ABSTRACT
The endosomal sorting complexes required for transport (ESCRTs) I, -II and –III, and their associated factors are a collection of ∼20 proteins in yeast and ∼30 in mammals, responsible for severing membrane necks in processes that range from multivesicular body formation, HIV release and cytokinesis, to plasma and lysosomal membrane repair. ESCRTs are best known for ‘reverse-topology’ membrane scission, where they act on the inner surface of membrane necks, often when membranes are budded away from the cytosol. These events are driven by membrane-associated assemblies of dozens to hundreds of ESCRT molecules. ESCRT-III proteins form filaments with a variety of geometries and ESCRT-I has now been shown to also form helical structures. The complex nature of the system and the unusual topology of its action has made progress challenging, and led to controversies with regard to its underlying mechanism. This Review will focus on recent advances obtained by structural in vitro reconstitution and in silico mechanistic studies, and places them in their biological context. The field is converging towards a consensus on the broad outlines of a mechanism that is driven by a progressive ATP-dependent treadmilling exchange of ESCRT subunits, as well as compositional change and geometric transitions in ESCRT filaments.
KEY WORDS: ESCRT, Membrane scission, HIV, Endosome, Membrane biophysics, In vitro reconstitution, Giant unilamellar vesicle, Cryo-electron microscopy, Optical tweezers
Summary: New structural and biophysical studies have shed much light on the physical mechanism of ESCRT-mediated membrane remodeling and scission.
Introduction
The endosomal sorting complexes required for transport (ESCRTs) are an ancient, multi-component membrane remodeling complex capable of bringing membrane necks to abscission in a reverse-topology manner. In other words, ESCRTs can sever membranes that are budding away from the cytoplasm rather than towards the cytoplasm as in endocytosis (Hurley, 2015; Vietri et al., 2020). The ESCRTs originated in a subset of the Archaea, where they are involved in cytokinetic membrane constriction and abscission (Samson et al., 2008; Risa et al., 2020). In eukaryotes, the ESCRTs are involved in an enormous number of cellular processes (Hurley, 2015; Vietri et al., 2020). Most famously, the ESCRTs are essential for multivesicular body biogenesis (Babst, 2011), HIV egress (Sundquist and Kräusslich, 2012; Hurley and Cada, 2018), exosome production (Colombo et al., 2013), cytokinesis (Carlton and Martin-Serrano, 2007), as well as nuclear envelope surveillance and/or resealing (Webster et al., 2014; Olmos et al., 2015; Vietri et al., 2015; Raab et al., 2016). More recently identified functions include plasma-membrane repair (Jimenez et al., 2014; Scheffer et al., 2014), autophagosome closure (Takahashi et al., 2018) and lysosome repair (Skowyra et al., 2018; Radulovic et al., 2018). In addition to these reverse-topology scission functions, ESCRTs can also carry out normal topology scission (McCullough et al., 2015). ESCRTs, therefore, comprise a versatile membrane remodeling machinery with a vast array of functions in eukaryotes, and their mechanism of action has naturally attracted a great deal of investigative effort.
The ESCRTs constitute a system of ∼20 proteins in yeast and >30 in mammals, summarized in Table 1. They consist of the complexes ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III, the AAA+ATPase Vps4 as well as associated proteins that include ALG-2 interacting protein X (ALIX, officially known as PDCD6IP in humans)/ BCK1-like resistance to osmotic shock protein 1 (Bro1 in yeast), tyrosine-protein phosphatase non-receptor type 23 (PTPN23, also known as HD-PTP ) and others. Other reviews have extensively described the properties of the individual ESCRT proteins and sub-complexes (Schöneberg et al., 2017; McCullough et al., 2018). This Review is intended to be topical and focusses on recent advances in understanding the biophysical and structural mechanisms of membrane scission so, here, we present only a brief introduction to the system as a whole. ESCRT-0 proteins are best characterized in multi-vesicular body (MVB) biogenesis. ESCRT-0 is an endosomal clathrin adaptor that initiates ESCRT recruitment during MVB biogenesis but, despite its name, it is not part of the membrane remodeling machinery and will not be discussed further here. The ESCRT-I complex is in the shape of a long stalk with a head that recruits the ESCRT-II complex (Kostelansky et al., 2007; Boura et al., 2012). The ESCRT-II complex has a Y-shaped architecture (Hierro et al., 2004; Teo et al., 2004), with two binding sites for Vps20 (CHMP6 in humans) (Im and Hurley, 2008) that connect it to the ESCRT-III complex described below.
Table 1.
ESCRT complexes and their protein components in yeast and human
ESCRT-III proteins are highly basic proteins that bind readily to negatively charged membranes, and are capable of polymerizing into large assemblies of various architectures and curvatures, thought to drive membrane constriction (Lee et al., 2015; McCullough et al., 2018). There are a total of eight ESCRT-III proteins in yeast, and twelve in humans (see Table 1). In addition to Vps20/CHMP6, mechanistic research into membrane severing has focused mainly on Snf7/CHMP4, Vps24/CHMP3, Vps2/CHMP2, IST1 and Did2/CHMP1. Of the two other subunits, Chm7/CHMP7 exerts specialized functions in nuclear surveillance and repair (Olmos et al., 2016), and Vps60/CHMP5 remains poorly characterized. Structural work revealed the common architecture of all ESCRT-III subunits, which are small helical proteins (Muzioł et al., 2006). All ESCRT-III proteins exist in an autoinhibited, closed state within the cytosol (Zamborlini et al., 2006; Shim et al., 2007; Bajorek et al., 2009). However, some, including Snf7 in yeast and CHMP1B in human, adopt an open extended conformation on membranes (McCullough et al., 2015; Tang et al., 2015; McMillan et al., 2016). Finally, the AAA+ATPase Vps4 (Babst et al., 1998) is recruited to membranes in order to translocate and unfold (Yang et al., 2015) ESCRT-III components; this process is mediated through microtubule interacting and trafficking (MIT) domains binding to MIT-interacting motifs (MIMs) of ESCRT-III proteins (Stuchell-Brereton et al., 2007; Obita et al., 2007; Yang et al., 2008). Although the unfoldase activity of Vps4 on individual subunits is well understood (Yang et al., 2015), there is still an ongoing debate whether Vps4 has a mechanical role in membrane constriction and scission or whether it merely allows the ESCRT polymer to remodel itself by recycling individual subunits to the cytosol. The role of coupling between ESCRT-III and Vps4 in membrane scission is discussed in detail below.
Foundational studies of overexpressed (Hanson et al., 2008) and reconstituted (Lata et al., 2008) ESCRT-III complexes showed that ESCRTs are capable of forming helical tubes (Hanson et al., 2008; Lata et al., 2008) and spirals (Hanson et al., 2008; Cashikar et al., 2014) on membranes. An influential early model of ESCRT-III function stems from the observation that the ESCRT-III components CHMP2 and CHMP3 can assemble into tubes that end in a dome cap (Fabrikant et al., 2009; Effantin et al., 2013) and which can be disassembled by VPS4 (Lata et al., 2008). This proposed model suggests that the CHMP2–CHMP3 heteropolymer templates the membrane from the inside until the domed cap and protein-lipid adhesion energy brings the two leaflets close enough to allow fission in conjunction with VPS4 activity (Fabrikant et al., 2009).
ESCRTs can also carry out membrane scission in normal topology, in other words, scission towards or into the cytoplasm. In cells depleted of the ESCRT-III subunit IST1, recycling tubular endosomes cannot be scissioned, which impaired lysosomal trafficking (Allison et al., 2017, 2019). The AAA+ATPase spastin (SPAST), which is best known as a microtubule-severing enzyme (Roll-Mecak and Vale, 2005), also interacts strongly with IST1 and CHMP1B, two additional ESCRT-III subunits, and is involved in normal topology scission by ESCRTs (Allison et al., 2013). IST1, CHMP1B and spastin are important for trafficking of lipids between lipid droplets and peroxisomes in a yet unclear mechanism (Chang et al., 2019). ESCRTs are essential for the release of new peroxisomes from the endoplasmic reticulum (ER) (Mast et al., 2018), which seems likely to be a normal-topology scission reaction involved in peroxisome and lipid droplet biogenesis.
In this Review, we summarize recent data and insights on ESCRT-mediated mechanisms, with an emphasis on in vitro and in silico studies. We describe major findings that were afforded by advances in techniques, such as cryo-electron microscopy (EM) that made it possible to characterize the various large-scale assemblies formed by ESCRT-III subunits, synthetic membrane biochemistry to reconstitute ESCRT scission using minimal components and simplified membrane compositions, as well as the use of optical tweezers in reconstituted systems. We conclude with an attempt to converge the available data into what we consider a most-likely mechanistic model, and outline crucial further experiments that might yield important new insights in the future.
Bulk-membrane biochemical studies
Early in vitro studies reported that yeast ESCRT-III proteins can bud and sever intralumenal vesicles (ILVs) using giant unilamellar vesicles (GUVs) as substrates (Wollert et al., 2009; Wollert and Hurley, 2010). This reaction corresponds to the role of ESCRTs in MVB biogenesis, where ESCRTs are responsible both for creating membrane buds and severing them into the lumen. This process is distinct from most other ESCRT functions in the cell, where the membrane neck is created by other machineries (Hurley, 2015; Vietri et al., 2020). In in vitro studies, GUVs with a negative charge density sufficient to recruit ESCRTs were incubated with various ESCRT components that had been added either simultaneously or sequentially; thereafter, ILVs were observed in the lumen of the GUV (Wollert et al., 2009). Experiments based on adding a membrane-impermeable dye to the GUV mix before initiating the ESCRT reaction showed that ILV formation depends on the presence of Vps20, Snf7, Vps24 and Vps2 (Wollert et al., 2009). The ATPase Vps4 is unnecessary for the initial round of budding and scission events under these conditions (Wollert et al., 2009). These findings are contradicted, however, by a series of subsequent cell-imaging studies that, consistently, found that Vps4 is recruited to sites of ESCRT action prior to scission (Baumgärtel et al., 2011; Jouvenet et al., 2011; Bleck et al., 2014; Adell et al., 2017; Mierzwa et al., 2017; Johnson et al., 2018).
Recent studies reproduced and extended these observations. A lipidated, chimeric yeast Snf7–Vps2 construct was shown to be capable of generating ILVs in GUVs, although complete scission of the ILVs into the lumen of the GUV with such a minimal ESCRT functionality was inefficient (Marklew et al., 2018). Similar findings were reported in a study of Entamoeba histolytica ESCRT-III proteins on GUVs (Avalos-Padilla et al., 2018). In addition, yeast ESCRT-III proteins were used to probe the effects of membrane tension and on the relative amounts of different subunits on ILV formation in GUVs, and found that membrane tension but not the relative concentrations of each subunit, is a key factor determining successful ILV formation (Booth et al., 2019). The former findings are consistent with in vivo observations that a decrease in tension on endosomes triggers ESCRT recruitment and function on the membrane (Mercier et al., 2020). These results support an intriguing feedback model, i.e. low membrane tension favors ESCRT-mediated formation of intraluminal vesicles, which causes a loss of membrane area with a consequent increase of tension in the GUV or endosome, and this tension increase then further inhibits ESCRT function (Booth et al., 2019). Since GUVs, typically, have lower bending rigidities and lower tension values than cellular membranes (Dimova et al., 2006), it is possible that the previously observed apparent Vps4 independence of scission in GUVs relates to the deformability of GUVs. Collectively, although the GUV system ultimately proved to be too permissive – i.e. it is easier to achieve scission in the GUV system than in cells – to completely reflect the biology, these experiments, nevertheless, establish the important point that the core ESCRT-III proteins Snf7, Vps24 and Vps2 comprise, in principle, the minimal machinery for membrane scission.
Membrane nanotube experiments
Membrane nanotubes that are pulled from GUVs provide a more-controlled setting for reconstitution and real-time imaging of single ESCRT-mediated scission events. In this modality, streptavidin-coated beads held in an optical trap are used to pull membrane tubes of tens of nm in diameter from GUVs under conditions of controlled membrane tension and force (Sorre et al., 2009; Prévost et al., 2017). Indeed, recent experiments from our lab and others have used such a setup to probe ESCRT function on small membrane tubes pulled from GUVs by dielectric beads held in an optical trap (Schöneberg et al., 2018; Pfitzner et al., 2020). This pulled nanotube comprises Gaussian-curvature geometry of the membrane neck that is similar to the physiological substrates of the ESCRTs. In work published by our group, we encapsulated Snf7, Vps24, Vps2, Vps4 and caged ATP (cATP) within GUVs during swelling of lipid films (Fig. 1A) (Schöneberg et al., 2018). The use of cATP prevented premature activation of Vps4. Prior to uncaging of ATP in response to illumination with UV light, ESCRT proteins, including Vps4, were observed exclusively in the lumen of the GUV. GUVs were aspirated onto a micropipette to control membrane tension and a tube was pulled from the GUV (Schöneberg et al., 2018). After uncaging of ATP, ESCRT-III proteins and Vps4 began to accumulate in puncta along the length of the tube. A progressively increasing force on the trapped bead, associated with membrane constriction, was measured over timescales of minutes (Schöneberg et al., 2018). The kinetics are slower than for biological scission reactions, and are likely to reflect reduced efficiency of the minimal system as opposed to the entire complement of ESCRT proteins. Snf7 and Vps4 were seen to steadily accumulate together at up to a few hundred subunits each (Schöneberg et al., 2018), which is roughly consistent with quantification data from live-cell imaging (Adell et al., 2017). The increase in force is concurrent with an increase in the fluorescence intensity of the puncta and a progressive decrease in the tube radius to <10nm1 (Schöneberg et al., 2018). This culminates in the scission of the tube from the GUV (Schöneberg et al., 2018).
Fig. 1.
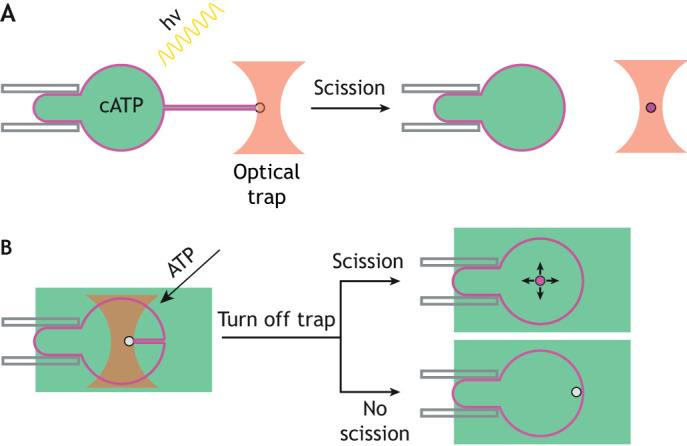
Experimental methods. (A) After a membrane nanotube is formed, UV illumination uncages any caged ATP (cATP) ATP present and ESCRT activity is observed. As the force to hold the bead rises, the membrane nanotube either scissions or pulls the bead out of the optical trap. (B) A membrane nanotube is pulled into the lumen of the GUV, with the ESCRT proteins outside. ESCRT proteins nucleate inside the membrane nanotube, and the addition of ATP triggers ESCRT activity. Scission is assayed by releasing the bead and observing whether it undergoes free diffusion, signifying scission, or a rapid pullback to the membrane, indicating that the membrane nanotube was still attached.
A subsequent study reproduced these results and extended them by including the additional components Did2 and Ist1, demonstrating that, as expected, scission is more efficient when more of the ESCRT components are included (Pfitzner et al., 2020). In order to add ESCRTs externally and avoid the technical demands of encapsulating the ESCRTs within GUVs, this study used tubes that were pushed into the lumen of the GUV, such that ESCRTs could be injected onto their surface (Fig. 1B). The authors further concluded that the precise composition of the ESCRT-III assembly is crucial for regulating deformation of the tube and its scission, with particularly important roles for Did2 and Ist1. Did2 appears to promote buckling and tubulation of ESCRT assemblies, whereas Ist1 facilitates constriction but blocks scission. On the basis of these findings, the authors suggest that remodeling of the ESCRT-III filament by Did2 causes initial deformation, after which Ist1 induces the final constriction. The subsequent removal of Ist1 from the filament by Vps4 then allows scission to occur (Pfitzner et al., 2020). However, ist1Δ and did2Δ yeast cells have only slight defects in regard to sorting of the cargoes carboxypeptidase S (CPS) (Dimaano et al., 2008; Rue et al., 2007) and mating factor α receptor Ste2 to the lumen of the vacuole (Dimaano et al., 2008), which suggests that they are not essential for efficient scission. Therefore, it remains to be reconciled how these in vitro findings relate to the biological function in vivo.
ESCRT and membrane mechanics
In silico simulations and modeling are becoming increasingly important in understanding key steps of mechanical events at the nanoscale, such as ESCRT-mediated scission, which are intractable to experimental observation. The earliest model of ESCRT scission to be analyzed in detail in silico was a continuum membrane mechanics calculation investigating whether an adhesive protein dome can sufficiently deform a membrane to scission. (Fabrikant et al., 2009). A subsequent reanalysis, also using continuum membrane mechanics (Agudo-Canalejo and Lipowsky, 2018), found that the first calculation had overestimated by several-fold the energy available from adhesion of the putative ESCRT dome to the membrane (Agudo-Canalejo and Lipowsky, 2018), thereby calling its feasibility into question. The ability of ESCRTs to undergo buckling transitions from flat spirals to helical tubes and, so, to deform membranes was implied from cellular EM studies (Hanson et al., 2008) that were analyzed by using a continuum mechanical model (Lenz et al., 2009) and observed experimentally (Chiaruttini et al., 2015), raising the possibility that the sudden buckling of a helical spiral in the neck of a membrane to a flat spiral pulls the membrane. By using a continuum model of the forces imparted by polymerizing proteins on an elastic membrane at the neck of a pre-formed vesicle, cone-like assemblies of adhesive proteins were shown to both require less energy to constrict the neck and to be capable of imparting greater forces for constriction than dome-like assemblies (Agudo-Canalejo and Lipowsky, 2018). The energy barrier to membrane scission can be lowered further when axial symmetry is broken (Vasan et al., 2020), suggesting that ESCRTs function with optimal efficiency if they constrict the membrane asymmetrically.
The most-detailed mechanical analysis, thus far, of how ESCRT-III polymerization generates membrane deformation and constriction made use of coarse-grained molecular simulations (Harker-Kirschneck et al., 2019). Here, a coarse-grained approximation of individual ESCRT-III subunits as a triplet of beads connected by springs was applied onto a one-bead-per-lipid approximation of an initially flat lipid bilayer. In the simulation, the filaments were driven to alternately tilt and flatten. By adding a model membrane-bound cargo capable of interacting with ESCRT beads to the simulation, Harker-Kirschneck and colleagues showed that the combination of cargo stabilizing the bud curvature, in competition with the tilting and flattening – i.e. buckling – of ESCRT filaments, is sufficient to progressively constrict and sever the membrane neck. Importantly, this model recapitulates the buckling observed in an experiment involving massive ESCRT assemblies (≥1000 subunits of Snf7) (Chiaruttini et al., 2015; Carlson et al., 2015), but with tens rather than thousands of subunits being involved. This brings this model into closer alignment with subunit numbers measured in vivo (Adell et al., 2017) and in in vitro scission reconstitution (Schöneberg et al., 2018). Taken together, these in silico studies are consistent with the idea that transitions in filament geometry at the membrane neck, including buckling and symmetry breaking, drive membrane scission.
Compositional control of scission
ESCRTs act at membrane necks that have negative Gaussian curvature, yet individual ESCRT-III subunits do not prefer this geometry. Indeed, filaments of pure Snf7 (yeast) or CHMP4 (human), the most abundant structural component of ESCRT-III, form flat spirals (Shen et al., 2014; Chiaruttini et al., 2015). In addition, CHMP1B alone and in combination with IST1 (McCullough et al., 2015), and CHMP2 in combination with CHMP3 (Bertin et al., 2019) preferentially bind to positively curved membranes. However, combinations of Snf7 with both Vps24 and Vps2, or of CHMP2 with both CHMP3 and CHMP4, preferentially localize to bud necks (Schöneberg et al., 2018; Bertin et al., 2019). In fact, the presence of Vps2 and Vps24 in Snf7 filaments radically alters their geometry to three-dimensional helices compared with the flat spiral geometry of Snf7-only filaments (Henne et al., 2012; Bertin et al., 2019; Moser von Filseck et al., 2020). When bound to membranes, the combinations of Snf7–Vps24–Vps2 or CHMP2–CHMP3–CHMP4 deform liposomes into helical tubes that are coated with these ESCRT subunits on their exterior positively curved surface (Bertin et al., 2019; Moser von Filseck et al., 2020). Although the link between these unusual helical tubular structures and biological ESCRT-III-mediated reactions is unclear, the dramatic remodeling events observed in vitro indicate that changes in the composition of ESCRT-III subunits profoundly alters its membrane-remodeling properties.
High-resolution structural data of CHMP1B and IST1 revealed that these subunits act together to remodel and, presumably, sever positively-curved tubular endosomes (McCullough et al., 2015). A recent cryo-EM study has shown that CHMP1B alone forms a single-start helical coat of 28 nm diameter around lipid tubes (Nguyen et al., 2020). Addition of IST1 leads to the coat to constrict to an outer diameter of 18 nm, reducing the lumen to just 4 nm (Nguyen et al., 2020), which, presumably, is sufficient for membrane scission to take place. Constriction is accomplished by flexing of the helical core of CHMP1B about an elbow joint between helices α3 and α4, bringing the helices closer together when flexed and, consequently, constricting the filament. (Nguyen et al., 2020). This work, thus, presents an important advance in the field, as it allows us to understand normal-topology scission by CHMP1B and IST1 at the structural level. It is conceivable that the constriction event in reverse-topology scission also depends on a compositional change and elbow-joint flexing. To solve this question requires cryo-EM experiments, which are likely to be more demanding because it is not obvious how to engineer constriction from within a tube in a manner that provides a large number of well-aligned molecules for the necessary averaging. Structural details of constriction in reverse-topology scission are, therefore, still lacking.
Membrane constriction can be triggered in an in vitro system by changing the ESCRT-III composition, e.g. by adding a new component. In cells, a change in ESCRT-III composition is enabled by constant Vps4-mediated removal of subunits from the membrane followed by rebinding. This was first observed for ESCRT-III during cytokinetic abscission by using rapid fluorescence recovery after photobleaching (FRAP) (Mierzwa et al., 2017). This finding is important in two respects. First, it provides a mechanism for a change in the composition of ESCRT-III during the course of membrane remodeling (Fig. 2). This can happen because the relative rates of Vps4-mediated removal and spontaneous rebinding can differ for different subunits (Mierzwa et al., 2017). Second, it provides insight into the function of the Vps4 ATPase in membrane scission. Vps4 does not mechanically power membrane rupture in a direct manner – at least, it is not essential for this – as evidenced by the ability of ESCRT-III to sever membranes in the absence of Vps4 in vitro (Wollert et al., 2009; Marklew et al., 2018; Avalos-Padilla et al., 2018). Rather, Vps4 facilitates changes in the relative subunit composition of ESCRT-III over the course of the reaction, leading to structural changes in the ESCRT coat and, so, to membrane constriction and scission.
Fig. 2.
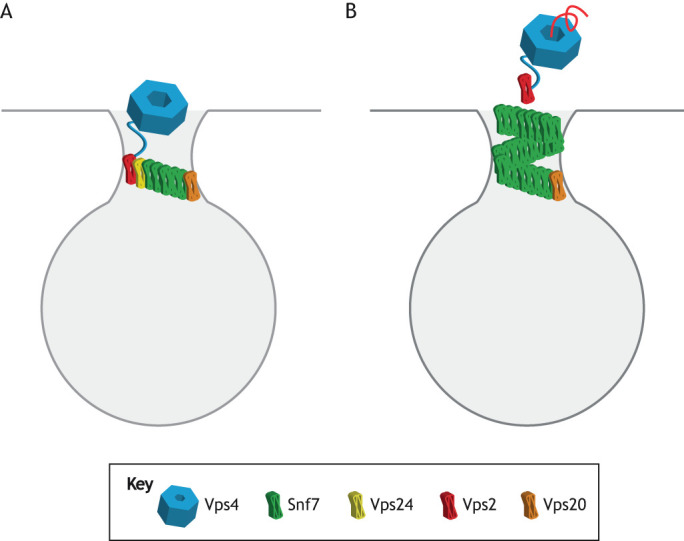
Compositional control of scission. (A) An ESCRT-III assembly whose growth is initially inhibited by Vps2 capping of the growing Snf7 polymer. Vps4 is recruited by Vps2 through an interaction between the MIT domain of Vps4 and the high-affinity MIT-interaction motif (MIM) of Vps2 (Stuchell-Brereton et al., 2007). (B) Vps4-mediated recycling of Vps2 by unfolding and translocation of the Vps2 subunit through the Vps4 central pore (Yang et al., 2015) uncaps Snf7 and allows for the polymer to continue growing.
Vps4 is the only ATPase demonstrated to mediate ESCRT-III subunit exchange but it may be that spastin can also promote exchange. CHMP1B and IST1 are unique in that they bind to both Vps4 and its cousin spastin. For CHMP1B and IST1 only, but not other ESCRT-III proteins, spastin is likely to contribute to treadmilling on the basis of its close similarity to Vps4, i.e. its ability to bind tightly to these proteins (Reid et al., 2005; Yang et al., 2008) and its role in the scission of endosomal recycling tubes (Allison et al., 2013, 2017, 2019). However, this remains to be tested directly. If true, this may have profound implications for the biological function of spastin, whose only biological function is commonly assumed to be that of severing microtubules (Roll-Mecak and Vale, 2005).
Geometry of scission
In in vitro systems that consist of bare lipid membranes and purified ESCRTs, formation of a bud neck is sufficient to recruit the set of core ESCRT-III proteins (Snf7, Vps24 and Vps2 in yeast; CHMP2, CHMP3 and CHMP4 in humans) involved in scission (Schöneberg et al., 2018; Bertin et al., 2019). In cells, where many proteins compete for space on membrane, ESCRT-III recruitment is tightly regulated and depends not just on membrane shape but also on the presence of the appropriate upstream factors. These upstream factors include – among many others – ESCRT-0 in multivesicular body biogenesis, HIV-1 Gag in HIV-1 release and CEP55 in mammalian cytokinesis (Hurley, 2015; Vietri et al., 2020). With the main exception of nuclear membrane reformation, most ESCRT-mediated pathways involve the recruitment and nucleation of ESCRT-III by ESCRT-I and ESCRT-II, as well as ALIX (Hurley, 2015; Vietri et al., 2020). Recently, ESCRT-I was found to be capable of polymerizing into helical structures containing ≤11 subunits (Hoffman et al., 2019). The outer diameter of this structure is 60 nm, which matches the inner diameter of a typical membrane bud neck that would be an ESCRT scission substrate as, for example, observed in HIV-1 release (Flower et al., 2020). We found that inhibition of this polymerization disrupts both HIV-1 egress and autophagosome closure (Flower et al., 2020), suggesting that such polymerization is required in multiple ESCRT-dependent scission pathways. Uniquely, cytokinesis, in which the ESCRTs nucleate at the outer rim of the midbody to form spirals with a diameter of >1 μm (Elia et al., 2011; Guizetti et al., 2011; Goliand et al., 2018), involves ESCRT assembly on a scale larger than any of the other known ESCRT-dependent processes and, therefore, has unique features that do not fit easily into this scheme. Coarse-grained simulations of HIV budding has shown that ESCRT-I polymerization could serve as a geometrical checkpoint for viral egress by recognizing the correct neck geometry and initiating downstream ESCRT-III polymerization (Flower et al., 2020). This would be consistent with the spoke-like assemblies in HIV-1 budding profiles (Ladinsky et al., 2014) and the gradual accumulation of ESCRT-I at HIV-1 budding sites, whereas ESCRT-III rapidly arrives immediately before scission (Johnson et al., 2018). In this model ESCRT-I gradually accumulates until the membrane neck reaches a geometry that is compatible with ESCRT-I polymerization. The polymerization of ESCRT-I then triggers rapid nucleation of ESCRT-III proteins that bring the membrane to scission (Flower et al., 2020).
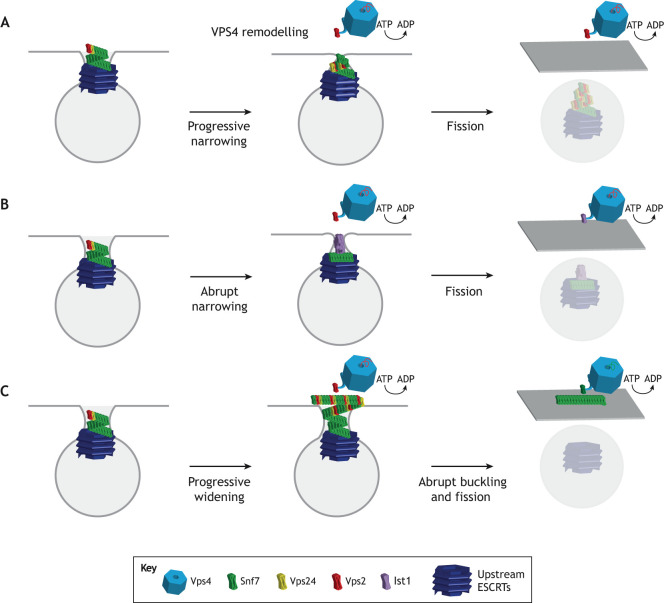
This new observation might hint at one of the other unanswered questions in the field: the location of the bulk of ESCRT molecules following scission. Fig. 3 illustrates three scenarios that follow from the premise of ESCRT-I nucleating the scission machinery from within the bud (Flower et al., 2020). Although conically tapered ESCRT-III structures have been observed and are likely to drive scission (Dobro et al., 2013; Cashikar et al., 2014; McCullough et al., 2015), it is currently unknown whether they grow from wide to narrow (Fig. 3A), narrow to wide (Fig. 3B) or either – depending on the situation. This is important because the dimension of the nucleating ESCRT-I ring corresponds to the narrow end of the cone, implying that the cones might grow from narrow to wide – which is counterintuitive for a constriction mechanism. The paradox would be resolved, however, if the widening cone buckled, leading to constriction and scission. The inward-tapering (wide-to-narrow) model gained some support from a recent EM study showing that VPS4 can remodel CHMP2–CHMP3 tubes into dome-capped structures in a transition that could bring the membrane to scission (Maity et al., 2019). If, as for HIV-1 release, ESCRT proteins nucleate from within the bud, the inward tapering model implies that all ESCRTs are be retained in the buds and that, for example, HIV-1 virions, contain hundreds of ESCRT proteins. Outwards growth, i.e. narrow to wide, from the bud neck followed by buckling (Fig. 3C) is the one most consistent with membrane and filament mechanics models (Agudo-Canalejo and Lipowsky, 2018; Harker-Kirschneck et al., 2019). It is also the only scenario compatible with both nucleation from within the bud and recycling of the bulk of the ESCRT-III complexes into the cytosol following scission, as depicted in Fig. 3.
Fig. 3.
Proposed models for ESCRT-mediated constriction and scission. Upstream ESCRTs, depicted in dark blue, initially recruit the ESCRT-III subunits to the membrane neck, after which three models of constriction and scission are proposed: (A) VPS4 initiates remodeling of the ESCRT-III polymer. Compositional rearrangement leads to gradual constriction and, ultimately, scission. (B) Compositional rearrangement of the polymer leads to specific geometric changes that lead to an abrupt structural change and so constrict the membrane neck. (C) Outward spiral growth causes a buildup of tension, leading the polymer to suddenly buckle from a helix to a flat spiral. This allows the ESCRT-III (but not upstream ESCRTs) to be recycled to the cytosol, in contrast to models A and B where presumably the bulk of the ESCRTs are encapsulated in the nascent vesicle.
Conclusions and perspectives
In this Review, we attempted to present the most-recent developments in biophysical reconstitution, structural biology and in silico simulations of ESCRT function, with in vivo data providing context and constraining the possible models. The field is now converging on a consensus mechanistic model, in which a spiral assembly undergoes progressive remodeling and compositional rearrangement and, thus, constricts the underlying membrane to the point of scission. That said, many key questions remain open. Although it is largely accepted that the role of VPS4 is to enable compositional changes, the thermodynamic determinants of composition are still unclear. A description of membrane scission at atomic resolution is only available for the two-component normal-topology scission by CHMP1B and IST1 (McCullough et al., 2015; Nguyen et al., 2020) but not yet for the, more characteristic, reverse-topology process. In normal-topology scission involving CHMP1B and IST1, the relative roles of VPS4 and spastin need to be clarified. Cryo-EM of scission-capable complexes in the reverse-topology process is still urgently needed to fully understand the ESCRT mechanism. Time-resolved studies with sufficient spatial resolution to address the directionality of filament growth are also crucial. The unique aspects of the large-scale structures formed in cytokinesis need further study. Given the surprising insight that ESCRT-I can form helical structures, unsuspected until recently, it is important to determine how ESCRT-II and ALIX fit into the picture, and how the upstream assemblies are regulated.
Molecular and cell biology textbooks generally portray membrane scission by the ESCRTs as a black box. The picture has become clear enough, though, and there is enough agreement in the field, that it is now reasonable to depict ESCRT action in the textbooks as a consensus version of the models shown in Fig. 3. Researchers in the ESCRT field should be proud of the recent progress, and optimistic that answers to the remaining questions will be forthcoming in the next few years.
Acknowledgements
We thank Kevin Larsen, Thomas Flower and King Cada for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
M.R.P. is a National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) fellow. ESCRT research in J.H.H.’s lab is funded by the National Institutes of Health (NIH) (grant number: R37 AI112442). Deposited in PMC for release after 12 months.
In membrane nanotube experiments, the quoted radii correspond to the distance from the center of the tube to the center of the limiting membrane. With typical biological lipid mixtures, a radius of 4 nm would correspond to scission. Tube radii are measured by comparing the intensity of the fluorescence of a lipid dye to a calibration standard. As tubes become very narrow, the signal decreases and it is difficult to quantitate accurately. When we speak of the radius dropping to ≤10 nm in a nanotube-imaging experiment, we essentially mean that it is entering a size regime where scission is possible.
References
- Adell M. A. Y., Migliano S. M., Upadhyayula S., Bykov Y. S., Sprenger S., Pakdel M., Vogel G. F., Jih G., Skillern W., Behrouzi R. et al. (2017). Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 6, e31652 10.7554/eLife.31652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo-Canalejo J. and Lipowsky R. (2018). Domes and cones: adhesion-induced fission of membranes by ESCRT proteins. PLOS Comput. Biol. 14, e1006422 10.1371/journal.pcbi.1006422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R., Lumb J. H., Fassier C., Connell J. W., Ten Martin D., Seaman M. N. J., Hazan J. and Reid E. (2013). An ESCRT–spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol. 202, 527-543. 10.1083/jcb.201211045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R., Edgar J. R., Pearson G., Rizo T., Newton T., Günther S., Berner F., Hague J., Connell J. W., Winkler J. et al. (2017). Defects in ER–endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216, 1337-1355. 10.1083/jcb.201609033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R., Edgar J. R. and Reid E. (2019). Spastin MIT domain disease-associated mutations disrupt lysosomal function. Front. Neurosci. 13, 1179 10.3389/fnins.2019.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos-Padilla Y., Knorr R. L., Javier-Reyna R., García-Rivera G., Lipowsky R., Dimova R. and Orozco E. (2018). The conserved ESCRT-III machinery participates in the phagocytosis of entamoeba histolytica. Front. Cell. Infect. Microbiol. 8, 53 10.3389/fcimb.2018.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. (2011). MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 23, 452-457. 10.1016/j.ceb.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J. and Emr S. D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982-2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M. and Sundquist W. I. (2009). Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360-1373. 10.1091/mbc.e08-05-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P. W., Kräusslich H.-G., Bräuchle C., Müller B. and Lamb D. C. (2011). Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13, 469-474. 10.1038/ncb2215 [DOI] [PubMed] [Google Scholar]
- Bertin A., de Franceschi N., de la Mora E., Maity S., Alqabandi M., Miguet N., di Cicco A., Roos W., Mangenot S., Weissenhorn W. et al. (2019). Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun. 11, 2663 10.1101/847319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleck M., Itano M. S., Johnson D. S., Thomas V. K., North A. J., Bieniasz P. D. and Simon S. M. (2014). Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc. Natl. Acad. Sci. USA 111, 12211-12216. 10.1073/pnas.1321655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A., Marklew C. J., Ciani B. and Beales P. A. (2019). In vitro membrane remodeling by ESCRT is regulated by negative feedback from membrane tension. iScience 15, 173-184. 10.1016/j.isci.2019.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E., Różycki B., Chung H. S., Herrick D. Z., Canagarajah B., Cafiso D. S., Eaton W. A., Hummer G. and Hurley J. H. (2012). Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure 20, 874-886. 10.1016/j.str.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L.-A., Shen Q.-T., Pavlin M. R. and Hurley J. H. (2015). ESCRT filaments as spiral springs. Dev. Cell 35, 397-398. 10.1016/j.devcel.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Carlton J. G. and Martin-Serrano J. (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908-1912. 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Cashikar A. G., Shim S., Roth R., Maldazys M. R., Heuser J. E. and Hanson P. I. (2014). Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3, e02184 10.7554/eLife.02184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-L., Weigel A. V., Ioannou M. S., Pasolli H. A., Xu C. S., Peale D. R., Shtengel G., Freeman M., Hess H. F., Blackstone C. et al. (2019). Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J. Cell Biol. 218, 2583-2599. 10.1083/jcb.201902061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S. and Roux A. (2015). Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866-879. 10.1016/j.cell.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L. F., Théry C. and Raposo G. (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126, 5553-5565. 10.1242/jcs.128868 [DOI] [PubMed] [Google Scholar]
- De Franceschi N., Alqabandi M., Miguet N., Caillat C., Mangenot S., Weissenhorn W. and Bassereau P. (2019). The ESCRT protein CHMP2B acts as a diffusion barrier on reconstituted membrane necks. J. Cell Sci. 132, jcs217968 10.1242/jcs.217968 [DOI] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M. and Babst M. (2008). Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465-474. 10.1091/mbc.e07-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova R., Aranda S., Bezlyepkina N., Nikolov V., Riske K. A. and Lipowsky R. (2006). A practical guide to giant vesicles. Probing the membrane nanoregime via optical microscopy. J. Phys. Condens. Matter 18, S1151-S1176. 10.1088/0953-8984/18/28/S04 [DOI] [PubMed] [Google Scholar]
- Dobro M. J., Samson R. Y., Yu Z., McCullough J., Ding H. J., Chong P. L.-G., Bell S. D. and Jensen G. J. (2013). Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol. Biol. Cell 24, 2319-2327. 10.1091/mbc.e12-11-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G., Dordor A., Sandrin V., Martinelli N., Sundquist W. I., Schoehn G. and Weissenhorn W. (2013). ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell. Microbiol. 15, 213-226. 10.1111/cmi.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N., Sougrat R., Spurlin T. A., Hurley J. H. and Lippincott-Schwartz J. (2011). Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA 108, 4846-4851. 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G., Lata S., Riches J. D., Briggs J. A. G., Weissenhorn W. and Kozlov M. M. (2009). Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comput. Biol. 5, e1000575 10.1371/journal.pcbi.1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower T. G., Takahashi Y., Hudait A., Rose K., Tjahjono N., Pak A. J., Yokom A. L., Liang X., Wang H.-G., Bouamr F. et al. (2020). A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission. Nat. Struct. Mol. Biol. 27, 570-580. 10.1038/s41594-020-0426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliand I., Adar-Levor S., Segal I., Nachmias D., Dadosh T., Kozlov M. M. and Elia N. (2018). Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep. 24, 1756-1764. 10.1016/j.celrep.2018.07.051 [DOI] [PubMed] [Google Scholar]
- Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T. and Gerlich D. W. (2011). Cortical constriction during abscission involves helices of ESCRT-III–dependent filaments. Science 331, 1616-1620. 10.1126/science.1201847 [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y. and Heuser J. E. (2008). Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389-402. 10.1083/jcb.200707031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker-Kirschneck L., Baum B. and Šarić A. (2019). Changes in ESCRT-III filament geometry drive membrane remodelling and fission in silico. BMC Biol. 17, 82 10.1186/s12915-019-0700-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Zhao Y. and Emr S. D. (2012). The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151, 356-371. 10.1016/j.cell.2012.08.039 [DOI] [PubMed] [Google Scholar]
- Hierro A., Sun J., Rusnak A. S., Kim J., Prag G., Emr S. D. and Hurley J. H. (2004). Structure of the ESCRT-II endosomal trafficking complex. Nature 431, 221-225. 10.1038/nature02914 [DOI] [PubMed] [Google Scholar]
- Hoffman H. K., Fernandez M. V., Groves N. S., Freed E. O. and van Engelenburg S. B. (2019). Genomic tagging of endogenous human ESCRT-I complex preserves ESCRT-mediated membrane-remodeling functions. J. Biol. Chem. 294, 16266-16281. 10.1074/jbc.RA119.009372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. (2015). ESCRTs are everywhere. EMBO J. 34, 2398-2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. and Cada A. K. (2018). Inside job: how the ESCRTs release HIV-1 from infected cells. Biochem. Soc. Trans. 46, 1029-1036. 10.1042/BST20180019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y. J. and Hurley J. H. (2008). Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell 14, 902-913. 10.1016/j.devcel.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A. J., Maiuri P., Lafaurie-Janvore J., Divoux S., Piel M. and Perez F. (2014). ESCRT machinery is required for plasma membrane repair. Science 343, 1247136 10.1126/science.1247136 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Bleck M. and Simon S. M. (2018). Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. eLife 7, e36221 10.7554/eLife.36221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Zhadina M., Bieniasz P. D. and Simon S. M. (2011). Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13, 394-401. 10.1038/ncb2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky M. S., Schluter C., Tam Y. Y. C., Lee S., Ghirlando R., Beach B., Conibear E. and Hurley J. H. (2007). Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell 129, 485-498. 10.1016/j.cell.2007.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky M. S., Kieffer C., Olson G., Deruaz M., Vrbanac V., Tager A. M., Kwon D. S. and Bjorkman P. J. (2014). Electron tomography of HIV-1 infection in gut-associated lymphoid tissue. PLoS Pathog. 10, e1003899 10.1371/journal.ppat.1003899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G. and Weissenhorn W. (2008). Helical structures of ESCRT-III are disassembled by VPS4. Science 321, 1354-1357. 10.1126/science.1161070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.-H., Kai H., Carlson L.-A., Groves J. T. and Hurley J. H. (2015). Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl. Acad. Sci. USA 112, 15892-15897. 10.1073/pnas.1518765113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz M., Crow D. J. G. and Joanny J.-F. (2009). Membrane buckling induced by curved filaments. Phys. Rev. Lett. 103, 38101 10.1103/PhysRevLett.103.038101 [DOI] [PubMed] [Google Scholar]
- Maity S., Caillat C., Miguet N., Sulbaran G., Effantin G., Schoehn G., Roos W. H. and Weissenhorn W. (2019). VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci. Adv. 5, eaau7198 10.1126/sciadv.aau7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklew C. J., Booth A., Beales P. A. and Ciani B. (2018). Membrane remodelling by a lipidated endosomal sorting complex required for transport-III chimera, in vitro. Interface Focus 8, 20180035 10.1098/rsfs.2018.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F. D., Herricks T., Strehler K. M., Miller L. R., Saleem R. A., Rachubinski R. A. and Aitchison J. D. (2018). ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217, 2087-2102. 10.1083/jcb.201706044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clippinger A. K., Talledge N., Skowyra M. L., Saunders M. G., Naismith T. V., Colf L. A., Afonine P., Arthur C., Sundquist W. I. et al. (2015). Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548-1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Frost A. and Sundquist W. I. (2018). Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 34, 85-109. 10.1146/annurev-cellbio-100616-060600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan B. J., Tibbe C., Jeon H., Drabek A. A., Klein T. and Blacklow S. C. (2016). Electrostatic interactions between elongated monomers drive filamentation of Drosophila shrub, a metazoan ESCRT-III protein. Cell Rep. 16, 1211-1217. 10.1016/j.celrep.2016.06.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier V., Larios J., Molinard G., Goujon A., Matile S., Gruenberg J. and Roux A. (2020). Endosomal membrane tension regulates ESCRT-III-dependent intra-lumenal vesicle formation. Nat. Cell Biol. 22, 947-959. 10.1038/s41556-020-0546-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B. E., Chiaruttini N., Redondo-Morata L., Moser von Filseck J., König J., Larios J., Poser I., Müller-Reichert T., Scheuring S., Roux A. et al. (2017). Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787 10.1038/ncb3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck J., Barberi L., Talledge N., Johnson I. E., Frost A., Lenz M. and Roux A. (2020). Anisotropic ESCRT-III architecture governs helical membrane tube formation. Nat. Commun. 11, 1516 10.1038/s41467-020-15327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H. and Weissenhorn W. (2006). Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821-830. 10.1016/j.devcel.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Nguyen H. C., Talledge N., McCullough J., Sharma A., Moss F. R., Iwasa J. H., Vershinin M. D., Sundquist W. I. and Frost A. (2020). Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat. Struct. Mol. Biol. 27, 392-399. 10.1038/s41594-020-0404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D. and Williams R. L. (2007). Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735-739. 10.1038/nature06171 [DOI] [PubMed] [Google Scholar]
- Olmos Y., Hodgson L., Mantell J., Verkade P. and Carlton J. G. (2015). ESCRT-III controls nuclear envelope reformation. Nature 522, 236-239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y., Perdrix-Rosell A. and Carlton J. G. (2016). Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 26, 2635-2641. 10.1016/j.cub.2016.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner A.-K., Mercier V., Jiang X., Moser von Filseck J., Baum B., Šarić A. and Roux A. (2020). An ESCRT-III Polymerization Sequence Drives Membrane Deformation and Fission. Cell 1140-1155. 10.1016/j.cell.2020.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost C., Tsai F. C., Bassereau P. and Simunovic M. (2017). Pulling membrane nanotubes from giant unilamellar vesicles. J. Vis. Exp. 130, e56086 10.3791/56086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.-R., Vargas P., Jimenez A. J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A. M., Manel N. et al. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359-362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Radulovic M., Schink K. O., Wenzel E. M., Nähse V., Bongiovanni A., Lafont F. and Stenmark H. (2018). ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, e99753 10.15252/embj.201899753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., Connell J., Edwards T. L., Duley S., Brown S. E. and Sanderson C. M. (2005). The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14, 19-38. 10.1093/hmg/ddi003 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A. and Vale R. D. (2005). The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15, 650-655. 10.1016/j.cub.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S. and Emr S. D. (2007). Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475-484. 10.1091/mbc.e07-07-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. Y., Obita T., Freund S. M., Williams R. L. and Bell S. D. (2008). A role for the ESCRT system in cell division in archaea. Science 322, 1710-1713. 10.1126/science.1165322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer L. L., Sreetama S. C., Sharma N., Medikayala S., Brown K. J., Defour A. and Jaiswal J. K. (2014). Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5, 5646 10.1038/ncomms6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Lee I.-H., Iwasa J. H. and Hurley J. H. (2017). Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5-17. 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Pavlin M. R., Yan S., Righini M., Lee I.-H., Carlson L.-A., Bahrami A. H., Goldman D. H., Ren X., Hummer G. et al. (2018). ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362, 1423-1428. 10.1126/science.aat1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.-T., Schuh A. L., Zheng Y., Quinney K., Wang L., Hanna M., Mitchell J. C., Otegui M. S., Ahlquist P., Cui Q. et al. (2014). Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 206, 763-777. 10.1083/jcb.201403108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A. and Hanson P. I. (2007). Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068-1079. 10.1111/j.1600-0854.2007.00584.x [DOI] [PubMed] [Google Scholar]
- Skowyra M. L., Schlesinger P. H., Naismith T. V. and Hanson P. I. (2018). Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 10.1126/science.aar5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B., Callan-Jones A., Manneville J.-B., Nassoy P., Joanny J.-F., Prost J., Goud B. and Bassereau P. (2009). Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA 106, 5622-5626. 10.1073/pnas.0811243106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S. and Sundquist W. I. (2007). ESCRT-III recognition by VPS4 ATPases. Nature 449, 740-744. 10.1038/nature06172 [DOI] [PubMed] [Google Scholar]
- Sundquist W. I. and Kräusslich H.-G. (2012). HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924 10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., He H., Tang Z., Hattori T., Liu Y., Young M. M., Serfass J. M., Chen L., Gebru M., Chen C. et al. (2018). An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9, 2855 10.1038/s41467-018-05254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Henne W. M., Borbat P. P., Buchkovich N. J., Freed J. H., Mao Y., Fromme J. C. and Emr S. D. (2015). Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife 4, e12548 10.7554/eLife.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrason Risa G., Hurtig F., Bray S., Hafner A. E., Harker-Kirschneck L., Faull P., Davis C., Papatziamou D., Mutavchiev D. R., Fan C. et al. (2020). The proteasome controls ESCRT-III–mediated cell division in an archaeon. Science 369, eaaz2532 10.1126/science.aaz2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H., Veprintsev D. B. and Williams R. L. (2004). Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 279, 28689-28696. 10.1074/jbc.M400023200 [DOI] [PubMed] [Google Scholar]
- Vasan R., Rudraraju S., Akamatsu M., Garikipati K. and Rangamani P. (2020). A mechanical model reveals that non-axisymmetric buckling lowers the energy barrier associated with membrane neck constriction. Soft Matter 16, 784-797. 10.1039/C9SM01494B [DOI] [PubMed] [Google Scholar]
- Vietri M., Schink K. O., Campsteijn C., Wegner C. S., Schultz S. W., Christ L., Thoresen S. B., Brech A., Raiborg C. and Stenmark H. (2015). Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231-235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Vietri M., Radulovic M. and Stenmark H. (2020). The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25-42. 10.1038/s41580-019-0177-4 [DOI] [PubMed] [Google Scholar]
- Webster B. M., Colombi P., Jäger J. and Lusk C. P. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388-401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T. and Hurley J. H. (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864-869. 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J. and Hurley J. H. (2009). Membrane scission by the ESCRT-III complex. Nature 458, 172-177. 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C. and Hurley J. H. (2008). Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278-1286. 10.1038/nsmb.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Stjepanovic G., Shen Q., Martin A. and Hurley J. H. (2015). Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492 10.1038/nsmb.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G. and Göttlinger H. (2006). Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA 103, 19140-19145. 10.1073/pnas.0603788103 [DOI] [PMC free article] [PubMed] [Google Scholar]