Abstract
A major challenge in hepatitis C research is the detection of early potential for progressive liver disease. MicroRNAs (miRNAs) are small RNAs that regulate gene expression and can be biomarkers of pathological processes. In this study, we compared circulating miRNAs identified in hepatitis C virus (HCV)-infected patients presenting two extremes of liver disease: mild/moderate fibrosis and cirrhosis. The patients in the cirrhosis group subsequently developed hepatocellular carcinoma (HCC). We identified 163 mature miRNAs in the mild/moderate fibrosis group and 171 in the cirrhosis group, with 144 in common to both groups. Differential expression analysis revealed 5 upregulated miRNAs and 2 downregulated miRNAs in the cirrhosis group relative to the mild/moderate fibrosis group. Functional analyses of regulatory networks (target gene and miRNA) identified gene categories involved in cell cycle biological processes and metabolic pathways related to cell cycle, cancer, and apoptosis. These results suggest that the differentially expressed circulating miRNAs observed in this work (miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, miR-885-5p, miR-26b-5p and miR -197-3p) may be candidates for biomarkers in the prognosis of liver disease.
Keywords: mirRNAome, Cirrhosis, Biomarkers, HCV, Massively parallel sequencing
Highlights
-
•
Circulating miRNome was performed in patients infected with HCV-1a or 1b.
-
•
Mature miRNAs were identified in patients with mild/moderate fibrosis and cirrhosis.
-
•
Five upregulated and two downregulated miRNAs were observed in the cirrhosis group.
-
•
Regulatory networks identified gene categories involved in cell cycle.
-
•
A routine baseline circulating biomarkers detection can have a prognostic value.
1. Introduction
Hepatitis C virus is responsible for a high rate of chronic infection (70–85%), since only 15–30% of cases have spontaneous resolution of viral infection. Chronic hepatitis C (CHC) evolves in 20% of cases to liver cirrhosis after progressive stages of fibrosis (Center for Diseases Control and Prevention - CDC, 2019). The most serious complication of chronic hepatitis C is the development of liver cancer, especially hepatocellular carcinoma (HCC). CHC is a major risk factor for the development of cirrhosis and subsequent HCC. Factors present in chronic patients that may contribute to disease progression to more severe stages, such as cirrhosis and HCC, are age and concomitant liver diseases, such as steatosis and steatohepatitis [1]. Early identification of progressive liver disease is important for efficient clinical management and improved quality of life. MicroRNAs (miRNAs) are being investigated for their diagnostic potential value in viral hepatitis, hepatic fibrosis, and HCC [2,3]. Non-coding RNAs correspond to 98% of RNA transcripts, a small part of them acts in a complex regulatory network and plays a relevant role in the development of various pathologies [4]. Among non-coding RNAs, miRNAs are the best studied. MiRNAs consist of small RNA molecules with 22 highly conserved nucleotides that regulate expression of target genes through antisense RNA-RNA interactions with the messenger RNA (mRNA) 3′UTR region. It is estimated that at least 60% of all mammalian genes may be under miRNA control [5]. The biological function of circulating miRNAs is still poorly known in HCV infection. However, it may provide a means of communication to recipient neighboring cells by influencing gene expression in target cells. The potential use of miRNAs as biomarkers was tested in patients with HCV-associated liver disease. The stability of miRNAs in the circulation make them ideal potential biomarkers [6]. Therefore, the identification of noninvasive or less invasive biomarkers, such as specific miRNAs, may aid prognostic assessment and clinical and therapeutic management. Importantly, preventive measures, monitoring and treatment of patients significantly lower the cost of medical and nursing care, hospitalizations, among others, generating lower expenses for the public health system. Preliminary studies of miRNAs in CHC-related HCC have identified promising candidates [[7], [8], [9]], with the development of miRNA panels that are now being explored to improve diagnostic accuracy. Nevertheless, few studies have been done with miRNome approach. In this work, we present an evaluation of miRNome in two groups of chronic hepatitis C patients that present liver stage disease of fibrosis or cirrhosis. Differential expression of the circulating miRNA between these two groups of chronical HCV patients suggests that the miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, miR-885-5p, miR-26b-5p and miR -197-3p may be indicative of worsening of liver disease and therefore are potential biomarkers.
2. Methods
2.1. Patients
Patients (101 individuals) above 18 years old with chronic HCV infection, genotypes 1a or 1b, attending the Hepatology Service at the Federal University Hospital Clementino Fraga Filho (HUCFF), Rio de Janeiro, Brazil, were selected for this work. These patients were diagnosed with chronic hepatitis C, confirmed by histopathology and serum HCV RNA detection by PCR. We excluded pregnant or nursing women; patients using immunosuppressants or antineoplastic therapy; and patients with hepatitis B virus (HBV) or human immunodeficiency virus (HIV) co-infection. Patients were treated for 48 weeks with PEG-IFN α2b (1,5 μg/kg once weekly) and RBV (1000–1200 mg/day); or IFN α2b (3 million units 3 times/week) and RBV (750–1000 mg/day). Patients were classified as responders (serum viral levels not detected at the end of treatment and sustained for additional 24 weeks); relapses (serum viral levels not detected at the end of treatment and detected after 24 weeks); and non-responders (serum viral levels detected throughout treatment). A total of 109 patients with a minimum medical follow-up of 6 years were selected, allowing us to monitor the evolution of liver disease. Clinical data were compiled using the SPSS 14.0 (Statistical Package for Social Sciences v.14.0) program based on medical records. Outcome and treatment response in this cohort have been previously described [10,11].
2.2. Ethical considerations
This work has been approved by the HUCFF Research Ethics Committee (CEP) and registered with CIC under the numbers: CEP- 166/05 and CIC 148/05.
2.3. Liver disease staging
The stage of liver fibrosis was assessed by biopsy or transient elastography (Fibroscan®) according to medical records. All patients were submitted to liver biopsy during staging of liver disease, Fibroscan was used during the follow-up. The severity of fibrosis was classified according to the Metavir score as: mild/moderate (F1– F2) and advanced (F3–F4); F4 indicatet liver cirrhosis [12]. The development of HCC was evaluated using medical records when a hypervascular nodule with washout was detected by Computer Tomography or magnetic resonance. Based on liver disease staging, a total of 8 patients were selected in a paired manner: 4 patients in the mild/moderate fibrosis group, and 4 patients in the cirrhosis group.
2.4. Total RNA extraction
Serum from patients at baseline, pre-treatment, were previously collected and frozen at −70 °C. Total RNA, including miRNAs, was isolated from 200 μl of serum using silica column filtration, using the miRNeasy Serum/Plasma Kit isolation kit (Qiagen, Germany). After extraction, the qualitative and quantitative analysis of RNAs were performed using a Bioanalyzer (Agilent Technologies, Santa Clara, USA) and the Eukaryote Total RNA Pico kit according to the manufacturer's protocol.
2.5. Library preparation, clonal amplification, and sequencing
Library preparation for sequencing was performed using the Total Ion RNA-Seq kit v2, the Ion Xpress RNA - Seq Barcode 01–16 kit, and the Magnetic Bead Clean Up Module (Thermo Fisher, Waltham, Massachusetts, EUA). Quantification and size distribution of library fragments were evaluated using a Bioanalyzer and the DNA 1000 kit. Clonal amplification was performed on an Ion OneTouch ™ 2 System equipment (Thermo Fisher) with Ion PI ™ Hi-Q ™ OT2 200 Kit reagents (ThermoFisher, Waltham, Massachusetts, EUA) according to the manufacturer's instructions. Sequencing was performed simultaneously for all samples by semiconductor sequencing using the Ion PI ™ Hi - Q ™ Sequencing 200 Kit (ThermoFisher, Waltham, Massachusetts, EUA) on Ion Proton High Power Sequencer (Thermo Fisher, Waltham, Massachusetts, EUA) on chip PI v2.
2.6. Analysis of miRNA transcripts
The first stage of analysis was performed by the Torrent Suite software package (Thermo Fisher). After base calling, sequences smaller than 8 bp were discarded. The sequences were exported in FASTQ format for further analysis using the CLC Genomics Workbench software, version 8.5 (QIAGEN). The transcripts were trimmed - so that only those between 15 bp and 30 bp in size were kept, compatible with mature miRNAs-, and then annotated by comparing sequence similarity to miRBASE (http://www,mirbase,org). After identification of mature miRNAs, differential expression analysis of miRNAs between the mild fibrosis and cirrhosis groups was performed using Empirical Analysis of Differential Gene Expression (EDGE) [13], considering significant transcripts that varied at least twice (fold-change ≥ 2 and ≤ −2), with p value ≤ 0.05, present in at least two of the samples from each sample group. To verify the potential target genes regulated by the differentially expressed miRNAs identified between the two groups studied, the miRnet tool version 2.0 [14] was applied to identify the regulatory networks in which the miRNAs are involved.
3. Results
3.1. Host miRNA analysis and association with liver disease
Eight patients were selected in a paired and homogeneous manner, taking into consideration clinical and demographic characteristics. Pre-treatment serum of 8 patients were analyzed for identification of circulating miRNAs, 4 with mild/moderate fibrosis (Group F1/F2) and 4 with cirrhosis (Group F4). Data from the 8 selected patients are shown in Table 1. The patients had a mean age of 60.8 and 67.8 for the mild/moderate fibrosis and cirrhosis groups, respectively. Similarly, the body mass index (BMI) was 26.8 for the mild/moderate fibrosis group and 27.0 for the fibrosis group. Two patients were infected with HCV genotype 1a and two with HCV 1b in both groups. Patients in group F1/F2 evolved to advanced fibrosis (F3) during the follow-up period (average of 12 years), whereas patients in group F4 evolved to HCC
Table 1.
Clinical and demographic data of chronic hepatitis C patients selected for identification of circulating miRNAs.
| Group F1/F2 |
Group F4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | 15 | 17 | 57 | 73 | 22 | 48 | 77 | 100 | t-Test | ||
| Gender | M | M | F | M | M | F | F | M | |||
| Age | 52 | 55 | 69 | 67 | 60.8 ± 7.4 | 66 | 71 | 71 | 63 | 67.8 ± 3.4 | 0.241 |
| HCV genotype | 1a | 1a | 1b | 1b | 1a | 1b | 1a | 1b | |||
| BMI | 26 | 29 | 23 | 29 | 26.8 ± 2.5 | 28 | 31 | 22 | 27 | 27 ± 3.2 | 0.824 |
| Degree of fibrosis | F1 | F1 | F2 | F2 | F4 | F4 | F4 | F4 | |||
| Treatment outcome Peg-IFN + RBV | REL | NR | NR | NR | NR | NR | SVR | SVR | |||
| Follow up (years) | 14 | 13 | 12 | 10 | 12.2 ± 1.5 | 9 | 9 | 6 | 10 | 8.5 ± 1.5 | 0.065 |
| Degree of fibrosis/CHC at the end of follow up | F3 | F3 | F3 | F3 | HCC | HCC | HCC | HCC | |||
BMI: body mass index; Peg-IFN: pegylated interferon; RBV: ribavirin; REL: relapser, NR: non-responder; SVR: sustained viral responder; HCC: hepatocellular carcinoma.
During the follow-up period (average of 8 years).
An average of 24, 256, 692 bp (1,273,208 reads with 22 bp average size) and 26,231,142 bp (1,307,130 reads with 23 bp average size) were obtained after QC (≥Q20) for Group F1/F2 and Group F4, respectively (Supporting information.
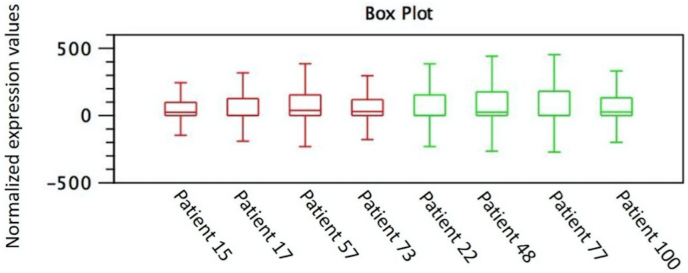
Table S1). The reads obtained, compatible with mature miRNAs, show that the fragments obtained in all libraries after filtering had an average size of approximately 25 and 26 bp, respectively. The list of transcripts previously obtained was subjected to a comparison analysis of the number of transcripts in each sample, showing that the expression values found in each group were similar (Fig. 1).
Fig. 1.
Box plot of normalized expression values of transcripts obtained in the sequencing of samples from the 4 patients of the Group F1/F2 (red) and Group F4 (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Selection, identification, and quantitative analysis of the serum miRNome.
We identified an average of 2249 small RNA transcripts in group F1/F2 and 2207 in group F4. Mature miRNAs annotated and grouped in serum samples from patients with chronic hepatitis C were categorized into an average of 315 and 322 unique mature miRNA species, respectively (Table 2). No significant difference was observed between the two groups.
Table 2.
Total transcripts identified, annotated, and grouped as mature miRNAs in serum samples from patients with chronic hepatitis C.
| Group F1/F2 |
Group F4 |
t-Test |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | 15 | 17 | 57 | 73 | 22 | 48 | 77 | 100 | |||
| Total reads (non-annotated) | 769.852 | 1.069.388 | 968.390 | 988.340 | 948,993 ± 110,125 | 812.417 | 1.048.843 | 1.109.741 | 1.043.486 | 1,003,622 ± 113,418 | 0,199 |
| Total reads (annotated) | 43.305 | 25.973 | 28.002 | 36.225 | 33,376 ± 6899 | 21.119 | 42.756 | 18.714 | 40.144 | 30,683 ± 10,839 | 0,769 |
| Small RNAs (non-annotated) | 373.115 | 548.284 | 393.930 | 448.444 | 440,943 ± 67,803 | 403.489 | 481.763 | 515.791 | 525.474 | 481,629 ± 47,946 | 0,387 |
| Small RNAs (Annotated) | 2.385 | 1.925 | 2.242 | 2.442 | 2249 ± 200 | 1.717 | 2.842 | 1.917 | 2.350 | 2207 ± 432 | 0,909 |
| Grouped (mature miRs) | 334 | 283 | 319 | 325 | 315 ± 19 | 273 | 379 | 291 | 343 | 322 ± 42 | 0,865 |
3.2. Qualitative evaluation of the serum miRNome
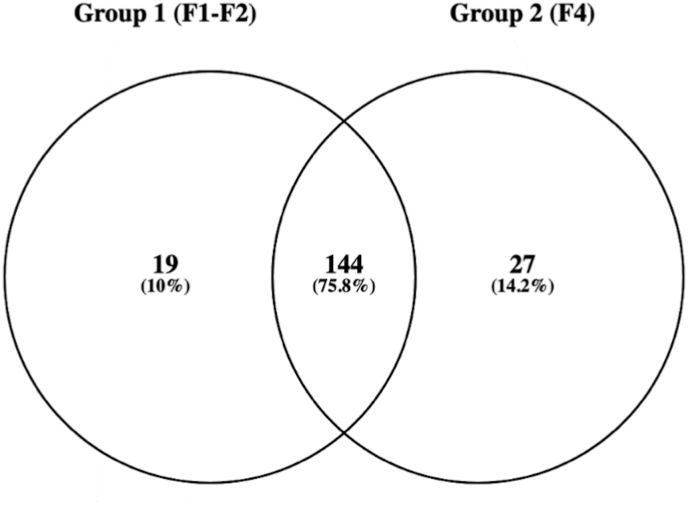
To evaluate the distribution of the identified miRNAs between the two groups of patients (Table 2), the miRNAs were filtered for those present in the four samples from each group. Annotated mature miRNAs corresponded to 163 species in group F1/F2 and 171 in group F4. Of these, 144 (75.8%) were common to both groups, as shown in the Venn Diagram in Fig. 2. The exclusively expressed miRNAs (19 and 27 in groups F1/F2 and F4, respectively) had very low expression level (ranging from 2 to 1 reads), owing to stochastic low-level expression detection.
Fig. 2.
Venn diagram indicating the number of unique mature miRNA species identified in the four patients from each group with one or more copies, and the mature miRNAs identified in both groups.
3.3. MiRNA differential expression analysis
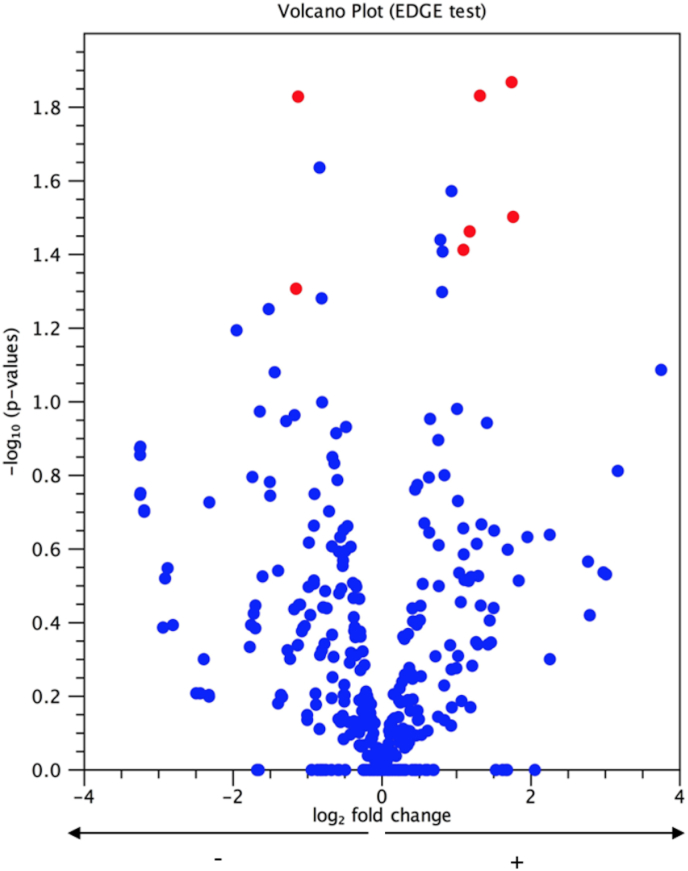
Differentially expressed miRNAs are shown relative to group F1/F2 because it represents less advanced liver disease. The results of this analysis are shown in a volcano plot in Fig. 3. Seven miRNAs with differential expression were identified: miR-26b-5p and miR-197-3p with reduced expression in group F4 compared to group F1/F2 (Table 3), and miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, and miR-885-5p with increased expression.
Fig. 3.
Differential expression of circulating microRNAs in the F1/F2 and F4 groups (volcano plot). Positive values are those with increased expression and negative values with reduced expression. Red dots indicate the seven differentially expressed miRNAs (fold change ≥ 2 and ≤ −2 and p-value <0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Reduced and increased expression of circulating miRNAs in group F4 relative to group F1/F2 identified by EDGE.
| MiRNA | Group F1/F2 (average of normalized expression values) | Group F4 (average of normalized expression values) | Fold-change | p Value | |
|---|---|---|---|---|---|
| Reduced expression | miR-26b-5p | 817,994 | 478,654944 | −2.19 | 0.01 |
| miR-197-3p | 2485,886 | 1348,49152 | −2.23 | 0.05 | |
| Increased expression | miR-215-5p | 86,92843 | 387,709343 | 3.34 | 0.01 |
| miR-483-5p | 317,504 | 896,002951 | 2.49 | 0.01 | |
| miR-193b-3p | 82,58706 | 315,116486 | 3.39 | 0.03 | |
| miR-34a-5p | 172,8424 | 480,041071 | 2.26 | 0.03 | |
| miR-885-5p | 34,5688 | 69,2780401 | 2.15 | 0.04 | |
Among the differentially expressed miRNAs, it was observed that the miR-215-5p had significant variation (fold-change: 3.34 and p-value: 0.01) compared to the other miRNAs, and miR-193b-3p had the largest variation (fold-change of 3.39).
Analysis of gene regulatory networks of differentially expressed miRNAs.
The biological interpretation of miRNA differential expression data is a major challenge owing to the fact that one miRNA can regulate multiple targets, and the same mRNA can be regulated by multiple miRNAs. One way to identify possible patterns is to obtain regulatory networks between miRNAs and their targets, making it possible to visualize identify clusters generated by miRNA-gene interactions.
3.4. Cirrhosis-decreased miRNA regulation networks
From the list of miRNAs with decreased expression in group F4, a regulatory network was constructed (Fig. 4). We observed the formation of a complex network in which a total of 2234 genes were identified. miR-197-3p regulated 409 genes and miR-26b-5p regulated 1874 genes. Among the genes found, only 49 (2,19%) were common to both miRNAs (Supporting information Fig. S1). Genes regulated by both miRNAs are shown in the Supporting information Table S2.
Fig. 4.
Regulatory network identified by miRNAs with decreased expression in group F4 (hsa-mir-26b-5p and hsa-mir-197-3p) relative to group F1/F2. Each blue square represents one miRNA and each red dot represents a target gene. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Cirrhosis-associated miRNA regulation networks
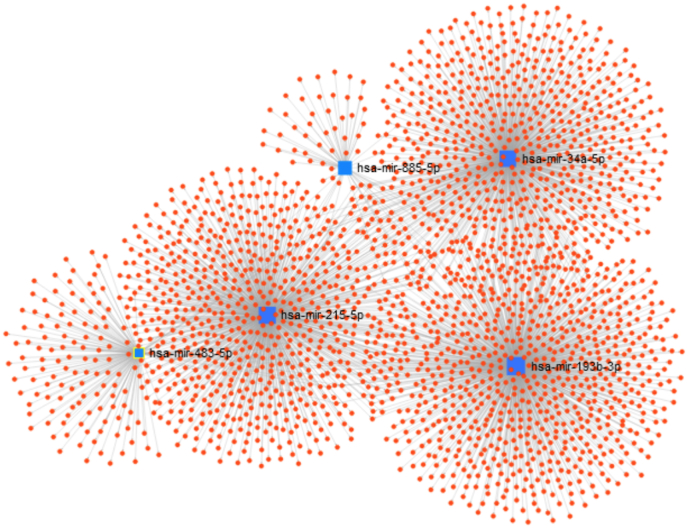
From miRNAs with increased expression in group F4, a network was constructed (Fig. 5). We observed the formation of a complex network in which 2312 target genes were identified. Of these, 853 genes are regulated by miR-193b-3p, 755 by miR-215-5p, 736 by miR-34a-5p, 176 by miR-483-3p, and 64 by miR-483-3p miR-885-5p. The number of predicted genes in the network of targets that are uniquely regulated and common to the five miRNAs with increased expression are shown in Supporting information Fig. S2. We observed 10 target genes common to miR-215-5p, miR-34a-5p, and miR-193b-3p, which are miRNAs with the highest number of targets and interaction within the formed network. Similarly, 74 target genes were found in common with miR-193b-3p and miR-34a-5p, and 34 target genes in common with miR-34a-5p and miR-215-5p. The target genes of these miRNAs are shown in Supporting information Table S3. MiR-215-5p shares the largest number of target genes with the other cirrhosis-associated miRNAs. No common target genes were identified for all miRNAs.
Fig. 5.
Regulatory network identified by miRNAs with increased expression in group F4 relative to group F1/F2. Each blue square represents one miRNA and each red dot represents a target gene. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Enriched gene ontology terms of the regulatory network
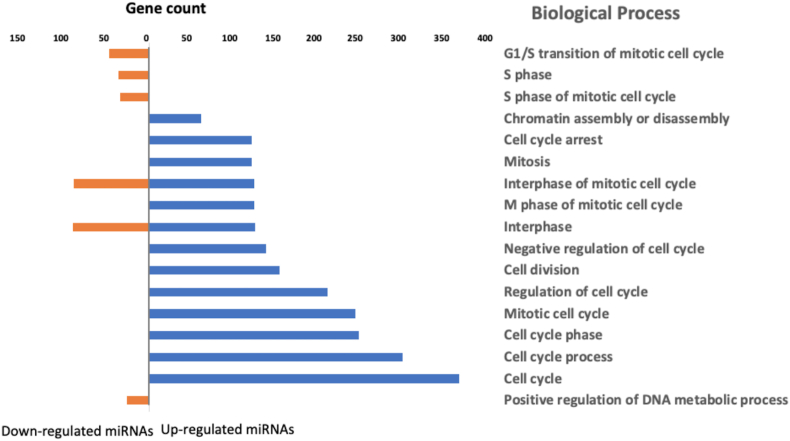
Gene ontology (GO) and biological process (BP) category analysis were performed to identify the enriched GO terms in the enhanced expression miRNA network targets. Enrichment analysis for metabolic pathways, deposited in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database of the regulatory network target genes, identified several genes (Supporting information Table S4). Fig. 6 shows the main enriched GO terms for BP based on the p value obtained with hypergeometric tests (p < 0.05). The main terms found are related to cell cycle processes.
Fig. 6.
Gene ontology terms in the enriched biological process categories of identified target genes network for differentially expressed miRNAs. Hypergeometric tests were performed (p < 0.05).
4. Discussion
4.1. MiRNA expression
A single miRNA can negatively suppress a large number of mRNAs from related target genes, thereby controlling various metabolic pathways. On the other hand, the same gene can be controlled by several miRNAs [15]. Owing to these characteristics, miRNA research is of great interest for a better understanding of gene expression control mechanisms in several diseases, with great potential for diagnosis, prognosis, and new therapeutic targets [[16], [17], [18]]. MiRNAs are involved in gene regulation of various biological processes such as HCV infection and progression to HCC [19,20]. The identification of miRNA signatures in HCC is of great value for its early diagnosis in patients with HCV infection. Hepatitis C virus modulates the expression of cellular miRNAs in the hepatocytes, favoring tumorigenesis [21]. Furthermore, miRNAs dysregulation has been associated with CHC initiation and progression [22]. In our work, serum miRNome analysis from eight patients with CHC identified seven miRNAs differentially expressed accordingly to the severity of liver fibrosis. Patients in both groups were treated with PEG-IFN and ribavirin at similar doses. Although interferon is considered as an antifibrogenic drug, long term longitudinal observation showed no effect on fibrosis progression [50].However, the role of miRNAs expression in HCV-associated HCC is still poorly understood. The increase and decrease in specific miRNA levels are associated with HCV infection and disease progression [21]. The efficacy of hepatitis C antiviral therapies [23] on morbidity and mortality depends on the identification of cirrhosis owing to CHC and its complications, including HCC. SVR has been shown to improve liver function in cirrhotic patients, although the risk of complications such as HCC remains [24]. The number of circulating miRNAs previously described in liver and serum samples from patients with CHC, using the Illumina platform, was 658 miRNAs (301 after normalization) in liver tissue and 549 miRNAs (397 after normalization) in serum [25], consistent with the results obtained in our study for circulating miRNAs. A study comparing serum miRNA expression levels of 33 patients with mild/moderate fibrosis level (F0–F2) and 45 patients with advanced fibrosis (F3) or cirrhosis (F4) reported 34 miRNAs differentially expressed between the two groups, p-values < 0.05. but none was significant after the Benjamini-Hochberg correction test for multiple comparisons [25].
4.2. Differentially miRNA expression
During HCV infection, miRNAs are released into the extracellular space by various means including their association with circulating argonaute-2 (Ago2), exosomes, or high-density lipoproteins (HDL) [26,27]. When HCV infects hepatocytes, a unique interaction occurs between the host miR-122 and the HCV 5′UTR region together with the Ago2, GW182 and heat shock (HSPs) proteins, which are part of the RNA-induced silencing complex (RISC), which resulting in increased viral replication. Other host factors, such as cyclin G1 (an miR-122 target), are also involved in increased HCV replication in the presence of alcohol. During HCV infection, other host miRNAs are modulated, leading to an increase in miR-155 levels and a decrease in miR-499 levels in hepatocytes, activating Notch and Wnt signaling pathways, which can lead from inflammation to cancer development. HCV infection causes damage to hepatocytes by releasing molecules that activate immune cells, increasing local and systemic inflammation. Activation of peripheral monocytes causes an increase in miR-155 levels, which subsequently increases the release of TNF into the circulation. Several miRNAs such as miR-122, miR-155, miR-34a, miR-21, miR-146a and miR-125b are increased in the circulation of HCV-infected patients and may contribute to the pathogenesis of the disease [28]. Therefore, circulating miRNAs have great potential to be used as biomarkers of liver disease progression [29]. In another study, miRNA profiles were identified in the serum of HCV infected individuals. Serum MiRNAs levels (miR-134, miR-320c, and miR-483-5p) were significantly increased in HCV-infected patients [9]. Serum from patients with HCC and/or other etiologies associated with liver injury had elevated serum levels of miR-122 and miR-192 when compared to sera from healthy patients [7]. Our data showed that miR-26b-5p and miR-197-3p are approximately two-fold down-regulated in group F4. The miRNAs, miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, and miR-885-5p were two to three-fold up-regulated in group F4. Previous study demonstrated elevated serum/plasma miR-20a and miR-92a levels in HCV-infected patients compared with healthy subjects, suggesting that these circulating miRNAs may serve as potential biomarkers of HCV infection. They also demonstrated elevated serum expression of miR-20a in early and late stage fibrosis in HCV-infected patients, but not in non-HCV-infected patients, suggesting its potential as a predictive biomarker for HCV-mediated liver disease progression [30].
4.3. Regulatory networks
We used the miRnet tool [14] to identify regulatory networks in which differentially expressed miRNAs are involved in the two groups of patients. The down-regulated microRNAs (miR-26b-5p and miR-197-3p), in group F4, have been related to liver cancer development in HCV-infected patients [31]. Some studies suggest that miR-26b-5p has tumor suppressor function in osteosarcoma through suppression of the glycolytic pathway [32] and its downregulation was correlated with the reduced apoptosis rate in HCC tissues [33]. In mice, the increase in miR-26 reduces cell proliferation and increases apoptosis in liver tumor, suggesting that this miRNA may be promising for the development of anticancer therapies and as a prognostic marker for HCC [34,35]. Also, decreased expression of miR-26 and miR-29 was observed in patients with HCC compared with healthy individuas [19]. Recent study reported decrease expression of miR-197-3p in HCC tissues [36]. The low level of miR-197-3p expression was associated with tumor aggressiveness, indicating that miR-197-3p may be a predictor of prognosis in patients with HCC [36]. In addition, miR-197-3p inhibited metastasis in HCC cells in vitro and in vivo [36]. These studies demonstrate that miR-26b and miR-197 have tumor suppressor function. Thus, its reduced expression may be related to the development of HCC, which corroborates our results showing that patients with cirrhosis presented a decreased expression of miR-26b and miR-197 in relation to the mild/moderate fibrosis group. In addition, all cirrhotic patients developed HCC over the years of follow-up. It should be noted that our work was performed using serum instead of tissue samples, therefore one cannot infer that intracellular and extracellular levels of gene expression of these miRNAs are the same. On the other hand, our findings of up-regulated miRNAs (miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, and miR-885-5p), in group F4, are associated with cell cycle and related functions. Studies have shown differential regulation of these five miRNAs in the pathophysiology of liver disease and in HCC. ROY et al., 2015 showed a deregulation of miR-193 and miR-30c levels in serum of the patients with chronic liver disease [37].Since TGF-β2 and SNAIL1 genes, important regulators of extracellular matrix formation, are potential targets for miR-193 and miR-30c, the authors suggested that these miRNAs are involved in the process of hepatofibrogenesis [37]. The increase in miR-193b expression level may also regulate apoptosis by modulating the expression of the anti-apoptotic protein Mcl in HCV-positive cells [38]. Furthermore, miR-193b regulates the expression of CCND1 and ETS1 oncogenes, thus regulating proliferation, migration and invasion in HCC cells [39]. MiR-34a is described in the literature as a liver tumor suppressor by inhibiting cell proliferation, targeting the SATB2 gene in HCC [40], inhibiting liver cancer cell metastasis [41] and progression to HCC by repressing hexokinase 1 [42]. In HCC-human tissue, miR-34a expression is decreased when compared to normal tissue, demonstrating that miRNA may play a role in tumor suppression through apoptosis and cell senescence. This indicates that decreased expression of this miRNA in tissue may be related to metastasis and may be a predictor of HCC prognosis [43,44]. Among all differentially expressed miRNAs in our study, miR-215 was the one with the largest expression variation, and the greatest interaction with other miRNAs in the analysis of regulatory networks. Other study report increased expression of miR-215 in the serum of patients with CHC, cirrhosis and/or HCC [45] suggesting its role as potential biomarkers. Also, GUI et al. (2011) showed increased expression of five miRNAs (miR-885-5p, miR-574-3p, miR-224, miR-215 and miR-146a) in serum from patients with cirrhosis and HCC, compared to control subjects without liver disease [46]. Of these, two miRNAs (miR-885-5p and miR-215-5p) corroborate our work. MiR-215 may be involved not only in the development of liver disease but also in the HCV life cycle infection [47]. Primary malignant tumors in the liver represent the sixth cause of cancer and the fourth cause of cancer death in the worldwide hepatitis B and C accounting for 80% of HCC cases [48]. HCV is the etiology most associated with HCC, and most cases are cirrhotic [49]. Differential expression of the circulating miRNAs of chronical HCV patients indicate that the miR-215-5p, miR-483-5p, miR-193b-3p, miR-34a-5p, miR-885-5p, miR-26b-5p and miR -197-3p may be indicative of worsening of liver disease and therefore a potential biomarker. Our main purpose was to explore serum miRNAome profile as a potential source of molecular risk biomarkers for risk prediction in HCV patients. Despite the small sample size, the massively parallel sequencing approach allows a broad and integrative analysis for potential interest-network identification. Unfortunately, cost limitations are a frequent issue determining sample size in massively parallel sequencing.
5. Conclusions
The present study compared the circulating miRnome of HCV-infected patients with mild/moderate fibrosis and cirrhosis, identifying the general expression of miRNAs in serum, without previous selection. Although the majority of circulating miRNAs (73.4%) were common in both groups. five miRNAs (miR-215-5p; miR-483-5p; miR-193b-3p; miR-34a-5p and miR-885-5p) had increased expression and two miRNAs (miR-26b-5p and miR-197- 3p) had decreased expression in patients with cirrhosis compared to patients with mild/moderate fibrosis. The enriched gene groups in the interaction network of differentially expressed miRNAs fell into categories related to cell division, cell cycle, and mitotic process regulation. Taken together, our results suggest that circulating miRNAs may be indicative of disease progression and, therefore, potential biomarkers.
Author contributions
RS, LH, DSF Conceptualization; PFC, ALAR, CANV, HSMC, BCAC Data curation;; RS, LH, DSF, BCAC Formal analysis; RS, Funding acquisition; BCAC Investigation; TPU Methodology; RS Project administration; CANV, HSMC, PFC, ALAR Resources; RS, LH, Supervision; TPU Validation; RS, DSF,LH, BCAC Visualization; Roles/Writing - BCAC original draft; RS, LH, DSF, TPU Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by research grants from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ - E-26/202.847/2018) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq -304156/2016-7).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100814.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Castera L., Hezode C., Roudot-Thoraval F., Bastie A., Zafrani E.-S., Pawlotsky J.-M. Worsening of steatosis is an independent factor of fibrosis progression in untreated patients with chronic hepatitis C and paired liver biopsies. Gut. 2003;52:288–292. doi: 10.1136/gut.52.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Al A., El-Ahwany E., Zoheiry M., Hassan M., Ouf A., Abu-Taleb H. miRNA-221 and miRNA-222 are promising biomarkers for progression of liver fibrosis in HCV Egyptian patients. Virus Res. Elsevier. 2018;253:135–139. doi: 10.1016/j.virusres.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Rashad N.M., El-Shal A.S., Shalaby S.M., Mohamed S.Y. vol. 447. Springer US; 2018. pp. 125–136. (Serum miRNA-27a and miRNA-18b as Potential Predictive Biomarkers of Hepatitis C Virus-Associated Hepatocellular Carcinoma. Mol Cell Biochem). [DOI] [PubMed] [Google Scholar]
- 4.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. Cold Spring Harbor Lab. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid G., Kirschner M.B., van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 2011;80:193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Cermelli S., Ruggieri A., Marrero J.A., Ioannou G.N., Beretta L. Circulating MicroRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. In: Tavis J.E., editor. vol. 6. 2011. p. e23937. (PLoS ONE). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bihrer V., Friedrich-Rust M., Kronenberger B., Forestier N., Haupenthal J., Shi Y. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 9.Shwetha S., Gouthamchandra K., Chandra M., Ravishankar B., Khaja M.N., Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci. Rep. 2013;3:1555. doi: 10.1038/srep01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann L., Ramos J.A., Souza EV de, Araújo Ramos AL de, Villela-Nogueira C.A., Urményi T.P. Dynamics of resistance mutations to NS3 protease inhibitors in a cohort of Brazilian patients chronically infected with hepatitis C virus (genotype 1) treated with pegylated interferon and ribavirin: a prospective longitudinal study. Virol. J. 2013;10:57. doi: 10.1186/1743-422X-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos J.A., Ramos AL. de A., Hoffmann L., Perez R. de M., Coelho H.S.M., Urményi T.P. A single nucleotide polymorphism, rs129679860, in the IL28B locus is associated with the viral kinetics and a sustained virological response in a chronic, monoinfected hepatitis C virus genotype-1 Brazilian population treated with pegylated interferon-ribavirin. Mem. Inst. Oswaldo Cruz. 2012;107:888–892. doi: 10.1590/S0074-02762012000700008. [DOI] [PubMed] [Google Scholar]
- 12.Bedossa P., Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 13.Robinson M.D., McCarthy D.J., Smyth G.K. vol. 26. Oxford University Press; 2010. pp. 139–140. (edgeR: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–W141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes C., Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. IJMS. 2016;17 doi: 10.3390/ijms17030280. 280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirola C.J., Fernández Gianotti T., Castaño G.O., Mallardi P., San Martino J., Mora Gonzalez Lopez Ledesma M. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumida Y., Nakajima A., Itoh Y. vol. 20. Baishideng Publishing Group Inc; 2014. pp. 475–485. (Limitations of Liver Biopsy and Non-invasive Diagnostic Tests for the Diagnosis of Nonalcoholic Fatty Liver Disease/nonalcoholic Steatohepatitis. World J Gastroenterol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widera C., Gupta S.K., Lorenzen J.M., Bang C., Bauersachs J., Bethmann K. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J. Mol. Cell. Cardiol. 2011;51:872–875. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Ura S., Honda M., Yamashita T., Ueda T., Takatori H., Nishino R. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 20.Varnholt H., Drebber U., Schulze F., Wedemeyer I., Schirmacher P., Dienes H.-P. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2007;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 21.Shrivastava S., Steele R., Ray R., Ray R.B. MicroRNAs: role in hepatitis C virus pathogenesis. Genes & Diseases. 2015;2:35–45. doi: 10.1016/j.gendis.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S., He X. The role of microRNAs in liver cancer progression. Br. J. Canc. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Association for the Study of the Liver Electronic address: easloffice@easloffice.eu, European association for the study of the liver. EASL recommendations on treatment of hepatitis C 2018. J. Hepatol. 2018:461–511. doi: 10.1016/j.jhep.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Foster G.R., Irving W.L., Cheung M.C.M., Walker A.J., Hudson B.E., Verma S. Impact of direct acting antiviral therapy in patients with chronic hepatitis C and decompensated cirrhosis. J. Hepatol. 2016;64:1224–1231. doi: 10.1016/j.jhep.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 25.Van Keuren Jensen K.R., Malenica I., Courtright A.L., Ghaffari L.T., Starr A.P., Metpally R.P. vol. 36. John Wiley & Sons, Ltd; 2016. pp. 334–343. (microRNA Changes in Liver Tissue Associated with Fibrosis Progression in Patients with Hepatitis C. Liver International). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. vol. 11. Nature Publishing Group; 2014. pp. 145–156. (Clinical Relevance of Circulating Cell-free microRNAs in Cancer). Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 27.Lemoinne S., Thabut D., Housset C., Moreau R., Valla D., Boulanger C.M. vol. 11. Nature Publishing Group; 2014. pp. 350–361. (The Emerging Roles of Microvesicles in Liver Diseases). [DOI] [PubMed] [Google Scholar]
- 28.Szabo G., Bala S. vol. 10. 2013. pp. 542–552. (MicroRNAs in Liver Disease). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderburg C., Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J. Hepatol. 2014;61:1434–1437. doi: 10.1016/j.jhep.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Shrivastava S., Petrone J., Steele R., Lauer G.M., Di Bisceglie A.M., Ray R.B. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863–871. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huan L., Liang L.H., medicine X.H.C.B. Role of microRNAs in inflammation-associated liver cancer. Cancer Biol Med. 2016 doi: 10.20892/.2095-3941.2016.0071. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J.Y., Wang L.F., Wang Q., reports L.Y.O. miR-26b inhibits proliferation, migration, invasion and apoptosis induction via the downregulation of 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3 driven glycolysis in osteosarcoma cells. Oncol. Rep. 2015;33:1890–1898. doi: 10.3892/or.2015.3797. 2105. [DOI] [PubMed] [Google Scholar]
- 33.Zhao N., Wang R., Zhou L., Zhu Y., Gong J., Zhuang S.-M. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol. Canc. 2014;13:35. doi: 10.1186/1476-4598-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kota J., Chivukula R.R., O'Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.-W. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llovet J.M., Ricci S., journal V.M.N.E. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. 2008. [DOI] [PubMed] [Google Scholar]
- 36.Ni J.S., Zheng H., Huang Z.P., Hong Y.G., Ou Y.L., Tao Y.P. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17:2317–2327. doi: 10.3892/ol.2018.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy S., Benz F., Vargas Cardenas D., Vucur M., Gautheron J., Schneider A. miR-30c and miR-193 are a part of the TGF-β-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J Dig Dis. 2015;16:513–524. doi: 10.1111/1751-2980.12266. [DOI] [PubMed] [Google Scholar]
- 38.Braconi C., Valeri N., Gasparini P., Huang N., Taccioli C., Nuovo G. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin. Canc. Res. 2010;16:957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu C., Liu S., Fu H., Li S., Tie Y., Zhu J. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur. J. Canc. 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 40.Wu G., Li Z., Wang Y., Ju X., Huang R. miR-34a inhibits cell proliferation by targeting SATB2 in hepatocellular carcinoma. BioMed Res. Int. 2018;2018:2863902. doi: 10.1155/2018/2863902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei W., Tang H., Tang L. MicroRNA-34a inhibits metastasis in liver cancer cells. Oncol Lett. 2018;16:6960–6965. doi: 10.3892/ol.2018.9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Liu K., Liu Y., Tan L. MicroRNA-34a inhibit hepatocellular carcinoma progression by repressing hexokinase-1. J. Cell. Biochem. 2018;120(5):7147–7153. doi: 10.1002/jcb.27988. [DOI] [PubMed] [Google Scholar]
- 43.Budhu A., Jia H.L., Forgues M., Liu C.G., Goldstein D., Lam A. vol. 47. John Wiley & Sons, Ltd; 2008. pp. 897–907. (Identification of Metastasis-Related microRNAs in Hepatocellular Carcinoma. Hepatology). [DOI] [PubMed] [Google Scholar]
- 44.Dang Y., Luo D., Rong M., Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PloS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z.-Q., Meng H., Wang N., Liang L.-N., Liu L.-N., Lu S.-M. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol. BioMed Central. 2014;9:135–137. doi: 10.1186/1746-1596-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gui J., Tian Y., Wen X., Zhang W., Zhang P., Gao J. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishida H., Tatsumi T., Hosui A., Nawa T., Kodama T., Shimizu S. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem. Biophys. Res. Commun. 2011;412:92–97. doi: 10.1016/j.bbrc.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 48.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 49.Ferenci P., Fried M., Labrecque D. World gastroenterology organisation global guideline. Hepatocellular carcinoma (HCC): a global perspective. J. Clin. Gastroenterol. 2010;44:239–245. doi: 10.1097/MCG.0b013e3181d46ef2. [DOI] [PubMed] [Google Scholar]
- 50.Ellis E, L, Mann D., A J. Hepatol. 2012;56:1171–1180. doi: 10.1016/j.jhep.2011.09.024. 23, In this issue. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.