Abstract
Natural killer (NK) cells are an important arm of the innate immune system. They constitutively express the NKp30 receptor. NKp30‐mediated responses are triggered by the binding of specific ligands e.g. tumour cell‐derived B7‐H6 and involve the secretion of cytotoxic mediators including TNF‐α, IFN‐γ, perforins and granzymes. The latter two constitute a target cell‐directed response that is critical in the process of immunosurveillance. The structure of NKp30 is presented, focusing on the ligand‐binding site, on the ligand‐induced structural changes and on the experimental data available correlating structure and binding affinity. The translation of NKp30 structural changes to disease progression is also reviewed. NKp30 role in immunotherapy has been explored in chimeric antigen receptor T‐cell (CAR‐T) therapy. However, antibodies or small ligands targeting NKp30 have not yet been developed. The data reviewed herein unveil the key structural aspects that must be considered for drug design in order to develop novel immunotherapy approaches.
Keywords: antigen recognition receptors, ligands, cancer, and carcinogenesis; cancer pharmacology; immunopharmacology
Abbreviations
- ACT
adoptive cell transfer
- APC
antigen preseting cell
- B7‐H6
B7 homolog 6
- BAG6
BCL2‐associated athanogene 6
- BAT3
HLA‐B associated transcript 3
- BCL‐2
B cell lymphoma 2 protein
- CAM
cell adhesion molecule
- CAR‐T
chimeric antigen receptor T cell
- CCL5
chemokine C‐C motif ligand 5
- CD
cluster of differentiation
- CTL
cytotoxic T lymphocyte
- CTLA‐4
CTL‐associated protein 4
- DAP10
HCST, haematopoietic cell signal transducer
- DNAM‐1
DNAX Accessory Molecule‐1
- GM‐CSF
granulocyte‐macrophage colony‐stimulating factor
- HA
haemagglutinin
- HCMV
human cytomegalovirus
- HLA‐B
major histocompatibility complex, class I, B
- ILT2
Ig‐like transcript 2
- ITAM
immunoreceptor tyrosine‐based activation motif
- ITIM
immunoreceptor tyrosine‐based inhibitory motif
- ITT
inducible translocation trap
- KAR
killer activation receptors
- KIR
killer‐cell immunoglobulin‐like receptors
- NKR‐P1A
NK cell receptor protein 1A (NKR‐P1) (CD161)
- NKG2A
NK cell inhibitory receptor G
- LAK
lymphocyte‐activated killer cells
- LFA
lymphocyte function‐associated antigen
- MHC
major histocompatibility complex
- MIP
chemokine macrophage inflammatory proteins
- MTOC
microtubule‐organizing centre
- NCAM
neural cell adhesion molecule
- NCR
natural cytotoxicity receptor
- NKp30
natural cytotoxicity triggering receptor 3
- NKp44
natural cytotoxicity triggering receptor 2
- NKp46
natural cytotoxicity triggering receptor 1
- NKp80
natural cytotoxicity triggering receptor, 80 kDa
- PD‐1
programmed cell death protein 1
- PfEMP‐1
Plasmodium falciparum erythrocyte membrane protein 1
- SNP
single nucleotide polimorphism
- SSEA‐1
stage‐specific embryogenic antigen 1
- STAT4
signal transducer and activator of transcription 4
- TAA
tumour‐associated antigens
- TCR
T cell receptor
- Th1
T helper type 1 cells
- TIL
tumour infiltrating lymphocytes
1. INTRODUCTION
Cancer is one of the major global health problems (Siegel, Miller, & Jemal, 2019). Each year, the number of cancer cases and associated deaths increases worldwide. Nonetheless, despite the increase in cancer incidence over the past 30 years, the number of deaths has fallen short of the estimated values. Several milestones in cancer therapy are at the basis of the steadily increasing survival rates and of particular interest are immunotherapy regimens (DeVita & Rosenberg, 2012).
Immunotherapy was recently added as a major cancer treatment approach, along with surgery, radiotherapy,and chemotherapy. In the early 1960s, it became evident that the immune system was able to prevent the onset of several cancers, as pointed out by some ground‐breaking experiments (Brondz, 1964; Granger & Weiser, 1964; Hellstrom, Hellstrom, Pierce, & Bill, 1968). The discovery of IL‐2 further stimulated the studies of the immune response of lymphocytes to cancer cells. The pioneering work of Rosenberg et al. (1985) showed that immune modulation with IL‐2 could be used to cause the regression of invasive metastases in patients suffering from various types of cancers, including malignant melanoma, colorectal cancer, sarcoma, renal cell cancer, adenocarcinoma and oesophageal cancer. This led to the approval of IL‐2 for the treatment of renal carcinoma and metastatic melanoma in 1992 and 1998, respectively (Jiang, Zhou, & Ren, 2016). Further studies led to the application of other cytokines in cancer treatment. First approved in 1986, recombinant human IFN‐α type 2b (IFN‐α2b) is currently used as a monotherapy or in combination with antineoplastic drugs (Asmana Ningrum, 2014).
The notion that the immune system could play an important role in cancer treatment led to the development of antibodies for use as immunomodulatory agents. Patients treated with ipilimumab or rituximab, two monoclonal antibodies designed to promote the immune system response against melanoma and lymphoma, respectively, showed an improvement in overall survival (Hodi et al., 2010; Molina, 2008). These rates increase significantly when these immunomodulators are used in combination with conventional chemotherapeutic regimens (Castro, Sandoval‐Sus, Bole, Rassenti, & Kipps, 2008) and other immunomodulators (Shanafelt et al., 2019).
These discoveries stimulated the development of new treatments based on harvesting the immune system potential to fight cancer. The field of immuno‐oncology grew significantly in the last 30 years and today it can be divided into two major strategies: passive and active immunotherapy. Passive immunotherapy comprises the use of antibodies or immunomodulatory molecules, as well as the adoptive transfer of activated immune cells. This last approach includes the isolation and expansion of patient lymphocyte‐activated killer (LAK) cells that are primed with IL‐2 and expanded ex vivo, resulting in highly active T cells that can be reinfused to the patient, with significant results in tumour regression (Nagasawa et al., 2012; Quattrocchi et al., 1999; Tsurushima et al., 1999). A similar approach has been successfully used with tumour‐infiltrating lymphocytes (TILs) and circulating T cells, supporting the therapeutic potential of tumour‐specific T cells (Sharpe & Mount, 2015). However, as tumours use immune escape mechanisms based on major histocompatibility complex (MHC) expression alteration and tumour‐associated antigen (TAA) processing, a fair part of these adoptive cell transfer (ACT) therapies fails to produce significant results. To overcome these problems, genetically engineered T cells have emerged as an alternative (Fousek & Ahmed, 2015).
The branch of active immunotherapy comprises the methodologies that aim to induce a response by stimulating the immune system. This differs from passive immunotherapy as the molecules/cells used in this case are not produced by the host. In active immunotherapy, the immune system is boosted, with cytokines, immunoadjuvants or vaccines to elicit generic or specific responses (Baxter, 2014).
All these techniques have been proven effective in cancer treatment. However, none of these are free from pitfalls. All the immunotherapy regimens currently approved or under study perform extremely well in some cancers and patients but fail completely in others. This was observed early in the immuno‐oncology era with IL‐2‐based treatments that have a response of only ~7% (Rosenberg et al., 1985). One drawback is the fact that some patients present little or no immune response to their tumours, with the tumour micro‐environment being devoid of infiltrating lymphocytes, rendering cytokine‐infiltrating, antibody‐infiltrating, cytotoxic T lymphocyte (CTL)‐infiltrating and TIL‐based therapies useless. This is nowadays recognized as the next big challenge in cancer immunotherapy (Gajewski, 2015).
It is important to mention the economic impact of the available treatments per patient. The cost of standard treatment with one checkpoint inhibitor antibody is around €100,000 per year (Fellner, 2012), while a single chimeric antigen receptor T cell (CAR‐T) treatment can reach €430,000 (Hay & Cheung, 2019). For these reasons, new cost‐effective immuno‐oncology treatments are necessary to increase the number of patients receiving such regimens and their efficacy.
Many authors have focused recently on a subset of lymphocytes, natural killer (NK) cells, that possess the innate ability to detect and kill transformed cells, proposing them as the next “major target in cancer immunotherapy” (Lorenzo‐Herrero et al., 2018; Souza‐Fonseca‐Guimarães, Cursons, & Huntington, 2019). In fact, in recent years, several clinical trials employing NK cells in cancer therapy have been launched, with many reporting cases of complete remissions, further supporting the use of these cells in future cancer treatment regimens (Shimasaki, Jain, & Campana, 2020).
2. NATURL KILLER (NK) CELLS
NK cells are a part of the innate immune system. This subset of lymphocytes was first identified in the 1970s as a set of cells that were neither B nor T and that could kill target cells without prior sensitization (Greenberg, 1994). The term natural killer was devised by Kiessling, Klein and Wigzell (1975) to describe these naturally occurring lymphocytes with lytic activity against both syngeneic and allogenic cells.
2.1. NK cell receptors
NK cells are large granular lymphocytes with a diameter ranging from 7 to 12 μm, depending on the activation state. Most phenotypical characteristics of NK cells are not exclusive, with many being shared with activated cytotoxic T cells. The unequivocal identification of NK cells must, therefore, rely on the absence and presence of specific cell surface markers. For the correct identification of the NK phenotype, CD3 must be absent and CD56 (neural cell adhesion molecule 1 [NCAM]) and CD16A (FcγRIII) must be present. Moreover, NK cells also lack surface Igs and constitutively express IL‐2 receptors (Nagler, Lanier, & Phillips, 1990; Whiteside & Herberman, 1994). Many other surface markers are present in NK cells, as well in other lymphocytes. However, the unique combination of the CD3−CD56+CD16A+ phenotype defines human NK cells (Abakushina, 2015). Additionally, two subsets of NK cells may be defined, depending on the relative expression of CD16A and CD56. These are called CD56brightCD16Adim/− and CD56dimCD16A+, where bright and dim are associated with high and low levels of expression, respectively. The first subset is outnumbered by the second in circulation but constitutes the majority of NK cells in lymphoid tissues. These subsets present different expression levels of other markers and receptors and different cytotoxic activities, with CD56bright being significantly less cytotoxic than CD56dim cells (Poli et al., 2009). It has been hypothesized that CD56bright cells are an immature form of NK cells that differentiate into CD56dim NK cells, which participate in natural and antibody‐mediated cell cytotoxicity (Chan et al., 2007). On the other hand, CD56bright NK cells express higher levels of cytokine receptors, such as the IL‐2 receptors αβγ and βγ (IL‐2Rαβγ and IL‐2Rβγ, respectively), as well as higher levels of some activating and inhibiting receptors and chemokine receptors. These cells produce higher levels of cytokines upon stimulation, namely IL‐10, an immunosuppressive cytokine, and therefore are thought to have immunoregulatory properties (Cooper et al., 2001).
NK cells descend from the same lineage as T cells, thus sharing some of the characteristics of these lymphocytes, such as cytolytic activity and lymphokine (lymphocyte‐produced cytokines) production. However, NK cells do not undergo T‐cell receptor gene rearrangement to create diverse receptors as part of the adaptive immune system (Caligiuri, 2008; Moretta, Ciccone, Mingari, Biassoni, & Moretta, 1994). The mechanisms through which NK cells recognize and kill target cells rely on a set of consistently expressed receptors with activation, inhibition or mixed activities (Biassoni, 2008; Whiteside & Herberman, 1994).
NK cells preferably lyse cells missing or under‐expressing MHC class I (MHC‐I) molecules, a characteristic that has been designated as the “missing‐self” hypothesis. Some viruses have the capacity to cause a down‐regulation in MHC‐I expression, preventing the infected cell from presenting fragments of abnormal or viral proteins. In this way, T lymphocytes are not triggered and the infected cell escapes recognition by T cell receptors. This happens also as a result of the immunoediting process during carcinogenesis (Garrido, Romero, Aptsiauri, & Garcia‐Lora, 2016). However, NK cells have evolved in such a way that cells missing MHC‐I molecules are promptly recognized as targets, while cells presenting MHC‐I molecules are regarded as “self” and are not targeted. These inhibitory signals are delivered to NK cells by C‐type lectin receptors and killer cell Ig‐like receptors (KIRs). Both classes possess immunoreceptor tyrosine‐based inhibitory motifs (ITIMs) in the cytoplasm that are responsible for the inhibitory signal transduction. Some of the known human NK cell inhibitory receptors are KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, KIR3DL2, NKR‐P1A, CD49/NKG2A and ILT2. Engaging these receptors produces an inhibitory effect in the cytotoxic activity of NK cells that prevents lysis of the ligand‐presenting cells (Paul & Lal, 2017). It is important to understand that NK cells do not detect the absence of MHC‐I molecules but rather their loss. In an environment lacking MHC‐I molecules, NK cells are not triggered. However, in an environment with cells presenting MHC‐I molecules, NK cells will target the cells missing MHC‐I. This means that, upon engaging, the inhibitory receptors deliver an inhibitory signal that does not disarm the cells but instead renders them responsive to an activation signal, a process known as licencing (Yokoyama & Kim, 2006). Paradoxically, inhibitory receptors are thus critical in the activation of NK cell responses (Long, Kim, Liu, Peterson, & Rajagopalan, 2013).
Non‐engagement of inhibitory receptors by cells lacking MHC‐I molecules is not, by itself, sufficient to induce cytolytic NK cell responses. Although some degree of response can be seen with MHC‐I‐missing cells, full activation of NK cells requires the binding of killer activation receptors (KARs) (Paul & Lal, 2017). This is defined as the “induced self” hypothesis and is based on the up‐regulation of stress‐induced ligands by the target cells. These interact with the activating receptors of NK cells, triggering a full cytolytic response that results in target cell lysis. This model is further complemented by the “altered‐self” hypothesis that is based on the capacity of killer activation receptors to recognize a large span of peptide–MHC combinations, thereby prompting the NK cell response (Boyington & Sun, 2002; Carrillo‐Bustamante, de Boer, & Kesmir, 2018).
Besides the activation and inhibition receptors, NK cells also display a broad range of cell adhesion molecules that are essential for their cytotoxic activities, as these participate in interactions between NK cells, target cells and accessory cells to generate effective immune responses. Some of these cell adhesion molecules are up‐regulated upon activation, including lymphocyte function‐associated antigen 1 (LFA‐1; CD11a/CD18), LFA‐3 (CD58) and intracellular adhesion molecule‐1 (CD54). Interestingly, upon in vitro stimulation with IL‐2, the cell surface levels of all these cell adhesion molecules increase (Mäenpää, Jääskeläinen, Carpén, Patarroyo, & Timonen, 1993), as do the levels of CD56 (Dybkaer et al., 2007). Even more interesting is the fact that, upon this increase in cell adhesion molecule expression, the cytotoxic activity of NK cells towards NK‐sensitive target cells also increases and cells that were previously non‐susceptible to NK‐killing become targets (Hood et al., 2019; Mäenpää et al., 1993; Robertson, Caligiuri, Manley, Levine, & Ritz, 1990).
One other interesting aspect in the NK cell receptor plethora is the existence of certain molecules with both activation and adhesion functions. DNAM‐1 (DNAX accessory molecule 1), a receptor part of the Ig superfamily, was found to be important in NK‐mediated tumour cell lysis. In fact, Iguchi‐Manaka et al. (2008) demonstrated, using an in vivo model, that this receptor is crucial in the immunosurveillance process. It has also been demonstrated that, besides the long‐known adhesion receptor activity (Shibuya et al., 1996), DNAM‐1 can also act as a killer activation receptor through signal transduction by its intracellular inducible translocation trap (ITT)‐like domain (Zhang et al., 2015).
2.2. The cytotoxic response of NK cells
The cytotoxic response of NK cells upon activation is divided into four steps: formation of the immunological synapse, microtubule reorganization, lysosome docking to the membrane, and lysosome fusion with the membrane (Paul & Lal, 2017).
In the first, an interface between the NK cell and the target cell is formed, deemed immunological synapse. This occurs when the NK cell approaches the target, either accidentally or “intentionally” due to chemotactic signalling.
Upon contact, CD2 molecules present on the surface of NK cells recognize the presence of stage‐specific embryogenic antigen 1 (SSEA‐1, also known as CD15 or Sialyl‐LewisX). At this point, if the target cell presents markers for NK cell inhibition, the formation of the immunological synapse is halted. Conversely, if activating receptors are present, tight adhesion between the two cells is promoted by receptor–ligand interactions of high affinity. LFA‐1 and MHC‐I molecules present on NK cells bind ligands present on the target cell surface. The formation of these receptor‐ligand complexes is enough to activate the cytolytic response, to some extent. However, these processes are considered more relevant in maintaining the immunological synapse than in triggering NK cell cytotoxicity. Full activation of the cytotoxic response depends on the engagement of specific receptors, such as the natural cytotoxicity receptors (NCRs), present on the surface of NK cells. The immunological synapse is shaped in a way that both cells form a ring‐shaped interface. Within this ring, cytotoxic granules and other cytotoxicity mediators are released directly and in a controlled fashion towards the target cell surface (Orange, 2008; Stinchcombe & Griffiths, 2007). The secretory lysosome exocytosis requires reorganization of the cell cytoskeleton. In this step, the microtubule‐organizing centre (MTOC) becomes polarized and the secretory lysosomes are transported along the microtubules towards the synapse. Upon reaching the cell membrane, the granules dock and fuse, releasing the contents towards the target, formally constituting the degranulation process (Paul & Lal, 2017; Topham & Hewitt, 2009).
2.3. The molecular mechanisms of target cell killing
As referred before, contrarily to T cells, NK cells do not rely on the somatic rearrangement of receptor genes to accommodate the expression of a variety of receptors. Instead, NK cell receptors are germ‐line encoded and consistently expressed (Biassoni, 2008; Whiteside & Herberman, 1994). The homeostasis of NK cell activity is ensured by a specific set of receptors with activating and inhibitory activities, some of which overlap. Inhibitory receptors contribute to self‐tolerance of NK cells, preventing the lysis of normal healthy cells. On the other hand, activating receptors trigger the lytic activity of these cells, prompting the destruction of cells that present activating ligands.
Upon activation, NK cells release a series of lytic enzymes through the degranulation process already described, namely perforins and granzymes. Granzymes induce target cell apoptosis, but their actions depend on being appropriately delivered by perforins (Boivin, Cooper, Hiebert, & Granville, 2009). Together with these enzymes, a wide variety of cytokines is also released, including IFN‐γ, TNF‐α, GM‐CSF, IL‐10, IL‐5, and IL‐13, chemokine macrophage inflammatory proteins 1α and 1β (MIP‐1α, CCL3 and MIP‐1β, CCL4) IL‐8 (CXCL8) and chemokine (C–C motif) ligand 5 (CCL5, also known as RANTES) (Paul & Lal, 2017).
Besides the direct effect of granzymes (and perforins), cytokines such as IFN‐γ also play an important role in inducing target cell death. IFN‐γ is an important activator of macrophages, as well of antigen‐presenting cells (APCs). Stimulation of the latter up‐regulates the expression of cytokine IL‐12, as well as IL‐18 and co‐stimulatory CD86 molecules that enhance Th1 differentiation and cytotoxic T lymphocyte function. IFN‐γ can also exert antiproliferative effects on tumour cells by enhancing the expression of the cell cycle inhibitor proteins p27Kip, p16 or p2 (Ni & Lu, 2018). TNF‐α is implicated in the collapse of tumour vasculature but can additionally induce tumour regression by triggering apoptosis, T cell activation (by Tregs' blockage) and neutrophil and monocyte chemoattraction to the tumour region and by down‐regulating the immunosuppressive phenotypes of tumours (Josephs et al., 2018).
Colony‐stimulating factor (GM‐CSF) is part of the inflammatory cascade, recruiting monocytes and inducing the differentiation, proliferation and migration of granulocytes (neutrophils, eosinophils and basophils) to the inflammation site. It is also essential for the differentiation of dendritic cells that are responsible for processing and presentation of tumour antigens and for activating cytotoxic T lymphocytes (Yan, Shen, Tien, Chen, & Liu, 2017).
IL‐5 and IL‐13 are associated with B cell growth, eosinophil activation and regulation of inflammatory and immune responses (Minty et al., 1993). IL‐10, on the other hand, seems to have a pleiotropic effect on the immune response. It down‐regulates Th1 responses, cytokine secretion, such as TNF‐α, IFN‐γ, and IL‐12 and CD4+ T cell activation (de Waal, Abrams, Bennett, Figdor, & de Vries, 1991; de Waal et al., 1991). However, IL‐10 can also induce the secretion of granzymes and perforins by CD8+ T cells and potentiate T cell receptor‐dependent IFN‐γ secretion (Emmerich et al., 2012). Lastly, MIP‐1α/1β, IL‐8 and CCL‐5 have chemotactic activities towards granulocytes, neutrophils and T cells, eosinophils and basophils (Kohidai & Csaba, 1998; Wolpe et al., 1988).
Through the secretion of these cytokines and chemokines, NK cells are able to recruit other immune cells, namely dendritic cells that can infiltrate the tumour tissue and trigger robust and sustained immune responses (Bottcher et al., 2018).
2.4. Balance between activation and inactivation of NK cells
NK cells express a wide variety of receptors with activating and inhibiting functions that allow for the fine discrimination between healthy and ailing cells in a matter of seconds. These activating and inhibitory signalling pathways involved are shown in Figure 1. Most of the inhibitory receptors detect the absence of MHC‐I molecules, while activation receptors probe for the presence of specific ligands that flag the target cells as harmful (Leung, 2014). Because NK cells are extremely cytotoxic when active and can very quickly deliver their lytic response without prior sensitization, contrarily to other lymphocytes, a complex activation/inactivation system tightly regulates their responses. Activation depends on tipping the balance between stimulatory and inhibitory signals, an equilibrium that is dictated by the engagement of different receptors. After cell‐to‐cell contact, NK cells integrate, within seconds, the signals from their activating and inhibiting surface receptors. Normal cells, expressing normal MHC‐I molecules, will engage the inhibitory receptors, rendering NK cells unresponsive, without compromising their functionality (licencing). Cells lacking MHC‐I and presenting surface activating ligands will trigger the cytolytic response. A dynamic equilibrium is reached when the target cells present both MHC‐I molecules and activating ligands. In this case, no response is triggered as the positive and negative feedback loops cancel each other. If more activating ligands are engaged, the activation signal dominates and target cell lysis occurs (Figure 2) (Vivier, Ugolini, Blaise, Chabannon, & Brossay, 2012).
FIGURE 1.
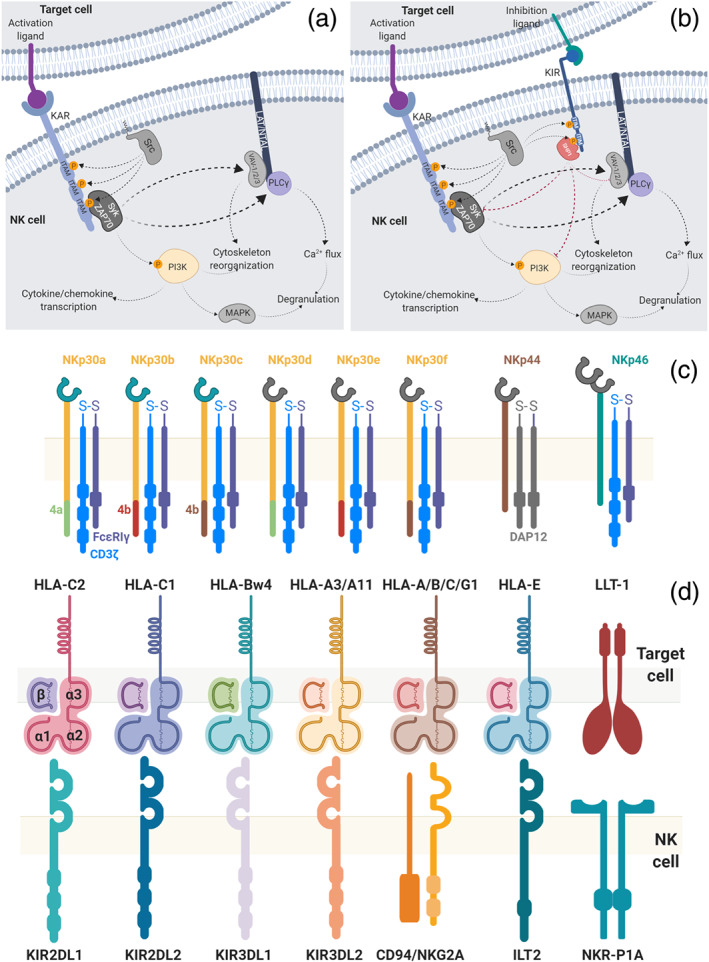
Natural killer (NK) cell signalling pathways. (a) Upon binding of the killer activation receptor (KAR), the ITAM motifs of the intracellular portion are phosphorylated by a member of the Src family. Syk and ZAP70 are activated and further phosphorylate the PI3K complex and activate VAV‐1/2/3, PLCγ1 and PLCγ2. This will further trigger cytoskeleton reorganization and affect the calcium flux into the cell, leading to an increase in Ca2+ levels. This cascade results in the cytotoxic/cytolytic response of NK cells, with the release of the cytotoxic granules (degranulation) and production of cytokines/chemokines. (b) Killing‐inhibiting receptors (KIRs), present on the surface of NK cells, may also be engaged by the corresponding inhibiting ligands. These ITIM‐bearing receptors are subsequently phosphorylated by Src kinases. This leads to the recruitment of SHP1/2 kinases that terminate the activation signalling emanating from ITAMs, rendering the NK cells inactive. The ultimate activation/inactivation result of these pathways is directly correlated with the number of receptors engaged. Target cells expressing more activation than inactivation ligands will engage more activation receptors resulting in the transduction of more activation signals, triggering the response of NK cells (Koch, Steinle, Watzl, & Mandelboim, 2013; Linnartz, Wang, & Neumann, 2010; Linnartz‐Gerlach, Kopatz, & Neumann, 2014; Paul & Lal, 2017; Shemesh et al., 2016; Watzl & Long, 2010). (c) Natural cytotoxicity receptors (NCR) protein isoforms. The six isoforms of NKp30 are presented in detail, in particular the exons 4a, 4b, and 4c, together with the overall organization of NKp44 and NKp46, which have three and five isoforms respectively. A heterodimeric intracellular CD3ζ/FcεRIγ adaptor protein is shown, but homodimers of each monomer are also possible. (d) Representation of the inhibitory receptors on NK cells and the corresponding ligands expressed in normal healthy cells. KIRs and ILT2 molecules are Ig‐like receptors, while NKR‐P1A and CD94/NKG2A are C‐type lectin receptors. HLA (human leukocyte antigen)‐type ligands are MHC‐I molecules comprising the polypeptide chains α (divided into three subunits: α1, α2 and α3) and β2 microglobulin, here abbreviated as β. KIRs have been shown to interact with the α1 and α2 subunits (Storkus et al., 1991). The NKR‐P1A receptor recognizes the lectin‐like transcript‐1 (LLT‐1) and the heterodimer CD94/NKG2A recognizes the non‐classical MHC‐I molecule HLA‐E (Paul & Lal, 2017)
FIGURE 2.
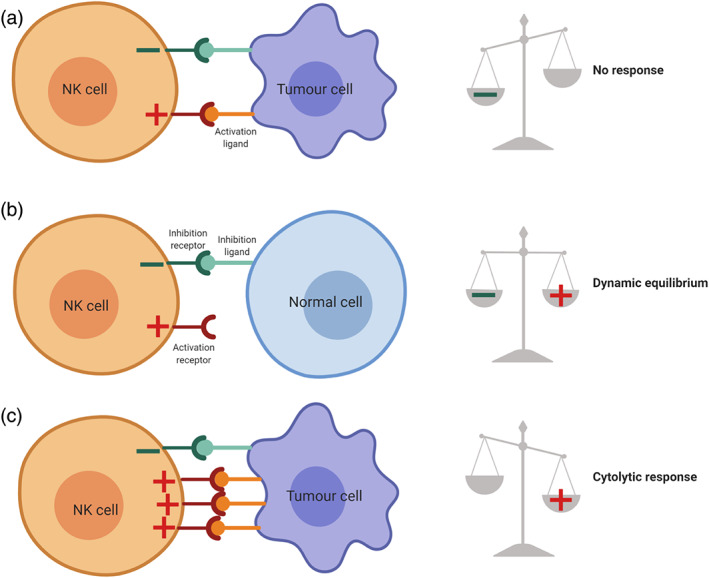
Natural killer (NK) cell activation/inhibition equilibrium. The relative number of engaged receptor types dictates the response. (a) Normal cells, expressing normal MHC‐I molecules (inhibitory ligands), do not trigger the cytolytic response of NK cells. (b) Stressed cells, such as tumour cells, engaging the same relative number of activating and inhibiting receptors of NK cells, induce a dynamic equilibrium that results in no cytolytic response of NK cells. (c) If the equilibrium is tipped over by engaging more activating receptors on NK cells, a cytolytic response will be triggered against the cells presenting the corresponding ligands
The engagement of different receptors is translated into activating and inactivating signals through immunoreceptor tyrosine‐based activation motif (ITAM) and immunoreceptor tyrosine‐based inhibitory motifs (ITIM) present in either the receptors themselves or in adaptor proteins. The ITAM and ITIM signalling cascades are combined intracellularly to determine the response of NK cells. Upon binding of activating ligands, the ITAM motifs are phosphorylated, triggering the recruitment of Syk‐family kinases and the subsequent activation of cytolytic responses. However, engagement of inhibitory receptors also leads to the phosphorylation of ITIM motifs on these receptors, a process that is promoted by the same Src kinase responsible for ITAM phosphorylation. The phosphorylated ITIMs recruit the tyrosine phosphatases SHP1 or SHP2, which can in turn terminate intracellular signals emanating from ITAM signalling receptors via their phosphatase activity, rendering NK cells inactive (Figure 2) (Linnartz‐Gerlach et al., 2014).
As mentioned, most of the inhibitory signals are triggered by ITIM motifs. Activation signals are transduced by ITAM motifs located in the cytoplasmic portion of the receptor or of associated molecules that include DAP12. The activating receptor NKG2D, constitutively expressed by all NK cells, bears no ITAM motif. Therefore, in order to transduce stimulatory signals, NKG2D is associated with an adaptor subunit designated DAP10 (Farag, Fehniger, Ruggeri, Velardi, & Caligiuri, 2002). DAP10 does not contain ITAM motifs either but is able to recruit PI3K activity upon tyrosine phosphorylation of its YINM motif (Lanier, 2009).
NK cells also present receptors that can either activate or inhibit the response, namely, the receptors 2B4 and NKR‐P1. The 2B4 receptor has been classified as a multifunctional receptor with an activity (activation or inhibition) that depends on the stage of NK cell maturation. Also, two isoforms of this receptor have been identified, with different intracellular domains (an ITIM and an immunoreceptor tyrosine‐based switching motif [ITSM]), of which only one transduces stimulatory signals. Similarly, the NKR‐P1 receptor family contains several members, of which two contain ITIM motifs and one is associated with a high‐affinity IgE receptor (FcεRI) that bears ITAM motifs, thus transducing an activation signal.
Tumour cells are generally under a constant state of cellular stress due to hypoxia, chronic proliferative signals and genome instability. Not surprisingly, cells under these conditions up‐regulate killer activation receptor ligands, becoming susceptible to NK cell killing. Some mutations, characteristic of carcinogenesis, also down‐regulate the presentation of MHC‐I molecules, rendering tumour cells more susceptible to attachment by NK cells. To survive, cancer cells develop mechanisms to evade NK cell killing. One of the proposed evasion mechanisms hypothesizes that tumour cells secrete soluble forms of the activating ligands. Certain human tumours can release soluble forms of MICA and MICB (MHC‐I polypeptide‐related sequences A and B, respectively), the natural activating ligands of NKG2D. As NKG2D‐dependent signalling requires dimerization of the ligands, by saturating the tumour environment with monomeric ligands, tumour cells are able to downmodulate NK cell responses (Groh, Wu, Yee, & Spies, 2002). On the other hand, it has been suggested that highly specific NKG2D soluble ligands can reverse completely the desensitization of NK cells and increase tumour regression (Deng et al., 2015).
Other direct evasion mechanisms rely on the secretion of immunosuppressive factors such as IL‐10 or TGF‐β1 or up‐regulation of MHC‐I molecules to counteract the stimulating signals of activating ligands. Indirect evasion mechanisms require the activation or inhibition of Tregs, killing of dendritic cells and even the use of monocytes and macrophages as NK cell desensitizers. Monocytes and macrophages can be reprogrammed by tumour‐derived signals and start to express inhibitory molecules, such as TGF‐β1, inhibiting the activity of the adjacent NK cells (Peng et al., 2017; Sabry & Lowdell, 2013).
3. NATURAL CYTOTOXICITY‐TRIGGERING RECEPTORS
Among the known activating receptors, NKG2D is by far the best characterized. It has been argued that, because NKG2D has a signalling pathway that does not overlap with the inhibitory signalling pathway of KIRs, its triggering is less susceptible to inhibitory ligands. At the same time, several studies have highlighted the importance of natural cytotoxicity receptors (NCRs) as part of the main mechanisms by which NK cells kill tumour targets (Pegram, Andrews, Smyth, Darcy, & Kershaw, 2011). In fact, one study demonstrated that lymphoma cells are able to grow in vivo in the absence of NCR+ NK cells in mutant mice, but in wild‐type mice, tumours were completely rejected over time (Halfteck et al., 2009).
Natural cytotoxicity receptors are expressed by NK cells and are extremely important in their activation. Besides NK cells, NCRs are also expressed in other cell types, although sometimes in non‐functional states. NKp44 and NKp46 have been found in innate lymphoid cells (ILCs), which are primarily tissue‐resident cells (Cella et al., 2009; Cella, Otero, & Colonna, 2010; Cortez et al., 2017; Diefenbach, Colonna, & Koyasu, 2014; Fuchs et al., 2013; Verrier et al., 2016). Additionally, NKp30, NKp44 and NKp46 have been found in different subpopulations of CD8+ cytotoxic T lymphocytes (Correia et al., 2018; Meresse et al., 2006) and in γδ T cells (Correia et al., 2011), two subtypes of T cells also involved in tumour cell lysis, as well as in IL‐15‐cultured umbilical cord blood T cells (Tang et al., 2008).
Of the four natural cytotoxicity receptors currently known, three are constitutively expressed (NKp30, NKp46 and NKp80) in NK cells, while NKp44 is only found on the surface of activated NK cells (Barrow, Martin, & Colonna, 2019; Moretta et al., 2001). Engagement of these receptors by their ligands transduces a strong activation signal to the cell. Signal transduction by natural cytotoxicity receptors happens through ITAM motifs of the associated proteins DAP12, in NKp44, and FcεRIγ and CD3ζ, in NKp30 and NKp46 (Kruse, Matta, Ugolini, & Vivier, 2014; Moretta et al., 2001). NKp80 has been described only recently and its signalling is transduced through a hemITAM motif (Bartel, Bauer, & Steinle, 2013). Interestingly, NKp44 presents three alternative mRNA splice variants, one of which bears an ITIM motif in its intracellular portion. The different levels of expression of these variants define the signalling arising from NKp44 engagement (Parodi et al., 2019).
The surface density of natural cytotoxicity receptors varies among individuals and is directly correlated with the ability to actively eliminate tumour cells. Moreover, lysis of certain NK‐sensitive tumours can be averted by simultaneously blocking the activity of NKp30, NKp44 and NKp46, showcasing the relevance of these molecules (Biassoni et al., 2001; Moretta et al., 2001). Contrarily to what happens to NKG2D, natural cytotoxicity receptors, NKp30 and NKp46 are not up‐regulated upon cytokine treatment. This demonstrates that the natural cytotoxicity receptor density at the NK cell surface is remarkably stable and its under expression (termed NCRdull) has been correlated with poor prognosis in cases of leukaemia (Fauriat et al., 2007) and non‐small‐cell lung cancer (Charrier et al., 2019).
Natural cytotoxicity receptors have been shown to recognize a multitude of ligands, including bacterial‐, viral‐ and parasite‐derived proteins, as well as stress‐induced proteins (Kruse et al., 2014). NKp30 interacts with a broad range of ligands without an obvious structural similarity, including viral, parasitic and tumoural proteins (Kruse et al., 2014). NKp30, for instance, has been shown to interact with B7‐H6 and BAT3, two proteins that share no homology (Pende et al., 1999). Binding of heparin and heparan sulphate sequences is also a puzzling observation. These sequences are widely expressed in all cells, but natural cytotoxicity receptors seem to be able to distinguish between the ones presented by tumoural and normal cells (Kruse et al., 2014).
NKp30 has been regarded as a possible target for future immunotherapies due to its importance in mediating the antitumour effects of NK cells in both solid tumours and haematological malignancies (Correia et al., 2011; Delahaye et al., 2011; Zhang, Wu, & Sentman, 2012).
4. NKp30 AS A MEDIATOR OF NK ACTIVITY
4.1. The structure of NKp30
The work of Pende et al. (1999), in which NKp30 was first identified, demonstrated the importance of this receptor in the lysis of several tumour cell lines. More recently, its structure has been obtained by X‐ray crystallography, revealing features distinct from the other known natural cytotoxicity receptors (Joyce et al., 2011).
The ectodomain of NKp30 contains an Ig‐like fold, connected to a transmembrane α‐helix via a short stalk domain. The Ig‐like domain is composed of eight β‐strands, forming two antiparallel β‐sheets, linked by a disulphide bond between Cys39 and Cys108 (Figure 3) (Joyce et al., 2011; Li et al., 2011; Pende et al., 1999).
FIGURE 3.
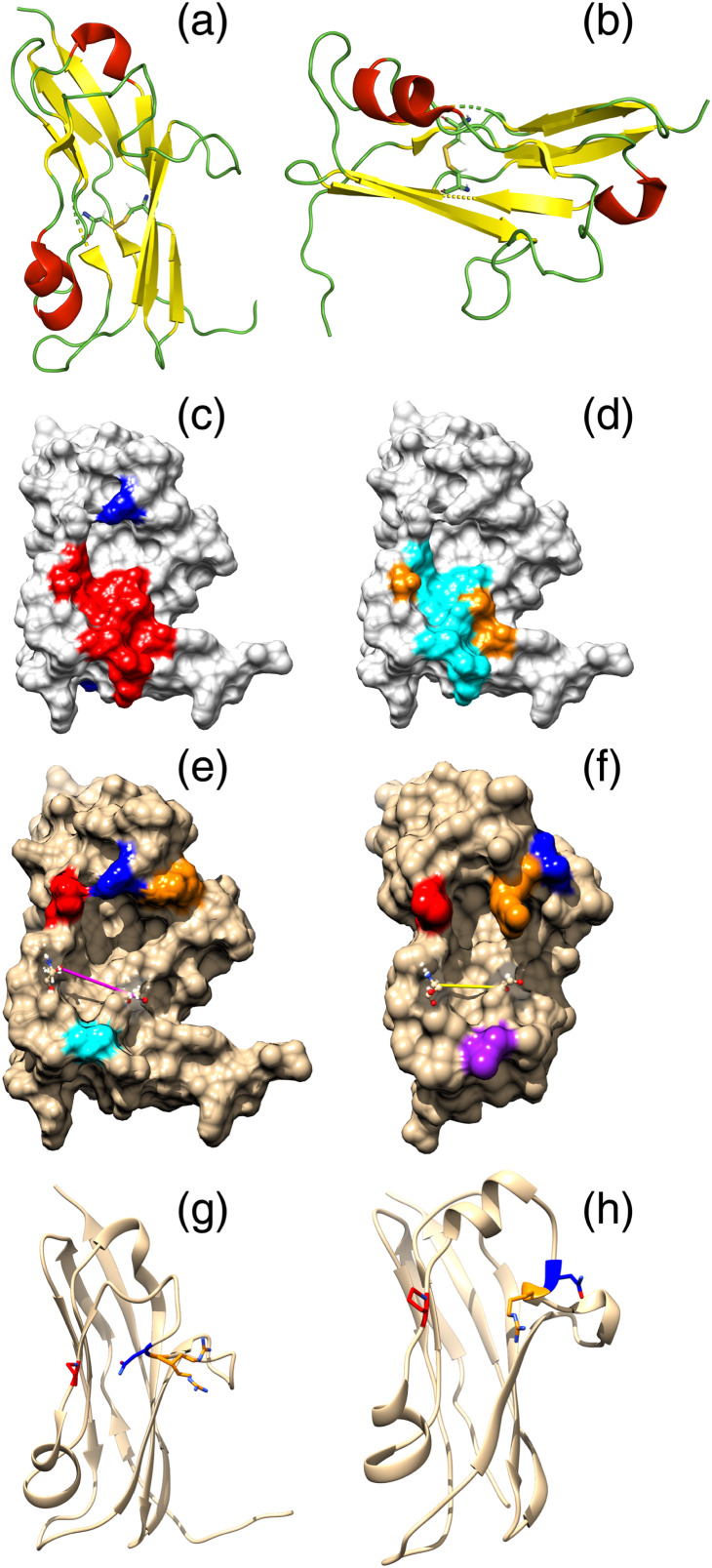
Structure representation of NKp30. The two antiparallel β‐sheets (in yellow) and the two α‐helixes (in red) are shown in opposing views (a, b). The two β‐sheets are bridged by a disulphide bond between residues Cys39 and Cys108. The stalk domain and the transmembrane α‐helix are not represented. The NKp30 molecular surface is also shown, with (a) B7‐H6‐contacting residues (red) and N‐glycosylation sites (blue) highlighted. Only one possibly N‐glycosylated residue (Asp68) is located near the identified binding site, which comprises residues Ile50, Gly51, Ser52, Val53, Leu80, Ser82, Phe85, Leu86, Glu111, Leu113 and Gly114, marked in red. (b) B7‐H6‐contacting residues differentiated by type of amino acid residue: polar residues (yellow) and hydrophobic residues (cyan). Image generated with PyMOL (Schrödinger, LLC, 2015) from the published X‐ray structure of unbound NKp30 (PDB ID: 3NOI) (Joyce et al., 2011). NKp30 structure representation in its unbound (c, e, g) and B7‐H6‐bound forms (d, f, h) are also shown. Isoleucine 50 is labelled as a positioning reference. Conformation differences between the two forms are observed in the distance between residues 52 and 82, labelled in yellow, which differs by about 1 Å and in the relative positions of arginine 67 and asparagine 68. The loop composed by arginine 67 (blue) and asparagine 68 (orange) seems to bend upon binding. Asparagine 68 moves away from proline 79 (in red) and arginine 67 bends down towards the pocket, approaching valine 53 (not highlighted). Images were generated from the published X‐ray structures of unbound NKp30 (PDB 3NOI) and B7‐H6‐bound NKp30 (PDB ID: 3PV6) (Li, Wang, & Mariuzza, 2011)
Although initially overlooked, the stalk domain has been shown to play an important role in the receptor activity, as it increases ligand‐binding affinity (Alter et al., 2011). Originally thought to include five to six residues (Pende et al., 1999), the stalk domain considered here is a longer 15‐residue domain (129KEHPQLGAGTVLLLR143) (Memmer et al., 2016). This domain is very sensitive to point mutations that lead to impaired binding and activity. The last residue of this domain is a cationic arginine. Arg143 is located on the membrane extracellular interface, but upon ligand binding, it is translocated into the membrane, strengthening the interaction with the CD3ζ and FCεRIγ Asp residues (Li et al., 2011; Memmer et al., 2016).
The initial structural elucidation of NKp30 identifies it to be monomeric in the crystal phase, in agreement with its behaviour in size exclusion chromatography and on the cell surface, but later work identified it as a dimer (Joyce et al., 2011; Pende et al., 1999). However, the authors also point out that many similarities between NKp30 and the homodimeric NKG2D receptor suggest that NKp30 can exist as a homodimer in which the binding site is formed by the two identical subunits (Li et al., 2011).
NKp30's closest human homologues are CTLA‐4 (von Strandmann et al., 2006), a homodimeric member of the CD28 family and PD‐1, an immune checkpoint protein that guards against autoimmunity (Francisco, Sage, & Sharpe, 2010; Joyce et al., 2011); a simple homology modelling approach based on the known structure of CTLA‐4 originated a homodimeric NKp30 structure, in which the subunit interaction is dominated by a network of salt bridges and hydrogen bonds between Glu8, Arg10, Glu13, Arg106 and Glu110 of both monomers (von Strandmann et al., 2006). However, this dimer is formed by a top‐top approach of the subunits and is unlikely to be relevant in vivo. It has recently been shown that the formation of NKp30 ectodomain dimers leads to a higher affinity towards the B7‐H6 ligand. This ectodomain dimerization appears to be driven by the association of the stalk domains of both polypeptide chains (Kasahara et al., 2018).
Superposition of the NKp30 and the NKp44 structures revealed common features as well as distinct conformations. The core regions of NKp30 and NKp44 are quite similar, but significant differences are found at the interacting region between the two β‐sheets, which in the case of NKp30 presents six more H‐bonds. An additional α‐helix is found between strands in NKp30 that is absent in NKp44. Furthermore, NKp30 lacks an extensive positively charged region that is found in NKp44. Sequence homology is about 30% between NKp30 and NKp44, while there is no significant overlapping with NKp46 (Foster, Colonna, & Sun, 2003; Joyce et al., 2011; Li et al., 2011).
As previously mentioned, signal transduction occurs through the ITAM‐bearing adaptor proteins CD3ζ and FCεRIγ. Like the other natural cytotoxicity receptors, association with the adaptor proteins is promoted by opposing charge residues. NKp30 and NKp46 contain positively charged Arg residues that interact with Asp residues in CD3ζ and FCεRIγ. On the other hand, NKp44 contains positively charged Lys residues that interact with Asp residues in DAP12 (Koch et al., 2013). Interestingly, NKp30 has been shown to be expressed in several variants (NKp30a, NKp30b, NKp30c, NKp30d, NKp30e and NKp30f) resultant from alternative splicing. Cells transfected with the NKp30a and NKp30b variants were found to produce high amounts of IFN‐γ upon NKp30 engagement. Conversely, cells transfected with the NKp30c variant produced small amounts of IFN‐γ but high levels of IL‐10, the immunosuppressor IL. This variant seems to interact poorly with the CD3ζ adaptor protein, in contrast with the a and b variants, which could justify the dual immunomodulatory effect of NKp30 (Delahaye et al., 2011; Siewiera et al., 2015). Moreover, one work has demonstrated that expression of higher levels of the c variant is directly correlated with poor prognosis in gastrointestinal tumour patients (Delahaye et al., 2011).
The role of the positively charged Arg143 residue in the end of the stalk domain in the association of the receptor with CD3ζ, together with the observation that CD3ζ increases cell membrane expression of NKp30, has led to the proposal that this receptor exists in two different assemblies—a signalling‐incompetent one and a ligand‐induced signalling‐competent complex (Memmer et al., 2016), which is consistent with the licencing hypothesis for NK cell response (Yokoyama & Kim, 2006).
It is also important to mention that NKp30 is present in the extracellular surface of the cell as a glycoprotein and that different glycosylation patterns affect the affinity towards its ligands. For instance, glycosylation of Asn42 increases the binding affinity of BAT‐3 but severely hampers the interaction with B7‐H6 (Hartmann et al., 2012). These structural features, which may vary between individuals, may be the cause of some pathologies and the reason why some patients develop aggressive malignancies even when their NK cell population is normal.
The NKp30 receptor is the product of the NCR3 gene expression, which can be transcribed as four exons that undergo alternative splicing (Delahaye et al., 2011; Tornroos, Hagerstrand, & Lindqvist, 2019). Up to this date, only isoforms NKp30a to NKp30c, resulting from alternative splicing of exon 4 and corresponding to different intracellular terminal domains, have been studied. NKp30a and NKp30b present a stronger association to the intracellular adaptor CD3ζ (and also to FcεRI‐γ), while NKp30c exhibits a lower association to both (Delahaye et al., 2011; von Strandmann et al., 2006).
NKp30a and NKp30b are typically associated with an immunostimulatory activity of NK cells and are associated with overall improved survival rate and with better prognosis in gastrointestinal stromal tumours (Delahaye et al., 2011; Fogel, Sun, Geurs, Carayannopoulos, & French, 2013) and in hepatocellular carcinoma (Jarahian et al., 2011), in contrast to a higher morbidity observed in patients that predominantly express isoform NKp30c. A similar trend is observed in the case of some viral infections, namely, in chronic hepatitis virus C infection (Tam, Martinson, Doligosa, & Klingemann, 2003), while no clear association between NKp30 expression status and outcome could be found in HIV‐1 patients (Rahmoune & Guest, 2018).
This isoform‐prognostic correlation is particularly evident in paediatric neuroblastoma patients, where the pattern of isoform expression is highly correlated to a 10‐year event‐free survival in multiple cohorts (Longo et al., 2012; Yang et al., 2019). However, this requires further investigation, as a later clinical study addressing paediatric neuroblastoma response to imatinib treatment failed to identify any correlation between NKp30 isoforms and clinical response (Phillips, Romeo, Bitsaktsis, & Sabatino, 2016).
While isoforms NKp30a, NKp30b and NKp30c present a V‐type Ig‐like extracellular domain, splice variants NKp30d, NKp30e and NKp30f encode a different C‐type Ig extracellular domain, coming from an exon 2 alternative splicing, with amino acid residues 66 to 90 being absent (Corry, Ampel, Christian, Locksley, & Galgiani, 1996; Neville & Campbell, 1999; Tornroos et al., 2019; von Strandmann et al., 2006). The intracellular domains, resulting from exon 4 expression, are shared between isoforms a and e, between isoforms b and d, and between isoforms c and f (Tornroos et al., 2019). A seventh, computer‐generated, isoform that matches experimental evidence at protein level, but corresponds only to the extracellular Ig domain of NKp30, has been proposed (Uniprot Accession Number A0A0G2JKT7) but has not been confirmed yet.
4.2. NKp30 polymorphisms in disease
The outcome variability associated with immunotherapy regimens is most likely of multifactorial origin and a contributing factor is the variability of the immune cell receptors involved in recognition and signalling. A large number of single nucleotide polymorphisms of the NKp30‐coding NCR3 gene have been identified, but only a very small number of them have been studied, particularly regarding population resistance to parasite infection (Berrien‐Elliott, Wagner, & Fehniger, 2015; Ghofrani, Lucar, Dugan, Reeves, & Jost, 2019).
A susceptibility locus for mild malaria, a major cause of morbidity and mortality in many developing countries, is located in the MHC region in chromosome 6p21, where TNF polymorphisms have been associated with mild malaria. The NCR3 gene is located just 15 kb distal to TNF and early studies have shown that the NCR3 ‐ 412G>C single nucleotide polymorphism is associated with increased risk and frequency of mild malaria (Ghofrani et al., 2019), when compared with the standard NCR3 −412G allele. A later study confirmed this observation and also showed that this association only occurs for mild malaria cases and not for severe malaria cases (Godoy‐Ramirez, Franck, Mahdavifar, Andersson, & Gaines, 2004). Noticeably, STAT4, a transcription factor essential for IL‐12‐mediated cytotoxicity and IFN‐γ production in mouse and human NK cells, binds this promoter region with higher affinity for the G allele (De Falco et al., 2017). In agreement with this, purified NK cells were found to lyse Plasmodium falciparum‐parasitized erythrocytes, through a direct interaction of NKp30 with P. falciparum erythrocyte membrane protein‐1 (PfEMP‐1) (Mavoungou, Held, Mewono, & Kremsner, 2007). A more recent study with healthy human volunteers found that parasitaemia levels and NK cell activation follow the same trend during the first 18 days post‐challenge by the bites of five P. falciparum 3D7 strain‐infected Anopheles mosquitoes, in what is the first study identifying the role of the NKp30 protein against P. falciparum (Katial, Sachanandani, Pinney, & Lieberman, 1998).
When considering the role of NKp30 on Trypanosoma cruzi infections, Hermann et al. found that cord blood NK cells from newborns congenitally infected with T. cruzi had a reduced expression of NKp30, which could be due to earlier NK cell activation or to NK cell activity down‐regulation by T. cruzi itself (Hermann et al., 2006). Later work has shown that NK cells were activated in utero, during the first exposure to the parasite and that the foetus response to T. cruzi is an adult‐like one, based on IFN‐γ production by CD8+ T cells and through an IL‐12‐dependent monocyte pathway (Berrien‐Elliott et al., 2015; Fassy, Tsalkitzi, Goncalves‐Maia, & Braud, 2017; Lapteva et al., 2012; Porrata, Inwards, Lacy, & Markovic, 2001).
Two other NKp30 single nucleotide polymorphisms in the promoter region, −201G>A and −163G>C, have been studied in healthy Japanese subjects, but no transcription binding factors are known at these sites; a second set of single nucleotide polymorphisms, c.111G>A and c.156C>T, were found in the same population, but these are synonymous substitutions and so the phenotype remained unchanged (Sato et al., 2001).
4.3. Ligands for NKp30
The NKp30 receptor may be engaged by several ligand types. Interestingly, and even though NKp30 is considered an activating receptor, some ligands have been associated with the opposite response. This is the case of poxvirus haemagglutinin (HA) that was found to bind NKp30, acting as antagonist. On the other hand, these same antigens are able to trigger the NKp46‐mediated response (Jarahian et al., 2011). Human cytomegalovirus tegument protein pp65 (HCMV pp65) is also able to block the triggering signal of NKp30 engaging. This protein interacts directly and specifically with NKp30 without blocking the interaction of this receptor with its activating ligands. Despite being able to form NKp30‐activating ligand complexes, the killing action of NK cells is suppressed by pp65, which suggests a downstream effect of this protein on the stimulatory signal transduction. It was demonstrated by Arnon et al. (2005) that pp65 disrupts the interaction of NKp30 with the adaptor protein CD3ζ, directly reducing the activation state of NK cells. The other known ligands for NKp30 have activation effects and their engaging results in cytokine production and perforin and granzyme release. For instance, it was found that molecules expressed on the surface of P. falciparum‐infected erythrocytes can be recognized by NKp30 and NKp46. The P. falciparum erythrocyte membrane protein 1 (PfEMP‐1) was identified as a strong activating ligand of NKp30 and NKp46 that, upon engagement, results in the lysis of the infected cells (Mavoungou et al., 2007).
Despite the importance of these virus‐ and parasite‐infected cell destruction activities, researchers have focused primarily on NK cell tumour targeting. One crucial finding stimulating this research field was the discovery of heparin and heparan sulphates as activating ligands for natural cytotoxicity receptors. An initial research paper described, in 2004, that membrane‐associated heparan sulphate proteoglycans were involved in the recognition of cellular targets by NK cells. In fact, the NKp44 natural cytotoxicity receptor bears an extensive positively charged surface that may constitute a binding site for sulphated carbohydrate structures (Bloushtain et al., 2004). Heparin and heparan sulphate sequences have formal negative charges in their sulphate groups that would account for strong interactions. Binding specificities of NKp30, NKp44 and NKp46 to these carbohydrates were further investigated, revealing that different receptors bind different monomer sequences (Hecht et al., 2009). However, since NKp30 does not possess a positively charged region, contrary to NKp44 and, as stated before, there is no structural homology between the receptors, the existence of other binding sites in the receptors or the cooperation of ligands to induce the activation response seems plausible. In fact, some evidence indicates that heparan sulphates are able to bind natural cytotoxicity receptors but unable to induce cytolysis on their own. On the other hand, the diversity of heparan sulphate molecules with different sequences and chemical moieties may account for the inconsistent results that have been presented, as these sequences might also interact with inhibitory receptors (Hershkovitz et al., 2008). Besides the wide diversity of heparan sulphate sequences, it is also worth to mention that different natural cytotoxicity receptor glycosylation states may play an important role in the binding of different molecules, especially large molecules that assume secondary and tertiary conformations, like polysaccharides (Brusilovsky, Radinsky, Yossef, Campbell, & Porgador, 2014; Hershkovitz et al., 2008).
Other molecules expressed by tumour cells that act as activating ligands of NKp30 are the BAT3 and B7‐H6 proteins, as seen before. BAT3 (HLA‐B‐associated transcript 3, also identified as BAG6, BCL2‐associated athanogene 6) is a protein chaperone that becomes overexpressed by tumour cells in response to stress signals. BAT3 transcription is strongly elevated in various tumour cell lines (Marchesi et al., 2013). The protein has been shown to be secreted by heat‐shocked 293T kidney cells and HeLa cells (Pogge von Strandmann et al., 2007) and soluble BAT3 has been identified in the serum of Hodgkin lymphoma and chronic lymphocyte leukaemia patients (Reiners et al., 2013; Reiners et al., 2013). BAT3 promotes exosome‐mediated metastization in melanoma patients (Daßler‐Plenker et al., 2016; Schuldner et al., 2019) and has been identified as a prognosis‐worth biomarker in hepatocellular carcinoma (Zhang, Wang, Liu, & Li, 2019), gastrointestinal stromal tumour (Rusakiewicz et al., 2017), and non‐small‐cell (Etokebe et al., 2015) and adenocarcinoma (Renieri et al., 2014) lung cancer patients.
Under these conditions, the protein is translocated to the cell membrane and released from the cells, engaging the NKp30 receptor. Interestingly, cytokine release is only experimentally attained using tumour‐released BAT3 and no activation of NK cells is observed when a recombinant protein is used. This suggests that BAT3 only induces the cell response when it is part of vesicles, such as exosomes (Pogge von Strandmann et al., 2007). More recently, the binding site of NKp30 on BAT3 has been localized to a BAT3686–936 sequence. Recombinant BAT3686–936 alone does not engage NKp30 but is able to form homodimers that inhibit NKp30‐mediated signalling, confirming that the activity of BAT3 depends on how it is presented to NK cells. The differences between soluble and membrane‐associated BAT3 activities are clearly due to different oligomerization states and consequently distinct affinities to the receptor (Binici & Koch, 2014; Marchesi et al., 2013), a regulatory motif often observed in NK cell activity.
On the other hand, the other known tumour‐derived ligand of NKp30, B7‐H6, is able to induce cytokine release both in its soluble and membrane‐bound forms (Gutierrez‐Franco et al., 2018; Phillips et al., 2016). Membrane expression of B7‐H6 has been widely implicated in the sensitivity of cancer cells to NK cell‐mediated cytolysis (Cao et al., 2015). Conversely, NK cells chronically stimulated with soluble forms of B7‐H6 have been shown to down‐regulate the expression of NKp30, contributing to tumour immune escape (Pesce et al., 2015). It seems that, besides the normal regulation mechanisms presented by NK cells, there is yet another layer of regulation dictating the activity of NK cells. Nevertheless, among all the known ligands for NKp30, B7‐H6 has been regarded as a possible target for novel cancer therapies. The basis of the B7‐H6‐NKp30 recognition has been widely studied in several works, in an effort to highlight the molecular mechanisms behind the strong NK cell response elicited by tumours presenting B7‐H6. Protein glycosylation also plays a role, as it has been shown that single, double and triple mutations, where residues Asn42, Asn68 or Asn121 were mutated to Gln, lead to increased K D values for B7‐H6 binding, with the exception of the Asn121Gln, which practically does not affect K D values (Hartmann et al., 2012).
Tumour cells also express galectin‐3 (Gal‐3), a β‐galactoside binding protein that specifically binds NK cells via the NKp30 receptor. Gal‐3 is highly expressed in many types of cancer cells in different locations, namely, within the nucleus, on the cell surface and in the extracellular compartment (Lahm et al., 2001; Perillo, Marcus, & Baum, 1998; Wang et al., 2014). While intracellularly Gal‐3 acts a cell cycle control agent, promoting tumour growth metastasis and protecting from apoptosis, extracellular Gal‐3 promotes cell adhesion and invasion and has been studied as a biomarker for early‐stage cancer detection (Hara et al., 2020; Salimi et al., 2016; Wang et al., 2014). Gal‐3 has also been recently shown to mediate immune escape by interacting with the NKp30 ectodomain N‐glycosylated Asn42 and Asn121 sites. This interaction interferes with ligand binding (Salimi et al., 2016; Wang et al., 2014) and reduces the diffusion of IFN‐γ by binding the extracellular matrix glycans and glycosylated cytokines, thus contributing to promoting tumour survival (Gordon‐Alonso, Hirsch, Wildmann, & van der Bruggen, 2017).
It has also been demonstrated that immature dendritic cells can induce the activity of NK cells via NKp30 signalling, promoting their expansion and increasing their cytotoxicity to levels comparable to those obtained with IL‐2 (Ferlazzo et al., 2002). The dendritic cell‐primed NK cells were found to promptly kill immature dendritic cells (Ferlazzo et al., 2002) that accumulate in the tumour micro‐environment, which hampers their differentiation. As these immature dendritic cells have impaired antigen‐presenting function, their accumulation on the lesion site contributes to immune evasion of cancer cells (Ma, Shurin, Peiyuan, & Shurin, 2013). By killing these cells, while sparing mature antigen‐presenting dendritic cells, NK cells promote a concerted immune response in the tumour environment (Ferlazzo et al., 2004; Harizi, 2013).
4.4. NKp30's B7‐H6 binding site
Despite the relevance of NKp30 receptor immunologically, the molecular details of ligand binding and receptor recognition have not been studied in detail for ligands other than B7‐H6. To understand the basis of tumour recognition by NKp30, several authors have tried to accurately describe the interactions between NKp30 and its natural ligands. As there is no significant sequence homology between the known NKp30 ligands, or similarity in their predicted structure folding, it is not possible to determine a plausible binding site by identifying common features in ligand structures. Site‐directed mutagenesis was used to alter the surface charge and/or hydrophobicity of specific areas of NKp30, allowing the identification of a relatively small region where at least one of the known ligands—B7‐H6—interacts (Joyce et al., 2011). The B7‐H6 binding region of NKp30 is comprised mostly of non‐polar amino acid residues that account for the hydrophobic interactions between the two proteins. Two hydrogen bonds are also involved, as well as a salt bridge. B7‐H6 affinity is extremely sensitive to changes in the protein structure, as the interactions of the two proteins are restricted to a small region of NKp30 (residues Ile50–Val53, Leu80, Ser82, Phe85, Leu86, Glu111, Leu113 and Gly114), as seen in Figure 3. In fact, the selective mutation of a few residues results in marked changes in the receptor affinity towards the ligand. Ser52Arg and Ser52Ala mutants display a 150‐fold and 14‐fold affinity reduction, respectively and Ile50Ala mutants display a 14‐fold affinity reduction. Also the Phe85Ala and Leu86Ala mutants display a 53‐fold reduction in the binding affinity (Li et al., 2011).
The substitution of Ser52 by an Arg residue has a tremendous impact on the binding affinity of B7‐H6. In this mutation, a hydrophobic amino acid residue is replaced by a positively charged one, which completely changes the polarity of the protein surface, dramatically increasing the dissociation constant (K D). Replacement of Ser52 with another hydrophobic amino acid (e.g. Ala) has a much more reduced impact on the binding of B7‐H6. Replacement of Phe85 with a non‐aromatic amino acid residue causes an accentuated decrease in the pocket hydrophobicity. The B7‐H6 region that interacts with this portion of NKp30 comprises the Thr59 and Pro128 residues, making 7 van der Waals contacts with NKp30, which account for 44% of all contacts between the two proteins (Li et al., 2011). This region of B7‐H6 forms a projection that seems to bind at the groove formed between Ser52 and Ser82 of NKp30 (see Figure 3), which may be a key feature involved in triggering signal transduction by NKp30.
Further work yielded the X‐ray structure of NKp30 bound to B7‐H6, providing important information about the interaction of these two molecules. Interestingly, B7‐H6 interacts with just 11 NKp30 residues, mainly through hydrophobic interactions. The three potentially N‐glycosylated residues identified by other authors (Asn residues 42, 68 and 121) are located outside the interface and therefore should not interfere with binding (Figure 3) (Li et al., 2011). However, it was argued that different glycosylation patterns may affect the overall three‐dimensional (3D) structure of the protein, increasing or decreasing binding affinity (Hartmann et al., 2012).
Marked NKp30 conformational changes occur upon B7‐H6 binding, as observed by comparison of the two available structures (bound and unbound, Figure 3). A key change is the reduction of the distance between residues 52 and 82, which define the ridge where B7‐H6 docks. Moreover, the loop composed by Arg67 and Asn68, located above the ridge, seems to rotate, pushing Asn68 backwards and pulling the Arg67 residue down towards the centre of the ridge. Although the available structures do not allow the identification of an H bond, the NH moiety of the side chain of Arg67 approaches the C=O of Val53 at about 4 Å.
Interestingly, Asn68 is one of the three identified glycosylation acceptor sites. Although all these sites are located outside the binding site, the glycosylation of Asn42 and Asn68 is essential for B7‐H6 binding; their glycosylation is likely to induce subtle NKp30 conformational changes, helping shape the ligand‐binding pocket (Hartmann et al., 2012). On the other hand, BAT‐3 binding depends essentially on Asn68 glycosylation, whereas Asn42 and Asn121 have a lower impact (Hartmann et al., 2012). This indicates that Asn68 is critical for correct ligand binding and subsequent signal transduction.
Although no experimental information exists regarding these conformational changes, it is clear that some degree of modification is required for NKp30‐based triggering of NK cells. One particular work demonstrated that, upon binding of B7‐H6, conformational changes in NKp30 enable the translocation of Arg143 to a position deeper in the phospholipid bilayer. As a result, the positively charged arginine residue aligns with the negatively charged aspartate in CD3ζ or FCεRIγ adaptor proteins, which triggers the NK cell response (Memmer et al., 2016).
4.5. Targeting NKp30
B7‐H6 is expressed by approximately 20% of available human tumour cell lines and by some primary human tumours (Brandt et al., 2009). It has been widely demonstrated that B7‐H6 expression is directly correlated with susceptibility to NK‐killing, as engaging of NKp30 by this protein triggers a strong cytolytic response. It has also been suggested that some cancer therapies may increase the stress levels on tumour cells and up‐regulate the expression of B7‐H6, increasing the susceptibility to NK cells (Cao et al., 2015). Moreover, it has been demonstrated that artificially coating cells with B7‐H6 promotes NK cell cytotoxicity, providing a proof of concept that NKp30 engagement may represent a new strategy in cancer therapy (Kellner et al., 2012). One recent work also noted that the consistent expression of NKp30 on NK cells makes it a very relevant marker for the detection of tumour‐infiltrating NK cells (Shaffer, Gambhir, Aalipour, & Schurch, 2020), which further supports the suitability of this receptor as a pharmacological target in the development of new therapies.
One example of this strategy focused on triggering the response of NK cells with B7‐H6‐derived small peptides. In this approach, the peptide sequence present in the projection of B7‐H6 that binds to the ridge of NKp30 was reproduced as a soluble fragment. Treatment of NK cells with this molecule induced the release of TNF‐α, proving that NKp30 may be targeted using small peptides. This work, however, failed to achieve a complete response of NK cells, as no release of IFN‐γ was triggered by the designed peptides and no data regarding the cytolytic activity of NK cells were reported (Phillips et al., 2016).
Another approach described in the literature reports the use of fusion proteins bearing B7‐H6 (Peipp et al., 2015). In this work, B7‐H6 was fused to a single‐chain fragment variable (scFv) targeting the human EGF receptor 2 (HER2), which is displayed by various solid tumours. As a result, the activity of NK cells towards cancer cells was increased by the bispecific fusion protein, once again demonstrating the suitability of NKp30 as target.
Despite the advances in NK cell characterization and in the identification of the triggering mechanisms, few therapies using NK cells are available for cancer treatment. In some ongoing clinical trials, autologous NK cells are expanded ex vivo and reinfused after stimulation with cytokines, namely IL‐2. In another approach, allogenic NK cells are directly administered to the patients and an immortalized NK cell line (NK‐92) is used in some instances. However, most of the ongoing trials rely on the use of specific antibodies aimed at triggering the cytolytic responses of NK cells against cancer cells, by targeting NK cell receptors and cancer cell markers. Some of the therapies were found to be effective and to significantly increase the life quality of the patients, while others presented serious side effects (Dianat‐Moghadam, Rokni, Marofi, Panahi, & Yousefi, 2018).
Only a few works have focused on NKp30 as a target for the activation of NK cells and as a possible route towards the development of new immuno‐oncology therapies. With all the benefits of immunotherapies and as part of the personalized medicine approaches, it is expected that, in the near future, new drugs and treatments focused on harvesting the power of the immune system through NK cells will be developed.
5. CONCLUSION
The immune activity of NK cells is mediated by a large number of cell surface receptors that recognize different ligands and mediate different signalling pathways. NK cells are one of the arms of the innate immune system and are particularly interesting because they are able to differentiate between self and non‐self cells, not by the absence of MHC‐I‐presented molecules but rather by their loss. This finely regulated activity comes from a balance between activating and inhibiting receptors that constantly sense the environment around the cell and generate a response that considers all the stimuli present.
NKp30 is a natural cytotoxicity trigger and its role on NK cell response to several stimuli is well explored, particularly in tumour settings. The usage of efficient NKp30 agonists would constitute an invaluable tool in immuno‐oncology because it would simultaneously make use of both the specificity and the potency conferred by this receptor.
NKp30 can be, in the future, targeted with antibodies in immunotherapy regimens. However, due to the inherent characteristics of the process, antibody production and usage is far from being a straightforward option when compared with the use of smaller and more affordable drugs. The development of specific and potent drugs, targeting the binding site of NKp30 and generating the same response, is therefore of utmost importance.
The structure of NKp30 has been resolved both in its free‐ and ligand‐bound forms, and the ligand‐binding region is well characterized. However, important aspects of NKp30‐mediated NK cell activation arise not only from direct ligand binding but also from other affected residues. All structural data available in the literature, reviewed herein, indicate that the design of specific and effective drugs requires not only binding to the adequate site but also the ability to replicate ligand‐induced conformational changes, in particular Arg67 displacement.
Targeting NKp30 with small molecules, such as peptides or low MW ligands, should be considered as a new strategy to trigger immune responses against cancer prompted by NK cells. NKp30‐targeted therapies have been proven efficient, as demonstrated by CAR‐T cells expressing chimeric NKp30 receptors that were found able to destroy B7‐H6+ cells (Zhang et al., 2012). This strategy, however, presents major drawbacks regarding the production and cost of CAR‐T cells. The work of Phillips et al. (2016) provided important insights regarding the activation of NK cell response with small peptides, but more affordable and comprehensive strategies should be developed in the future as part of a new branch of cancer immunotherapy.
5.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY http://www.guidetopharmacology.org and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
CONFLICT OF INTEREST
The authors declare that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGEMENTS
This work was partially funded by research grants from Fundação para a Ciência e a Tecnologia (FCT) (PTDC/QUI‐QAN/32242/2017) and from FCT and COMPETE2020 actions (SAICTPAC/0019/2015) and by a Terry Fox grant from Liga Portuguesa Contra o Cancro (LPCC/NRS‐Terry Fox grant 2015‐2017). P.F.P. was an FCT grantee (SFRH/BD/110945/2015). Centro de Química Estrutural is funded by FCT (UIDB/00100/2020).
Pinheiro PF, Justino GC, Marques MM. NKp30 ‐ A prospective target for new cancer immunotherapy strategies. Br J Pharmacol. 2020;177:4563–4580. 10.1111/bph.15222
REFERENCES
- Abakushina, E. V. (2015). The technique of flow cytometry in evaluation of NK‐cells and their activity. Klinicheskaia Laboratornaia Diagnostika, 60, 37–44. [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , … Southan, C . (2019). The Concise Guide to PHARMACOLOGY 2019/20: Introduction and other protein targets. British Journal of Pharmacology, 176, S1–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter, G. , Jost, S. , Rihn, S. , Reyor, L. L. , Nolan, B. E. , Ghebremichael, M. , … Lauer, G. M. (2011). Reduced frequencies of NKp30+NKp46+, CD161+, and NKG2D+ NK cells in acute HCV infection may predict viral clearance. Journal of Hepatology, 55, 278–288. 10.1016/j.jhep.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, T. I. , Achdout, H. , Levi, O. , Markel, G. , Saleh, N. , Katz, G. , … Mandelboim, O. (2005). Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nature Immunology, 6, 515–523. 10.1038/ni1190 [DOI] [PubMed] [Google Scholar]
- Asmana Ningrum, R. (2014). Human interferon alpha‐2b: A therapeutic protein for cancer treatment. Scientifica (Cairo), 2014, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow, A. D. , Martin, C. J. , & Colonna, M. (2019). The natural cytotoxicity receptors in health and disease. Frontiers in Immunology, 10, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, Y. , Bauer, B. , & Steinle, A. (2013). Modulation of NK cell function by genetically coupled C‐type lectin‐like receptor/ligand pairs encoded in the human natural killer gene complex. Frontiers in Immunology, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, D. (2014). Active and passive immunization for cancer. Hum Vacc Immunother, 10, 2123–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrien‐Elliott, M. M. , Wagner, J. A. , & Fehniger, T. A. (2015). Human cytokine‐induced memory‐like natural killer cells. Journal of Innate Immunity, 7, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biassoni, R. (2008). Natural killer cell receptors. Advances in Experimental Medicine and Biology, 640, 35–52. 10.1007/978-0-387-09789-3_4 [DOI] [PubMed] [Google Scholar]
- Biassoni, R. , Cantoni, C. , Pende, D. , Sivori, S. , Parolini, S. , Vitale, M. , … Moretta, A . (2001). Human natural killer cell receptors and co‐receptors. Immunological Reviews, 181, 203–214. [DOI] [PubMed] [Google Scholar]
- Binici, J. , & Koch, J. (2014). BAG‐6, a jack of all trades in health and disease. Cellular and Molecular Life Sciences, 71, 1829–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloushtain, N. , Qimron, U. , Bar‐Ilan, A. , Hershkovitz, O. , Gazit, R. , Fima, E. , … Porgador, A. (2004). Membrane‐associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. Journal of Immunology, 173, 2392–2401. 10.4049/jimmunol.173.4.2392 [DOI] [PubMed] [Google Scholar]
- Boivin, W. A. , Cooper, D. M. , Hiebert, P. R. , & Granville, D. J. (2009). Intracellular versus extracellular granzyme B in immunity and disease: Challenging the dogma. Laboratory Investigation, 89, 1195–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher, J. P. , Bonavita, E. , Chakravarty, P. , Blees, H. , Cabeza‐Cabrerizo, M. , Sammicheli, S. , … e Sousa, C. R . (2018). NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell, 172, 1022–1037. 10.1016/j.cell.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington, J. C. , & Sun, P. D. (2002). A structural perspective on MHC class I recognition by killer cell immunoglobulin‐like receptors. Molecular Immunology, 38, 1007–1021. [DOI] [PubMed] [Google Scholar]
- Brandt, C. S. , Baratin, M. , Yi, E. C. , Kennedy, J. , Gao, Z. , Fox, B. , … Levin, S. D. (2009). The B7 family member B7‐H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. The Journal of Experimental Medicine, 206, 1495–1503. 10.1084/jem.20090681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondz, B. D. (1964). In vivo and in vitro studies on the cytotoxic effect of cellular and humoral antibodies. II. Relation of immune lymphocytes to homologous normal and tumor tissues. Voprosy Onkologii, 10, 64–71. [PubMed] [Google Scholar]
- Brusilovsky, M. , Radinsky, O. , Yossef, R. , Campbell, K. S. , & Porgador, A. (2014). Carbohydrate‐mediated modulation of NK cell receptor function: Structural and functional influences of heparan sulfate moieties expressed on NK cell surface. Frontiers in Oncology, 4, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri, M. A. (2008). Human natural killer cells. Blood, 112, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, G. , Wang, J. , Zheng, X. , Wei, H. , Tian, Z. , & Sun, R. (2015). Tumor therapeutics work as stress inducers to enhance tumor sensitivity to natural killer (NK) cell cytolysis by up‐regulating NKp30 ligand B7‐H6. The Journal of Biological Chemistry, 290, 29964–29973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Bustamante, P. , de Boer, R. J. , & Kesmir, C. (2018). Specificity of inhibitory KIRs enables NK cells to detect changes in an altered peptide environment. Immunogenetics, 70, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, J. E. , Sandoval‐Sus, J. D. , Bole, J. , Rassenti, L. , & Kipps, T. J. (2008). Rituximab in combination with high‐dose methylprednisolone for the treatment of fludarabine refractory high‐risk chronic lymphocytic leukemia. Leukemia, 22, 2048–2053. 10.1038/leu.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M. , Fuchs, A. , Vermi, W. , Facchetti, F. , Otero, K. , Lennerz, J. K. , … Colonna, M . (2009). A human natural killer cell subset provides an innate source of IL‐22 for mucosal immunity. Nature, 457, 722–725. 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M. , Otero, K. , & Colonna, M. (2010). Expansion of human NK‐22 cells with IL‐7, IL‐2, and IL‐1β reveals intrinsic functional plasticity. Proceedings of the National Academy of Sciences of the United States of America, 107, 10961–10966. 10.1073/pnas.1005641107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, A. , Hong, D. L. , Atzberger, A. , Kollnberger, S. , Filer, A. D. , Buckley, C. D. , … Bowness, P. (2007). CD56bright human NK cells differentiate into CD56dim cells: Role of contact with peripheral fibroblasts. Journal of Immunology, 179, 89–94. 10.4049/jimmunol.179.1.89 [DOI] [PubMed] [Google Scholar]
- Charrier, M. , Mezquita, L. , Lueza, B. , Dupraz, L. , Planchard, D. , Remon, J. , … Chaput, N. (2019). Circulating innate immune markers and outcomes in treatment‐naive advanced non‐small cell lung cancer patients. European Journal of Cancer, 108, 88–96. 10.1016/j.ejca.2018.12.017 [DOI] [PubMed] [Google Scholar]
- Cooper, M. A. , Fehniger, T. A. , Turner, S. C. , Chen, K. S. , Ghaheri, B. A. , Ghayur, T. , … Caligiuri, M. A . (2001). Human natural killer cells: A unique innate immunoregulatory role for the CD56bright subset. Blood, 97, 3146–3151. [DOI] [PubMed] [Google Scholar]
- Correia, D. V. , Fogli, M. , Hudspeth, K. , da Silva, M. G. , Mavilio, D. , & Silva‐Santos, B. (2011). Differentiation of human peripheral blood Vδ1+ T cells expressing the natural cytotoxicity receptor NKp30 for recognition of lymphoid leukemia cells. Blood, 118, 992–1001. [DOI] [PubMed] [Google Scholar]
- Correia, M. P. , Stojanovic, A. , Bauer, K. , Juraeva, D. , Tykocinski, L.‐O. , Lorenz, H.‐M. , … Cerwenka, A . (2018). Distinct human circulating NKp30+FcεRIγ+CD8+ T cell population exhibiting high natural killer‐like antitumor potential. P Natl Acad Sci USA, 115, E5980–E5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry, D. B. , Ampel, N. M. , Christian, L. , Locksley, R. M. , & Galgiani, J. N. (1996). Cytokine production by peripheral blood mononuclear cells in human coccidioidomycosis. The Journal of Infectious Diseases, 174, 440–443. [DOI] [PubMed] [Google Scholar]
- Cortez, V. S. , Ulland, T. K. , Cervantes‐Barragan, L. , Bando, J. K. , Robinette, M. L. , Wang, Q. , … Colonna, M. (2017). SMAD4 impedes the conversion of NK cells into ILC1‐like cells by curtailing non‐canonical TGF‐β signaling. Nature Immunology, 18, 995–1003. 10.1038/ni.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daßler‐Plenker, J. , Reiners, K. S. , van den Boorn, J. G. , Hansen, H. P. , Putschli, B. , Barnert, S. , … Coch, C. (2016). RIG‐I activation induces the release of extracellular vesicles with antitumor activity. Oncoimmunology, 5, e1219827–e1219827. 10.1080/2162402X.2016.1219827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco, G. , Terlizzi, M. , Sirignano, M. , Commodo, M. , D'Anna, A. , Aquino, R. P. , … Sorrentino, R . (2017). Human peripheral blood mononuclear cells (PBMCs) from smokers release higher levels of IL‐1‐like cytokines after exposure to combustion‐generated ultrafine particles. Scientific Reports, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal, M. R. , Abrams, J. , Bennett, B. , Figdor, C. G. , & de Vries, J. E. (1991). Interleukin 10 (IL‐10) inhibits cytokine synthesis by human monocytes: An autoregulatory role of IL‐10 produced by monocytes. The Journal of Experimental Medicine, 174, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal, M. R. , Haanen, J. , Spits, H. , Roncarolo, M. G. , te Velde, A. , Figdor, C. , … De Vries, J. E . (1991). Interleukin 10 (IL‐10) and viral IL‐10 strongly reduce antigen‐specific human T cell proliferation by diminishing the antigen‐presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. The Journal of Experimental Medicine, 174, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye, N. F. , Rusakiewicz, S. , Martins, I. , Menard, C. , Roux, S. , Lyonnet, L. , … Minard‐Colin, V . (2011). Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nature Medicine, 17, 700–707. [DOI] [PubMed] [Google Scholar]
- Deng, W. , Gowen, B. G. , Zhang, L. , Wang, L. , Lau, S. , Iannello, A. , … Raulet, D. H . (2015). A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science, 348, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita, V. T. , & Rosenberg, S. A. (2012). Two hundred years of cancer research. The New England Journal of Medicine, 366, 2207–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianat‐Moghadam, H. , Rokni, M. , Marofi, F. , Panahi, Y. , & Yousefi, M. (2018). Natural killer cell‐based immunotherapy: From transplantation toward targeting cancer stem cells. Journal of Cellular Physiology, 234, 259–273. [DOI] [PubMed] [Google Scholar]
- Diefenbach, A. , Colonna, M. , & Koyasu, S. (2014). Development, differentiation, and diversity of innate lymphoid cells. Immunity, 41, 354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybkaer, K. , Iqbal, J. , Zhou, G. , Geng, H. , Xiao, L. , Schmitz, A. , … Chan, W. C . (2007). Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: Gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics, 8, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich, J. , Mumm, J. B. , Chan, I. H. , LaFace, D. , Truong, H. , McClanahan, T. , … Oft, M. (2012). IL‐10 directly activates and expands tumor‐resident CD8+ T cells without de novo infiltration from secondary lymphoid organs. Cancer Research, 72, 3570–3581. 10.1158/0008-5472.CAN-12-0721 [DOI] [PubMed] [Google Scholar]
- Etokebe, G. E. , Zienolddiny, S. , Kupanovac, Z. , Enersen, M. , Balen, S. , Flego, V. , … Haugen, A . (2015). Association of the FAM46A gene VNTRs and BAG6 rs3117582 SNP with non small cell lung cancer (NSCLC) in Croatian and Norwegian populations. PLoS ONE, 10, e0122651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag, S. S. , Fehniger, T. A. , Ruggeri, L. , Velardi, A. , & Caligiuri, M. A. (2002). Natural killer cell receptors: New biology and insights into the graft‐versus‐leukemia effect. Blood, 100, 1935–1947. [DOI] [PubMed] [Google Scholar]
- Fassy, J. , Tsalkitzi, K. , Goncalves‐Maia, M. , & Braud, V. M. (2017). A real‐time cytotoxicity assay as an alternative to the standard chromium‐51 release assay for measurement of human NK and T cell cytotoxic activity. Current Protocols in Immunology, 118, 7.42.41–7.42.12. [DOI] [PubMed] [Google Scholar]
- Fauriat, C. , Just‐Landi, S. , Mallet, F. , Arnoulet, C. , Sainty, D. , Olive, D. , & Costello, R. T . (2007). Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood, 109, 323–330. 10.1182/blood-2005-08-027979 [DOI] [PubMed] [Google Scholar]
- Fellner, C. (2012). Ipilimumab (Yervoy) prolongs survival in advanced melanoma: Serious side effects and a hefty price tag may limit its use. Pharm Ther, 37, 503–530. [PMC free article] [PubMed] [Google Scholar]
- Ferlazzo, G. , Thomas, D. , Lin, S.‐L. , Goodman, K. , Morandi, B. , Muller, W. A. , … Münz, C . (2004). The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig‐like receptors and become cytolytic. Journal of Immunology, 172, 1455–1462. [DOI] [PubMed] [Google Scholar]
- Ferlazzo, G. , Tsang, M. L. , Moretta, L. , Melioli, G. , Steinman, R. M. , & Münz, C. (2002). Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. The Journal of Experimental Medicine, 195, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel, L. A. , Sun, M. M. , Geurs, T. L. , Carayannopoulos, L. N. , & French, A. R. (2013). Markers of nonselective and specific NK cell activation. Journal of Immunology, 190, 6269–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, C. E. , Colonna, M. , & Sun, P. D. (2003). Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. The Journal of Biological Chemistry, 278, 46081–46086. [DOI] [PubMed] [Google Scholar]
- Fousek, K. , & Ahmed, N. (2015). The evolution of T‐cell therapies for solid malignancies. Clinical Cancer Research, 21, 3384–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco, L. M. , Sage, P. T. , & Sharpe, A. H. (2010). The PD‐1 pathway in tolerance and autoimmunity. Immunological Reviews, 236, 219–242. 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, A. , Vermi, W. , Lee, J. S. , Lonardi, S. , Gilfillan, S. , Newberry, R. D. , … Colonna, M . (2013). Intraepithelial type 1 innate lymphoid cells are a unique subset of IL‐12‐ and IL‐15‐responsive IFN‐γ‐producing cells. Immunity, 38, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski, T. F. (2015). The next hurdle in cancer immunotherapy: Overcoming the non‐T‐cell‐inflamed tumor microenvironment. Seminars in Oncology, 42, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido, F. , Romero, I. , Aptsiauri, N. , & Garcia‐Lora, A. M. (2016). Generation of MHC class I diversity in primary tumors and selection of the malignant phenotype. International Journal of Cancer, 138, 271–280. [DOI] [PubMed] [Google Scholar]
- Ghofrani, J. , Lucar, O. , Dugan, H. , Reeves, R. K. , & Jost, S. (2019). Semaphorin 7A modulates cytokine‐induced memory‐like responses by human natural killer cells. European Journal of Immunology, 49, 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy‐Ramirez, K. , Franck, K. , Mahdavifar, S. , Andersson, L. , & Gaines, H. (2004). Optimum culture conditions for specific and nonspecific activation of whole blood and PBMC for intracellular cytokine assessment by flow cytometry. Journal of Immunological Methods, 292, 1–15. [DOI] [PubMed] [Google Scholar]
- Gordon‐Alonso, M. , Hirsch, T. , Wildmann, C. , & van der Bruggen, P. (2017). Galectin‐3 captures interferon‐gamma in the tumor matrix reducing chemokine gradient production and T‐cell tumor infiltration. Nature Communications, 8, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, G. A. , & Weiser, R. S. (1964). Homograft target cells: Specific destruction in vitro by contact interaction with immune macrophages. Science, 145, 1427–1429. [DOI] [PubMed] [Google Scholar]
- Greenberg, A. H. (1994). The origins of the NK cell, or a Canadian in King Ivan's court. Clinical and Investigative Medicine, 17, 626–631. [PubMed] [Google Scholar]
- Groh, V. , Wu, J. , Yee, C. , & Spies, T. (2002). Tumour‐derived soluble MIC ligands impair expression of NKG2D and T‐cell activation. Nature, 419, 734–738. [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Franco, J. , Hernandez‐Gutierrez, R. , Bueno‐Topete, M. R. , Haramati, J. , Navarro‐Hernandez, R. E. , Escarra‐Senmarti, M. , … del Toro‐Arreola, S. (2018). Characterization of B7H6, an endogenous ligand for the NK cell activating receptor NKp30, reveals the identity of two different soluble isoforms during normal human pregnancy. Immunobiology, 223, 57–63. 10.1016/j.imbio.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Halfteck, G. G. , Elboim, M. , Gur, C. , Achdout, H. , Ghadially, H. , & Mandelboim, O. (2009). Enhanced in vivo growth of lymphoma tumors in the absence of the NK‐activating receptor NKp46/NCR1. Journal of Immunology, 182, 2221–2230. [DOI] [PubMed] [Google Scholar]
- Hara, A. , Niwa, M. , Noguchi, K. , Kanayama, T. , Niwa, A. , Matsuo, M. , … Tomita, H . (2020). Galectin‐3 as a next‐generation biomarker for detecting early stage of various diseases. Biomolecules, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harizi, H. (2013). Reciprocal crosstalk between dendritic cells and natural killer cells under the effects of PGE2 in immunity and immunopathology. Cellular & Molecular Immunology, 10, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, J. , Tran, T. V. , Kaudeer, J. , Oberle, K. , Herrmann, J. , Quagliano, I. , … Wollscheid, B . (2012). The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. The Journal of Biological Chemistry, 287, 31527–31539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A. E. , & Cheung, M. C. (2019). CAR T‐cells: Costs, comparisons, and commentary. Journal of Medical Economics, 22, 613–615. [DOI] [PubMed] [Google Scholar]
- Hecht, M. L. , Rosental, B. , Horlacher, T. , Hershkovitz, O. , De Paz, J. L. , Noti, C. , … Seeberger, P. H . (2009). Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. Journal of Proteome Research, 8, 712–720. 10.1021/pr800747c [DOI] [PubMed] [Google Scholar]
- Hellstrom, I. E. , Hellstrom, K. E. , Pierce, G. E. , & Bill, A. H. (1968). Demonstration of cell‐bound and humoral immunity against neuroblastoma cells. Proc Natl Acad Sci USA, 60, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, E. , Alonso‐Vega, C. , Berthe, A. , Truyens, C. , Flores, A. , Cordova, M. , … Carlier, Yves (2006). Human Congenital Infection With Trypanosoma cruzi Induces Phenotypic and Functional Modifications of Cord Blood NK Cells. Pediatric Research, 60(1), 38–43. 10.1203/01.pdr.0000220335.05588.ea [DOI] [PubMed] [Google Scholar]
- Hershkovitz, O. , Jarahian, M. , Zilka, A. , Bar‐Ilan, A. , Landau, G. , Jivov, S. , … Porgador, A. (2008). Altered glycosylation of recombinant NKp30 hampers binding to heparan sulfate: A lesson for the use of recombinant immunoreceptors as an immunological tool. Glycobiology, 18, 28–41. 10.1093/glycob/cwm125 [DOI] [PubMed] [Google Scholar]
- Hodi, F. S. , O'Day, S. J. , McDermott, D. F. , Weber, R. W. , Sosman, J. A. , Haanen, J. B. , … Akerley, W. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. The New England Journal of Medicine, 363, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, S. P. , Foulds, G. A. , Imrie, H. , Reeder, S. , McArdle, S. E. B. , Khan, M. , & Pockley, A. G. (2019). Phenotype and function of activated natural killer cells from patients with prostate cancer: Patient‐dependent responses to priming and IL‐2 activation. Frontiers in Immunology, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi‐Manaka, A. , Kai, H. , Yamashita, Y. , Shibata, K. , Tahara‐Hanaoka, S. , Honda, S. , … Shibuya, A. (2008). Accelerated tumor growth in mice deficient in DNAM‐1 receptor. The Journal of Experimental Medicine, 205, 2959–2964. 10.1084/jem.20081611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarahian, M. , Fiedler, M. , Cohnen, A. , Djandji, D. , Hammerling, G. J. , Gati, C. , … Hengel, H. (2011). Modulation of NKp30‐ and NKp46‐mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathogens, 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T. , Zhou, C. , & Ren, S. (2016). Role of IL‐2 in cancer immunotherapy. Oncoimmunology, 5, e1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs, S. F. , Ichim, T. E. , Prince, S. M. , Kesari, S. , Marincola, F. M. , Escobedo, A. R. , & Jafri, A. (2018). Unleashing endogenous TNF‐alpha as a cancer immunotherapeutic. Journal of Translational Medicine, 16, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, M. G. , Tran, P. , Zhuravleva, M. A. , Jaw, J. , Colonna, M. , & Sun, P. D. (2011). Crystal structure of human natural cytotoxicity receptor NKp30 and identification of its ligand binding site. Proc Natl Acad Sci USA, 108, 6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, Y. , Shin, C. , Kubo, N. , Iwabuchi, H. , Imamura, M. , Saitoh, A. , & Imai, C. (2018). Superior activity of NKp44‐CAR‐T cells in comparison with ex vivo expended activated NK cells or NKp30‐CAR‐T cells. Pediatric Blood & Cancer, 65, S24–S24. [Google Scholar]
- Katial, R. K. , Sachanandani, D. , Pinney, C. , & Lieberman, M. M. (1998). Cytokine production in cell culture by peripheral blood mononuclear cells from immunocompetent hosts. Clinical and Diagnostic Laboratory Immunology, 5, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner, C. , Maurer, T. , Hallack, D. , Repp, R. , van de Winkel, J. G. , Parren, P. W. , … Peipp, M. (2012). Mimicking an induced self phenotype by coating lymphomas with the NKp30 ligand B7‐H6 promotes NK cell cytotoxicity. Journal of Immunology, 189, 5037–5046. [DOI] [PubMed] [Google Scholar]
- Kiessling, R. , Klein, E. , & Wigzell, H. (1975). “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. European Journal of Immunology, 5, 112–117. [DOI] [PubMed] [Google Scholar]
- Koch, J. , Steinle, A. , Watzl, C. , & Mandelboim, O. (2013). Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends in Immunology, 34, 182–191. [DOI] [PubMed] [Google Scholar]
- Kohidai, L. , & Csaba, G. (1998). Chemotaxis and chemotactic selection induced with cytokines (IL‐8, RANTES and TNF‐α) in the unicellular Tetrahymena pyriformis . Cytokine, 10, 481–486. [DOI] [PubMed] [Google Scholar]
- Kruse, P. H. , Matta, J. , Ugolini, S. , & Vivier, E. (2014). Natural cytotoxicity receptors and their ligands. Immunology and Cell Biology, 92, 221–229. [DOI] [PubMed] [Google Scholar]
- Lahm, H. , André, S. , Hoeflich, A. , Fischer, J. R. , Sordat, B. , Kaltner, H. , … Gabius, H. J. (2001). Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT‐PCR and its implications for diagnostic and therapeutic procedures. Journal of Cancer Research and Clinical Oncology, 127, 375–386. 10.1007/s004320000207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, L. L. (2009). DAP10‐ and DAP12‐associated receptors in innate immunity. Immunological Reviews, 227, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapteva, N. , Durett, A. G. , Sun, J. , Rollins, L. A. , Huye, L. L. , Fang, J. , … Ando, J. (2012). Large‐scale ex vivo expansion and characterization of natural killer cells for clinical applications. Cytotherapy, 14, 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, W. (2014). Infusions of allogeneic natural killer cells as cancer therapy. Clinical Cancer Research, 20, 3390–3400. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, Q. , & Mariuzza, R. A. (2011). Structure of the human activating natural cytotoxicity receptor NKp30 bound to its tumor cell ligand B7‐H6. The Journal of Experimental Medicine, 208, 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz, B. , Wang, Y. , & Neumann, H. (2010). Microglial immunoreceptor tyrosine‐based activation and inhibition motif signaling in neuroinflammation. International Journal of Alzheimer's Disease, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz‐Gerlach, B. , Kopatz, J. , & Neumann, H. (2014). Siglec functions of microglia. Glycobiology, 24, 794–799. 10.1093/glycob/cwu044 [DOI] [PubMed] [Google Scholar]
- Long, E. O. , Kim, H. S. , Liu, D. , Peterson, M. E. , & Rajagopalan, S. (2013). Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annual Review of Immunology, 31, 227–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, D. M. , Louie, B. , Wang, E. , Pos, Z. , Marincola, F. M. , Hawtin, R. E. , & Cesano, A. (2012). Inter‐donor variation in cell subset specific immune signaling responses in healthy individuals. Am J Clin Exp Immunol, 1, 1–11. [PMC free article] [PubMed] [Google Scholar]
- Lorenzo‐Herrero, S. , Lopez‐Soto, A. , Sordo‐Bahamonde, C. , Gonzalez‐Rodriguez, A. P. , Vitale, M. , & Gonzalez, S. (2018). NK cell‐based immunotherapy in cancer metastasis. Cancers, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Shurin, G. V. , Peiyuan, Z. , & Shurin, M. R. (2013). Dendritic cells in the cancer microenvironment. Journal of Cancer, 4, 36–44. 10.7150/jca.5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäenpää, A. , Jääskeläinen, J. , Carpén, O. , Patarroyo, M. , & Timonen, T. (1993). Expression of integrins and other adhesion molecules on NK cells; impact of IL‐2 on short‐ and long‐term cultures. International Journal of Cancer, 53, 850–855. [DOI] [PubMed] [Google Scholar]
- Marchesi, M. , Andersson, E. , Villabona, L. , Seliger, B. , Lundqvist, A. , Kiessling, R. , & Masucci, G. V. (2013). HLA‐dependent tumour development: A role for tumour associate macrophages? Journal of Translational Medicine, 11, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavoungou, E. , Held, J. , Mewono, L. , & Kremsner, P. G. (2007). A Duffy binding‐like domain is involved in the NKp30‐mediated recognition of Plasmodium falciparum‐parasitized erythrocytes by natural killer cells. The Journal of Infectious Diseases, 195, 1521–1531. [DOI] [PubMed] [Google Scholar]
- Memmer, S. , Weil, S. , Beyer, S. , Zoller, T. , Peters, E. , Hartmann, J. , … Koch, J. (2016). The stalk domain of NKp30 contributes to ligand binding and signaling of a preassembled NKp30‐CD3ζ complex. The Journal of Biological Chemistry, 291, 25427–25438. 10.1074/jbc.M116.742981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meresse, B. , Curran, S. A. , Ciszewski, C. , Orbelyan, G. , Setty, M. , Bhagat, G. , … Winchester, R. J. (2006). Reprogramming of CTLs into natural killer‐like cells in celiac disease. The Journal of Experimental Medicine, 203, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty, A. , Chalon, P. , Derocq, J. M. , Dumont, X. , Guillemot, J. C. , Kaghad, M. , … Minty, C. (1993). Interleukin‐13 is a new human lymphokine regulating inflammatory and immune responses. Nature, 362, 248–250. [DOI] [PubMed] [Google Scholar]
- Molina, A. (2008). A decade of rituximab: Improving survival outcomes in non‐Hodgkin's lymphoma. Annual Review of Medicine, 59, 237–250. 10.1146/annurev.med.59.060906.220345 [DOI] [PubMed] [Google Scholar]
- Moretta, A. , Bottino, C. , Vitale, M. , Pende, D. , Cantoni, C. , Mingari, M. C. , … Moretta, L. (2001). Activating receptors and coreceptors involved in human natural killer cell‐mediated cytolysis. Annual Review of Immunology, 19, 197–223. 10.1146/annurev.immunol.19.1.197 [DOI] [PubMed] [Google Scholar]
- Moretta, L. , Ciccone, E. , Mingari, M. C. , Biassoni, R. , & Moretta, A. (1994). Human natural killer cells: Origin, clonality, specificity, and receptors. Advances in Immunology, 55, 341–380. 10.1016/s0065-2776(08)60513-1 [DOI] [PubMed] [Google Scholar]
- Nagasawa, D. T. , Fong, C. , Yew, A. , Spasic, M. , Garcia, H. M. , Kruse, C. A. , & Yang, I. (2012). Passive immunotherapeutic strategies for the treatment of malignant gliomas. Neurosurgery Clinics of North America, 23, 481–495. 10.1016/j.nec.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagler, A. , Lanier, L. L. , & Phillips, J. H. (1990). Constitutive expression of high affinity interleukin 2 receptors on human CD16‐natural killer cells in vivo. The Journal of Experimental Medicine, 171, 1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville, M. J. , & Campbell, R. D. (1999). A new member of the Ig superfamily and a V‐ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC. Journal of Immunology, 162, 4745–4754. [PubMed] [Google Scholar]
- Ni, L. , & Lu, J. (2018). Interferon gamma in cancer immunotherapy. Cancer Medicine, 7, 4509–4516. 10.1002/cam4.1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange, J. S. (2008). Formation and function of the lytic NK‐cell immunological synapse. Nature Reviews. Immunology, 8, 713–725. 10.1038/nri2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi, M. , Favoreel, H. , Candiano, G. , Gaggero, S. , Sivori, S. , Mingari, M. C. , … Cantoni, C. (2019). NKp44‐NKp44 ligand interactions in the regulation of natural killer cells and other innate lymphoid cells in humans. Frontiers in Immunology, 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S. , & Lal, G. (2017). The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Frontiers in Immunology, 8, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegram, H. J. , Andrews, D. M. , Smyth, M. J. , Darcy, P. K. , & Kershaw, M. H. (2011). Activating and inhibitory receptors of natural killer cells. Immunology and Cell Biology, 89, 216–224. 10.1038/icb.2010.78 [DOI] [PubMed] [Google Scholar]
- Peipp, M. , Derer, S. , Lohse, S. , Staudinger, M. , Klausz, K. , Valerius, T. , … Kellner, C. (2015). HER2‐specific immunoligands engaging NKp30 or NKp80 trigger NK‐cell‐mediated lysis of tumor cells and enhance antibody‐dependent cell‐mediated cytotoxicity. Oncotarget, 6, 32075–32088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende, D. , Parolini, S. , Pessino, A. , Sivori, S. , Augugliaro, R. , Morelli, L. , … Bottino, C. (1999). Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. The Journal of Experimental Medicine, 190, 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L.‐S. , J‐y, Z. , Y‐s, T. , Y‐l, Z. , Wang, T.‐t. , Mao, F.‐y. , … Duan, M. (2017). Tumor‐associated monocytes/macrophages impair NK‐cell function via TGFβ1 in human gastric cancer. Cancer Immunology Research, 5, 248–256. [DOI] [PubMed] [Google Scholar]
- Perillo, N. L. , Marcus, M. E. , & Baum, L. G. (1998). Galectins: Versatile modulators of cell adhesion, cell proliferation, and cell death. Journal of Molecular Medicine (Berlin, Germany), 76, 402–412. 10.1007/s001090050232 [DOI] [PubMed] [Google Scholar]
- Pesce, S. , Tabellini, G. , Cantoni, C. , Patrizi, O. , Coltrini, D. , Rampinelli, F. , … Marcenaro, E. (2015). B7‐H6‐mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. Oncoimmunology, 4, e1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, M. , Romeo, F. , Bitsaktsis, C. , & Sabatino, D. (2016). B7H6‐derived peptides trigger TNF‐α‐dependent immunostimulatory activity of lymphocytic NK92‐MI cells. Biopolymers, 106, 658–672. [DOI] [PubMed] [Google Scholar]
- Pogge von Strandmann, E. , Simhadri, V. R. , von Tresckow, B. , Sasse, S. , Reiners, K. S. , Hansen, H. P. , … McKinnon, P. J. (2007). Human leukocyte antigen‐B‐associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity, 27, 965–974. [DOI] [PubMed] [Google Scholar]
- Poli, A. , Michel, T. , Theresine, M. , Andres, E. , Hentges, F. , & Zimmer, J. (2009). CD56bright natural killer (NK) cells: An important NK cell subset. Immunology, 126, 458–465. 10.1111/j.1365-2567.2008.03027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrata, L. F. , Inwards, D. J. , Lacy, M. Q. , & Markovic, S. N. (2001). Immunomodulation of early engrafted natural killer cells with interleukin‐2 and interferon‐α in autologous stem cell transplantation. Bone Marrow Transplantation, 28, 673–680. 10.1038/sj.bmt.1703203 [DOI] [PubMed] [Google Scholar]
- Quattrocchi, K. B. , Miller, C. H. , Cush, S. , Bernard, S. A. , Dull, S. T. , Smith, M. , … Varia, M. A. (1999). Pilot study of local autologous tumor infiltrating lymphocytes for the treatment of recurrent malignant gliomas. Journal of Neuro‐Oncology, 45, 141–157. [DOI] [PubMed] [Google Scholar]
- Rahmoune, H. , & Guest, P. C. (2018). Studies of isolated peripheral blood cells as a model of immune dysfunction. Methods in Molecular Biology, 1735, 221–229. 10.1007/978-1-4939-7614-0_12 [DOI] [PubMed] [Google Scholar]
- Reiners, K. S. , Kessler, J. , Sauer, M. , Rothe, A. , Hansen, H. P. , Reusch, U. , … Von Strandmann, E. P. (2013). Rescue of impaired NK cell activity in Hodgkin lymphoma with bispecific antibodies in vitro and in patients. Molecular Therapy, 21, 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners, K. S. , Topolar, D. , Henke, A. , Simhadri, V. R. , Kessler, J. , Sauer, M. , … Krönke, M. (2013). Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti‐tumor activity. Blood, 121, 3658–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renieri, A. , Mencarelli, M. A. , Cetta, F. , Baldassarri, M. , Mari, F. , Furini, S. , … Frullanti, E. (2014). Oligogenic germline mutations identified in early non‐smokers lung adenocarcinoma patients. Lung Cancer, 85, 168–174. [DOI] [PubMed] [Google Scholar]
- Robertson, M. J. , Caligiuri, M. A. , Manley, T. J. , Levine, H. , & Ritz, J. (1990). Human natural killer cell adhesion molecules. Differential expression after activation and participation in cytolysis. Journal of Immunology, 145, 3194–3201. [PubMed] [Google Scholar]
- Rosenberg, S. A. , Lotze, M. T. , Muul, L. M. , Leitman, S. , Chang, A. E. , Ettinghausen, S. E. , … Reichert, C. M. (1985). Observations on the systemic administration of autologous lymphokine‐activated killer cells and recombinant interleukin‐2 to patients with metastatic cancer. The New England Journal of Medicine, 313, 1485–1492. 10.1056/NEJM198512053132327 [DOI] [PubMed] [Google Scholar]
- Rusakiewicz, S. , Perier, A. , Semeraro, M. , Pitt, J. M. , Pogge von Strandmann, E. , Reiners, K. S. , … Zitvogel, L. (2017). NKp30 isoforms and NKp30 ligands are predictive biomarkers of response to imatinib mesylate in metastatic GIST patients. Oncoimmunology, 6, e1137418 10.1080/2162402X.2015.1137418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabry, M. , & Lowdell, M. W. (2013). Tumor‐primed NK cells: Waiting for the green light. Frontiers in Immunology, 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi, M. , Xue, L. , Jolin, H. , Hardman, C. , Cousins, D. J. , McKenzie, A. N. J. , & Ogg, G. S. (2016). Group 2 innate lymphoid cells express functional NKp30 receptor inducing type 2 cytokine production. Journal of Immunology, 196, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Ohashi, J. , Tsuchiya, N. , Tadokoro, K. , Juji, T. , Hanaoka, K. , … Yabe, T. (2001). Identification of novel single nucleotide substitutions in the NKp30 gene expressed in human natural killer cells. Tissue Antigens, 58, 255–258. [DOI] [PubMed] [Google Scholar]
- Schrödinger, LLC . (2015). The PyMOL Molecular Graphics System, Version 1.8.
- Schuldner, M. , Dörsam, B. , Shatnyeva, O. , Reiners, K. S. , Kubarenko, A. , Hansen, H. P. , … von Strandmann, E. P. (2019). Exosome‐dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300‐dependent acetylation of p53. Theranostics, 9, 6047–6062. 10.7150/thno.36378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, T. , Gambhir, S. S. , Aalipour, A. , & Schurch, C. (2020). PET imaging of the natural killer cell activation receptor NKp30. Journal of Nuclear Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanafelt, T. D. , Wang, X. V. , Kay, N. E. , Hanson, C. A. , O'Brien, S. , Barrientos, J. , … Coutre, S. E. (2019). Ibrutinib–rituximab or chemoimmunotherapy for chronic lymphocytic leukemia. The New England Journal of Medicine, 381, 432–443. 10.1056/NEJMoa1817073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, M. , & Mount, N. (2015). Genetically modified T cells in cancer therapy: Opportunities and challenges. Disease Models & Mechanisms, 8, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh, A. , Kugel, A. , Steiner, N. , Yezersky, M. , Tirosh, D. , Edri, A. , … Campbell, K. S. (2016). NKp44 and NKp30 splice variant profiles in decidua and tumor tissues: A comparative viewpoint. Oncotarget, 7, 70912–70923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, A. , Campbell, D. , Hannum, C. , Yssel, H. , Franz‐Bacon, K. , McClanahan, T. , … Phillips, J. H. (1996). DNAM‐1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity, 4, 573–581. [DOI] [PubMed] [Google Scholar]
- Shimasaki, N. , Jain, A. , & Campana, D. (2020). NK cells for cancer immunotherapy. Nature Reviews. Drug Discovery, 19, 200–218. 10.1038/s41573-019-0052-1 [DOI] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2019). Cancer statistics, 2019. CA: A Cancer Journal for Clinicians, 69, 7–34. [DOI] [PubMed] [Google Scholar]
- Siewiera, J. , Gouilly, J. , Hocine, H. R. , Cartron, G. , Levy, C. , Al‐Daccak, R. , & Jabrane‐Ferrat, N. (2015). Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. Nature Communications, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza‐Fonseca‐Guimarães, F. , Cursons, J. , & Huntington, N. D. (2019). The emergence of natural killer cells as a major target in cancer immunotherapy. Trends in Immunology, 40, 142–158. [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J. C. , & Griffiths, G. M. (2007). Secretory mechanisms in cell‐mediated cytotoxicity. Annual Review of Cell and Developmental Biology, 23, 495–517. [DOI] [PubMed] [Google Scholar]
- Storkus, W. J. , Salter, R. D. , Alexander, J. , Ward, F. E. , Ruiz, R. E. , Cresswell, P. , & Dawson, J. R. (1991). Class I‐induced resistance to natural killing: Identification of nonpermissive residues in HLA‐A2. Proceedings of the National Academy of Sciences of the United States of America, 88, 5989–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, Y. K. , Martinson, J. A. , Doligosa, K. , & Klingemann, H. G. (2003). Ex vivo expansion of the highly cytotoxic human natural killer‐92 cell‐line under current good manufacturing practice conditions for clinical adoptive cellular immunotherapy. Cytotherapy, 5, 259–272. [DOI] [PubMed] [Google Scholar]
- Tang, Q. , Grzywacz, B. , Wang, H. , Kataria, N. , Cao, Q. , Wagner, J. E. , … Verneris, M. R. (2008). Umbilical cord blood T cells express multiple natural cytotoxicity receptors after IL‐15 stimulation, but only NKp30 is functional. Journal of Immunology, 181, 4507–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham, N. J. , & Hewitt, E. W. (2009). Natural killer cell cytotoxicity: How do they pull the trigger? Immunology, 128, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornroos, H. , Hagerstrand, H. , & Lindqvist, C. (2019). Culturing the human natural killer cell line NK‐92 in interleukin‐2 and interleukin‐15—Implications for clinical trials. Anticancer Research, 39, 107–112. 10.21873/anticanres.13085 [DOI] [PubMed] [Google Scholar]
- Tsurushima, H. , Liu, S. Q. , Tuboi, K. , Matsumura, A. , Yoshii, Y. , Nose, T. , … Ohno, T. (1999). Reduction of end‐stage malignant glioma by injection with autologous cytotoxic T lymphocytes. Japanese Journal of Cancer Research, 90, 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier, T. , Satoh‐Takayama, N. , Serafini, N. , Marie, S. , Di Santo, J. P. , & Vosshenrich, C. A. (2016). Phenotypic and functional plasticity of murine intestinal NKp46+ group 3 innate lymphoid cells. Journal of Immunology, 196, 4731–4738. [DOI] [PubMed] [Google Scholar]
- Vivier, E. , Ugolini, S. , Blaise, D. , Chabannon, C. , & Brossay, L. (2012). Targeting natural killer cells and natural killer T cells in cancer. Nature Reviews. Immunology, 12, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Strandmann, E. P. , Simhadri, V. R. , von Tresckow, B. , Sasse, S. , Reiners, K. S. , Hansen, H. P. , … Engert, A. (2006). Tumor cell‐derived HLA‐B‐associated transkript 3 (BAT3) is a ligand for NKp30 and activates NK cells. Blood, 108, 194a–194a. [Google Scholar]
- Wang, W. , Guo, H. , Geng, J. , Zheng, X. , Wei, H. , Sun, R. , & Tian, Z. (2014). Tumor‐released Galectin‐3, a soluble inhibitory ligand of human NKp30, plays an important role in tumor escape from NK cell attack. The Journal of Biological Chemistry, 289, 33311–33319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzl, C. , & Long, E. O. (2010). Signal transduction during activation and inhibition of natural killer cells. Current Protocols in Immunology, 90, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside, T. L. , & Herberman, R. B. (1994). Role of human natural killer cells in health and disease. Clinical and Diagnostic Laboratory Immunology, 1, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpe, S. D. , Davatelis, G. , Sherry, B. , Beutler, B. , Hesse, D. G. , Nguyen, H. T. , … Cerami, A. (1988). Macrophages secrete a novel heparin‐binding protein with inflammatory and neutrophil chemokinetic properties. The Journal of Experimental Medicine, 167, 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W. L. , Shen, K. Y. , Tien, C. Y. , Chen, Y. A. , & Liu, S. J. (2017). Recent progress in GM‐CSF‐based cancer immunotherapy. Immunotherapy, 9, 347–360. [DOI] [PubMed] [Google Scholar]
- Yang, H. G. , Kang, M. C. , Kim, T. Y. , Hwang, I. , Jin, H. T. , Sung, Y. C. , … Kim, S. W. (2019). Discovery of a novel natural killer cell line with distinct immunostimulatory and proliferative potential as an alternative platform for cancer immunotherapy. Journal for Immunotherapy of Cancer, 7, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, W. M. , & Kim, S. (2006). Licensing of natural killer cells by self‐major histocompatibility complex class I. Immunological Reviews, 214, 143–154. 10.1111/j.1600-065X.2006.00458.x [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Wu, M. R. , & Sentman, C. L. (2012). An NKp30‐based chimeric antigen receptor promotes T cell effector functions and antitumor efficacy in vivo. Journal of Immunology, 189, 2290–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, Y. , Liu, H. , & Li, B. (2019). Six genes as potential diagnosis and prognosis biomarkers for hepatocellular carcinoma through data mining. Journal of Cellular Physiology, 234, 9787–9792. 10.1002/jcp.27664 [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Wu, N. , Lu, Y. , Davidson, D. , Colonna, M. , & Veillette, A. (2015). DNAM‐1 controls NK cell activation via an ITT‐like motif. The Journal of Experimental Medicine, 212, 2165–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]


