Abstract
Liver cancer is the fastest growing cause of cancer deaths in the United States due to its aggressiveness and lack of effective therapies. The current preclinical study examines valeric acid (pentanoic acid [C5H10O2]), one of the main compounds of valerian root extract, for its therapeutic use in liver cancer treatment. Anticancer efficacy of valeric acid was tested in a series of in vitro assays and orthotopic xenograft mouse models. The molecular target of valeric acid was also predicted, followed by functional confirmation. Valeric acid has a broad spectrum of anticancer activity with specifically high cytotoxicity for liver cancer in cell proliferation, colony formation, wound healing, cell invasion, and 3D spheroid formation assays. Mouse models further demonstrate that systematic administration of lipid-based nanoparticle-encapsulated valeric acid significantly reduces the tumor burden and improves survival rate. Histone deacetylase (HDAC)-inhibiting functions of valeric acid are also revealed by a structural target prediction tool and HDAC activity assay. Further transcriptional profiling and network analyses illustrate that valeric acid affects several cancer-related pathways that may induce apoptosis. In summary, we demonstrate for the first time that valeric acid suppresses liver cancer development by acting as a potential novel HDAC inhibitor, which warrants further investigation on its therapeutic implications.
Keywords: valeric acid, hepatocellular carcinoma, lipid nanoparticle, HDACi, apoptosis, cell proliferation, colony formation, cell migration, 3D formation, mouse model
Graphical Abstract
This study demonstrates the viability of an active compound from valerian root extract as a novel chemotherapeutic for the treatment of hepatocellular carcinoma. We show that valeric acid suppresses liver cancer development in a series of in vitro assays and in orthotopic xenograft mouse models, possibly by acting as a novel histone deacetylase inhibitor.
Introduction
Hepatocellular carcinoma (HCC) is the dominant type of liver cancer that ranks as the sixth-most common malignant tumors and cancer-caused deaths.1 HCC is the fastest growing cancer type in the United States, and its incidence has tripled during the past 20 years.2 HCC has a high mortality and poor prognosis3 due to its aggressiveness and lack of effective treatments. There is a tremendous unmet need for the development of novel therapeutics for liver cancer.
Valerian (Valeriana officinalis), a member of Valerianaceae family, is a perennial herb4 and has been authorized by the US Food and Drug Administration (FDA) as a complementary and alternative medicine (CAM) to treat insomnia and other sleep-related disorders.5 Valerian has been found to have anticancer effect for liver cancer,6 and its extraction compounds, such as iridoids, Valepotriates, and F3, have been shown as promising antitumor agents in many types of cancer,7, 8, 9 which warrants further exploration to discover novel active compounds from valerian for cancer treatment.10
Valeric acid (VA), or pentanoic acid (C5H10O2), another major active chemical ingredient of valerian, has been reported to have therapeutic effects on diseases, like insomnia and seizures.11 Recent evidence has also shown that VA can improve immunity against cancer.12 In addition, as an organic acid, VA shares a high structural similarity with the known histone deacetylase inhibitor (HDACi) valproic acid (C8H16O2),13 which is used as a treatment for seizure disorders, and a FDA-approved suberoylanilide hydroxamic acid (SAHA) (C14H20N2O3), which is used to treat T cell lymphoma.14 However, the antitumor activity of VA has not been examined in previous publications.
In this study, we aimed to explore the anticancer effect of VA with a focus on liver cancer. In vitro assays, including cell proliferation, migration, invasion, colony formation, and even 3D formation, were performed. Lipid nanoparticles were used as a delivery vehicle for the systemic administration of VA in animal studies. Functional experiments were also carried out to confirm the HDACi role of VA and its global transcriptional impact on cancer-related pathways.
Results
Anticancer Effect of VA in Cell Proliferation Assay
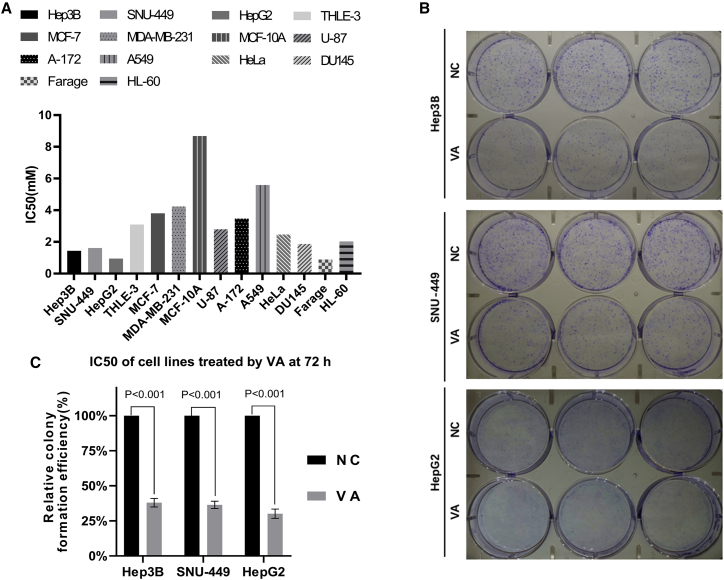
We first used the MTS assay to measure cell proliferation to test the anticancer effect of VA in 12 cancer and 2 normal cell lines. Significant anticancer effects with a positive dose-dependent effect relationship were observed in all tested cell lines. Figure S1 shows cell proliferation results of VA at 5 different concentrations (0.5, 1, 2, 4, and 8 mM) for all 14 cell lines from 4 time points (24, 48, 72, and 96 h). The half-maximal inhibitory concentration (IC50) at 72 h was presented in Figure 1A for all cell lines tested. Three liver cancer cell lines were among the group with the lowest IC50 (Farage: 0.89 mM; HepG2: 0.948 mM; Hep3B: 1.439 mM: and SNU-449: 1.612 mM). However, the IC50 for liver normal cell line THLE-3 (3.097) was 1.92, 2.15, and 3.27 times higher than that for liver cancer cells SNU-449, Hep3B, and HepG2, respectively. A similar trend was also observed for breast cancer and normal cell lines, which exhibited an overall higher IC50 when compared to liver cells. Based on these data, we chose to further investigate the anticancer efficacy of VA for liver cancer. We used the VA concentration of 0.85 mM for the rest of in vitro assays because it displayed a low inhibitory effect (<10%) on normal liver cells while maintaining a high inhibitory effect (>30%) on liver cancer cells.
Figure 1.
Anticancer Effect of Valeric Acid (VA) in Cell Proliferation and Colony Formation Assays
(A) The IC50 values were calculated based on dose-response data (cell absorbance) at 72 h. Liver cancer cell lines Hep3B, SNU-449, and HepG2 were sensitive to the treatment of VA. (B) Images from the colony formation assay using Hep3B, SNU-449, and HepG2 cells. (C) The relative colony formation efficiency showed a significant reduction (∼70%) of colonies formed after VA treatment in all 3 liver cancer cell lines tested. Data are presented as the mean ± standard deviation (SD); NS, not significant; NC, negative control.
VA Suppresses Colony Formation, Migration, and Invasion of Liver Cancer Cells
We then carried out the colony formation assay to assess the anticancer effect of VA over a relatively long time period of 10 days. Figure 1B showed the images of the assay from 3 liver cancer cell lines. Compared to the negative control (NC) group, VA significantly reduced the number of cell colonies formed with the 67.99% ± 2.51% (p < 0.001) difference in cell colony number for Hep3B, 63.56% ± 2.11% (p < 0.001) for SNU-449, and 69.83% ± 2.71% (p < 0.001) for HepG2 (Figure 1C).
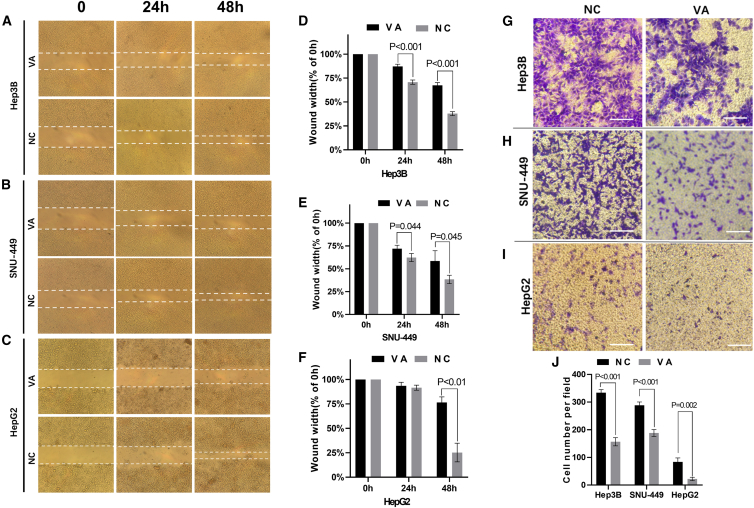
The wound healing assay also showed that cells (Hep3B, SNU-449, HepG2) treated with VA had significantly slower healing rates compared to the control groups (Figures 2A–2C). For Hep3B, the average width of wound gaps in the VA group was 87.22% ± 1.91% of its initial width compared to 67.49% ± 2.38% of the NC group (p < 0.001) at 48 h. Similar significant differences were observed for liver cancer cells SNU-449 and HepG2 (Figures 2D–2F).
Figure 2.
VA Inhibited Migration and Invasion of Liver Cancer Cells
(A–C) Images from the wound healing assay using 3 liver cancer cell lines, Hep3B (A), SNU-449 (B), and HepG2 (C). (D–F) Pictures were taken at 0, 24, and 48 h, and the bar graphs present the percentage of wound recovery in Hep3B (D), SNU-449 (E), and HepG2 (F). The assay was performed in triplicate for each cell line. The wound in the NC groups closed significantly faster than VA groups, respectively. (G–I) Images taken at 24 h from the Transwell invasion assay using liver cancer cell lines Hep3B (G), SNU-449 (H), and HepG2 (I). The average cell number was calculated from counting three randomly chosen different fields. (J) A significantly smaller number of cells was observed in VA-treated groups. Data are presented as the mean ± SD; NS, not significant; NC, negative control.
The cell invasion assays showed that the VA-treated cells displayed significantly weaker invasive abilities than the control group (Figures 2G–2I). For Hep3B cell, the average counts of invading cells in the VA group were 156.67 ± 12.04 cells compared to 334 ± 9.2 cells from the control group (p < 0.001). Similar significant differences were observed for liver cancer cells SNU-449 and HepG2 (Figure 2J).
VA Suppresses 3D Spheroid Formation of Liver Cancer Cells
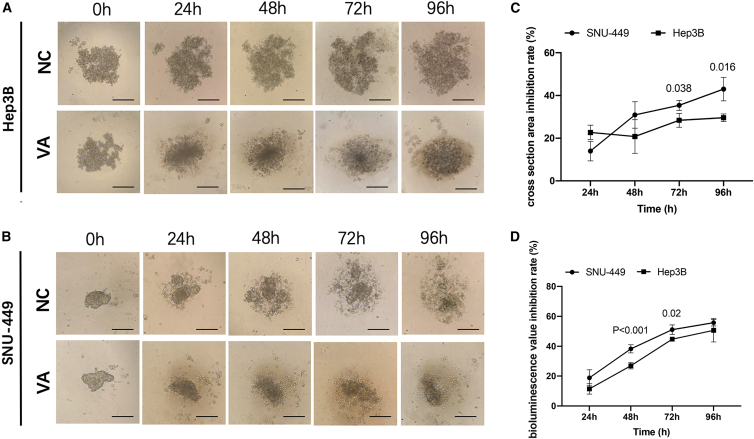
To investigate the effect of VA on 3D spheroid formation of liver cancer cells, Hep3B and SNU-449 cells, containing the luciferase reporter gene, were both cultured using a hanging drop method. The dynamic changes of the cross-section of 3D spheroid formation were displayed in Figures 3A and 3B. 3D formation efficiency calculated from the cell cross-section area showed a significantly larger 3D spheroid formed (p < 0.01) in the control group compared to the VA group at all time points measured (24 h, 48 h, 72 h, and 96 h). Inhibition rates were further calculated for both cell lines, which were 13.96% ± 4.57% at 24 h, 35.36% ± 2.31% at 72 h, and 42.97% ± 5.52% at 96 h for SNU-449. A similar trend was observed for Hep3B cells but with relatively lower inhibition rates compared to SNU-449 cells (Figure 3C).
Figure 3.
VA Restrained the 3D Formation Ability of Liver Cancer Cells
(A and B) The representative 3D spheroid models of Hep3B (A) and SNU-449 (B) cells treated by VA and NC. The relative 3D formation efficiency calculated from both the cross-section area and fluorescence values showed a significant reduction at all time points (p < 0.01) in VA-treated groups compared to control groups. (C and D) Relative inhibition rates of SNU-449 and Hep3B in response to VA were calculated by comparing the cross-section area (C) and fluorescence values (D) of VA groups to NC at 24, 48, 72, and 96 h, respectively. All assays were performed in triplicate.
Measurements from the luciferase reporter gene assay for the 3D spheroids formed showed similar results, as calculated above using the cross-section area. The inhibition rates of VA raised from 11.42% ± 2.8% to 49.3% ± 3.9% (24 h to 96 h) for Hep3B cells, and cells in the VA group climbed steadily from 18.87% ± 2.8% to 55.07% ± 1.8% (24 h to 96 h) for SNU-449 cells (Figure 3D).
LV Increases the Inhibitory Effect for Liver Cells
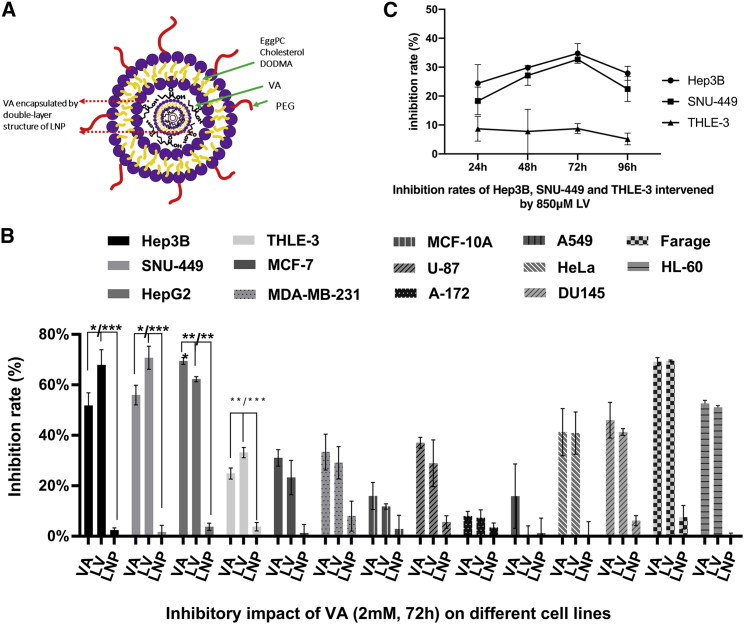
We further tested whether VA encapsulated by a cationic lipid nanoparticle (LNP) can increase its efficacy against liver cancer cells using the MTS assay. Figure 4A showed a brief structure of the LNP-encapsulated VA (LV). Figure S2 showed cell proliferation results of LV at 5 different concentrations (0.5, 1, 2, 4, and 8 mM) for all 14 cell lines from 4 time points (24, 48, 72, and 96 h).
Figure 4.
LNP Increases Anticancer Efficacy of VA in Liver Cancer
(A) A diagram of lipid nanoparticle-encapsulated VA (LV) with double-layer structure composed by PEG bad egg/cholesterol/DODMA. (B) Compared to the VA-treated group, the increased anti-cell proliferation effect was only detected for HCC cell lines Hep3B and SNU-449 but not for hepatoblastoma cell HepG2. (C) Relative inhibition rates were calculated by comparing the OD value of LV to NC at the concentration of 850 μM of LV, which gives over 30% inhibitory rates for liver cancer cells Hep3B and SNU-449 and less than 10% for normal liver cell THLE-3. Data are presented as the mean ± SD; NS, not significant; NC, negative control. ∗p < 0.05; ∗∗p < 0.01;∗∗∗p < 0.001.
Our results showed that LV only increased the inhibitory effect for liver cells compared to VA tested in the same concentration at all 4 time points. Examples of these results were presented in Figure 4B using data collected at 72 h for all 14 testing cell lines treated by 2 mM of VA and LV. For Hep3B, the inhibition rate of LV (67.82% ± 6.06%) was over 16% higher than that of VA (51.75% ± 5.06%, p = 0.024). For SNU-449, the inhibition rate of LV was 70.71% ± 4.57% compared to 55.93% ± 3.88% for VA with a 15% increase (p = 0.013). For normal liver cells (THLE-3), the inhibition rate was 33.15% ± 2.04% for LV and 24.86% ± 2.21% for VA (p = 0.009). However, for HepG2 cells, the inhibition rate significantly dropped from 69.46% ± 1.36% to 62.23% ± 0.99% (p = 0.002) after LV encapsulation.
0.85 mM concentration of LV was further used to treat Hep3B, SNU-449, and THLE-3 cells at different time points (24 h, 48 h, 72 h, 96 h) because the same dose of VA was used in all above in vitro assays due to its high inhibitory rate for liver cancer cells and low toxicity on normal liver cells. Figure 4C illustrated that the inhibition rates in Hep3B and SNU-449 were significantly higher (∼>30%) than their own counterparts in THLE-3 (∼<9%) at 48, 72, and 96 h (all p values less than 0.01).
Anticancer Effect of LV in Xenograft Mouse Models
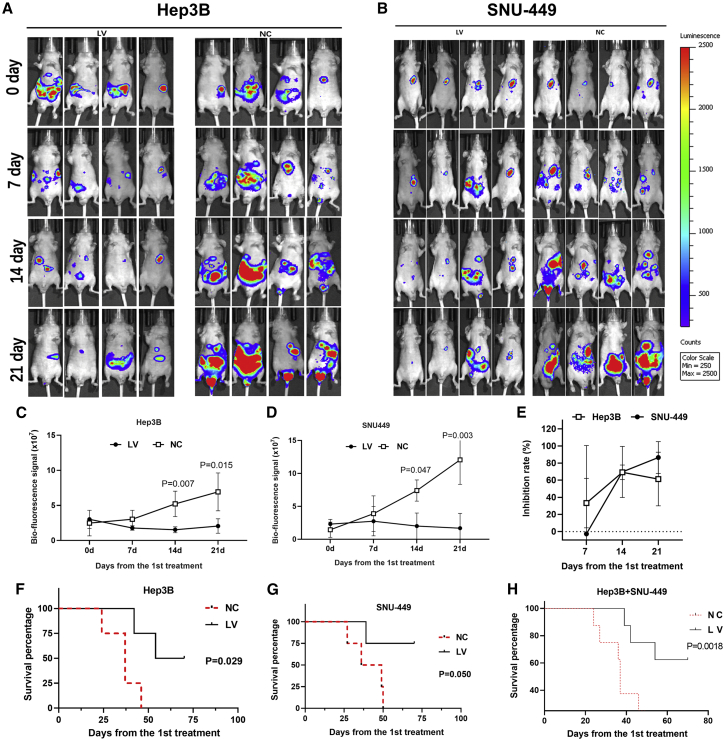
Systematic delivery of LV via tail-vein injection was applied in mouse models implanted by 2 liver cancer cell lines Hep3BLuc and SNU-449Luc. Tumor sizes represented by bioluminescence signals from all testing mice were shown in Figures 5A and 5B. The Hep3B tumors treated by LV measured on day 14 (14d) (1.53 ± 0.42) × 107 and 21d (2.05 ± 1.03) × 107 were significantly smaller compared to the control group (14d: (5.22 ± 1.82) × 107, p = 0.007; 21d: (6.93 ± 2.7) × 107, p = 0.015) (Figure 5C). Similar findings were observed in SNU-449 cell line models as well (Figure 5D). The SNU-449 tumors of LV-treated mice (2.0 ± 1.99) × 107] started to show significant difference compared to the NC group (7.4 ± 1.6) × 107, p = 0.047] on 14d of treatment. On 21d, the bioluminescence value of LV group was (1.69 ± 2.24) × 107 compared to (12.06 ± 3.73) × 107 of the control group (p = 0.003). The inhibition rates of LV were further calculated and shown in Figure 5E, which demonstrated a reduction of 70% and 61% for Hep3B tumors and 70% and 86% for SNU-449 tumors on 14d and 21d of treatment, respectively.
Figure 5.
LV Suppressed HCC Development and Improved the Survival Rate in the Mouse Study
(A and B) The images of bioluminescence of HCC implanted in xenograft mice of Hep3B (A) or SNU-449 (B), from 0 to the 21st day of treatment, were displayed. (C and D) The bioluminescence value was significantly lower in the LV group compared to NC, at 14 and 21 days of treatment, in both Hep3B (C) and SNU-449 (D) cell line mice models. (E) The inhibition rates were calculated by comparing the biofluorescence signal of the LV group to that of NC. (F and G) LV also improved the survival rate of mice implanted with Hep3B (F) or SNU-449 (G) cells compared to NC groups. (H) Combined survival curve of both Hep3B and SNU-449 implanted mice. Data are presented as the mean ± SD; NS, not significant; NC, negative control.
Significant differences in mice survivorship were also detected between LV-treated and control groups (p = 0.029 for the Hep3B model; p = 0.050 for the SNU-449 model). In the Hep3B model, the first mouse in the NC group died on 24d of the experiment, whereas the first mouse in the LV group died on 42d after starting treatment (Figure 5F). After the 70d, the survival rate was 50% in the LV group compared to 0% in the NC group. In the SNU-449 model, mice in the NC group died on the 27d, 36d, 49d, and 50d postinitiation of treatment, whereas only one mouse succumbed on 39d, and the rest of the mice all survived to the end of 70d in the LV-treated group (Figure 5G), which yielded a 75% survival rate for the LV-treated group and 0% survival rate for the NC group. When survival data from both Hep3B and SNU-449 models were combined (Figure 5H), the Mantel-Cox log rank test generated a highly significant difference between LV-treated and control groups (p = 0.0018).
Mouse weight was monitored daily, and we did not find significant difference between treatment and control groups. We observed the redness and desquamation in the tail-injection site in 2 mice, 2 weeks after starting the treatment. The redness and desquamation completely disappeared about 2 weeks after the last treatment.
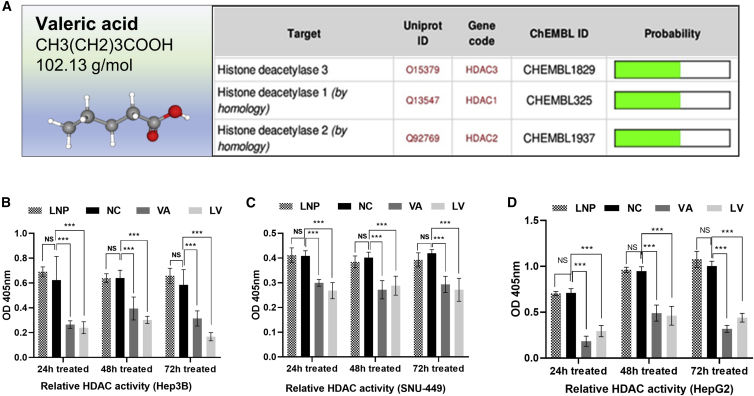
VA Functions as a Potential HDACi
The molecular formula, weight, and 3D structure of VA were illustrated in Figure 6A. The top proteins predicted as VA targets by the SwissTargetPrediction tool were 5 solute carrier family members and 3 HDAC enzymes (HDAC1, HDAC2, and HDAC3) (Figure 6A). The impact of VA on HDAC activity was tested using the HDAC activity assay, and results showed that both VA and LV significantly decreased HDAC activity in all liver cancer cells at 24 h, 48 h, and 72 h after treatment. For example, in the Hep3B experiment, the normalized HDAC activity of the NC group was 0.623 ± 0.156, calculated by optical density (OD) values, and measured higher compared to 0.265 ± 0.025 in the VA-treated group (p < 0.001) and 0.24 ± 0.039 in the LV-treated group (p < 0.001) (Figure 6B). Similar results were observed in experiments using liver cancer cells SNU-449 (Figure 6C) and HepG2 (Figure 6D). Furthermore, LV inhibited HDAC activity (0.168 ± 0.027) significantly more than VA (0.315 ± 0.05) in Hep3B cells (p = 0.021) at 72 h. A similar trend was also detected in SNU-449 and HepG2 experiments.
Figure 6.
VA Targeted HDAC Enzymes and Reduced Their Activities
(A) Three members of HDAC (HDAC1, HDAC2, and HDAC3) were predicted to be the potential targets of VA by the Swiss Target Prediction Tool. (B–D) The relative HDAC activity values in the VA- or LV-treated groups were significantly lower than the activity values in NC at 24, 48, and 72 h in Hep3B (B), SNU-449 (C), and HepG2 (D) cells. No significant differences were observed between the LNP and NC groups. Data are presented as the mean ± SD; NS, not significant; NC, negative control, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Impact of VA on Transcriptome Associated with Cancer-Related Pathways
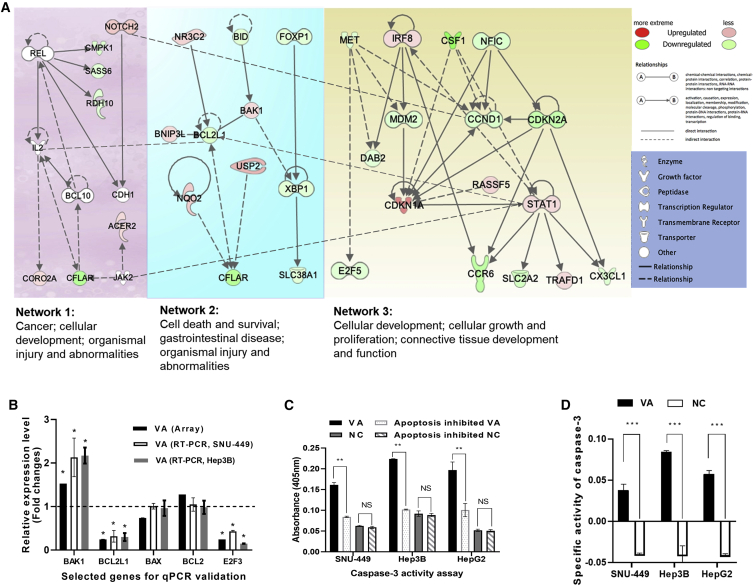
A gene expression array (Affymetrix) was performed to better understand potential anticancer mechanisms of VA in liver cancer. Treatment of VA caused expression changes of 2,003 genes in liver cancer cells (Hep3B); of these, 880 genes were downregulated (<−2-fold, Bonferroni p < 0.05), and 1,123 genes were overexpressed (>2-fold, Bonferroni p < 0.05). Input of these genes into the Ingenuity Pathway Analysis (IPA) tool (Ingenuity Systems; https://digitalinsights.qiagen.com/) generated top networks with functional relevance to “Cell death and survival; gastrointestinal disease; organismal injury and abnormalities.” Figure 7A summarized key features from these networks, which included multiple cancer- and apoptosis-related pathways that are significantly modulated by the VA treatment. For example, the FOXP1/XBP1 pathway was repressed and resulted in the downregulation of oncogene SLC38A1 (Network 2 in Figure 7A). Moreover, MDM2 (a negative regulator of p53) was downregulated, followed by the suppression of the MET/MDM2, which further affected genes downstream of the p21/CDKN1A pathway and downregulated oncogene CCND1 (Network 3).
Figure 7.
Global Transcriptional Impact of VA on Cancer-Related Networks
(A) A combined network from genes significantly affected by VA. These networks include “Cancer; cellular development; organismal injury and abnormalities,” Cell death and survival, gastrointestinal disease; organismal injury and abnormalities, and “Cellular development; cellular growth and proliferation; connective tissue development and function.” (B) Consistent expression results from both array and qPCR analyses for 5 selected genes. (C) Significantly increased CASP3 activities were detected in the VA-treated group compared to NC in all cell lines. (D) In SNU-449, Hep3B, and HepG2 cells, VA-treated groups have significantly higher SA values compared to the control groups, respectively (p < 0.001). Values represent the mean ± SD. NS, nonsignificant; NC, negative control. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure 7B illustrated that expression levels of 5 selected genes were all consistent between the array date and qPCR results, which confirmed that one gene was overexpressed (BAK1), 2 were downregulated (BCL2L1 and E2F3), and 2 did not have significant expression changes (BCL2 and BAX) after VA treatment.
The caspase-3 (CASP3) activity was measured to examine and confirm the predicted impact of VA on apoptosis. Figure 7C showed that the CASP3 activity in the VA-treated group was significantly higher compared to all 3 control groups (inhibited apoptosis + VA, NC, and inhibited apoptosis + NC) in all 3 cell lines tested (all p values < 0.001). The CASP3 specific activity (SA) also showed a similar trend: the VA-treated group had significantly higher SA values compared to the control group (p < 0.001) (Figure 7D).
Discussion
The anticancer efficacy of VA, a small compound and a major ingredient of valerian, has been examined in this study. Our results demonstrated for the first time that VA has a broad spectrum of anticancer activity with specifically high cytotoxicity for liver cancer by serving as a novel HDACi. The IC50 data generated from a series of VA concentrations in the cell proliferation assay showed that VA has the strongest tumor-suppressing role for liver cancer cells. More importantly, its anti-cell proliferation activity was less evident for normal liver cells, suggesting that VA could be a promising agent for liver cancer treatment.
This anticancer efficacy of VA for liver cancer is further supported by results from other in vitro assays. Compared to the cell proliferation assay, the colony formation assay allows a longer time period (10–14 days) to evaluate cell survival in response to various treatment conditions. As expected, ∼30% inhibition rate was observed in the cell proliferation assay, and approximately a 60% inhibition rate was detected in the colony formation assay when treated with the same dose of VA. Moreover, VA significantly reduced (>50%) migration and cell-cell interaction of liver cancer cells, as observed in the wound healing and Transwell invasion assays. Results from the 3D spheroid formation assay demonstrated more physiologically relevant information about the tumor-suppressing activity of VA and may provide more predictive data for in vivo tests.
Before starting animal studies, we first tested lipid nanoparticle as a delivery vehicle for the systemic administration of VA. The LNP used in this study has been modified as a drug carrier specifically targeting liver cancer cells.15 LNP, at different concentrations (1 mM, 2 mM, 4 mM, 8 mM), showed no impact on cell proliferation as compared to water controls (data not shown), which indicated the safety of this LNP as reported in previous studies.16 More importantly, LV showed a significantly stronger antiproliferative effect than naked VA only for liver cancer Hep3B, SNU-449, and normal liver THLE-3 cells but not for liver cancer HepG2 and other cancer cell lines tested. This is probably due to the different affinities possessed by the LNP to different cell types. Both Hep3B and SNU-449 are HCC and represent the most common type of primary liver cancer. However, HepG2 is considered to be a hepatoblastoma, a rare type of liver cancer in adults but common in children.17 Our results from the cell proliferation assay also indicate that LV at 1 mM has over a 30% inhibition rate for HCC cells but less than 10% for normal liver cells, which further suggested that LV is a great potential agent for liver cancer treatment because of its HCC-related high cytotoxicity and normal cell-related low toxicity.
LV, via tail-vein injection, was used in our animal studies to treat orthotopic xenograft mice, established by injecting tumor cells directly into mouse liver. Compared to HCC subcutaneous models, the orthotopic model closely mimics the development of human HCC by imitating similar collagen distribution and blood supply in tumor growth and the ability to take up LNP more efficiently.18 Our results show that mice that received LV treatment experienced a significantly decreased tumor burden (>90% after 21d) but also had a significantly improved survival rate in both Hep3B and SNU-449 models, indicating the high effectiveness and feasibility of LV in HCC treatment. One of the main reasons for high HCC mortality is the cancer’s high metastatic potential.19 Data from the orthotopic HCC model showed that LV may also prevent liver cancer cells from metastasizing. In our animal experiment, LV might not have achieved the maximum therapeutic potential because the blood supply in tumors of the xenograft model is usually insufficient due to the hindrance of angiogenesis by the fibrotic tissues deposited in the surrounding region of implanted tumor cells.18 Taken together, results from both in vitro and in vivo assays suggest LV as a new and effective therapeutic agent for liver cancer treatment.
Possible molecular mechanisms accounting for the anticancer effect of VA were also investigated by first searching for its predicted protein targets based on chemical structure similarities. Three HDAC enzymes (HDAC1, HDAC2, and HDAC3) are among the top hits of predicted targets, suggesting VA as a potential HDACi. This prediction was confirmed by findings from the HDAC activity assay. HDAC is a hallmark in cancer and plays a crucial role in gene-transcription regulation, controlling the proliferation, cell survival, differentiation, and genetic stability.20 Therefore, HDACs are among the most promising therapeutic targets for cancer treatment. So far, there are 4 FDA-approved HDACi that are all used to treat lymphoma and over one dozen HDACi being tested to treat solid tumors in different phases of clinical trials (by 2018).21 Studies have demonstrated that HDACs strongly suppress cancer cell apoptosis by decreasing the activity of apoptosis-key effector CASP3.22 Previous studies have reported that inhibition of HDAC can increase the activity of CASP3 and promote apoptosis,23 which was exactly what we observed in our study.
HDACs are a superfamily with 18 members,24 and HDACs have been implicated in multiple types of cancer with different expression levels. For example, HDAC1, -2, and -3 are frequently upregulated in primary human HCC.20 HDAC4, -5, -7, and -9 are overexpressed in glioma.25 In breast cancer, the common overexpressed HDACs have been found to be HDAC1, -2, -6, and -8.26 Researchers have reported that HDAC3, a predicted target of VA, was required for the self-renewal of liver cancer stem cells.27 These differential expressions of HDAC genes may help explain the high therapeutic effect of VA on HCC cells because all 3 overexpressed HDACs in HCC are targets of VA. Therefore, VA could function as a novel HDACi and an emerging approach for HCC treatment.
The global impact of VA on the human transcriptome revealed more cancer-related pathways and networks affected by VA treatment. The intrinsic apoptotic pathway is reported to be the main mechanism for HDACi to induce apoptosis in cancer cells.28 Bcl-2 family members, such as BCL-2, BCL-XL, BAX, BID, and BAK1, are frequently observed to be significantly regulated by HDACi via an intrinsic apoptotic pathway, therefore activated CASP3, CASP7, and eventually cell apoptosis.28 Moreover, STAT1- and p53-related MDM2/MET29 and p2130-related pathways are reported to take part in HDACi-induced apoptosis in cancer cells.31,32 However, apoptotic pathways, like CDKN2A/CCND, have not been reported to be affected by HDACi before, which may provide a new direction for further research.33,34
In summary, we demonstrate for the first time that VA suppresses liver cancer development possibly by acting as a novel HDACi in this study. We also show the success of using LNPs as a delivery vehicle to increase treatment feasibility of VA. These findings represent the key preclinical steps in the development of a new chemotherapeutic with HDACi functionality. Further investigation of LV is warranted to explore its therapeutic potential for the treatment of HCC and cancer in general.
Materials and Methods
Cell Lines and Culture Medium
All human cell lines were purchased from American Type Culture Collection (ATCC; USA) and cultured in ATCC-suggested medium and conditions. Cell lines included in the study were HCC cells Hep3B and SNU-449, hepatoblastoma cell HepG2, normal liver cell THLE-3, breast cancer cells MCF-7 and MDA-MB-231, normal breast cell MCF-10A, lung cancer cell A549, glioblastoma cell U-87 and A172, cervix adenocarcinoma cell HeLa, lymphoma cell Farage, prostate cancer cell DU145, and acute promyelocytic leukemia cell HL-60.
Transduction of Luciferase Lentivirus
To better image tumor sizes, both Hep3B and SNU-449 cells containing cytomegalovirus (CMV)-Firefly luciferase-internal ribosome entry site (IRES)-Puro lentivirus (Cellomics, USA) were constructed according to the standard protocol. After a 12-day selection on medium with Puro (1 μg/mL), stable fluorescence signals of both Hep3B-luciferase (Luc) and SNU-449-Luc were then confirmed by adding luciferin substrate and measured by the Microplate Luminometer (Promega, USA).
Preparation of LV
VA was purchased from Sigma-Aldrich (USA) and diluted in culture medium to a working concentration for all in vitro assays. The cationic liposomes were prepared with minor modifications.16 A prewarmed 100% ethanolic lipid solution composed of DODMA/EggPC/Chol/PEG-lipid at 45:15:35:5 (molar ratio) was mixed with 20 mM HEPES (pH 7.4) solution at 60°C in a water bath. Ethanol was removed by dialysis using a molecular weight cutoff (MWCO) 10,000-dalton Float-A-Lyzer (Spectrum Laboratories, Rancho Dominguez, CA, USA) and then sterilized by passing the mixture through a 0.22-μm syringe filter (Millipore, Billerica, MA, USA). LV was prepared by mixing cationic liposomes with an equal volume of VA in 20 mM HEPES buffer at room temperature for 15 min and centrifuged at 2,100 × g rpm for 15 min. For in vitro experiments, the LV mixture was diluted in culture medium to testing concentrations. For tail-vein injection in animal experiments, LV was centrifuged and concentrated to the final volume of 100 μL using the Amicon 50K centrifugal filter (Merck Millipore, USA). The LNP-encapsulated ddH2O were used as an NC.
MTS Cell Proliferation Assay
MTS cell proliferation assay was used to determine the anticancer ability of VA and LV for 14 cell lines. The assay was performed by adding 20 μL of MTS solution (Promega, USA) into each well in a dark hood at different incubation time points (24 h, 48 h, 72 h, 96 h). A Microplate Spectrophotometer (BioTek, USA) was used to determine the absorbance at the wavelength of 450 nm. The assay was performed in triplicate. The proliferation inhibition rate was calculated based on the formula: inhibition rate = (1 − absorbance of treated sample/absorbance of control sample) × 100%. IC50s were also calculated based on dose-response data (cell absorbance) by using GraphPad Prism (version 8.2.1).
Colony Formation Assay
Approximately 2,000 cells of Hep3B, SNU-449, or HepG2 were seeded into each well of 6-well cell-culture plates (Falcon, USA) in 2 mL of the completed medium. The cells were treated with either VA with final concentration of 850 μM (treatment group) or the same amount of ddH2O (control group) and incubated in a humidified incubator at 37°C with 5% CO2 for 10 days. Cells were then fixed by 4% paraformaldehyde and dyed with crystal violet (Sigma-Aldrich, USA). This assay was performed in triplicate, and the number of colonies with more than 30 cells was counted.
Wound Healing Assay
Approximately 1 × 106 cells of Hep3B, SNU-449, or HepG2 were seeded into each well of 6-well tissue-culture plates in triplicate. When the cell monolayer reached 90% confluence, a straight wound scratch in each well was gently made by a 100-μL pipette tip. 2 mL culture medium with VA at concentration of 850 μM was added to pregrown plates to replace the original medium (treatment group). Medium with the same volume of ddH2O as VA were added to control plates. The plates were then incubated at 37°C for another 48 h. Images were taken at 0 h, 24 h, and 48 h after the wound for the average of wound-closure measurement.
Transwell Invasion Assay
1 × 104 of Hep3B, SNU-449, or HepG2 cells were seeded in triplicate into each upper layer of the culture insert of a 3.0-μm pore-size Transwell chamber (Falcon, USA) with 100 μL medium containing 20% Matrigel (Corning, USA), 850 μM VA (or ddH2O), and 0.1% fetal bovine serum (FBS) in each well. After 24 h incubation at 37°C, cells invaded through the membrane were fixed by 4% paraformaldehyde and dyed with crystal violet. The surface of the upper layer of the membrane was cleaned, and cells in different fields of view were counted using an inverted microscope (Olympus, Japan) to get an average sum of cells.
3D Spheroid Formation Assay
Approximately 500 Hep3B-Luc and SNU-449-Luc cells were added in 30 μL of solution, which contained Corning Matrigel Matrix High Concentration (HC), Phenol Red-free, and Lactate Dehydrogenase-Elevating Virus (LDEV)-free (Corning, USA) and complete culture medium. VA (treatment group) or an equal amount of ddH2O (NC group) was added into the cell drops after 24 h incubation. The cross-section area was used to determine the 3D volume, which was calculated by ImageJ (version 1.52a; National Institutes of Health, USA). The cross-section area inhibition rate = (1 − cross-section area of treated sample/cross-section area control sample [NC]) × 100%. In vitro bioluminescence signals were also determined by transferring cells to a 96-well plate in the presence of D-luciferin (150 μL/mL) (PerkinElmer, USA). The assay was performed in triplicate, and the 3D spheroid-formation inhibition rate was calculated using the formula: inhibition rate = (1 − absorbance of treated sample [or mock sample]/absorbance of control sample [NC]) × 100%.
Mouse Models and In Vivo Fluorescence Imaging
Athymic nude male mice (N = 16) were purchased from The Jackson Laboratory (Sacramento, CA, USA). Experimental protocols were approved by the Yale University Institutional Animal Care and Use Committee (IACUC; number 2013-11504) and adhered to the NIH Guide for the Care and Use of Laboratory Animals. A freshly prepared mixture (50 μL complete culture medium, 5 × 105 Hep3B-Luc or SNU-449-Luc cells, 50 μL Corning Matrigel Matrix HC, Phenol Red-free, and LDEV-free) was directly injected into mouse liver under inhalation isoflurane anesthesia. The injection point was 2 mm below the angle, which formed by the xiphoid and the left costal margin of the mouse. All mice were then maintained for 15 days and examined by In Vivo Imaging System (IVIS) Lumina LT Imaging System (Xenogen/Caliper Life Sciences, USA) with Living Image 4.3.1 software (Caliper Life Sciences, USA). Intraperitoneal (i.p.) injection of D-luciferin was used to provide substrate bioluminescent imaging. All mice were then divided into an LV group (LNP-VA) (N = 4) and NC group (LNP-ddH2O) (N = 4). 100 μL LV at a concentration of 100 mg/kg was administered to each mouse by tail injection once a week for 4 consecutive weeks, and images of tumor sizes were collected every week. Mice were monitored daily for 70 days to collect survivorship data. Mice were sacrificed in a CO2 chamber when ethically necessary due to clinical symptoms or substantial loss in body weight. To compare survival differences between LV-treated and control groups, log rank (Mantel-Cox) test was used for survival analysis in GraphPad Prism (version 8.2.1).
Swiss Target Prediction Assay
Potential molecule targets of VA were predicted using the SwissTargetPrediction tool that used 2D and 3D structure of VA (http://www.swisstargetprediction.ch/).35
HDAC Colorimetric Activity Assay
Effects of VA on HDAC activity in Hep3B, SNU-449, and HepG2 cells were evaluated in triplicate using the Colorimetric HDAC Activity Assay Kit (BioVision, USA). Cells were treated with VA (final concentration of 850 μM), LV (final concentration of 850 μM), or the same volume of ddH2O (NC group). Absorbance was assessed by microplate spectrophotometer (BioTek, Winooski, VT, USA) at the wavelength of 405 nm. HDAC activity was presented as the relative OD value per microgram protein sample.
Gene Expression Profiling and Pathway-Based Network Analyses
The impact of VA on genome-wide gene expression was assessed using the Affymetrix Human Clariom S Assay (Thermo Fisher Scientific, USA). After 48 h treatment with either VA (final concentration of 850 μM) or ddH2O, total RNA was isolated from Hep3B cells using the RNeasy Mini Kit (QIAGEN, Germany). 5 μg of total RNA from each sample was sent to Yale Center for Genome Analysis for expression microarray analyses. Expression arrays for 3 VA-treated samples and 5 control samples in 2 batches were performed (GEO: GSE140280). Batch effects were adjusted by Affymatrix Transcriptome Analysis Console (TAC) Software. Genes with more than 2-fold expression changes were further investigated for network and functional inter-relatedness using the IPA (Ingenuity Systems; https://digitalinsights.qiagen.com/). Expressions of several selected genes (E2F1, E2F3, BAX, Bcl-2, and BclX) were measured using qPCR for confirmation. The housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was used as the control gene, and PCR primers were listed in Table S1. All qPCR reactions were conducted in triplicate. Relative expression levels of a target gene were calculated using the 2−ΔΔCt method.
CASP3 Activity Assay
The CaspACE Assay was conducted in a total volume of 100 μL in 96-well plates according to the manufacturer’s protocol. Briefly, 2 × 106 cells cultured in 2 mL medium were treated with either 850 μM VA or the same amount of 1 × PBS (NC) to create the induced apoptosis groups (72 h after treatment). 3 μL Z-VAD-FMK inhibitor was added at 72 h to create the inhibited apoptosis groups. Protein concentration of each sample was determined by the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, USA), and the p-Nitroaniline (pNA) Calibration Curves were also made by a colorimetric assay system. CASP3 SA was calculated as the following formulas: SA = (pmol pNA liberated per hour)/μg protein.
Author Contributions
Conception and Design, R.H., X.C., and Y.Z.; Development of Methodology, R.H. and Y.Z.; Acquisition of Data, R.H.; Analysis and Interpretation, R.H. and Y.Z.; Writing, Review, and/or Revision of the Manuscript, R.H., O.N., and Y.Z.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was partially supported by funds from Yale University and NIH research grant CA238100. R.H.’s visit to Yale University was supported by the China Scholarship Council (CSC).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.08.017.
Supplemental Information
References
- 1.Niino M., Matsuda T. Incidence rates of liver cancer in the world from the Cancer Incidence in Five Continents XI. Jpn. J. Clin. Oncol. 2019;49:693–694. doi: 10.1093/jjco/hyz092. [DOI] [PubMed] [Google Scholar]
- 2.Zhou M., Wang H., Zeng X., Yin P., Zhu J., Chen W., Li X., Wang L., Wang L., Liu Y. Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145–1158. doi: 10.1016/S0140-6736(19)30427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevention of Infection Related Cancer (PIRCA) Group, Specialized Committee of Cancer Prevention and Control, Chinese Preventive Medicine Association. Non-communicable & Chronic Disease Control and Prevention Society, Chinese Preventive Medicine Association. Health Communication Society, Chinese Preventive Medicine Association [Strategies of primary prevention of liver cancer in China: expert consensus (2018)] Zhonghua Yu Fang Yi Xue Za Zhi. 2019;53:36–44. doi: 10.3760/cma.j.issn.0253-3766.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Hobbs C. Phu: valerian and other anti-hysterics in European and American medicine (1733-1936) Pharm. Hist. 1990;32:132–137. [PubMed] [Google Scholar]
- 5.Zare A., Khaksar Z., Sobhani Z., Amini M. Analgesic Effect of Valerian Root and Turnip Extracts. World J. Plast. Surg. 2018;7:345–350. doi: 10.29252/wjps.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakehashi A., Kato A., Ishii N., Wei M., Morimura K., Fukushima S., Wanibuchi H. Valerian inhibits rat hepatocarcinogenesis by activating GABA(A) receptor-mediated signaling. PLoS ONE. 2014;9:e113610. doi: 10.1371/journal.pone.0113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian S., Wang Z., Wu Z., Wei Y., Yang B., Lou S. Valtrate from Valeriana jatamansi Jones induces apoptosis and inhibits migration of human breast cancer cells in vitro. Nat. Prod. Res. 2019:1–4. doi: 10.1080/14786419.2018.1548454. [DOI] [PubMed] [Google Scholar]
- 8.Tan Y.Z., Peng C., Hu C.J., Li H.X., Li W.B., He J.L., Li Y.Z., Zhang H., Zhang R.Q., Wang L.X., Cao Z.X. Iridoids from Valeriana jatamansi induce autophagy-associated cell death via the PDK1/Akt/mTOR pathway in HCT116 human colorectal carcinoma cells. Bioorg. Chem. 2019;87:136–141. doi: 10.1016/j.bioorg.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Lin S., Fu P., Chen T., Ye J., Su Y.Q., Yang X.W., Zhang Z.X., Zhang W.D. Minor valepotriates from Valeriana jatamansi and their cytotoxicity against metastatic prostate cancer cells. Planta Med. 2015;81:56–61. doi: 10.1055/s-0034-1383369. [DOI] [PubMed] [Google Scholar]
- 10.Li X., Chen T., Lin S., Zhao J., Chen P., Ba Q., Guo H., Liu Y., Li J., Chu R. Valeriana jatamansi constituent IVHD-valtrate as a novel therapeutic agent to human ovarian cancer: in vitro and in vivo activities and mechanisms. Curr. Cancer Drug Targets. 2013;13:472–483. doi: 10.2174/1568009611313040009. [DOI] [PubMed] [Google Scholar]
- 11.Torres-Hernández B.A., Del Valle-Mojica L.M., Ortíz J.G. Valerenic acid and Valeriana officinalis extracts delay onset of Pentylenetetrazole (PTZ)-Induced seizures in adult Danio rerio (Zebrafish) BMC Complement. Altern. Med. 2015;15:228. doi: 10.1186/s12906-015-0731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 13.Gurvich N., Tsygankova O.M., Meinkoth J.L., Klein P.S. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 14.Duvic M., Talpur R., Ni X., Zhang C., Hazarika P., Kelly C., Chiao J.H., Reilly J.F., Ricker J.L., Richon V.M., Frankel S.R. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baig B., Halim S.A., Farrukh A., Greish Y., Amin A. Current status of nanomaterial-based treatment for hepatocellular carcinoma. Biomed. Pharmacother. 2019;116:108852. doi: 10.1016/j.biopha.2019.108852. [DOI] [PubMed] [Google Scholar]
- 16.Hsu S.H., Yu B., Wang X., Lu Y., Schmidt C.R., Lee R.J., Lee L.J., Jacob S.T., Ghoshal K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine (Lond.) 2013;9:1169–1180. doi: 10.1016/j.nano.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Terrada D., Cheung S.W., Finegold M.J., Knowles B.B. Hep G2 is a hepatoblastoma-derived cell line. Hum. Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Wisse E., Jacobs F., Topal B., Frederik P., De Geest B. The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther. 2008;15:1193–1199. doi: 10.1038/gt.2008.60. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y.S., Kashida Y., Kulp S.K., Wang Y.C., Wang D., Hung J.H., Tang M., Lin Z.Z., Chen T.J., Cheng A.L., Chen C.S. Efficacy of a novel histone deacetylase inhibitor in murine models of hepatocellular carcinoma. Hepatology. 2007;46:1119–1130. doi: 10.1002/hep.21804. [DOI] [PubMed] [Google Scholar]
- 20.Buurman R., Sandbothe M., Schlegelberger B., Skawran B. HDAC inhibition activates the apoptosome via Apaf1 upregulation in hepatocellular carcinoma. Eur. J. Med. Res. 2016;21:26. doi: 10.1186/s40001-016-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suraweera A., O’Byrne K.J., Richard D.J. Combination Therapy With Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 2018;8:92. doi: 10.3389/fonc.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Z., Yang Y., Liu S., Lu J., Huang B., Zhang Y. HDAC inhibitor PAC-320 induces G2/M cell cycle arrest and apoptosis in human prostate cancer. Oncotarget. 2017;9:512–523. doi: 10.18632/oncotarget.23070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajji N., Wallenborg K., Vlachos P., Nyman U., Hermanson O., Joseph B. Combinatorial action of the HDAC inhibitor trichostatin A and etoposide induces caspase-mediated AIF-dependent apoptotic cell death in non-small cell lung carcinoma cells. Oncogene. 2008;27:3134–3144. doi: 10.1038/sj.onc.1210976. [DOI] [PubMed] [Google Scholar]
- 24.Seto E., Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laws M.T., Bonomi R.E., Kamal S., Gelovani D.J., Llaniguez J., Potukutchi S., Lu X., Mangner T., Gelovani J.G. Molecular imaging HDACs class IIa expression-activity and pharmacologic inhibition in intracerebral glioma models in rats using PET/CT/(MRI) with [18F]TFAHA. Sci. Rep. 2019;9:3595. doi: 10.1038/s41598-019-40054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller B.M., Jana L., Kasajima A., Lehmann A., Prinzler J., Budczies J., Winzer K.J., Dietel M., Weichert W., Denkert C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer--overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X.F., Cao X.Y., Zhu Y.J., Wu Z.R., Zhuang X., Shao M.Y., Xu Q., Zhou Y.J., Ji H.J., Lu Q.R. Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell Death Dis. 2018;9:398. doi: 10.1038/s41419-018-0428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Zhong Q. Histone deacetylase inhibitors and cell death. Cell. Mol. Life Sci. 2014;71:3885–3901. doi: 10.1007/s00018-014-1656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosenza M., Pozzi S. The Therapeutic Strategy of HDAC6 Inhibitors in Lymphoproliferative Disease. Int. J. Mol. Sci. 2018;19:2337. doi: 10.3390/ijms19082337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richon V.M., Sandhoff T.W., Rifkind R.A., Marks P.A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hrout A., Chaiboonchoe A., Khraiwesh B., Murali C., Baig B., El-Awady R., Tarazi H., Alzahmi A., Nelson D.R., Greish Y.E. Safranal induces DNA double-strand breakage and ER-stress-mediated cell death in hepatocellular carcinoma cells. Sci. Rep. 2018;8:16951. doi: 10.1038/s41598-018-34855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin A.R.M.R., Karpowicz P.A., Carey T.E., Arbiser J., Nahta R., Chen Z.G., Dong J.T., Kucuk O., Khan G.N., Huang G.S. Evasion of anti-growth signaling: A key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds. Semin. Cancer Biol. 2015;35(Suppl):S55–S77. doi: 10.1016/j.semcancer.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feitelson M.A., Arzumanyan A., Kulathinal R.J., Blain S.W., Holcombe R.F., Mahajna J., Marino M., Martinez-Chantar M.L., Nawroth R., Sanchez-Garcia I. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015;35(Suppl):S25–S54. doi: 10.1016/j.semcancer.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Kharrag R., Amin A., Hisaindee S., Greish Y., Karam S.M. Development of a therapeutic model of precancerous liver using crocin-coated magnetite nanoparticles. Int. J. Oncol. 2017;50:212–222. doi: 10.3892/ijo.2016.3769. [DOI] [PubMed] [Google Scholar]
- 35.Gfeller D., Michielin O., Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29:3073–3079. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.