Abstract
Gastric cancer is the second most common cause of cancer-associated death in Asia. The incidence and mortality rates of gastric cancer have markedly increased in the past few decades. Therefore, the identification of novel gastric cancer biomarkers are needed to determine prognosis. The role of serpin peptidase inhibitor clade A member 1 (SERPINA1) has been studied in several types of cancer; however, little is known about its mechanism in gastric cancer. The present study aimed to evaluate SERPINA1 as a potential prognostic biomarker in gastric cancer and to identify the possible mechanisms underlying its action. The expression levels of SERPINA1 in several gastric cancer datasets were assessed, and it was identified that high expression of SERPINA1 was associated to poor clinical outcomes. Furthermore, using histochemical analysis, western blotting, apoptotic analysis, gap closure and invasion assays in cell lines, it was reported that silencing of SERPINA1 inhibited the formation of cellular pseudopodia and did not affect apoptosis, but promoted cell cycle S-phase entry. In addition, overexpression of SERPINA1 increased the migration and invasion of gastric cancer cells, whereas knockdown of SERPINA1 decreased these functions. Moreover, SERPINA1 overexpression increased the protein levels of SMAD4, which is a key regulator of the transforming growth factor (TGF)-β signaling pathway. Taken together, the present data demonstrated that SERPINA1 promotes gastric cancer progression through TGF-β signaling, and suggested that SERPINA1 may be a novel prognostic biomarker from tumor tissue biopsy in gastric cancer.
Keywords: serpin peptidase inhibitor clade A member 1, gastric cancer, SMAD4, tumor progression, prognosis
Introduction
Gastric carcinoma is the fifth most frequently diagnosed cancer and the third leading cause of cancer-associated death worldwide (1). In 2018 alone, 1,033,701 new cases and 782,685 deaths from gastric cancer were expected globally (1). The 5-year overall survival rate of patients with metastatic gastric cancer is only 2%, with a median survival time of 8.6 months (2). Several studies have identified various oncogenes and tumor suppressors that regulate the tumorigenesis of gastric cancer (3,4). For instance, TP53 regulates target genes in response to cellular stress and BRCA2 is involved in DNA repair, which are both major genes that are frequently mutated in gastric cancer (5,6). However, no well-established targets besides human epidermal growth factor receptor 2 (7,8) have been shown to modify the outcomes of patients with gastric cancer. Therefore, it is essential to develop novel targets and therapeutic approaches.
Transforming growth factor (TGF)-β is one of the most extensively expressed cytokines in the tumor microenvironment, and it plays an important role in tumor initiation and progression (9). TGF-β is produced in large amounts by numerous tumor types and is known to be pro-oncogenic (10,11). The activated TGF-β receptor transduces its signal via the phosphorylation of SMAD2/3 and subsequent recruitment of SMAD4 intracellularly. This protein complex then enters the nucleus and initiates the transcription of the mesenchymal markers SNAI1, SNAI2, TWIST1 and ZEB1 (12–14), eventually promoting the migration and invasion of tumor cells (15). It has been reported that high expression of TGF-β1 decreases the overall survival rate of patients with gastric cancer (16).
Serpin peptidase inhibitor clade A member 1 (SERPINA1), a member of the protease inhibitor family of proteins, is primarily synthesized in the liver. It is also produced in certain neoplastic cells, such as those of colon, ovarian and lung cancer (17–19). Tumor cells synthesize and release SERPINA1, which plays a major role in physiological and pathological processes, such as angiogenesis, wound healing, and tumor invasion and metastasis (20). The expression of SERPINA1 has been reported to be correlated with poor prognoses in terms of metastasis among patients with lung, colon and skin cancer (18,19,21,22). However, details regarding the mechanism underlying the role of SERPINA1 in the progression and metastasis of gastric cancer remains unknown.
The present study aimed to provide important insights into the mechanism underlying the pathogenesis of gastric cancer and to evaluate SERPINA1 as a potential prognostic biomarker.
Materials and methods
Database analyses
The ONCOMINE database (https://www.oncomine.org) (23) was used to compare the transcriptional profiles of SERPINA1 in cancer tissues and adjacent normal tissues [cut-off values, P<0.01 and fold-change (FC)>1.5]. The Gene Expression Profiling Interactive Analysis (GEPIA2) bioinformatics tool (http://gepia2.cancer-pku.cn) (24) was used to assess the mRNA expression levels of SERPINA1 in gastric cancer tissues and normal tissues deposited in The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) and Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/) databases. The TCGA stomach adenocarcinoma (STAD) dataset (25) (408 tumor and 36 normal tissues), colon adenocarcinoma (COAD) dataset (26) (275 tumor and 41 normal tissues), esophageal carcinoma (ESCA) dataset (27) (182 tumor and 13 normal tissues) and pancreatic adenocarcinoma (PAAD) dataset (28) (179 tumor and 4 normal tissues), together with the GTEx database (29) (923 normal tissues), were used to determine the association between the mRNA levels of SERPINA1, as well as the overall stage (according to the TNM system) (30) of patients. Survival curves of patients with gastric cancer were generated using the Kaplan-Meier Plotter online tool (https://kmplot.com/) (31). Differentially expressed genes (DEGs) in the TCGA-STAD dataset were analyzed (cut-off values, |log2FC|>2 and q-value <0.01). Gene Set Enrichment Analysis (GSEA) was performed using online tools (http://software.broadinstitute.org/gsea) (32). The cBioPortal for Cancer Genomics (https://www.cbioportal.org/) (5,33) was used to identify genes associated with SERPINA1. Biological function networks were generated using Ingenuity Pathway Analysis (IPA) software (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) (34). The BioGRID (https://thebiogrid.org/) (35) and Wiki-Pi (https://hagrid.dbmi.pitt.edu/wiki-pi/) (36) databases were used to query the protein-protein interactions of SERPINA1.
Cell culture and transfection
The AGS gastric cancer cell line was purchased from the American Type Culture Collection and authenticated using short tandem repeat analysis (Beyotime Institute of Biotechnology) in 2020. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) and 10% fetal calf serum (both Invitrogen; Thermo Fisher Scientific, Inc.) in an incubator at 37°C with 5% carbon dioxide. For SERPINA1-knockdown analyses, AGS cells were transfected with 10 nM SERPINA1 small interfering (si)RNA (siSERPINA1) or non-targeting negative control siRNA (siCONTROL) using RNAiMAX transfection reagent (all Invitrogen; Thermo Fisher Scientific, Inc.). For SERPINA1-overexpression experiments, AGS cells were transfected with 1 µg SERPINA1 overexpression vector pcDNA3.1(+)-SERPINA1 (pSERPINA1) plasmid (GenScript) or 1 µg empty control vector pcDNA3.1(+)-SERPINA1_del (pCONTROL) plasmid (GenScript) and selected using 1,200 µg/ml of G418 for 4 weeks.
Histochemical staining
Cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at room temperature for 15 min and permeabilized in 0.1% Triton X-100 in PBS. Filamentous (F)-actin was labeled with Alexa Fluor 555 phalloidin (165 nM; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature and nuclei were stained with DAPI (1:10; PerkinElmer, Inc.) for 15 min at room temperature. Images were captured using the Mantra quantitative pathology imaging system (v1.03; PerkinElmer, Inc.).
Gap closure assay
An ibidi culture-insert (Ibidi GmBH) was placed in one well of a 6-well cell culture plate. Cells (70 µl/well; 1×106 cells/ml) were seeded into both wells and incubated at 37°C in 5% carbon dioxide. After 24 h, the insert was removed, creating a 500-µm cell-free gap, and subsequently 2 ml/well serum-free cell culture medium (DMEM) was added. Gap closure was tracked and images were captured using an inverted light microscope with a digital camera under ×10 magnification (Olympus Corporation). Quantification of gap closure was performed using ImageJ software (v1.52; National Institutes of Health).
Invasion assay
Matrigel (Corning, Inc.) was diluted with cold H2O to a final concentration of 0.15 µg/µl, and 50 µl diluted Matrigel was added to the upper Transwell chamber (Corning, Inc.). The chambers were left at room temperature overnight and after Matrigel had completely dried on the membranes, cells (100 µl/well; 5×105 cells/ml in DMEM containing 0.1% BSA; Sigma-Aldrich; Merck KGaA) were seeded into the upper chamber and DMEM medium containing 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) was added to the bottom well of the Transwell chamber (600 µl/well). After 24 h of incubation at 37°C, the membranes were fixed with 4% PFA in PBS for 15 min and stained with 0.2% crystal violet for 15 min at room temperature, and the cells that had invaded onto the lower surface of the porous membrane were captured and counted in four random squares using a light microscope under ×10 magnification with a digital camera (Olympus Corporation) and ImageJ software (v1.52; National Institutes of Health), respectively.
Analysis of apoptosis
Apoptosis was determined using Alexa Fluor 488-labeled Annexin V and the PI apoptosis detection kit (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were seeded in a 6-well plate (2 ml/well; 0.15×106 cells/ml) and cultured at 37°C for 48 h. Cells were then harvested and resuspended, stained with Annexin V and propidium iodide (PI), and analyzed using flow cytometry (BD LSRFortessa, BD Biosciences) and FlowJo software (v10.6.1; BD Biosciences) within 1 h.
Cell cycle analysis
Cells were harvested, resuspended, and fixed in 70% cold ethanol at 4°C for 12 h. The cells were then treated with DAPI (BD Biosciences) in PBS for 30 min at room temperature in the dark. The cell cycle was analyzed using the BD LSRFortessa flow cytometer and FlowJo software (v10.6.1; BD Biosciences).
Protein isolation and western blotting
Cultured cells were lysed in a radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.) containing a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Inc.). Lysates were sonicated at 20 kHz for 15 sec on ice and centrifuged at 10,000 × g for 10 min at 4°C. Whole-lysate proteins (15–25 µg) were loaded in each lane. Gel electrophoresis was performed using a 4–12% gradient polyacrylamide gel (Invitrogen; Thermo Fisher Scientific, Inc.) and the proteins were transferred to polyvinylidene difluoride membranes (Invitrogen; Thermo Fisher Scientific, Inc.) according to standard protocols (37). The membranes were then cut and incubated with SERPINA1 (1:1,500, cat. no. ab207303, Abcam), SMAD4 (1:1,500, cat. no. 46535S) or GAPDH (1:3,000, cat. no. 5174S) (both Cell Signaling Technology, Inc.) antibodies according to the known molecular weights of the proteins. HRP-coupled goat anti-rabbit secondary antibody (cat. no. 7074S, Cell Signaling Technology, Inc.) was used at 1:3,000 dilution. Enhanced chemiluminescence signals were recorded and quantified using the ChemiDoc MP imaging system and Image Lab v5.0 software (both Bio-Rad Laboratories, Inc.).
RNA preparation and reverse transcription-quantitative (RT-q) PCR
Total RNA was isolated from cultured cell lines using the RNeasy Plus Mini kit (Qiagen GmBH). RNA was reverse transcribed into cDNA using the SuperScript IV First-Strand Synthesis System kit (Invitrogen; Thermo Fisher Scientific, Inc.) at 50°C for 20 min and then 80°C for 10 min. RT-qPCR was performed using the QuantStudio 7 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the PowerUp SYBR Green Master mix (Invitrogen; Thermo Fisher Scientific, Inc.). The PCR primer sequences were as follows: SERPINA1, Forward: 5′-GAAGAGCGTCCTGGGTCAAC-3′ and reverse: 5′-TGGTCAGCACAGCCTTATGC-3′; SMAD4, forward: 5′-CCCATCCCGGACATTACTGG-3′ and reverse: 5′-TAGGGCAGCTTGAAGGAACC-3′; PAI-1, forward: 5′-GCAAGGCACCTCTGAGAACT-3′ and reverse: 5′-GGGTGAGAAAACCACGTTGC-3′; ACTB, forward: 5′-TGACATTAAGGAGAAGCTGTGCTA-3′ and reverse: 5′-GAGTTGAAGGTAGTTTCGTGGATG-3′. The relative expression of genes was assessed using the 2−ΔΔCq method (38), and the expression of ACTB was used as a reference.
Statistical analyses
Statistical analyses were carried out using GraphPad Prism version 7 (GraphPad Software, Inc.). Data are presented as the mean ± standard deviation and were compared using unpaired Student's t-tests. The association between SERPINA1 expression and tumor stages in digestive system cancer datasets was analyzed using one-way ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
Transcriptional levels of SERPINA1 are elevated in patients with digestive system cancer
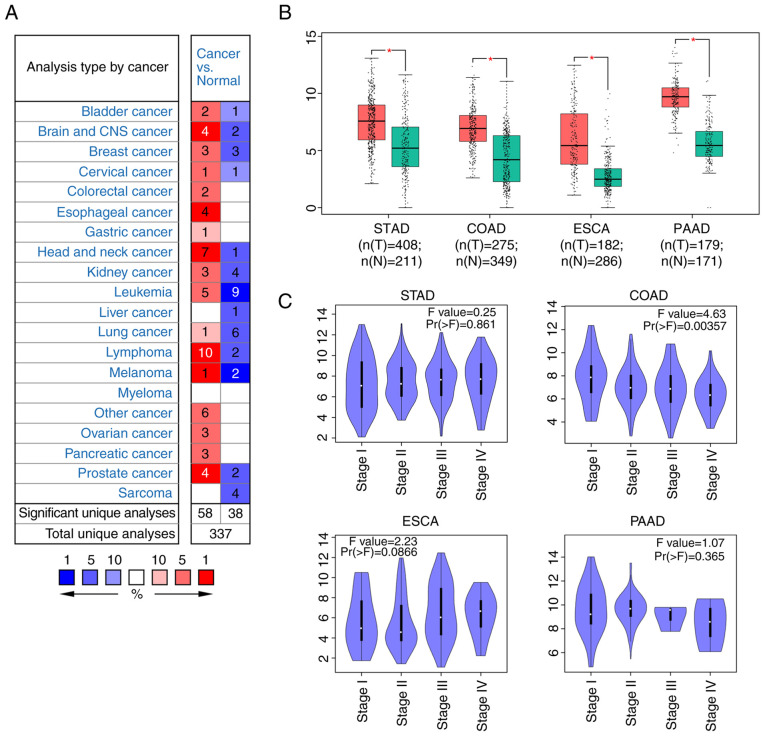
To understand the expression patterns of SERPINA1, the transcriptional levels of SERPINA1 in different cancer types were compared with those in normal samples using data from the ONCOMINE database (23). The mRNA expression levels of SERPINA1 were significantly upregulated in 58 analyses and downregulated in 38 analyses in different cancer types (cut-off values, P<0.01 and FC >1.5) (Fig. 1A). Notably, SERPINA1 was significantly upregulated in analyses of digestive system cancer types, such as gastric, colorectal, esophageal and pancreatic cancer (Fig. 1A). In the studies conducted by Skrzypczak et al (39) and Sabates-Bellver et al (40), SERPINA1 was found to be overexpressed in colon cancer compared with normal samples, with FCs of 3.273 and 2.592, respectively (Table SI). Kimchi et al (41) and Kim et al (42) reported that the mRNA levels of SERPINA1 were also elevated in esophageal adenocarcinoma, with FCs of 23.160 and 6.244, respectively, compared with those in normal esophageal tissues (Table SI). In a study conducted by Wang et al (43), SERPINA1 was found to have an increased FC of 3.319 in patients with gastric cancer compared with that in normal gastric tissues (Table SI). SERPINA1 overexpression was also found in pancreatic ductal adenocarcinoma, with an FC of 5.122 in a study conducted by Badea et al (44), and in pancreatic adenocarcinoma, with an increased FC of 6.752, in the study conducted by Logsdon et al (45) (Table SI).
Figure 1.
Expression profiles of SERPINA1 in different cancer types. (A) Summary of SERPINA1 expression in the ONCOMINE database. The numbers in each cell indicate the ONCOMINE profiles, with significant gene overexpression (red) or underexpression (blue) for each combination. Cell color is determined by the gene rank percentile for the analyses within the cell. (B) The mRNA expression of SERPINA1 in STAD, COAD, ESCA and PAAD. Box plots derived from gene expression data using Gene Expression Profiling Interactive Analysis comparing expression of tumor tissues and normal tissues. *P<0.05. (C) Association between SERPINA1 expression and tumor stage in STAD, COAD, ESCA and PAAD. In (B) and (C) the Y-axis represents the relative expression levels of SERPINA1 in terms of log2(Transcripts Per Million +1). SERPINA1, serpin peptidase inhibitor clade A member 1; STAD, stomach adenocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; PAAD, pancreatic adenocarcinoma.
Using GEPIA2 (24), the mRNA expression levels of SERPINA1 between STAD, ESCA, PAAD and COAD tissues and normal tissues were compared using TCGA and the GTEx databases. The results indicated that the expression levels of SERPINA1 were significantly higher in tissues of the aforementioned cancer types compared with those in normal tissues (all P<0.05; Fig. 1B). The expression of SERPINA1 was also analyzed based on clinical tumor stages for the four aforementioned cancer types. The STAD, ESCA and PAAD groups did not show any significant differences; however, in the COAD group, SERPINA1 expression was significantly associated with clinical stage (P<0.01; Fig. 1C). Overall, the present results suggested that SERPINA1 upregulation may be involved in digestive system cancer development.
Increased mRNA expression of SERPINA1 is associated with poor prognosis of patients with gastric cancer
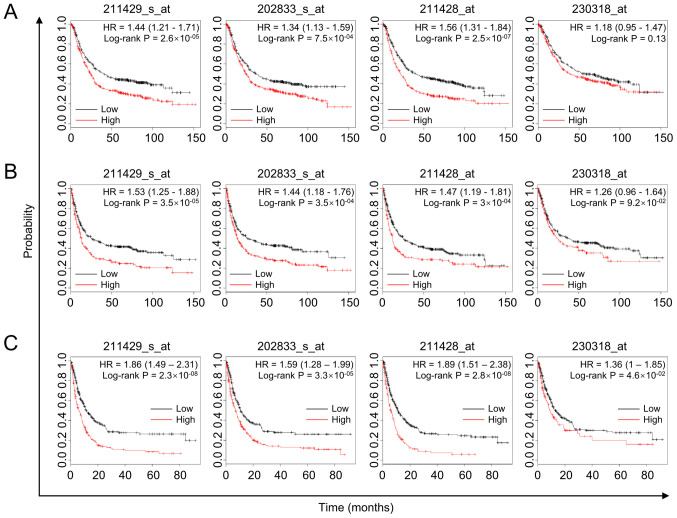
The effect of SERPINA1 on survival among patients with gastric cancer was further analyzed by examining the association between high SERPINA1 mRNA expression and poor prognosis using the Kaplan-Meier Plotter online survival analysis tool (31). The overall survival (OS) of 876 patients with gastric cancer showed that high SERPINA1 expression was associated with shorter survival time in probe 211429_s_at, 202833_s_at and 211428_at (log-rank P=2.6×10−5, 7.5×10−4 and 2.5×10−7, respectively; Fig. 2A). The progression-free survival (PFS) time of 646 patients with gastric cancer also showed the same outcomes in probe 202833_s_at, 211428_at and 211429_s_at (log-rank P=3.5×10−4, 3.0×10−4 and 3.5×10−5, respectively; Fig. 2B), as did the post-progression survival time of 499 patients in all probes (Fig. 2C). In the analyses of OS and PFS, the difference obtained for probe 230318_at did not reach statistical significance (P<0.05). However, a similar tendency was observed with the probes mentioned above (log-rank P=0.13 and 0.09, respectively; Fig. 2A and B). Overall, SERPINA1 is associated with clinical outcome of gastric cancer patients and high SERPINA1 expression indicates a short lifespan.
Figure 2.
The prognostic value of SERPINA1 expression in gastric cancer. (A) Kaplan-Meier plots showing overall survival time among patients with gastric cancer (n=876). (B) Survival curves plotted for progression-free survival time (n=646). (C) Survival curves plotted for post-progression survival time of 499 patients with gastric cancer. High SERPINA1 expression is indicated by the red line, and low SERPINA1 expression by the black line. SERPINA1, serpin peptidase inhibitor clade A member 1; HR, hazard ratio.
SERPINA1 is predicted to be involved in cellular movement
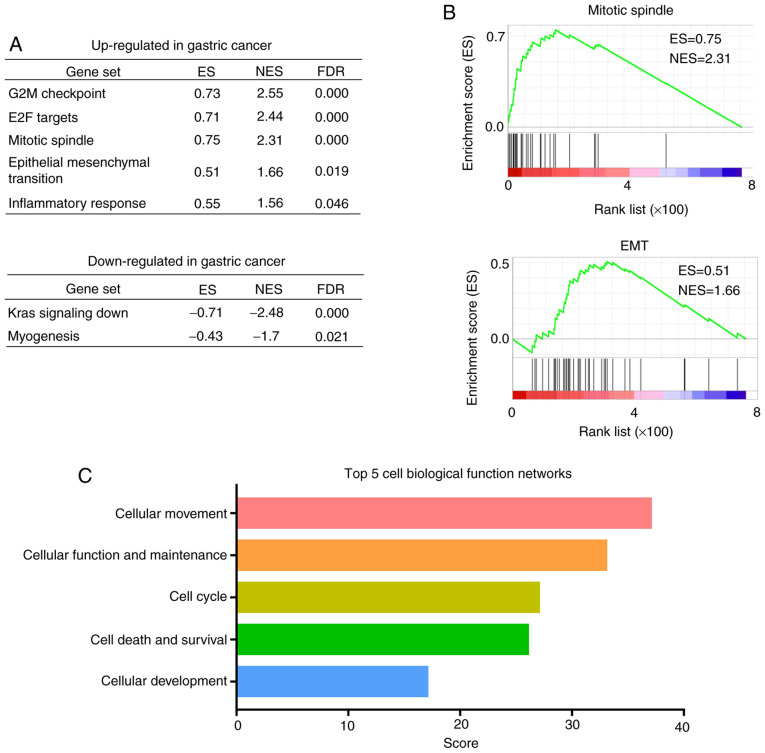
Employing GEPIA2, the DEGs in the gastric adenocarcinoma TCGA-STAD dataset were analyzed. SERPINA1 (Log2FC=2.374, adjusted P=3.34×10−19) was identified as one of 843 DEGs (Table SII). A subsequent GSEA (32) was performed that focused on hallmark gene sets representing 50 specific well-defined, large-scale biological processes and displaying coherent expression (46). By analyzing the TCGA-STAD dataset, five hallmark gene sets (G2M checkpoint, E2F targets, mitotic spindle, epithelial mesenchymal transition and inflammatory response) were identified in which DEGs were upregulated, and two hallmark gene sets (Kras signaling down and myogenesis) in which the DEGs were downregulated [all false discovery rate (FDR) q-value <0.05; Fig. 3A]. Furthermore, two of the upregulated gene sets, ‘mitotic spindle’ [normalized enrichment score (NES)=2.31] (Fig. 3B) and ‘epithelial-mesenchymal-transition’ (EMT; NES=1.66) (Fig. 3B) showed an association with tumor progression.
Figure 3.
Bioinformatics analysis of The Cancer Genome Atlas-stomach adenocarcinoma dataset. (A) Gene Set Enrichment Analysis of mRNA profiles from tumor tissues vs. normal tissues of patients with gastric cancer (FDR q-value <0.05). (B) Selected enrichment score plots of tumor progression related gene sets. (C) Top five cell biological function networks identified using Ingenuity Pathway Analysis. ES, enrichment score; NES, normalized ES; FDR, false discover rate.
Using the enrichment analysis function of cBioPortal for Cancer Genomics (5,33), an online TCGA data analysis platform, it was reported that the expressions of 480 genes were correlated with SERPINA1 expression (all P<0.001; Table SIII) in the TCGA-STAD dataset. To explore the potential functions of SERPINA1, IPA software (34) was then used to illustrate the biological pathways of these SERPINA1-enriched genes. IPA analysis revealed that the top five cell biological function networks were associated with cellular movement, cellular function and maintenance, cell cycle, cell death and survival and cellular development (Fig. 3C). The dominant biological function was cellular movement, which refers to cell migration and motility.
Silencing of SERPINA1 inhibits the formation of cellular pseudopodia, cell migration and invasion in vitro
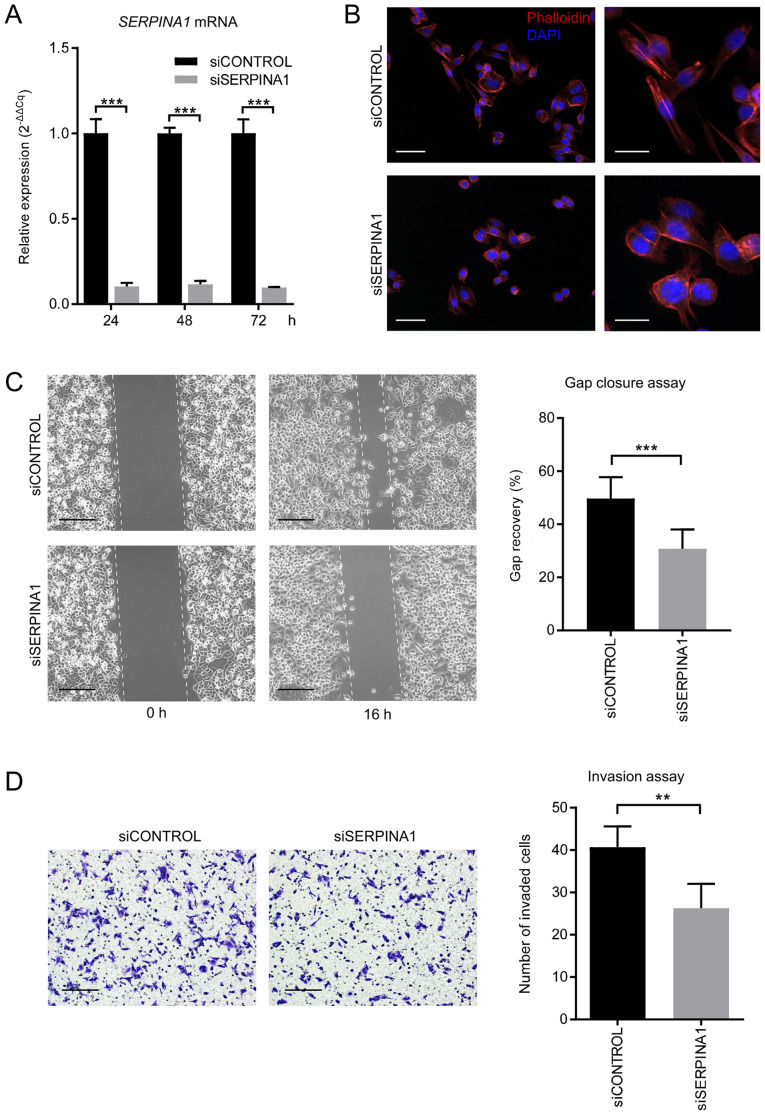
To obtain evidence supporting the role of SERPINA1 in cell movement in vitro, the gene expression of SERPINA1 was knocked down in the AGS gastric cancer cell line using siRNA targeting exon 3 of SERPINA1 mRNA. After 24 h of 10-nM siRNA transfection, the SERPINA1 expression levels in siSERPINA1-treated cells were reduced to 10% of that of the control siRNA-treated cells. This reduction persisted for at least 72 h (Fig. 4A). Following this, phalloidin, a high-affinity F-actin probe (47), was used to assess remodeling of the cytoskeleton. The AGS gastric cell line was stained with Alexa Fluor 555-labeled phalloidin after gene silencing of SERPINA1, and changes in cell morphology were assessed using light microscopy. The extension of long membranous pseudopodia in AGS cells was decreased in cells treated with 10 nM SERPINA1 siRNA compared to control siRNA-treated cells (Fig. 4B).
Figure 4.
Effect of siRNA interference on expression of SERPINA1 in AGS gastric cancer cells. (A) mRNA expression of SERPINA1 detected using reverse transcription-quantitative PCR at 24, 48 and 72 h after transfection with negative control siRNA (siCONTROL) or SERPINA1 siRNA (siSERPINA1). The relative levels of SERPINA1 mRNA were normalized using β-actin (n=3). (B) The morphology of siRNA-treated AGS cells. Actin fibers (filamentous actin) were visualized using a phalloidin-Alexa Fluor 555 probe (red), and nuclei were detected using DAPI (blue) at 72 h after siRNA transfection. Scale bar, 100 µm (left panels) and 20 µm (right panels). (C) Gap closure assay. The areas of the gaps were measured at 0 and 16 h post gap insert removal and % gap recovery of the initial gap areas were compared (n=8). Scale bar, 200 µm. (D) Invasion assay. The invading cells were stained with 0.2% crystal violet and counted in four random squares. n=4. Scale bar, 200 µm. **P<0.01, ***P<0.001. SERPINA1, serpin peptidase inhibitor clade A member 1; si, small interfering.
To assess whether silencing of SERPINA1 affected cell migration, a gap closure assay was performed to evaluate the motility of AGS cancer cells. SERPINA1-knockdown significantly decreased the cell migration area by 18.89% after 16 h compared with that of control siRNA-transfected cells (Fig. 4C). The invasion of the cells was assessed using a Matrigel assay. SERPINA1-knockdown significantly reduced the number of invasive cells from 40.7 cells/field to 26.3 cells/field (Fig. 4D). Overall, these observations further support that SERPINA1 may be involved in the migration and motility of cancer cells.
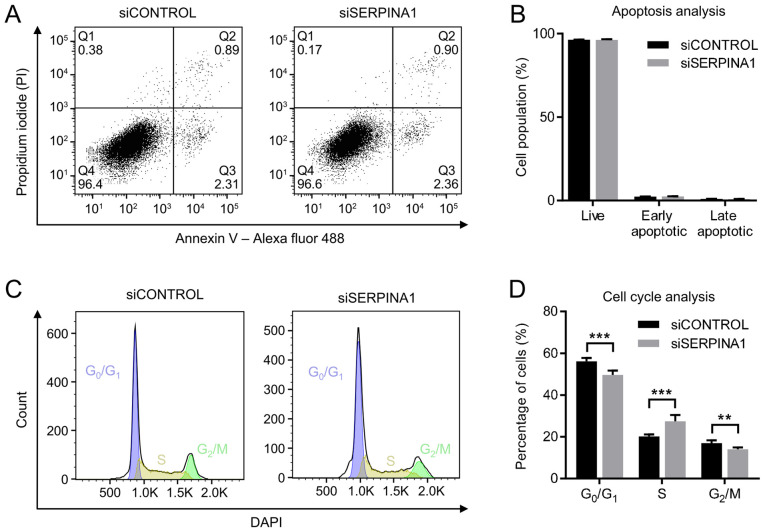
Knockdown of SERPINA1 does not impact apoptosis but promotes the S-phase entry of AGS cells
To exclude the possibility that the inhibition of cell migration and invasion was due to an increased rate of apoptosis and/or a state of proliferation arrest among cells, these effects in the AGS cell line were analyzed after SERPINA1-knockdown. Dual staining of the SERPINA1-downregulated AGS cells with PI and Annexin V-Alexa Fluor 488 was used to assess apoptosis. Flow cytometry revealed no significant difference between SERPINA1 siRNA-treated AGS cells and control siRNA-treated cells regarding the proportions of live cells, early apoptotic cells and late apoptotic cells (Fig. 5A and B). Cell cycle distribution was also evaluated by flow cytometric analysis using DAPI staining. No significant shifts in the G1 and G2 peak positions were observed among AGS cells after SERPINA1 siRNA transfection (Fig. 5C). However, a significantly increased proportion of S phase cells (from 20.2 to 27.4%), decreased G0/G1-phase (from 56.2 to 49.6%) and G2/M-phase (from 17.0 to 14.1%) cells were observed (Fig. 5D) after SERPINA1-knockdown. This suggested that gene silencing of SERPINA1 promoted S phase entry in the AGS cell line.
Figure 5.
Flow cytometry analysis of SERPINA1-knockdown AGS cells. (A) Cell apoptosis analysis was performed using the Annexin V-PI kit. (B) Apoptotic cells were measured at 72 h after siRNA transfection (n=6). (C) Cell cycle analysis was performed with DAPI after 72 h treatment with the indicated siRNAs. (D) Quantification of the cell cycle analysis of AGS cells, n=6. **P<0.01, ***P<0.001. SERPINA1, serpin peptidase inhibitor clade A member 1; si, small interfering.
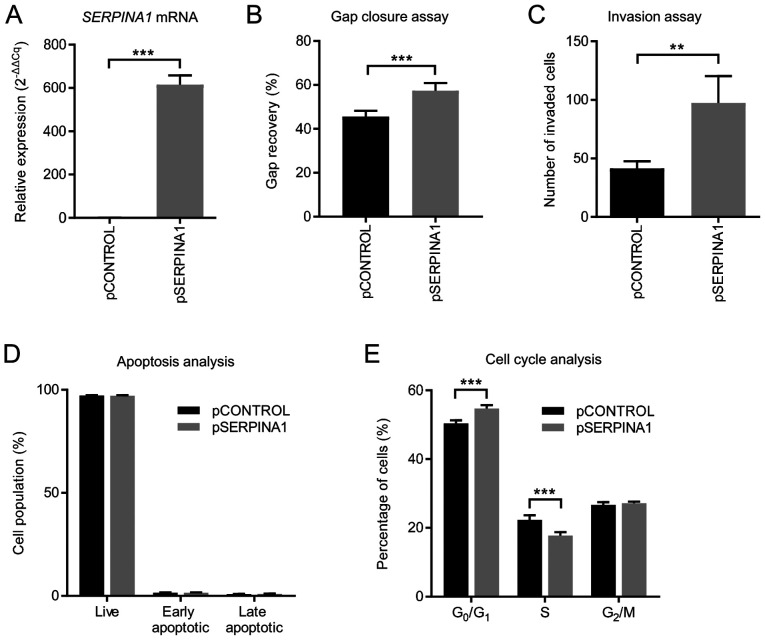
Overexpression of SERPINA1 in gastric cancer cell lines promotes tumor cell migration and invasion, but decreases the number of S phase cells
To confirm the earlier finding that knockdown of SERPINA1 inhibited cell migration and invasion, a SERPINA1-overexpression plasmid (pSERPINA1) and its control plasmid (pCONTROL) were generated. The AGS gastric cancer cell line was transfected with either of these vectors and selected with G418 for 4 weeks. SERPINA1 overexpression in these cells after transfection was confirmed using RT-qPCR (Fig. 6A). The gap closure assay showed that the average migration area of AGS cells transfected with SERPINA1 overexpression plasmid was significantly increased by 11.81% compared with that of cells transfected with control plasmid after 16 h (Fig. 6B). The number of invasive AGS cells significantly increased from 41.4 cells/field to 97.3 cells/field (Fig. 6C) after induced SERPINA1 overexpression. The rate of apoptosis remained the same after SERPINA1-upregulation (Fig. 6D). A decreased proportion of S phase cells (from 22.4 to 17.8%) and an increased proportion of G0/G1 phase cells (from 50.4 to 54.7%) were observed in AGS cells with SERPINA1-overexpression compared with control cells (Fig. 6E). Altogether, these results confirm that increased SERPINA1 expression promotes gastric cancer cell migration and invasion, but inhibits cell proliferation.
Figure 6.
Effect of SERPINA1 overexpression in gastric cancer cells. (A) mRNA expression of SERPINA1 in AGS cells after treatment with overexpression vector (pSERPINA1) or control vector (pCONTROL) was detected using reverse transcription-quantitative PCR after 4 weeks of G418 selection. The relative levels of SERPINA1 mRNA were normalized using β-actin as an internal control. n=3. (B) Gap closure assay. The areas of the gaps were quantified at 0 and 16 h post gap insert removal and % gap recovery of the initial gap areas were compared. n=8. (C) Invasion assay. The invading cells were stained with 0.2% crystal violet, and counted in four random squares. n=4. (D) Apoptotic cells were measured at 72 h after being seeded (n=6). (E) Cell cycle analysis was performed with DAPI after 72 h in culture. n=6. **P<0.01, ***P<0.001. SERPINA1, serpin peptidase inhibitor clade A member 1; si, small interfering.
SERPINA1 upregulates SMAD4, a regulator of the TGF-β signaling pathway
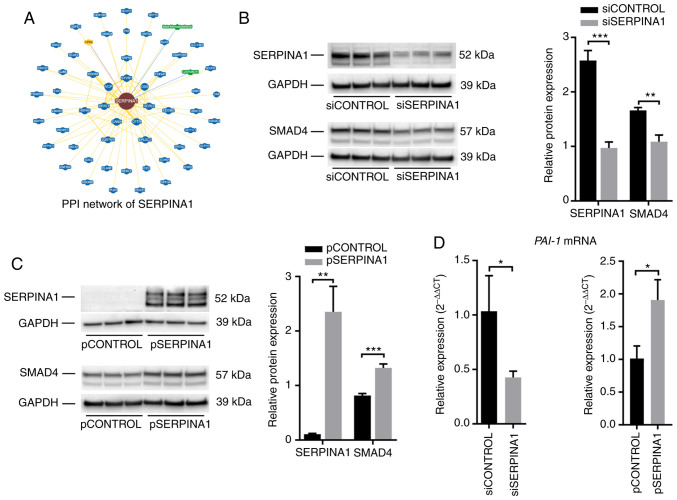
To elucidate the mechanism underlying the effects of SERPINA1, protein-protein interaction data of SERPINA1 were queried using the BioGRID (35) and Wiki-Pi (36) databases. Seventy-three interactions involving SERPINA1 were identified in the BioGRID database (Fig. 7A and Table SIV), and 26 interactions were reported in the Wiki-Pi database (Table SV). SMAD4, which plays a role in regulating TGF-β-mediated EMT in breast cancer (48), nasopharyngeal carcinoma (49) and squamous cell carcinoma of the head and neck (50), was found to interact with SERPINA1 in both databases.
Figure 7.
SERPINA1 regulates the TGF-β signaling pathway through SMAD4. (A) PPI network of SERPINA1 was queried from the BioGRID database. Green nodes represent chemicals, blue nodes represent proteins from the same organism and yellow nodes represent proteins from different organisms. Yellow edges represent protein interactions, blue edges represent chemical interactions and purple edges represent both protein and genetic interactions. (B) SERPINA1 and SMAD4 protein expression were evaluated using western blotting with lysates from AGS cells that were treated with a 72-h transfection of the indicated siRNAs (left panel). Quantification of SERPINA1 and SMAD4 protein. Columns indicate the ratio of SERPINA1 or SMAD4 intensity to GAPDH intensity (right panel, n=3). (C) Western blots of SERPINA1 and SMAD4 proteins in SERPINA1 overexpression vector or control vector-transfected AGS cells (left panel). The relative intensities of the bands were normalized using GAPDH (right panel, n=3). (D) The transcription levels of PAI-1 were measured using reverse transcription quantitative-PCR under the indicated conditions. β-actin was used as an internal control (n=3). *P<0.05, **P<0.01, ***P<0.001. SERPINA1, serpin peptidase inhibitor clade A member 1; SMAD4, SMAD family member 4; PPI, protein-protein interaction; si, small interfering; PAI-1, plasminogen activator inhibitor 1.
To further assess whether SERPINA1 regulated the TGF-β signaling pathway via SMAD4, the expression of SMAD4 protein in both SERPINA1-knockdown and SERPINA1-overexpression AGS cells was analyzed. Western blotting showed that the SMAD4 protein levels were downregulated after silencing SERPINA1 (Fig. 7B), whereas overexpression of SERPINA1 led to an upregulation of SMAD4 protein (Fig. 7C). In addition, changes in the mRNA levels of plasminogen activator inhibitor 1 (a SMAD4-dependent TGF-β signaling target gene) (51) (Fig. 7D) were consistent with changes in SMAD4 protein expression in AGS cells subjected to SERPINA1-knockout and overexpression. These findings suggested that SERPINA1 might regulate the TGF-β signaling pathway through interaction with SMAD4.
Discussion
In several Western Asian countries, gastric cancer is the most commonly diagnosed cancer and, in Eastern Asia, incidence rates have increased markedly in the past few decades; for example, the crude incidence rate in the Republic of Korea was 78.3 per 100,000 among males and 37.2 among females in 2012, compared with a crude incidence rate of 49.2 among males and 27.3 among females in 1999 (1,52,53). However, the underlying mechanisms driving poor clinical outcomes are not understood. Therefore, the identification of novel biomarkers and therapeutic targets for gastric cancer are essential to improve prognosis determination and treatment.
SERPINA1 is a member of the serpins superfamily of protease inhibitors, which play a crucial physiological role in hormone transport, blood clotting, corticosteroid binding and blood pressure regulation (54). However, serpins have also been found to function in tumorigenesis and cancer metastasis (54). Particularly, SERPINA1 has been reported to be overexpressed in various malignant tumors. High expression of SERPINA1 has been observed in prostate (55), lung (55) and colorectal (18) cancer. SERPINA1 was also reported to be upregulated in serum samples of patients with gastric cancer compared with healthy individuals (56). The present study demonstrated that SERPINA1 was overexpressed in colorectal, esophageal, gastric and pancreatic cancer, which indicated a strong association between a high SERPINA1 mRNA levels and digestive system tumorigenesis. In addition, it was reported that overexpression of SERPINA1 was associated with a shorter lifespan in four gastric cancer datasets. Consistent with the present results, Kwon et al (57) also reported an inverse correlation between SERPINA1 expression and survival time in the Korean population.
Despite its clinical relevance, the functional role of SERPINA1 in tumor cells remains unknown. Using bioinformatics analyses of the TCGA-STAD database, the present study revealed that the expression of SERPINA1 was significantly upregulated compared with normal tissues. Further GSEA revealed that the ‘mitotic spindle’ and ‘EMT’ gene sets were enriched in cancer tissues, which were associated with the function of the cytoskeleton. In addition, IPA analysis demonstrated that the top biological function network of SERPINA1-enriched genes was ‘cellular movement’. All these findings suggested that SERPINA1 may be involved in cellular movement, which is mediated by the cytoskeleton (58). To resolve the underlying molecular mechanisms of SERPINA1 in gastric cancer progression, the expression of the SERPINA1 gene was manipulated in vitro. A series of investigations revealed that SERPINA1 regulated cell morphology, migration and invasion and the number of S phase cells, but had no impact on apoptosis in the AGS gastric cell line. Similarly, the regulation of SERPINA1 in cell migration and invasion has also been reported in ovarian (17) and colon cancer (18) cell lines.
Although the mechanisms underlying the role of SERPINA1 in tumor cell migration/invasion and cell cycle have not been fully elucidated, it has been shown that fibronectin is upregulated by SERPINA1 (18). The upregulation of fibronectin promotes cell migration and invasion in colorectal cancer (18). Byon et al (59) and Hernanda et al (60) observed enhanced cell migration through SMAD4 in breast cancer cells and hepatoma cells, respectively. The present study demonstrated that overexpression of SERPINA1 upregulated the expression of SMAD4, and subsequently activated the SMAD4-dependent TGF-β signaling pathway. This promoted the migration and invasion of human gastric cancer cells and reduced the proportion of S phase cells. Based on the present data, it was hypothesized that SERPINA1 may protect the intracellular transport of SMAD4 from the cell membrane to the nucleus (61), as SERPINA1 inhibits the activity of protease and may prevent SMAD4 complex degradation (62). However, co-immunoprecipitation experiments are required to confirm the binding of SERPINA1 and SMAD4, and further signal transduction studies may be helpful to elucidate the precise mechanisms underlying the regulation of SMAD4 expression by SERPINA1.
Taken together, the results of the present study illustrated that SERPINA1 expression is elevated in digestive system cancer tissues and is associated with less favorable clinical prognoses in patients with gastric cancer from database analyses. These data provide evidence that SERPINA1 induces gastric cancer cell migration and invasion, possibly through the TGF-β pathway mediated by SMAD4, which might be a potential mechanism involved in tumor progression. These results suggested that SERPINA1 may be a novel biomarker for tumor metastasis and could be a novel therapeutic target.
Supplementary Material
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by Amgen Asia Research and Development Center.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LJ and LGH conceived and designed this study and wrote the manuscript. LJ performed the experiments and analyzed the data. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ben Kridis W, Marrekchi G, Mzali R, Daoud J, Khanfir A. Prognostic factors in metastatic gastric carcinoma. Exp Oncol. 2019;41:173–175. doi: 10.32471/exp-oncology.2312-8852.vol-41-no-2.13283. [DOI] [PubMed] [Google Scholar]
- 3.Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroentero. 2018;24:2818–2832. doi: 10.3748/wjg.v24.i26.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol. 2016;27:763–769. doi: 10.1093/annonc/mdw040. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, Yang D, Li X, Sun B, Song F, Cao W, Brat DJ, Gao Z, Li H, Liang H, et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112:1107–1112. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boku N. HER2-positive gastric cancer. Gastric Cancer. 2014;17:1–12. doi: 10.1007/s10120-013-0252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh DY, Bang YJ. HER2-targeted therapies-a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 9.Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, et al. A SNAIL1- SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 14.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu C, Xiao X, Wu K, Nie Y, Zhang H, Fan D. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol. 2013;139:1033–1042. doi: 10.1007/s00432-012-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawinkels LJ, Verspaget HW, van Duijn W, van der Zon JM, Zuidwijk K, Kubben FJ, Verheijen JH, Hommes DW, Lamers CB, Sier CF. Tissue level, activation and cellular localisation of TGF-beta1 and association with survival in gastric cancer patients. Br J Cancer. 2007;97:398–404. doi: 10.1038/sj.bjc.6603877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Normandin K, Péant B, Le Page C, de Ladurantaye M, Ouellet V, Tonin PN, Provencher DM, Mes-Masson AM. Protease inhibitor SERPINA1 expression in epithelial ovarian cancer. Clin Exp Metastas. 2010;27:55–69. doi: 10.1007/s10585-009-9303-6. [DOI] [PubMed] [Google Scholar]
- 18.Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK, Jo HJ, Kim HS, Oh N, Song GA, Park DY. Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget. 2015;6:20312–20326. doi: 10.18632/oncotarget.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ercetin E, Richtmann S, Delgado BM, Gomez-Mariano G, Wrenger S, Korenbaum E, Liu B, DeLuca D, Kuhnel MP, Jonigk D, et al. Clinical significance of SERPINA1 gene and its encoded alpha1-antitrypsin protein in NSCLC. Cancers (Basel) 2019;11:1306. doi: 10.3390/cancers11091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janciauskiene S. Conformational properties of serine proteinase inhibitors (serpins) confer multiple pathophysiological roles. Biochim Biophys Acta. 2001;1535:221–235. doi: 10.1016/s0925-4439(01)00025-4. [DOI] [PubMed] [Google Scholar]
- 21.Farshchian M, Kivisaari A, Ala-aho R, Riihilä P, Kallajoki M, Grénman R, Peltonen J, Pihlajaniemi T, Heljasvaara R, Kähäri VM. Serpin peptidase inhibitor Clade A member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol. 2011;179:1110–1119. doi: 10.1016/j.ajpath.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan HJ, Li H, Liu Z, Yuan YC, Mortimer J, Chen S. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget. 2015;6:25815–25827. doi: 10.18632/oncotarget.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45W:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C, Shen H, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, Shih J, Pedamallu CS, Cibulskis C, Dunford A, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–174. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raphael BJ, Hruban RH, Aguirre AJ, Moffitt RA, Yeh JJ, Stewart C, Robertson AG, Cherniack AD, Gupta M, Getz G, et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 31.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabo A, Gyorffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47D:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orii N, Ganapathiraju MK. Wiki-pi: A web-server of annotated human protein-protein interactions to aid in discovery of protein function. PLoS One. 2012;7:e49029. doi: 10.1371/journal.pone.0049029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim B. Western blot techniques. Methods Mol Biol. 2017;1606:133–139. doi: 10.1007/978-1-4939-6990-6_9. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 41.Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, Khodarev NN. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 42.Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One. 2010;5:e15074. doi: 10.1371/journal.pone.0015074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 44.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 45.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 46.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faulstich H, Zobeley S, Rinnerthaler G, Small JV. Fluorescent phallotoxins as probes for filamentous actin. J Muscle Res Cell Motil. 1988;9:370–383. doi: 10.1007/BF01774064. [DOI] [PubMed] [Google Scholar]
- 48.Deckers M, van Dinther M, Buijs J, Que I, Lowik C, van der Pluijm G, ten Dijke P. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 49.Huang G, Du MY, Zhu H, Zhang N, Lu ZW, Qian LX, Zhang W, Tian X, He X, Yin L. MiRNA-34a reversed TGF-β-induced epithelial-mesenchymal transition via suppression of SMAD4 in NPC cells. Biomed Pharmacother. 2018;106:217–224. doi: 10.1016/j.biopha.2018.06.115. [DOI] [PubMed] [Google Scholar]
- 50.Yu C, Liu Y, Huang D, Dai Y, Cai G, Sun J, Xu T, Tian Y, Zhang X. TGF-β1 mediates epithelial to mesenchymal transition via the TGF-β/Smad pathway in squamous cell carcinoma of the head and neck. Oncol Rep. 2011;25:1581–1587. doi: 10.3892/or.2011.1251. [DOI] [PubMed] [Google Scholar]
- 51.Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bray F, Colombet M, Mery L, Piñeros M, Znaor A, Zanetti R, Ferlay J, editors. XI. International Agency for Research on Cancer; Lyon: 2017. [Jul 20;2020 ]. Cancer Incidence in Five Continents. [Google Scholar]
- 53.Curado MP, Edwards B, Shin HR, Storm H, Ferlay J, Heanue M, Boyle P, editors. IX. International Agency for Research on Cancer; Lyon: 2007. [Jul 20;2020 ]. Cancer Incidence in Five Continents. [Google Scholar]
- 54.Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, Nebert DW, Vasiliou V. Update of the human and mouse SERPIN gene superfamily. Hum Genomics. 2013;7:22. doi: 10.1186/1479-7364-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Akawi ZJ, Al-Hindawi FK, Bashir NA. Alpha-1 antitrypsin (alpha1-AT) plasma levels in lung, prostate and breast cancer patients. Neuro Endocrinol Lett. 2008;29:482–484. [PubMed] [Google Scholar]
- 56.Yang J, Xiong X, Wang X, Guo B, He K, Huang C. Identification of peptide regions of SERPINA1 and ENOSF1 and their protein expression as potential serum biomarkers for gastric cancer. Tumour Biol. 2015;36:5109–5118. doi: 10.1007/s13277-015-3163-2. [DOI] [PubMed] [Google Scholar]
- 57.Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY, Jo HJ, Kim DH, Kim GH, Park DY. Serpin peptidase inhibitor clade A member 1 is a biomarker of poor prognosis in gastric cancer. Br J Cancer. 2014;111:1993–2002. doi: 10.1038/bjc.2014.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollard TD. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422:741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 59.Byon CH, Hardy RW, Ren C, Ponnazhagan S, Welch DR, McDonald JM, Chen Y. Free fatty acids enhance breast cancer cell migration through plasminogen activator inhibitor-1 and SMAD4. Lab Invest. 2009;89:1221–1228. doi: 10.1038/labinvest.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hernanda PY, Chen K, Das AM, Sideras K, Wang W, Li J, Cao W, Bots SJ, Kodach LL, de Man RA, et al. SMAD4 exerts a tumor-promoting role in hepatocellular carcinoma. Oncogene. 2015;34:5055–5068. doi: 10.1038/onc.2014.425. [DOI] [PubMed] [Google Scholar]
- 61.Heldin CH, Miyazono K, Ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 62.Komiyama T, Gron H, Pemberton PA, Salvesen GS. Interaction of subtilisins with serpins. Protein Sci. 1996;5:874–882. doi: 10.1002/pro.5560050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christensson A, Laurell CB, Lilja H. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine proteinase inhibitors. Eur J Biochem. 1990;194:755–763. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]
- 64.Korkmaz B, Attucci S, Hazouard E, Ferrandiere M, Jourdan ML, Brillard-Bourdet M, Juliano L, Gauthier F. Discriminating between the activities of human neutrophil elastase and proteinase 3 using serpin-derived fluorogenic substrates. J Biol Chem. 2002;277:39074–39081. doi: 10.1074/jbc.M202918200. [DOI] [PubMed] [Google Scholar]
- 65.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem. 2000;275:27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 66.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Yuan Y, Zhou Y, Guo L, Zhang L, Kuai X, Deng B, Pan Z, Li D, He F. Protein interaction data set highlighted with human Ras-MAPK/PI3K signaling pathways. J Proteome Res. 2008;7:3879–3889. doi: 10.1021/pr8001645. [DOI] [PubMed] [Google Scholar]
- 69.LaLonde DP, Bretscher A. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J Biol Chem. 2011;286:4892–4901. doi: 10.1074/jbc.M110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 2010;40:905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 71.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimori T, Kamiya Y, Nagata K, Kato K, Hosokawa N. Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant. FEBS J. 2013;280:1563–1575. doi: 10.1111/febs.12157. [DOI] [PubMed] [Google Scholar]
- 73.Schaafhausen A, Rost S, Oldenburg J, Muller CR. Identification of VKORC1 interaction partners by split-ubiquitin system and coimmunoprecipitation. Thromb Haemost. 2011;105:285–294. doi: 10.1160/TH10-07-0483. [DOI] [PubMed] [Google Scholar]
- 74.Mikami K, Yamaguchi D, Tateno H, Hu D, Qin SY, Kawasaki N, Yamada M, Matsumoto N, Hirabayashi J, Ito Y, Yamamoto K. The sugar-binding ability of human OS-9 and its involvement in ER-associated degradation. Glycobiology. 2010;20:310–321. doi: 10.1093/glycob/cwp175. [DOI] [PubMed] [Google Scholar]
- 75.Yamaguchi D, Hu D, Matsumoto N, Yamamoto K. Human XTP3-B binds to alpha1-antitrypsin variant null (Hong Kong) via the C-terminal MRH domain in a glycan-dependent manner. Glycobiology. 2010;20:348–355. doi: 10.1093/glycob/cwp182. [DOI] [PubMed] [Google Scholar]
- 76.Nagasawa K, Higashi T, Hosokawa N, Kaufman RJ, Nagata K. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep. 2007;8:483–489. doi: 10.1038/sj.embor.7400933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moussavi-Harami SF, Annis DS, Ma W, Berry SM, Coughlin EE, Strotman LN, Maurer LM, Westphall MS, Coon JJ, Mosher DF, Beebe DJ. Characterization of molecules binding to the 70 K N-terminal region of fibronectin by IFAST purification coupled with mass spectrometry. J Proteome Res. 2013;12:3393–3404. doi: 10.1021/pr400225p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex network: A systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jung CH, Kim YH, Lee K, Im H. Retarded protein folding of the human Z-type α1-antitrypsin variant is suppressed by Cpr2p. Biochem Biophys Res Commun. 2014;445:191–195. doi: 10.1016/j.bbrc.2014.01.156. [DOI] [PubMed] [Google Scholar]
- 80.Huang CH, Hsiao HT, Chu YR, Ye Y, Chen X. Derlin2 protein facilitates HRD1-mediated retro-translocation of sonic hedgehog at the endoplasmic reticulum. J Biol Chem. 2013;288:25330–25339. doi: 10.1074/jbc.M113.455212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan W, Cheng J, Zhang S, Liu X. Cloning and functions of the HBxAg-binding protein XBP1. Mol Med Rep. 2013;7:618–622. doi: 10.3892/mmr.2012.1232. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, Huo K, Ma L, Tang L, Li D, Huang X, Yuan Y, Li C, Wang W, Guan W, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feng L, Zhang J, Zhu N, Ding Q, Zhang X, Yu J, Qiang W, Zhang Z, Ma Y, Huang D, et al. Ubiquitin ligase SYVN1/HRD1 facilitates degradation of the SERPINA1 Z variant/α-1-antitrypsin Z variant via SQSTM1/p62-dependent selective autophagy. Autophagy. 2017;13:686–702. doi: 10.1080/15548627.2017.1280207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen JH, Wen H, Gibson-Corley KN, Glenn KA. FBG1 is the final arbitrator of A1AT-Z degradation. PLoS One. 2015;10:e135591. doi: 10.1371/journal.pone.0135591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dersh D, Jones SM, Eletto D, Christianson JC, Argon Y. OS-9 facilitates turnover of nonnative GRP94 marked by hyperglycosylation. Mol Biol Cell. 2014;25:2220–2234. doi: 10.1091/mbc.E14-03-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khodayari N, Wang RL, Marek G, Krotova K, Kirst M, Liu C, Rouhani F, Brantly M. SVIP regulates Z variant alpha-1 antitrypsin retro-translocation by inhibiting ubiquitin ligase gp78. PLoS One. 2017;12:e172983. doi: 10.1371/journal.pone.0172983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao Y, Han J, Wang Q, Mao Y, Wei M, Jia W, Wei L. A novel interacting protein SERP1 regulates the N-Linked glycosylation and function of GLP-1 receptor in the liver. J Cell Biochem. 2017;118:3616–3626. doi: 10.1002/jcb.26207. [DOI] [PubMed] [Google Scholar]
- 89.Kadowaki H, Nagai A, Maruyama T, Takami Y, Satrimafitrah P, Kato H, Honda A, Hatta T, Natsume T, Sato T, et al. Pre-emptive quality control protects the ER from protein overload via the proximity of ERAD components and SRP. Cell Rep. 2015;13:944–956. doi: 10.1016/j.celrep.2015.09.047. [DOI] [PubMed] [Google Scholar]
- 90.Pankow S, Bamberger C, Calzolari D, Martinez-Bartolome S, Lavallee-Adam M, Balch WE, Yates JR., III ∆F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kadowaki H, Satrimafitrah P, Takami Y, Nishitoh H. Molecular mechanism of ER stress-induced pre-emptive quality control involving association of the translocon, Derlin-1, and HRD1. Sci Rep. 2018;8:7317. doi: 10.1038/s41598-018-25724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giurato G, Nassa G, Salvati A, Alexandrova E, Rizzo F, Nyman TA, Weisz A, Tarallo R. Quantitative mapping of RNA-mediated nuclear estrogen receptor β interactome in human breast cancer cells. Sci Data. 2018;5:180031. doi: 10.1038/sdata.2018.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu S, Ito S, Wada I, Hosokawa N. ER-resident protein 46 (ERp46) triggers the mannose-trimming activity of ER degradation-enhancing α-mannosidase-like protein 3 (EDEM3) J Biol Chem. 2018;293:10663–10674. doi: 10.1074/jbc.RA118.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lamriben L, Oster ME, Tamura T, Tian W, Yang Z, Clausen H, Hebert DN. EDEM1's mannosidase-like domain binds ERAD client proteins in a redox-sensitive manner and possesses catalytic activity. J Biol Chem. 2018;293:13932–13945. doi: 10.1074/jbc.RA118.004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.