- RT-qPCR:
Real-time quantitative PCR
- GO:
Gene Ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- FA:
Fatty acid
- H3F3C:
H3 histone family 3C
- KMT2D:
Histone-lysine N-methyltransferase 2D
- PNISR:
PNN-interacting serine- and arginine-rich protein
- TNFRSF11A:
TNF receptor superfamily member 11a
- IDH1:
Isocitrate dehydrogenase (NADP (+)) 1
- SON:
SON DNA-binding protein
- PSAP:
Prosaposin
- NCOR1:
Nuclear receptor corepressor 1
- MTTP:
Microsomal triglyceride transfer protein
- APOA1:
Apolipoprotein A1
- PLA2G12B:
Secreted phospholipase A2 (SPLA2) G12B
- ABCG5/8:
ATP-binding cassette (ABC) transporter 5/8
- SELEDI:
Glycerophospholipid metabolism included selenoprotein I
- PLD1:
Phospholipase D1
- LPCAT3:
Lysophosphatidylcholine acyltransferase 3
- STRADA:
STE20-related kinase adaptor alpha
- PRKAB2:
Protein kinase AMP-activated noncatalytic subunit beta 2
- PI3KR1:
Phosphatidylinositol 3-kinase 1
- APO A3/4/5:
Apolipoprotein A3/4/5
- SCARB1:
Scavenger receptor class B member 1
- ACSL3/4/5:
Acyl-CoA synthetase long-chain family member 3/4/5
- ADH1C:
Alcohol dehydrogenase 1C
- CPT1A:
Carnitine palmitoyltransferase 1A
- CYP3A5:
Cytochrome P450 family 3 subfamily A member 5
- STS:
Steroid sulfatase
- CHO:
Cholesterol
- HLF:
Hawthorn leaf flavonoids
- CIEA:
Chrysanthemum indicum ethyl acetate
- WAT:
White adipose tissue
- AMPK:
AMP-activated protein kinase
- TFP:
Total flavonoids extracted from Polygonum perfoliatum L.
- SR-B1:
Scavenger receptor class B type 1
- PCs:
Polyunsaturated phosphatidylcholines
- HDL:
High-density lipoprotein
- OSM:
Oncostatin M
- DEGs:
Differentially expressed genes.
Data Availability
Answer: Yes. Comment:
Conflicts of Interest
The authors declare that they have no conflict of interest.
Authors' Contributions
Mi Wang and Yao Li participated in the design of the study and critically revised the first manuscript. Shanshan Wang and Linlin Ying provided some technical support for the experiment. Bo Wang, Yanjun Mao, and Han Lu performed the experiments and participated in the statistical analysis. Yao Li modified the manuscript and have given final approval of the version to be submitted.
References
- 1.Zhang M., Xie Z., Gao W., Pu L., Wei J., Guo C. Quercetin regulates hepatic cholesterol metabolism by promoting cholesterol-to-bile acid conversion and cholesterol efflux in rats. Nutrition Research. 2016;36(3):271–279. doi: 10.1016/j.nutres.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff S. C. Quercetin: potentials in the prevention and therapy of disease. Current Opinion in Clinical Nutrition and Metabolic Care. 2008;11(6):733–740. doi: 10.1097/MCO.0b013e32831394b8. [DOI] [PubMed] [Google Scholar]
- 3.Zhang R., Yao Y., Wang Y., Ren G. Antidiabetic activity of isoquercetin in diabetic KK-Ay mice. Nutrition & Metabolism (London) 2011;8(1):p. 85. doi: 10.1186/1743-7075-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirumbolo S. The role of quercetin, flavonols and flavones in modulating inflammatory cell function. Inflammation & Allergy Drug Targets. 2010;9(4):263–285. doi: 10.2174/187152810793358741. [DOI] [PubMed] [Google Scholar]
- 5.Jung C. H., Cho I. J., Ahn J., Jeon T. I., Ha T. Y. Quercetin reduces high-fat diet-induced fat accumulation in the liver by regulating lipid metabolism genes. Phytotherapy Research. 2013;27(1):139–143. doi: 10.1002/ptr.4687. [DOI] [PubMed] [Google Scholar]
- 6.Rivera L., Morón R., Sánchez M., Zarzuelo A., Galisteo M. Quercetin ameliorates metabolic syndrome and improves the inflammatory status in obese Zucker rats. Obesity. 2008;16(9):2081–2087. doi: 10.1038/oby.2008.315. [DOI] [PubMed] [Google Scholar]
- 7.Kobori M., Masumoto S., Akimoto Y., Oike H. Chronic dietary intake of quercetin alleviates hepatic fat accumulation associated with consumption of a Western-style diet in C57/BL6J mice. Molecular Nutrition & Food Research. 2011;55(4):530–540. doi: 10.1002/mnfr.201000392. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Zhao W., OuYang W. W., WANG M., JIN F. Effect of quercetin lipid metabolism in adipocytes. Journal of Northeast Agricultural University. 2013;44(3):58–65. [Google Scholar]
- 9.Panchal S. K., Poudyal H., Brown L. Quercetin ameliorates cardiovascular, hepatic, and metabolic changes in diet-induced metabolic syndrome in rats. The Journal of Nutrition. 2012;142(6):1026–1032. doi: 10.3945/jn.111.157263. [DOI] [PubMed] [Google Scholar]
- 10.Choi H. N., Jeong S. M., Huh G. H., Kim J. I. Quercetin ameliorates insulin sensitivity and liver steatosis partly by increasing adiponectin expression in ob/ob mice. Food Science and Biotechnology. 2015;24(1):273–279. doi: 10.1007/s10068-015-0036-9. [DOI] [Google Scholar]
- 11.Tang Z., Gao W., Yang J., Wei J., Meng B., Guo C. Effect of quercetin on cholesterol distribution in rats. Acta Nutrimenta Sinica. 2013;35(2):172–175. [Google Scholar]
- 12.Hurwitz S., Bar A., Katz M., Sklan D., Budowski P. Absorption and secretion of fatty acids and bile acids in the intestine of the laying fowl. The Journal of Nutrition. 1973;103(4):543–547. doi: 10.1093/jn/103.4.543. [DOI] [PubMed] [Google Scholar]
- 13.Tancharoenrat P., Ravindran V., Zaefarian F., Ravindran G. Digestion of fat and fatty acids along the gastrointestinal tract of broiler chickens. Poultry Science. 2014;93(2):371–379. doi: 10.3382/ps.2013-03344. [DOI] [PubMed] [Google Scholar]
- 14.Lemme A., Ravindran V., Bryden W. Ileal digestibility of amino acids in feed ingredients for broilers. World’s Poultry Science Journal. 2004;60(4):423–438. doi: 10.1079/WPS200426. [DOI] [Google Scholar]
- 15.Ajuyah A., Balnave D., Annison E. Determination of apparent and true dietary fatty acid digestibilities and metabolisable energy using ileal digesta and excreta from broiler chickens. Animal Feed Science and Technology. 1996;62(2-4):131–139. doi: 10.1016/s0377-8401(96)00996-0. [DOI] [Google Scholar]
- 16.Truong A. D., Hong Y. H., Lillehoj H. S. High-throughput sequencing reveals differing immune responses in the intestinal mucosa of two inbred lines afflicted with necrotic enteritis. Veterinary Immunology and Immunopathology. 2015;166(3-4):116–124. doi: 10.1016/j.vetimm.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Blech-Hermoni Y., Ladd A. N. Identification of transcripts regulated by CUG-BP, Elav-like family member1 (CELF1) in primary embryonic cardiomyocytes by RNA-seq. Genomics Data. 2015;6(C):74–76. doi: 10.1016/j.gdata.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Fan Y., Zhang Z., et al. Analysis of gene expression and regulation implicates C2H9orf152 has an important role in calcium metabolism and chicken reproduction. Animal Reproduction Science. 2017;176(1):1–10. doi: 10.1016/j.anireprosci.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Zhao C., Li J., Feng Y., Gong Y. Transcriptome analysis of the potential roles of FOXL2 in chicken pre-hierarchical and pre-ovulatory granulosa cells. Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics. 2017;21(3):56–66. doi: 10.1016/j.cbd.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Yang J. X., Maria T. C., Zhou B., et al. Quercetin improves immune function in arbor acre broilers through activation of NF-κB signaling pathway. Poultry Science. 2020;99(2):906–913. doi: 10.1016/j.psj.2019.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somerset S. M., Johannot L. Dietary flavonoid sources in Australian adults. Nutrition and Cancer. 2008;60(4):442–449. doi: 10.1080/01635580802143836. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Fu J., Wang B. D., Wang Y. B., Shan A. S. Effect of flavones of sea buckthorn on carcass characteristics and meat quality of arbor acres broilers. Chinese Journal of Animal and Veterinary Sciences. 2008;9:p. 13. [Google Scholar]
- 23.Ma J. S., Chang W. H., Liu G. H., et al. Effects of flavones of sea buckthorn fruits on growth performance, carcass quality, fat deposition and lipometabolism for broilers. Poultry Science. 2015;94(11):2641–2649. doi: 10.3382/ps/pev250. [DOI] [PubMed] [Google Scholar]
- 24.Musalrz H. H., Chen G. H., Chengl J. H., Li B. C., Mekkil D. M. Study on carcass characteristics of chicken breeds raised under the intensive condition. International Journal of Poultry Science. 2006;5(6):530–533. doi: 10.3923/ijps.2006.530.533. [DOI] [Google Scholar]
- 25.Cao F. L., Zhang X. H., Yu W. W., Zhao L. G., Wang T. Effect of feeding fermented Ginkgo biloba leaves on growth performance, meat quality, and lipid metabolism in broilers. Poultry Science. 2012;91(5):1210–1221. doi: 10.3382/ps.2011-01886. [DOI] [PubMed] [Google Scholar]
- 26.Ahmadipour B., Hassanpour H., Asadi E., Khajali F., Rafiei F., Khajali F. Kelussia odoratissima Mozzaf—a promising medicinal herb to prevent pulmonary hypertension in broiler chickens reared at high altitude. Journal of Ethnopharmacology. 2015;159:49–54. doi: 10.1016/j.jep.2014.10.043. [DOI] [PubMed] [Google Scholar]
- 27.Kishawy A. T. Y., Amer S. A., Abd El-Hack M. E., Saadeldin I. M., Swelum A. A. The impact of dietary linseed oil and pomegranate peel extract on broiler growth, carcass traits, serum lipid profile, and meat fatty acid, phenol, and flavonoid contents. Asian-Australasian Journal of Animal Sciences. 2019;32(8):1161–1171. doi: 10.5713/ajas.18.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung U. J., Kim H. J., Lee J. S., et al. Naringin supplementation lowers plasma lipids and enhances erythrocyte antioxidant enzyme activities in hypercholesterolemic subjects. Clinical Nutrition. 2003;22(6):561–568. doi: 10.1016/S0261-5614(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 29.Nepali S., Cha J. Y., Ki H. H., et al. Chrysanthemum indicum inhibits adipogenesis and activates the AMPK pathway in high-fat-diet-induced obese mice. The American Journal of Chinese Medicine. 2018;46(1):119–136. doi: 10.1142/S0192415X18500076. [DOI] [PubMed] [Google Scholar]
- 30.Dong P. Z., Pan L. L., Zhang X. T., et al. Hawthorn (Crataegus pinnatifida Bunge) leave flavonoids attenuate atherosclerosis development in apoE knock-out mice. Journal of Ethnopharmacology. 2017;198(23):479–488. doi: 10.1016/j.jep.2017.01.040. [DOI] [PubMed] [Google Scholar]
- 31.Jayachitra J., Nalini N. Effect of naringenin (citrus flavanone) on lipid profile in ethanol-induced toxicity in rats. Journal of Food Biochemistry. 2012;36(4):502–511. doi: 10.1111/j.1745-4514.2011.00561.x. [DOI] [Google Scholar]
- 32.Reidy S. P., Weber J. M. Leptin: an essential regulator of lipid metabolism. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2000;125(3):285–298. doi: 10.1016/S1095-6433(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 33.Combs T. P., Berg A. H., Obici W., Scherer P. E., Rossetti L. Endogenous glucose production is inhibited by the adipose-derived protein Acrp 30. The Journal of Clinical Investigation. 2001;108(12):1875–1881. doi: 10.1172/JCI14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pajvani U. B., Scherer P. E. Adiponectin: systemic contributor to insulin sensitivity. Current Diabetes Reports. 2003;3(3):207–213. doi: 10.1007/s11892-003-0065-2. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi T., Kamon J., Waki H., et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nature Medicine. 2001;7(8):941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 36.Jambocus N. G. S., Ismail A., Khatib A., et al. Morinda citrifolia L. leaf extract prevent weight gain in Sprague-Dawley rats fed a high fat diet. Food & Nutrition Research. 2017;61(1, article 1338919) doi: 10.1080/16546628.2017.1338919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia S., Gao Z., Yan S., et al. Anti-obesity and hypoglycemic effects of Poncirus trifoliata L. extracts in high-fat diet C57BL/6 mice. Molecules. 2016;21(4):p. 453. doi: 10.3390/molecules21040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484.RNA-Seq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozsolak F., Milos P. M. RNA sequencing: advances, challenges and opportunities. Nature Reviews Genetics. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermann M., Mahon M. G., Lindstedt K. A., Nimpf J., Schneider W. J. Lipoprotein receptors in extraembryonic tissues of the chicken. The Journal of Biological Chemistry. 2000;151(1):299–16844. doi: 10.1016/S0021-9150(00)81359-3. [DOI] [PubMed] [Google Scholar]
- 41.Powell K. A., Deans E. A., Speake B. K. Fatty acid esterification in the yolk sac membrane of the avian embryo. Journal of Comparative Physiology B. 2004;174(2):163–168. doi: 10.1007/s00360-003-0401-5. [DOI] [PubMed] [Google Scholar]
- 42.Coburn C. T., Hajri T., Ibrahimi A., Abumrad N. A. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. Journal of Molecular Neuroscience. 2001;16(2-3):117–122. doi: 10.1385/JMN:16:2-3:117. [DOI] [PubMed] [Google Scholar]
- 43.Yi G., Yuan J., Bi H., Yan W., Yang N., Qu L. In-depth duodenal transcriptome survey in chickens with divergent feed efficiency using RNA-Seq. PLoS One. 2015;10(9, article e136765) doi: 10.1371/journal.pone.0136765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X., Hou Y., Yang S., et al. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genomics. 2014;15(1):p. 226. doi: 10.1186/1471-2164-15-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coble D. J., Fleming D., Persia M. E., et al. RNA-seq analysis of broiler liver transcriptome reveals novel responses to high ambient temperature. BMC Genomics. 2014;15(1):p. 1084. doi: 10.1186/1471-2164-15-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Yang L., Wang H., et al. Growth hormone-regulated mRNAs and miRNAs in chicken hepatocytes. PLoS One. 2014;9(11, article e112896) doi: 10.1371/journal.pone.0112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan M., Qu L., Tan W., Chen L., Wong C. W. Hepatocyte nuclear factor-4 alpha regulates liver triglyceride metabolism in part through secreted phospholipase A2 GXIIB. Hepatology. 2011;53(2):458–466. doi: 10.1002/hep.24066. [DOI] [PubMed] [Google Scholar]
- 48.Peng Y., Xu J., Zeng Y., Chen L., Xu X. L. Polydatin attenuates atherosclerosis in apolipoprotein E-deficient mice: role of reverse cholesterol transport. Phytomedicine. 2019;62, article 152935 doi: 10.1016/j.phymed. [DOI] [PubMed] [Google Scholar]
- 49.Sun Q., Zhang W., Zhong W., Sun X., Zhou Z. Dietary fisetin supplementation protects against alcohol-induced liver injury in mice. Alcoholism, Clinical and Experimental Research. 2016;40(10):2076–2084. doi: 10.1111/acer.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M. M., Shi J., Dreizen P. Microsomal triglyceride transfer protein and its role in ApoB-lipoprotein assembly. Journal of Lipid Research. 2003;44(1):22–32. doi: 10.1194/jlr.R200014-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Hussain M. M., Rava P., Walsh M., Rana M., Iqbal J. Multiple functions of microsomal triglyceride transfer protein. Nutrition & Metabolism (London) 2012;9(1):p. 14. doi: 10.1186/1743-7075-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Guo Y., Ning D. Y., Peng Y., Yang Y., Liu D. Analysis of liver transcriptome in broilers with ascites and regulation by L-carnitine. The Journal of Poultry Science. 2013;50(2):126–137. doi: 10.2141/jpsa.0120124. [DOI] [Google Scholar]
- 53.Han C. C., Wang J. W., Pan Z. X., et al. Effect of liver X receptor activation on the very low density lipoprotein secretion and messenger ribonucleic acid level of related genes in goose primary hepatocytes. Poultry Science. 2011;90(2):402–409. doi: 10.3382/ps.2010-00995. [DOI] [PubMed] [Google Scholar]
- 54.Baroukh N., Bauge E., Akiyama J., et al. Analysis of apolipoprotein A5, c3, and plasma triglyceride concentrations in genetically engineered mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2004;24(7):1297–1302. doi: 10.1161/01.ATV.0000130463.68272.1d. [DOI] [PubMed] [Google Scholar]
- 55.Delgado-Lista J., Perez-Jimenez F., Ruano J., et al. Effects of variations in the APOA1/C3/A4/A5 gene cluster on different parameters of postprandial lipid metabolism in healthy young men. Journal of Lipid Research. 2009;51(1):63–73. doi: 10.1194/jlr.M800527-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haas M. J., Onstead-Haas L. M., Szafran-Swietlik A., et al. Induction of hepatic apolipoprotein A-I gene expression by the isoflavones quercetin and isoquercetrin. Life Sciences. 2014;110(1):8–14. doi: 10.1016/j.lfs.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Zhuo Z., Lamont S. J., Lee W. R., Abasht B. RNA-seq analysis of abdominal fat reveals differences between modern commercial broiler chickens with high and low feed efficiencies. PLoS One. 2015;10(8, article e0135810) doi: 10.1371/journal.pone.0135810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang H., Li Z., Huan C., Jiang X. C. Macrophage lysophosphatidylcholine acyltransferase 3 deficiency-mediated inflammation is not sufficient to induce atherosclerosis in a mouse model. Frontiers in Cardiovascular Medicine. 2019;5:p. 192. doi: 10.3389/fcvm.2018.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z., Jiang H., Ding T., et al. Deficiency in lysophosphatidylcholine acyltransferase 3 reduces plasma levels of lipids by reducing lipid absorption in mice. Gastroenterology. 2015;149(6):1519–1529. doi: 10.1053/j.gastro.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabir I., Li Z., Bui H. H., Kuo M. S., Gao G., Jiang X. C. Small intestine but not liver lysophosphatidylcholine acyltransferase 3 (Lpcat3) deficiency has a dominant effect on plasma lipid metabolism. The Journal of Biological Chemistry. 2016;291(14):7651–7660. doi: 10.1074/jbc.M115.697011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rong X., Wang B., Dunham M. M., et al. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. eLife. 2015;4, article e06557 doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng W., Tan S., Xu Y., et al. LC‑MS/MS metabolome analysis detects the changes in the lipid metabolic profiles of dMMR and pMMR cells. Oncology Reports. 2018;40(2):1026–1034. doi: 10.3892/or.2018.6510. [DOI] [PubMed] [Google Scholar]
- 63.Hur J. H., Park S. Y., Dall’Armi C., et al. Phospholipase D1 deficiency in mice causes nonalcoholic fatty liver disease via an autophagy defect. Scientific Reports. 2016;6(1):1–13. doi: 10.1038/srep39170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trujillo Viera J., el-Merahbi R., Nieswandt B., Stegner D., Sumara G. Phospholipases D1 and D2 suppress appetite and protect against overweight. PLoS One. 2016;11(6, article e0157607) doi: 10.1371/journal.pone.0157607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holloway G. P., Bezaire V., Heigenhauser G. J. F., et al. Mitochondrial long chain fatty acid oxidation, fatty acid translocase/CD36 content and carnitine palmitoyltransferase I activity in human skeletal muscle during aerobic exercise. The Journal of Physiology. 2006;571(1):201–210. doi: 10.1113/jphysiol.2005.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holloway G. P., Luiken J. J. F. P., Glatz J. F. C., Spriet L. L., Bonen A. Contribution of FAT/CD36 to the regulation of skeletal muscle fatty acid oxidation: an overview. Acta Physiologica. 2008;194(4):293–309. doi: 10.1111/j.1748-1716.2008.01878.x. [DOI] [PubMed] [Google Scholar]
- 67.Drover V. A., Abumrad N. A. CD36-dependent fatty acid uptake regulates expression of peroxisome proliferator activated receptors. Biochemical Society Transactions. 2005;33(1):311–315. doi: 10.1042/bst0330311. [DOI] [PubMed] [Google Scholar]
- 68.Pohl J., Ring A., Korkmaz U., Ehehalt R., Stremmel W. FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Molecular and Cellular Biology. 2005;16(1):24–31. doi: 10.1091/mbc.E04-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Febbraio M., Guy E., Coburn C., et al. The impact of overexpression and deficiency of fatty acid translocase (FAT)/CD36. Molecular and Cellular Biochemistry. 2002;239(1/2):193–197. doi: 10.1023/a:1020515210972. [DOI] [PubMed] [Google Scholar]
- 70.Bonen A., Campbell S. E., Benton C. R., et al. Regulation of fatty acid transport by fatty acid translocase/CD36. The Proceedings of the Nutrition Society. 2004;63(2):245–249. doi: 10.1079/pns2004331. [DOI] [PubMed] [Google Scholar]
- 71.Guo J., Shu G., Zhou L., et al. Selective transport of long-chain fatty acids by FAT/CD36 in skeletal muscle of broilers. Animal. 2013;7(3):422–429. doi: 10.1017/s1751731112001619. [DOI] [PubMed] [Google Scholar]
- 72.Cabanillas B. J., le Lamer A. C., Olagnier D., et al. Leishmanicidal compounds and potent PPARγ activators from Renealmia thyrsoidea (Ruiz & Pav.) Poepp. & Endl. Journal of Ethnopharmacology. 2014;157:149–155. doi: 10.1016/j.jep.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 73.Williams R., Berndt A., Miller S., Hon W. C., Zhang X. Form and flexibility in phosphoinositide 3-kinases. Biochemical Society Transactions. 2009;37(4):615–626. doi: 10.1042/BST0370615. [DOI] [PubMed] [Google Scholar]
- 74.Barbour L. A., Mizanoor Rahman S., Gurevich I., et al. Increased P85alpha is a potent negative regulator of skeletal muscle insulin signaling and induces in vivo insulin resistance associated with growth hormone excess. The Journal of Biological Chemistry. 2005;280(45):37489–37494. doi: 10.1074/jbc.m506967200. [DOI] [PubMed] [Google Scholar]
- 75.Brachmann S. M., Ueki K., Engelman J. A., Kahn R. C., Cantley L. C. Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit deletion have opposite effects on insulin sensitivity in mice. Molecular and Cellular Biology. 2005;25(5):1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campa D., Claus R., Dostal L., et al. Variation in genes coding for AMP-activated protein kinase (AMPK) and breast cancer risk in the European Prospective Investigation on Cancer (EPIC) Breast Cancer Research and Treatment. 2011;127(3):761–767. doi: 10.1007/s10549-010-1269-1. [DOI] [PubMed] [Google Scholar]
- 77.Ono M., Fujimori K. Antiadipogenic effect of dietary apigenin through activation of AMPK in 3T3-L1 cells. Journal of Agricultural and Food Chemistry. 2011;59(24):13346–13352. doi: 10.1021/jf203490a. [DOI] [PubMed] [Google Scholar]
- 78.Khalilpourfarshbafi M., Gholami K., Murugan D. D., Abdul Sattar M. Z., Abdullah N. A. Differential effects of dietary flavonoids on adipogenesis. European Journal of Nutrition. 2019;58(1):5–25. doi: 10.1007/s00394-018-1663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soupene E., Kuypers F. A. Mammalian long-chain acyl-CoA synthetases. Experimental Biology and Medicine. 2008;233(5):507–521. doi: 10.3181/0710-mr-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mashek D. G., Bornfeldt K. E., Coleman R. A., et al. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. Journal of Lipid Research. 2004;45(10):1958–1961. doi: 10.1194/jlr.E400002-JLR200. [DOI] [PubMed] [Google Scholar]
- 81.Bu S. Y., Mashek M. T., Mashek D. G. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. The Journal of Biological Chemistry. 2009;284(44):30474–30483. doi: 10.1074/jbc.M109.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cao A., Li H., Zhou Y., Wu M., Liu J. Long chain acyl-CoA synthetase-3 is a molecular target for peroxisome proliferator-activated receptor δ in hepg2 hepatoma cells. The Journal of Biological Chemistry. 2010;285(22):16664–16674. doi: 10.1074/jbc.M110.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruść A., Sieczkowska H., Krzęcio E., et al. The association between acyl-CoA synthetase (ACSL4) polymorphism and intramuscular fat content in (Landrace × Yorkshire) × Duroc pigs. Meat Science. 2011;89(4):440–443. doi: 10.1016/j.meatsci.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 84.Corominas J., Ramayo-Caldas Y., Castelló A., et al. Evaluation of the porcine ACSL4 gene as a candidate gene for meat quality traits in pigs. Animal Genetics. 2012;43(6):714–720. doi: 10.1111/j.1365-2052.2012.02335.x. [DOI] [PubMed] [Google Scholar]
- 85.Singh A. B., Kan C. F. K., Kraemer F. B., Sobel R. A., Liu J. Liver-specific knockdown of long-chain acyl-CoA synthetase 4 reveals its key role in VLDL-TG metabolism and phospholipid synthesis in mice fed a high-fat diet. American Journal of Physiology. Endocrinology and Metabolism. 2019;316(5):e880–e894. doi: 10.1152/ajpendo.00503.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li L. O., Grevengoed T. J., Paul D. S., et al. Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes. 2014;64(1):23–35. doi: 10.2337/db13-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowman T. A., O'Keeffe K. R., D'Aquila T., et al. Acyl CoA synthetase 5 (ACSL5) ablation in mice increases energy expenditure and insulin sensitivity and delays fat absorption. Molecular Metabolism. 2016;5(3):210–220. doi: 10.1016/j.molmet.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Senkal C. E., Salama M. F., Snider A. J., et al. Ceramide is metabolized to acylceramide and stored in lipid droplets. Cell Metabolism. 2017;25(3):686–697. doi: 10.1016/j.cmet.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bu S. Y., Mashek D. G. Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. Journal of Lipid Research. 2010;51(11):3270–3280. doi: 10.1194/jlr.m009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chiang C. P., Jao S. W., Lee S. P., et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human large bowel: association of the functional polymorphisms of ADH and ALDH genes with hemorrhoids and colorectal cancer. Alcoholism, Clinical and Experimental Research. 2012;46(1):37–49. doi: 10.1016/j.alcohol.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 91.Hines L. M., Hunter D. J., Stampfer M. J., et al. Alcohol consumption and high-density lipoprotein levels: the effect of ADH1C genotype, gender and menopausal status. Atherosclerosis. 2005;182(2):293–300. doi: 10.1016/j.atherosclerosis.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 92.Pennacchio L. A., Olivier M., Hubacek J. A., et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294(5540):169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 93.Wang J., Wang Y., Xu C., et al. Effects of total flavonoids extracted from Polygonum perfoliatum L. on hypolipidemic and antioxidant in hyperlipidemia rats induced by high-fat diet. International Journal of Clinical and Experimental Medicine. 2018;11(7):6758–6766. [Google Scholar]
- 94.Casaschi A., Wang Q., Dang K., Richards A., Theriault A. Intestinal apolipoprotein B secretion is inhibited by the flavonoid quercetin: potential role of microsomal triglyceride transfer protein and diacylglycerol acyltransferase. Lipids. 2002;37(7):647–652. doi: 10.1007/s11745-002-0945-8. [DOI] [PubMed] [Google Scholar]
- 95.Guardiola M., Oliva I., Guillaumet A., et al. Tissue-specific DNA methylation profiles regulate liver-specific expression of the APOA1/C3/A4/A5 cluster and can be manipulated with demethylating agents on intestinal cells. Atherosclerosis. 2014;237(2):528–535. doi: 10.1016/j.atherosclerosis.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 96.Ma R., Zhu X., Yan B. SCARB1 rs5888 gene polymorphisms in coronary heart disease: a systematic review and a meta-analysis. Gene. 2018;678:280–287. doi: 10.1016/j.gene.2018.08.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Answer: Yes. Comment:


