Abstract
Objective
The purpose of this study was to explore the effect of microRNA-6071 (miR-6071) on glioblastoma (GBM) and its potential mechanisms.
Methods
In this study, the expressions of miR-6071 and UL16 binding protein 2 (ULBP2) were measured by qRT-RCR in GBM tissues and cells. The prognostic values of miR-6071 and ULBP2 were evaluated by Kaplan–Meier methods using the data obtained from The Cancer Genome Atlas (TCGA) database. The cell clones, proliferation, apoptosis, migration and invasion in GBM cells were detected by colony formation assay, EdU assay, flow cytometry, wound-healing assay and transwell assay. The targeting relationship between miR-6071 and ULBP2 was predicted by Targetscan 7.2 and further verified by dual-luciferase reporter gene assay. Moreover, the expressions of Bax, caspase-3, Bcl-2, matrix metalloproteinases 2 (MMP-2), MMP-9, phosphatidylinositol 3′-kinase (PI3K), p-PI3K, protein kinase B (AKT), p-AKT, mammalian target of rapamycin (mTOR) and p-mTOR were measured by Western blot.
Results
miR-6071 was lowly expressed and ULBP2 was highly expressed in GBM tissues and cells. miR-6071 significantly repressed the proliferation, migration and invasion, and promoted apoptosis in GBM cells. Moreover, miR-6071 also inhibited the activation of PI3K/AKT/mTOR pathway in GBM cells. Additionally, miR-6071 has been shown to negatively regulate ULBP2 expression. We also confirmed that ULBP2 could reverse the effects of miR-6071 on GBM cells through regulating PI3K/AKT/mTOR pathway.
Conclusion
Our study demonstrated that miR-6071 could suppress cell proliferation, migration and invasion, as well as promote apoptosis through the inhibition of PI3K/Akt/mTOR pathway by binding to ULBP2 in GBM.
Keywords: glioblastoma, miR-6071, ULBP2, proliferation, metastasis, PI3K/Akt/mTOR pathway
Introduction
Glioblastoma (GBM) is one of the most malignant primary brain tumors in the human central nervous system and is associated with high morbidity and lethality.1,2 Although great efforts have been made towards the effective treatment of GBM in recent years, the overall survival rate of GBM patients is relatively low.3 Hence, exploring new markers for the diagnosis and treatment of GBM is critical.
MicroRNAs (miRNAs) are a class of evolutionally conserved small non-coding RNA molecules (19–25 nucleotides) that modulate gene expression via binding their target mRNA.4,5 miRNAs have been shown to be important in cellular processes, including cell proliferation, differentiation, apoptosis, metastasis, autophagy and cell death.6–9 In recent years, there has been increasing evidence that miRNAs play an important role in several kinds of cancers, including GBM.10,11 As an example is miR-154-5p, which has been reported to inhibit the proliferation and metastasis of GBM cells through targeting the Piwi like RNA-mediated gene silencing 1 gene.12 A study by Xu et al has indicated that miR-148a could suppress GBM cell proliferation and migration via targeting ITGA9.13 The analysis of The Cancer Genome Atlas (TCGA) dataset shows that the high expression of miR-6071 is associated with a better prognosis in GBM patients. However, the effect of miR-6071 on GBM and its related mechanisms have not yet been reported.
The phosphatidylinositol 3′-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway is one of the most frequently activated signal-transduction pathways in various cancers.14 Moreover, an increasing number of studies have confirmed that miRNAs play vital roles in GBM through modulating the PI3K/Akt/mTOR pathway. One example is miR-579, which potentially suppresses cell proliferation, migration and invasion in GBM via the regulation of the PI3K/AKT pathway.15 Kalhori et al16 have suggested that the overexpression of miR-548x and miR-4698 could inhibit proliferation of GBM cells by affecting the PI3K/AKT signaling pathway. However, whether miR-6071 affects the progression of GBM by modulating the PI3K/Akt/mTOR pathway is not clear.
In the present study, we explored the effects and molecular mechanisms of miR-6071 on GBM. Our data showed that miR-6071 could repress cell proliferation, migration and invasion, as well as promote apoptosis via the inhibition of the PI3K/Akt/mTOR pathway by binding to ULBP2 in GBM. Our findings may facilitate the understanding of GBM pathogenesis and identify a potential therapeutic for GBM treatment.
Materials and Methods
Human Tissue Samples
Thirty GBM tissue samples and 10 pair-matched normal brain tissues (NBTs) from the trauma or epilepsy patients were acquired from our hospital between July 2016 and June 2019, and then reserved at −80°C until usage. Before surgery, no patients received chemotherapy or radiotherapy. The study was approved by the Ethics Committee of Yan’an Hospital of Kunming and written informed consent was signed by all recruited patient.
Cell Cultures
Four human glioma cell lines (A172, U251, U87 and LN229) and normal human astrocyte (NHA) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, USA) with 10% fetal bovine serum (FBS, GE Healthcare Life Sciences, USA) and 1% penicillin/streptomycin (HyClone, USA) at 37°C in an atmosphere with 5% CO2.
Cell Transfection
The miR-6071 mimics, miR-6071 inhibitor, UL16 binding protein 2 (ULBP2) siRNA (si-ULBP2) and their corresponding negative controls were transfected into U251 and A172 cells by using Lipofectamine® 3000 Reagent (Invitrogen, USA) following to the manufacturer’s protocols. The transfected U251 and A172 cells were randomly divided into 9 groups: Control group (no treatment), mimics NC group (transfected with miR-6071 mimics negative control), miR-6071 mimics group (transfected with miR-6071 mimics), inhibitor NC group (transfected with miR-6071 inhibitor negative control), miR-6071 inhibitor group (transfected with miR-6071 inhibitor), inhibitor NC + si-NC group (transfected with inhibitor NC and ULBP2 siRNA negative control), inhibitor NC + si-ULBP2 group (transfected with inhibitor NC and ULBP2 siRNA), miR-6071 inhibitor + si-NC group (transfected with miR-6071 inhibitor and si-NC) and miR-6071 inhibitor + si-ULBP2 group (transfected with miR-6071 inhibitor and si-ULBP2). Then, all the cells were cultured in 37°C incubator for 48 h.
Colony Formation Assay
The ability of cell clones was measured by using colony formation assay. Simply, the transfected U251 and A172 cells were seeded onto a six-well plate and cultured for 14 days. Afterwards, the colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma, USA), respectively. Finally, the stained colonies were counted by using a light microscope.
5-Ethynyl-20-Deoxyuridine (EdU) Proliferation Assay
The proliferation ability of U251 and A172 cells was determined by using EdU assay kit (RiboBio, Guangzhou, China). In brief, the transfected U251 and A172 cells were planted into a 96-well plant and then incubated with 50 μm EdU for two hours. After that, the 4′,6-diamidino-2phenylindole (DAPI, Sigma, USA) was used to stain the cell nuclei of U251 and A172 cells. In the end, the EdU-positive cells were observed by using a fluorescence microscope.
Apoptosis Assay
The apoptosis ability of U251 and A172 cells was detected by Annexin-V-fluorescein isothiocyanate (FITC) cell apoptosis assay kit (Biovision, USA) following to the manufacturer’s protocols. In brief, the transfected U251 and A172 cells were collected, washed twice with cold PBS buffer and resuspended in binding buffer. Afterwards, U251 and A172 cells were incubated with Annexin V-FITC (5 μL) and propidium iodide (PI, 10 μL) for 15 min. The apoptotic cells were analysed by using flow cytometry within 1 h.
Wound-Healing Assay
The transfected U251 and A172 cells were cultured in a 6-well plate for 24 h to form a confluent monolayer. Then, the cell monolayer was scratched in a straight line with a sterile pipette tip. After that, the U251 and A172 cells were washed with PBS for three times to remove the debris, and then serum-free medium was added into the well plates for continuous incubation. The images were captured at 0 and 24 h after scratching under a light microscope, and then the wound healing rate was calculated and recorded.
Transwell Assay
The transfected U251 and A172 cells were resuspended in serum-free medium and then added to the top chamber of transwell chamber (Corning, Corning, NY, USA). For invasion assay, the upper chamber was pre-coated with Matrigel (BD Biosciences, USA). The lower chamber was filled with DMEM including FBS. After 48 h, the migrated cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 15 min and then stained by 0.1% crystal violet solution (Beyotime, Shanghai, China) for 15 min. The number of stained cells was observed under an inverted microscope (Olympus, Tokyo, Japan) in five randomly selected fields.
Dual-Luciferase Reporter Gene Assay
Targetscan 7.2 was used to predict the targeted relationship between miR-6071 and ULBP2. For dual-luciferase gene assay, U251 and A172 cells were co-transfected with PsiCHECK-2 luciferase plasmids (Promega, USA) carrying ULBP2-wildtype (ULBP2-Wt)/ULBP2-mutant (ULBP2-Mut) and miR-6071 mimics/miR-6071 NC by using Lipofectamine 3000 (Invitrogen, USA). After 48 h of transfection, the luciferase activity was detected by using a dual luciferase kit (Promega).
Quantitative Real Time-PCR (qRT-PCR)
Total RNA from tissue and cells was extracted with the use of TRIZOL kit (Invitrogen, USA). Reverse-transcribed complementary DNAs (cDNAs) were prepared according to the Revert aid first-strand cDNA synthesis kit (Thermo Fisher, USA), and qRT-PCR was conducted utilizing the SYBR Green One-Step RT-PCR Master Mixes (Thermo Fisher) on an ABI 7900 real-time PCR system (Applied Biosystems, USA). We used U6 and GAPDH as internal control. The primer sequences were as follows: miR-6071 (sense): 5′-TGGTACTGATGTGATGGACT-3′ (anti-sense): 5′-TCATATCACACAGCACCGAT-3′; U6 (sense): 5′-CTCGCTTCGGCAGCACA-3′ (anti-sense): 5′-AACGCTTCACGAATTTGCGT-3′; ULBP2 (sense): 5′-AAATGTCACAACGGCCTG-3′ (anti-sense): 5′-TGAGGGGTTCCTTGGG-3′; GAPDH (sense): 5′-CGAGCCACATCGCTCAGACA-3′ (anti-sense): 5′-GTGGTGAAGACGCCAGTGGA-3′. The relative gene expression levels were calculated using the 2−ΔΔCt method.
Western Blot Analysis
Total proteins were extracted from U251 and A172 cells using RIPA lysis buffer (Solarbio, Beijing, China) containing protease inhibitor. The same amount of protein from each group was separated by 10% SDS-PAGE and then transferred to a polyvinylidene difluoride membrane with the method of wet transfer. After sealed with 5% non-fat milk for 2 h at room temperature, the membrane was incubated with the specific primary antibody (anti-Bax, 1:1000, ab182733; anti-B-cell lymphoma-2 (Bcl-2), 1:500, ab32124; anti-caspase-3, 1:500, ab2302; anti-MMP-2, 1:500, ab97779; anti-MMP-9, 1:500, ab38898, Abcam, UK; anti-p-PI3K, 1:300, #17366; anti-PI3K, 1:1000, #4257; anti-p-AKT, 1:500, #4060; anti-AKT, 1:1000, #8596; anti-p-mTOR, 1:500, #2971; anti-mTOR, 1:1000, #2983, anti-GAPDH, 1:1500, #2118, Cell signaling, USA) overnight at 4°C. After washing three times, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit; 1:5000, Sigma) for 1 h at room temperature. The protein bands were visualized with ECL system (Thermo, USA) and then analyzed by using Image Lab™ Software (Bio-Rad, USA).
Statistical Analysis
The data were presented as mean ± SEM. All statistical analysis was performed using SPSS 22.0 statistical software. Statistical analysis was performed using Student’s t test or one-way ANOVA test. And the correlation between miR-6071 and ULBP2 was analyzed by Pearson correlation analysis. P < 0.05 expresses that the differences were statistically significant.
Results
miR-6071 Expression is Low in GBM and is Associated with Disease Progression
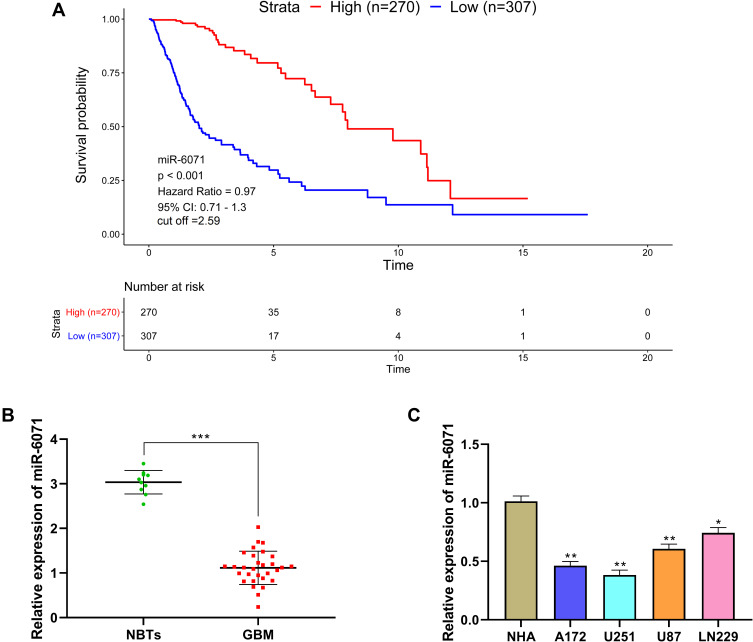
The analysis of The Cancer Genome Atlas (TCGA) dataset revealed that the high expression of miR-6071 was significantly associated with a better prognosis in GBM patients (Figure 1A). In addition, we first measured the expression of miR-6071 in GBM tissues and found that the expression of miR-6071 was markedly decreased in GBM tissues (P < 0.001) (Figure 1B). Similarly, the expression of miR-6071 was dramatically decreased in U87 (P < 0.01) and LN229 cells (P < 0.05), especially in U251 (P < 0.01) and A172 cells (P < 0.01) when compared to NHA cells (Figure 1C). Together, the data suggested that the expression of miR-6071 was downregulated in GBM and might play an important role in the progression of GBM.
Figure 1.
miR-6071 expression was low in GBM and was associated with disease progression. (A) Kaplan–Meier analyses of the overall survival of The Cancer Genome Atlas (TCGA) cohort patients with high miR-6071 expression and those with low miR-6071 expression. (B) The expression of miR-6071 in GBM tissues was detected by qRT-PCR. (C) The expression of miR-6071 in GBM cells was detected by qRT-PCR. ***P < 0.001, vs Normal brain tissues (NBTs) group (B); *P < 0.05, **P < 0.01, vs NHA cells group (C).
miR-6071 Inhibits the Proliferation Ability of U251 and A172 Cells
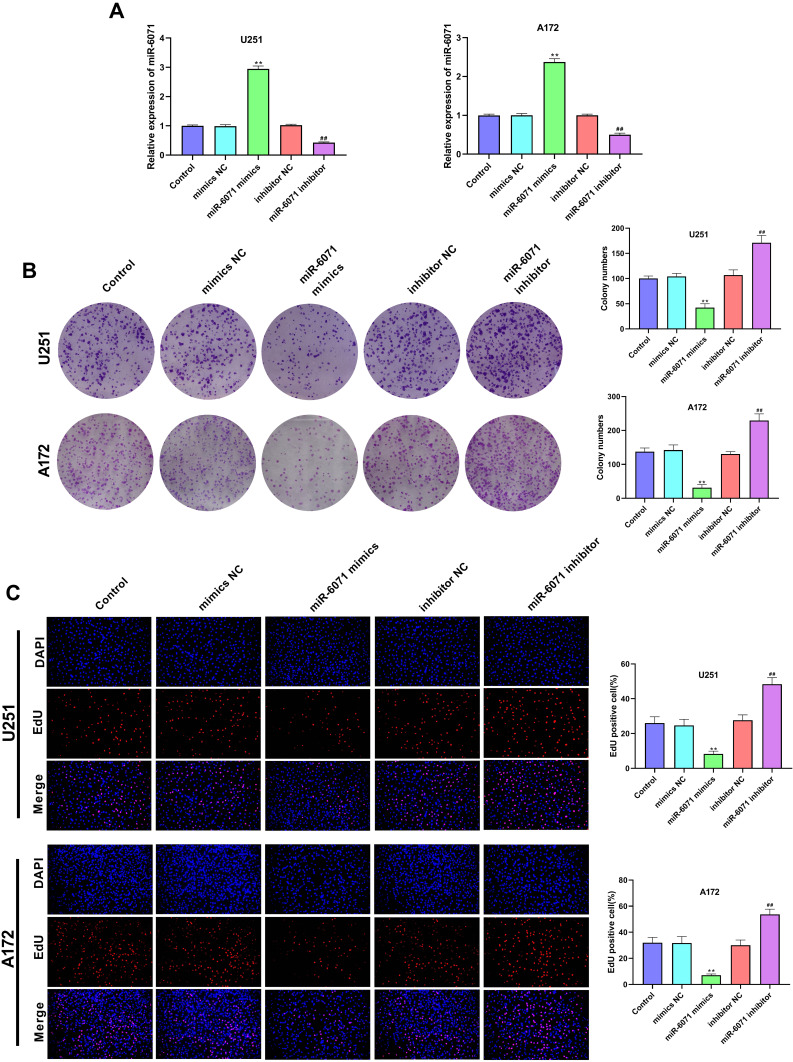
As shown in Figure 2A, the expression of miR-6071 in U251 and A172 cells was significantly increased in the miR-6071 mimics group compared to the Control and the mimics NC groups (P < 0.01). In addition, the expression of miR-6071 in miR-6071 inhibitor group was lower than that in Control and inhibitor NC groups (P < 0.01), suggesting that the transfection was successful. The results of the colony formation assay (Figure 2B) and EdU assay (Figure 2C) showed that the upregulation of miR-6071 significantly decreased cell clone numbers and cell proliferation in both U251 and A172 cells (P < 0.01). In contrast, the downregulation of miR-6071 markedly increased cell clone numbers and cell proliferation (P < 0.01). All the results suggested that miR-6071 could inhibit the proliferation of U251 and A172 cells.
Figure 2.
miR-6071 inhibited the proliferation ability of U251 and A172 cells. (A) The expression of miR-6071 in transfected U251 and A172 cells was detected by qRT-PCR. (B) The colonies number of U251 and A172 cells was detected by colony formation assay. (C) The proliferation of U251 and A172 cells was tested by EdU assay. **P < 0.01, vs Control and mimics NC group; ##P < 0.01, vs Control and inhibitor NC group.
miR-6071 Promotes the Apoptosis Ability of U251 and A172 Cells
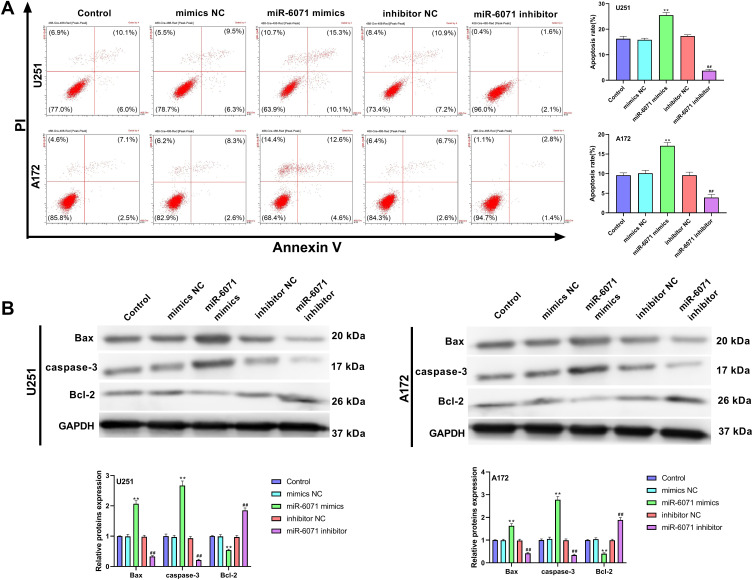
The results of flow cytometry revealed that the apoptosis of U251 and A172 cells was notably elevated in the miR-6071 mimics group in comparison with the Control and the mimics NC groups (P < 0.01) (Figure 3A). We also observed that the apoptosis of the cells in the miR-6071 inhibitor group was dramatically reduced relative to the Control and the inhibitor NC groups (P < 0.01) (Figure 3A). To further confirm the results, we detected the expressions of Bax, caspase-3 and Bcl-2 by Western blot (Figure 3B). Indeed, the results showed that the upregulation of miR-6071 dramatically decreased Bcl-2 expression in U251 and A172 cells relative to the Control and the mimics NC groups (P < 0.01), but increased the expression levels of Bax and caspase-3 (P < 0.01). In comparison, the downregulation of miR-6071 significantly elevated the expression of Bcl-2 compared with the Control and the inhibitor NC groups (P < 0.01), but reduced the expression levels of Bax and caspase-3 (P < 0.01). These results indicated that miR-6071 could promote the apoptosis ability of U251 and A172 cells.
Figure 3.
miR-6071 promoted the apoptosis ability of U251 and A172 cells. (A) The apoptosis of U251 and A172 cells was detected by flow cytometry. (B) The expressions of Bax, caspase-3 and Bcl-2 in U251 and A172 cells were measured by Western blot. **P < 0.01, vs Control and mimics NC group; ##P < 0.01, vs Control and inhibitor NC group.
miR-6071 Inhibits the Ability of Migration and Invasion in U251 and A172 Cells
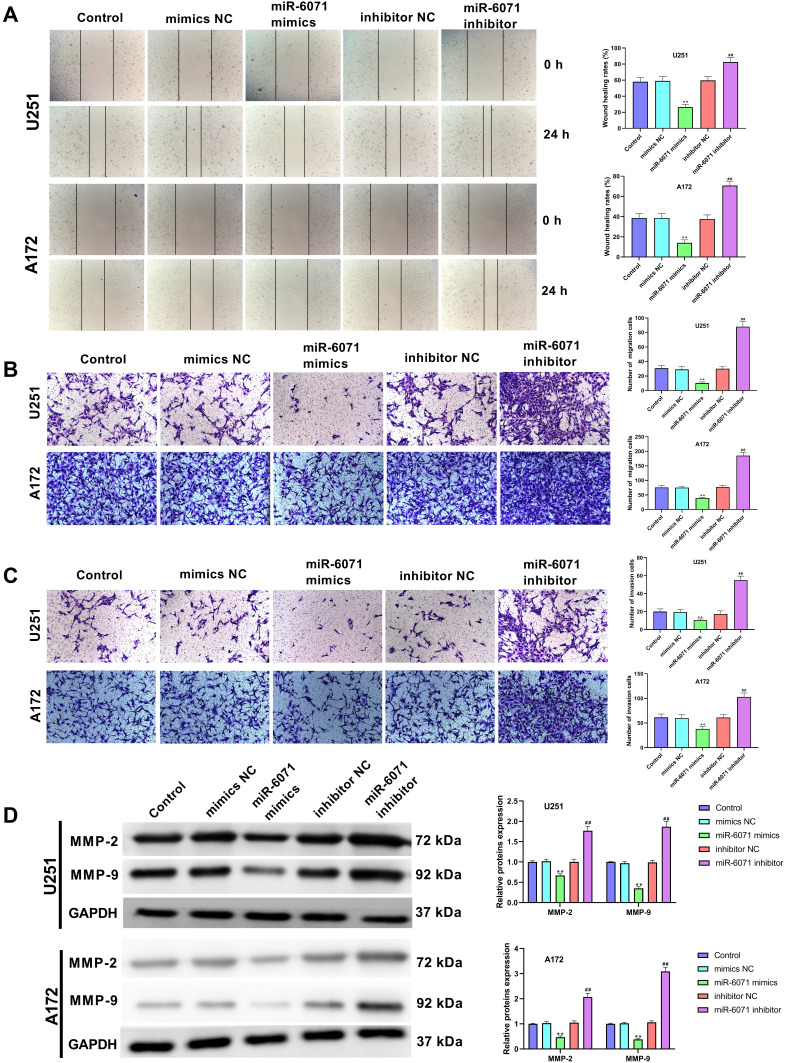
The ability of migration and invasion in U251 and A172 cells was determined by using wound-healing assay (Figure 4A) and transwell assay (Figure 4B and C). The results showed that the upregulation of miR-6071 significantly reduced the migration and invasion in U251 and A172 cells compared with the Control and the mimics NC groups (P < 0.01). Conversely, the downregulation of miR-6071 dramatically elevated the migration and invasion relative to the Control and the inhibitor NC groups (P < 0.01). To further confirm the results, we detected the expressions of MMP-2 and MMP-9 in U251 and A172 cells using Western blot (Figure 4D) and found that the expression of MMP-2 and MMP-9 was significantly decreased by the upregulation of miR-6071 (P < 0.01) and markedly increased by the downregulation of miR-6071 (P < 0.01). Our data indicated that miR-6071 could inhibit the ability of migration and invasion in U251 and A172 cells.
Figure 4.
miR-6071 inhibited the ability of migration and invasion in U251 and A172 cells. (A) The migration of U251 and A172 cells was tested by wound-healing assay. (B) The migration of U251 and A172 cells was tested by transwell assay. (C) The invasion of U251 and A172 cells was tested by transwell assay. (D) The expressions of MMP-2 and MMP-9 in U251 and A172 cells were measured by Western blot. **P < 0.01, vs Control and mimics NC group; ##P < 0.01, vs Control and inhibitor NC group.
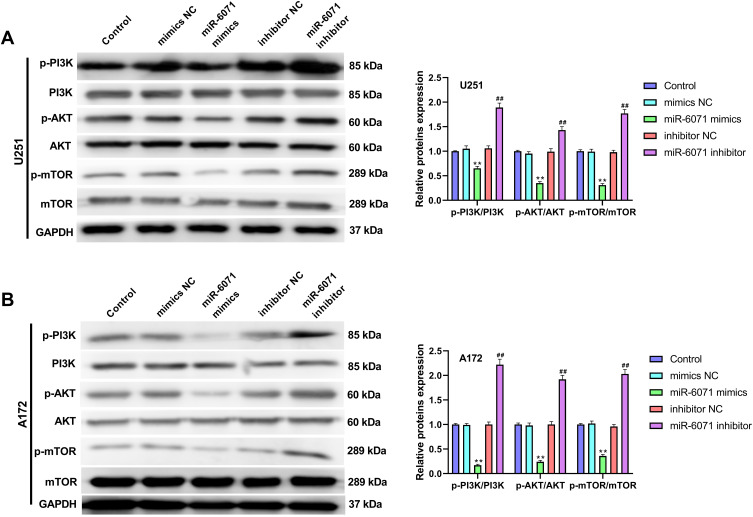
miR-6071 Inhibits the Activation of PI3K/AKT/mTOR Pathway in U251 and A172 Cells
As Figure 5A and B showed, the expressions of p-PI3K, p-AKT and p-mTOR in U251 and A172 cells were significantly inhibited by the upregulation of miR-6071 (P < 0.01), and markedly promoted by the downregulation of miR-6071 (P < 0.01). In addition, the expressions of PI3K, AKT and mTOR were not statistically significant between different groups (P > 0.05). Taken together, the results suggested that miR-6071 could inhibit the activation of PI3K/AKT/mTOR pathway in U251 and A172 cells.
Figure 5.
miR-6071 inhibited the activation of PI3K/AKT/mTOR pathway in U251 and A172 cells. (A) The expressions of p-PI3K, PI3K, p-AKT, AKT, p-mTOR and mTOR in U251 cells were measured by Western blot. (B) The expressions of p-PI3K, PI3K, p-AKT, AKT, p-mTOR and mTOR in A172 cells were measured by Western blot. **P < 0.01, vs Control and mimics NC group; ##P < 0.01, vs Control and inhibitor NC group.
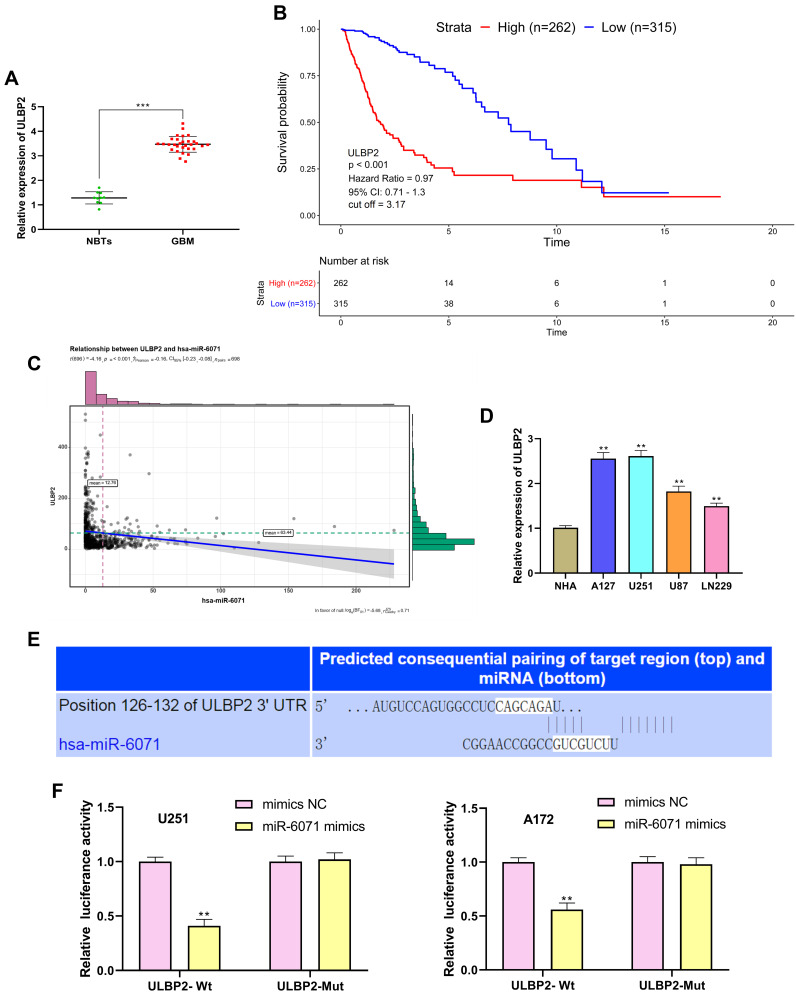
miR-6071 Negatively Modulates the Expression of ULBP2
The results of qRT-PCR showed that ULBP2 expression was markedly increased in GBM tissues in comparison with the NBTs group (P < 0.001) (Figure 6A). As shown in Figure 6B, the analysis of the TCGA dataset indicated that the low expression of ULBP2 was significantly associated with a better prognosis in GBM patients. In addition, the analysis of TCGA dataset also confirmed that the expression of miR-6071 was negatively correlated with the expression of ULBP2 in GBM tissues (P < 0.001) (Figure 6C). Subsequently, we detected the expression of ULBP2 in GBM cells and NHA cells, and found that ULBP2 expression was markedly elevated in U87 and LN229 cells, especially in U251 and A172 cells when compared with NHA cells (Figure 6D). As Figure 6E showed, a binding site of miR-6071 was predicted at 3ʹ-UTR of ULBP2 using Targetscan 7.2. Moreover, dual-luciferase reporter gene assay revealed that miR-6071 mimics dramatically reduced the fluorescence intensity in the ULBP2-Wt group (P < 0.01), but had no significant effect on the fluorescence intensity in the ULBP2-Mut group (P > 0.05) (Figure 6F). Collectively, these data demonstrated that miR-6071 could negatively modulate ULBP2 expression.
Figure 6.
miR-6071 negatively modulated the expression of ULBP2. (A) The expression of ULBP2 in GBM tissues was detected by qRT-PCR. (B) Kaplan–Meier analyses of the overall survival of TCGA cohort patients with high ULBP2 expression and those with low ULBP2 expression. (C) Correlation between miR-6071 and ULBP2 in clinical GBM samples from TCGA. (D) The expression of ULBP2 in GBM cells was detected by qRT-PCR. (E) The binding target of ULBP2 and miR-6071 was predicted by Targetscan 7.2 software. (F) The luciferase activity was determined by dual-luciferase reporter gene assay. ***P < 0.001, vs NBTs group (A); **P < 0.01, vs NHA cells group (D); **P < 0.01, vs mimics NC group (F).
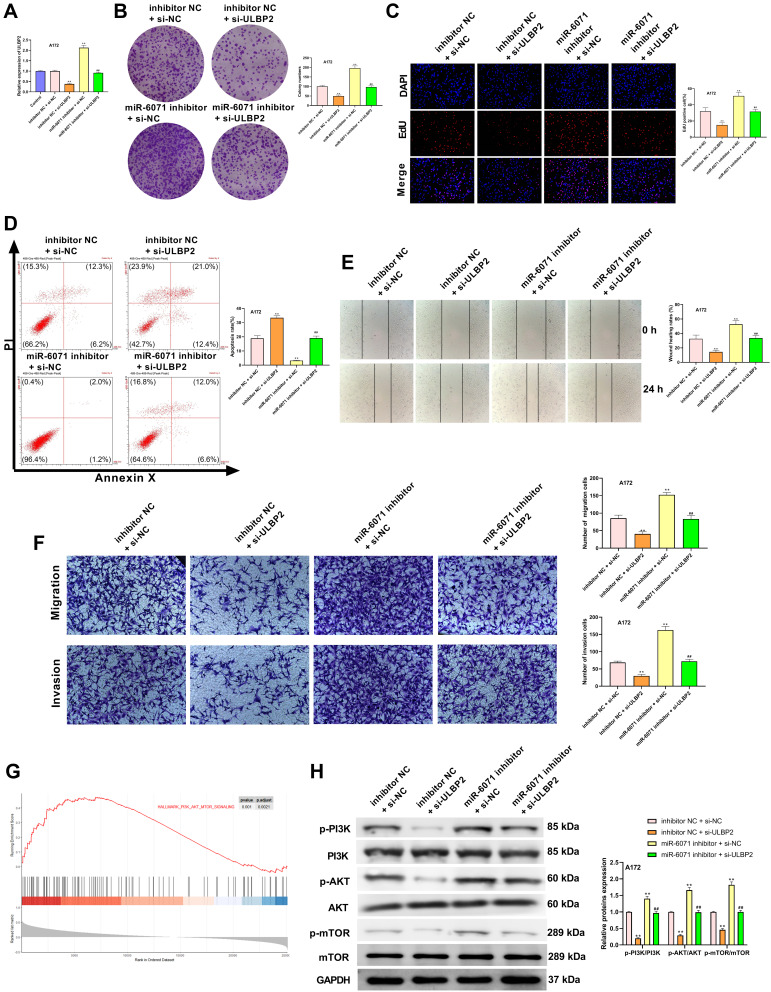
ULBP2 Reverses the Effects of miR-6071 on GBM Cells Through Regulating the PI3K/AKT/mTOR Pathway
As shown in Figure 7A, the expression of ULBP2 in A172 cells was significantly decreased in the inhibitor NC + si-ULBP2 group compared with the Control and the inhibitor NC + si-NC groups (P < 0.01), whereas its expression was markedly increased in the miR-6071 inhibitor + si-NC group (P < 0.01). When compared with the miR-6071 inhibitor + si-ULBP2 group, ULBP2 expression was reduced in the inhibitor NC + si-ULBP2 group (P < 0.01) and elevated in miR-6071 the inhibitor + si-NC group (P < 0.01), suggesting that the miR-6071 inhibitor and si-ULBP2 were successfully transfected. In addition, as shown in Figure 7B-F, silencing ULBP2 significantly decreased the rate of proliferation, migration and invasion, as well as increased cells apoptosis (P < 0.01). Meanwhile, silencing ULBP2 dramatically reversed miR-6071 inhibitor-medicated promotion of cell proliferation, invasion and migration, and inhibition of cell apoptosis in A172 cells (P < 0.01). To investigate the potential mechanisms by which ULBP2 promotes GBM progression, gene set enrichment analysis (GSEA) was performed on TCGA glioma datasets. As shown in Figure 7G, the high expression of ULBP2 was positively correlated with “PI3K-AKT-mTOR signaling”. To confirm the results, the related proteins of PI3K/AKT/mTOR signaling pathway were measured by Western blot (Figure 7H). When compared with the inhibitor NC + si-NC group, the expressions of p-PI3K, p-AKT and p-mTOR were significantly decreased in the inhibitor NC + si-ULBP2 group (P < 0.01), while their expressions were markedly increased in miR-6071 inhibitor + si-NC group (P < 0.01). On the contrary, the expressions of p-PI3K, p-AKT and p-mTOR were reduced in the inhibitor NC + si-ULBP2 group (P < 0.01) and elevated in the miR-6071 inhibitor + si-NC group (P < 0.01) compared with the miR-6071 inhibitor + si-ULBP2 group. These results suggested that miR-6071 could inhibit the progression of GBM through the inhibition of the PI3K/AKT/mTOR pathway by binding to ULBP2.
Figure 7.
ULBP2 reversed the effects of miR-6071 on GBM cells through regulating PI3K/AKT/mTOR pathway. (A) The expression of ULBP2 in A172 cells was detected by qRT-PCR. (B) The colonies number of A172 cells was detected by colony formation assay. (C) The proliferation of A172 cells was tested by EdU assay. (D) The apoptosis of A172 cells was detected by flow cytometry. (E) The migration of A172 cells was tested by wound-healing assay. (F) The migration and invasion of A172 cells was tested by transwell assay. (G) Gene set enrichment analysis of RNA-seq data for patients with high ULBP2 expression and low ULBP2 expression from the TCGA dataset. (H) The expressions of p-PI3K, PI3K, p-AKT, AKT, p-mTOR and mTOR in A172 cells were measured by Western blot. **P < 0.01, vs Control and inhibitor NC + si-NC group; ##P < 0.01, vs inhibitor NC + si-ULBP2 and miR-6071 inhibitor + si-NC group (A); **P < 0.01, vs inhibitor NC + si-NC group; ##P < 0.01, vs inhibitor NC + si-ULBP2 and miR-6071 inhibitor + si-NC group (B-F, H).
Discussion
Among solid tumors of the central nervous system, GBM is the most malignant and has shown to have poor prognosis all over the world.17,18 Currently, the effects of surgery, chemotherapy and radiotherapy on GBM are limited. Hence, there is an urgent need to explore novel therapeutic targets and molecular mechanisms for the treatment of GBM. In this research, we demonstrated that miR-6071 could repress cell proliferation, migration and invasion, as well as promote apoptosis via the inhibition of the PI3K/Akt/mTOR pathway by binding to ULBP2 in GBM.
There is growing evidence that miRNAs could play crucial roles in the progression of various cancers including GBM.19,20 In addition, the abnormal miRNAs expression is considered to be related to tumorigenesis. In the present study, we found that the high expression of miR-6071 was significantly associated with a better prognosis in GBM patients. In addition, the expression of miR-6071 was significantly decreased in GBM tissues and cells when compared to that in normal tissue and cells. There is increasing evidence that miRNAs participate in the growth, metastasis and apoptosis of tumor cells. For example, miR-21 could promote cell migration and invasion through the activation of Sox2 and the β-catenin pathway in glioma.21 Xia et al22 have confirmed that miR-1908 could facilitate cell proliferation and migration by suppressing PTEN tumor suppressor pathway in GBM. Liu et al23 have reported that miR-500a-5p could accelerate the proliferation, migration and invasion through targeting chromodomain helicase DNA binding protein 5 in GBM. In this study, we demonstrated that miR-6071 could significantly suppress proliferation, migration and invasion, and also facilitate apoptosis in U251 and A172 cells. Taken together, these findings suggested that miR-6071 might be a tumor-suppressed gene in GBM.
Natural killer (NK) cells-mediated clearance of tumor cells is considered an important intrinsic mechanism to inhibit cancer development.24 Natural killer group 2 member D (NKG2D), a receptor of NK cells and cytotoxic T lymphocytes, participates in the detection of abnormal self during infection or malignant transformation.25 In recent years, ULBP2, a ligand of NKG2D, is reported to be not expressed or at a low level in normal tissue cells and shows abnormal expression in cancer cells.26,27 Previous studies have indicated that ULBP2 is highly expressed in liver cancer,28 breast cancer,24 cervical cancer29 and melanoma.25 In the present study, the analysis of the TCGA dataset indicated that the low expression of ULBP2 was significantly associated with a better prognosis in GBM patients. In addition, we found that ULBP2 expression was significantly increased in GBM tissues and cells, and ULBP2 was the target gene of miR-6071. The data suggested that miR-6071 could negatively modulate the expression of ULBP2 in GBM. We then explored whether miR-6071 affects GBM by targeting ULBP2 and found that ULBP2 could reverse the function of miR-6071 on GBM cells, indicating that miR-6071 could inhibit proliferation, migration and invasion, as well as promote apoptosis via targeting ULBP2 in GBM.
Accumulating studies have indicated that PI3K/AKT/mTOR pathway could play a critical role in many human cancers through regulating various biological processes including cell proliferation, cell cycle, migration, invasion and apoptosis.30,31 In addition, various miRNAs have been considered to regulate the progression of GBM via modulating the PI3K/AKT/mTOR pathway. For instance, Li et al32 have suggested that miR-489 could reduce cell proliferation and cell cycle arrest, and increase cell apoptosis through targeting the SPIN1-mediated PI3K/AKT/mTOR pathway in glioma. In addition, miR-124-3p is reported to facilitate GBM cell growth via targeting NRP-1 through activating the PI3K/AKT pathway.33 In our study, we also demonstrated that miR-6071 could inhibit the activation of the PI3K/AKT/mTOR pathway in GBM. In addition, the results of GSEA on TCGA datasets showed that the effect of ULBP2 on promoting cancer was closely associated with the PI3K/AKT/mTOR pathway in GBM. Our data also confirmed that ULBP2 could significantly reverse the effect of miR-6071 on the PI3K/AKT/mTOR pathway.
In summary, we can conclude that miR-6071 has the potential to suppress cell proliferation, migration and invasion, as well as promote apoptosis through the inhibition of the PI3K/Akt/mTOR pathway by binding to ULBP2 in GBM. More importantly, the miR-6071/ULBP2/PI3K/AKT/mTOR axis may represent a promising therapeutic strategy for the treatment of GBM.
Acknowledgments
Yunyan Zhou and Hongwei An are co-first authors for this study.
Funding Statement
There is no funding to report.
Statement of Ethics
This study was authorized by the ethical committee of Yan’an Hospital of Kunming and all patients provided written informed consent. (No: 2018-057-01).
Disclosure
The authors report no potential conflicts of interest.
References
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer MA. Malignant gliomas in adults. N Engl J Med. 2008;359(17):1850;author reply 1850. [DOI] [PubMed] [Google Scholar]
- 3.Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol Res Pract. 2019;215(8):152476. doi: 10.1016/j.prp.2019.152476 [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. [DOI] [PubMed] [Google Scholar]
- 5.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 6.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367 [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- 8.Chen CH, Guo M, Hay BA. Identifying microRNA regulators of cell death in Drosophila. Methods Mol Biol. 2006;342:229–240. [DOI] [PubMed] [Google Scholar]
- 9.Fu LL, Wen X, Bao JK, Liu B. MicroRNA-modulated autophagic signaling networks in cancer. Int J Biochem Cell Biol. 2012;44(5):733–736. doi: 10.1016/j.biocel.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Wang Q, Yan XL, et al. miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition via EphA8. FEBS Lett. 2015;589(6):756–765. doi: 10.1016/j.febslet.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Pang B, Xin T, et al. Plasma miR-221/222 family as novel descriptive and prognostic biomarkers for glioma. Mol Neurobiol. 2016;53(3):1452–1460. doi: 10.1007/s12035-014-9079-9 [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Sun S, Tong X, et al. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res. 2017;1676:69–76. [DOI] [PubMed] [Google Scholar]
- 13.Xu TJ, Qiu P, Zhang YB, Yu SY, Xu GM, Yang W. MiR-148a inhibits the proliferation and migration of glioblastoma by targeting ITGA9. Hum Cell. 2019;32(4):548–556. [DOI] [PubMed] [Google Scholar]
- 14.Slattery ML, Mullany LE, Sakoda LC, et al. The PI3K/AKT signaling pathway: associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol Carcinog. 2018;57(2):243–261. doi: 10.1002/mc.22752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalhori MR, Irani S, Soleimani M, Arefian E, Kouhkan F. The effect of miR-579 on the PI3K/AKT pathway in human glioblastoma PTEN mutant cell lines. J Cell Biochem. 2019;120(10):16760–16774. doi: 10.1002/jcb.28935 [DOI] [PubMed] [Google Scholar]
- 16.Kalhori MR, Arefian E, Fallah Atanaki F, Kavousi K, Soleimani M. miR-548x and miR-4698 controlled cell proliferation by affecting the PI3K/AKT signaling pathway in Glioblastoma cell lines. Sci Rep. 2020;10(1):1558. doi: 10.1038/s41598-020-57588-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Y, Zhao J, Yi L, Jiang Y. microRNA-153 targets mTORC2 component rictor to inhibit glioma cells. PLoS One. 2016;11(6):e0156915. doi: 10.1371/journal.pone.0156915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Hung MC. Signaling-mediated regulation of MicroRNA processing. Cancer Res. 2015;75(5):783–791. doi: 10.1158/0008-5472.CAN-14-2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue J, Zhou A, Wu Y, et al. miR-182-5p induced by STAT3 activation promotes glioma tumorigenesis. Cancer Res. 2016;76(14):4293–4304. doi: 10.1158/0008-5472.CAN-15-3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo G, Luo W, Sun X, Lin J, Wang M, Zhang Y. MicroRNA‑21 promotes migration and invasion of glioma cells via activation of Sox2 and β‑catenin signaling. Mol Med Rep. 2017;15(1):187–193. doi: 10.3892/mmr.2016.5971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Xia X, Li Y, Wang W, et al. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol Cancer. 2015;14:154. doi: 10.1186/s12943-015-0423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Su D, Qi X, Ma J. MiR‑500a‑5p promotes glioblastoma cell proliferation, migration and invasion by targeting chromodomain helicase DNA binding protein 5. Mol Med Rep. 2018;18(3):2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breunig C, Pahl J, Küblbeck M, et al. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. 2017;8(8):e2973. doi: 10.1038/cddis.2017.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinemann A, Zhao F, Pechlivanis S, et al. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012;72(2):460–471. doi: 10.1158/0008-5472.CAN-11-1977 [DOI] [PubMed] [Google Scholar]
- 26.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235(1):267–285. doi: 10.1111/j.0105-2896.2010.00893.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paschen A, Sucker A, Hill B, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin Cancer Res. 2009;15(16):5208–5215. doi: 10.1158/1078-0432.CCR-09-0886 [DOI] [PubMed] [Google Scholar]
- 28.Weng J, Han X, Liu K, et al. CD44 3ʹ-untranslated region functions as a competing endogenous RNA to enhance NK sensitivity of liver cancer stem cell by regulating ULBP2 expression. Int J Biol Sci. 2019;15(8):1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang HX, Li YH. MiR-873, as a suppressor in cervical cancer, inhibits cells proliferation, invasion and migration via negatively regulating ULBP2. Genes Genomics. 2020. [DOI] [PubMed] [Google Scholar]
- 30.Tian L, Li W, Wang T. Therapeutic effects of silibinin on LPS-induced acute lung injury by inhibiting NLRP3 and NF-kappaB signaling pathways. Microb Pathog. 2017;108:104–108. [DOI] [PubMed] [Google Scholar]
- 31.Bruhn MA, Pearson RB, Hannan RD, Sheppard KE. AKT-independent PI3-K signaling in cancer - emerging role for SGK3. Cancer Manag Res. 2013;5:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Ma X, Wang Y, Li G. miR-489 inhibits proliferation, cell cycle progression and induces apoptosis of glioma cells via targeting SPIN1-mediated PI3K/AKT pathway. Biomed Pharmacother. 2017;93:435–443. doi: 10.1016/j.biopha.2017.06.058 [DOI] [PubMed] [Google Scholar]
- 33.Zhang G, Chen L, Khan AA, et al. miRNA-124-3p/neuropilin-1(NRP-1) axis plays an important role in mediating glioblastoma growth and angiogenesis. Int J Cancer. 2018;143(3):635–644. [DOI] [PubMed] [Google Scholar]