Abstract
Plant-microbe interactions are both symbiotic and antagonistic, and the knowledge of both these interactions is equally important for the progress of agricultural practice and produce. This review gives an insight into the recent advances that have been made in the plant-microbe interaction study in the post-genomic era and the application of those for enhancing agricultural production. Adoption of next-generation sequencing (NGS) and marker assisted selection of resistant genes in plants, equipped with cloning and recombination techniques, has progressed the techniques for the development of resistant plant varieties by leaps and bounds. Genome-wide association studies (GWAS) of both plants and microbes have made the selection of desirable traits in plants and manipulation of the genomes of both plants and microbes effortless and less time-consuming. Stress tolerance in plants has been shown to be accentuated by association of certain microorganisms with the plant, the study and application of the same have helped develop stress-resistant varieties of crops. Beneficial microbes associated with plants are being extensively used for the development of microbial consortia that can be applied directly to the plants or the soil. Next-generation sequencing approaches have made it possible to identify the function of microbes associated in the plant microbiome that are both culturable and non-culturable, thus opening up new doors and possibilities for the use of these huge resources of microbes that can have a potential impact on agriculture.
Keywords: Plant-microbe interaction, crop improvement, plant immune response, GWAS, plant growth-promoting bacteria, plant stress management
1. Introduction
The population of the world in December 2019, as reported by the United Nations through the Worldometer, is 7.8 billion, and it is increasing exponentially. Therefore, there is a dire need to meet the increasing demand for food, with enhancement in agricultural practices. This needs to be done in harmony with the ecological balance, which, in turn, translates to a reduction in the use of chemical fertilizers and pesticides. Agricultural research thus needs to focus on alternative options to enhance food production. Molecular study of plant-microbe interactions can be a better alternative for sustainable agriculture [1]. Plants live together with different microorganisms that survive in the rhizosphere below the ground and above in the phyllosphere [2, 3]. They are present within the plants as endophytes, as epiphytes attached to plant surface, and around the roots in the surrounding soil. These microorganisms may have positive, neutral or harmful effects on the health and development of plants [4, 5]. The mechanism behind plant-microbe interaction is still not completely known and there are many questions, which need to be answered. These queries are about plant immune response, signaling pathway (in both plants and microorganisms), beneficial and harmful interactions between plants and microorganisms, etc. These queries will help us understand the whole mechanism of plant-microbe interaction and also help to identify those microorganisms which can be used in the near future to increase crop yield [6]. In agriculture, the association of microbes with plants serves as a catalyst to spontaneously improve yield [7, 8]. Current farming activities, which rely heavily on the intensive use of high yielding agrochemicals, often cause environmental hazards [9]. The consequences of drastic global climate change, diminishing agricultural lands, rapid urbanization, and widespread use of agrochemicals, have had a devastating impact on crop production and ecology around the world, thus prioritizing the need for eco-friendly and sustainable development in agriculture [10]. An important strategy related to climate-smart agricultural practices is to harness the role of microorganisms in increasing plant nutrient quality and crop yield [11]. Beneficial plant-microbe interactions include Plant Growth Promotion (PGP), biotic and abiotic stress protection by plant immune system priming or activating plant defense mechanisms; variable ecosystem adaptation, mycorrhizal symbiosis, nutrient uptake, and plant-accessible transfer of inaccessible nutrient sources have been summarized previously [12].
Additionally, emerging plant diseases pose a serious threat to the world's agricultural industries, food safety, and plant species survival. Therefore, it is important to be able to quickly identify a new phytopathogen and explain- “What factor is responsible?, How it has evolved?, How do they interact with plant systems?”, to answer these questions, it is necessary to mine and annotate the genes involved in the plant-microbe associations from the genomes of both partners. In this regard, DNA and RNA, genomics data analysis, transcriptomics, metagenomics, metabolomics, NGS techniques, and proteomics methods have proved to be valuable tools for exploring plant-microbe interactions and their associations [13, 14].
The key proteins, that are involved in the growth and development of plants, and stress tolerance (both biotic and abiotic), play a vital role in the maintenance of cellular functions in the plants by controlling physiological and biochemical pathways [15, 16]. Recent research in the post-genomic era showed that modern “omics” technologies emerged as an important tool for the discovery of new genes responsible for encoding a functional protein, which will be helpful in many crop advancement programs [17]. In this present review, we have discussed the importance of plant-microbe interaction for crop improvement and stress management, focusing specifically on the advantages of NGS, along with Genome-Wide Association (GWA) mapping.
2. Plant-microbe Interaction For Crop Advancement
Sustainable crop production will be one of the key challenges for the twenty-first century. Due to changes in the environment, agriculture production has been severely affected and therefore, deployment of technologies for the enhancement of agricultural production is necessary to provide sufficient food for the growing population. Current agriculture production practices, such as the improper use of synthetic pesticides and fertilizer, create a long list of environmental and health problems, therefore, to optimize the use of plant-microbe interaction for crop production is one of the better alternatives [18]. Research has shown repeatedly over the last several decades that bacteria and fungi associate closely with their host plants and are capable of fostering plant growth as well as suppressing plant pathogens [19-21]. Besides these, plant biotechnology has contributed to the development of several new crop varieties by using molecular breeding and genetic engineering approach to transfer the resistance genes against pathogens with greater disease resistance, enhance nutrient availability and uptake, and promote biodiversity, this approach is most effective and environmentally friendly to counter microbial diseases as opposed to the use of chemical pesticides [22-25]. In addition, emerging, re-emerging, and endemic plant pathogens continue to affect agricultural production, hence, to tackle these challenges, strategic measures should be taken in the agricultural management system [26]. One of the strategies is the application of Plant Growth-Promoting Rhizobacteria (PGPR) in agriculture [27]. Large-scale application of PGPR to crops as inoculants has been proven to be beneficial in increasing crop yield, as it eventually leads to a reduction in the use of chemical fertilizers and pesticides, which pollute the environment and contaminate food [28]. The recent development of new techniques in the post-genomic era has opened new horizons for the development of superior or novel PGPR strains with enhanced plant growth promotion characteristics; also, the development of transgenic crop plants expressing the PGPR gene has been developed with increased biotic and abiotic stress resistance [29]. In recent years, various high-throughput ‘omics’ approaches have advanced biological science; these approaches include genomics, the study of the structural and functional aspects of genes; and comparison of the degree of gene expression in contrasting genotypes, transcriptomics that quantifies mRNA transcripts, proteomics that analyzes the protein composition and metabolomics which identifies and quantifies cellular metabolites [30-32]. ‘Omics’ platforms are widely used in understanding and selecting efficient endophytic or beneficial strains with various improved traits such as nutrient uptake, imparting abiotic and biotic stress tolerance during their interaction with plants [33, 34]. Comparative studies by large-scale genome and proteome analysis of hosts and pathogens have helped in the identification of various effector genes, key proteins and the nature of pathogenesis induced by pathogen and also the difference in defense mechanisms elicited by the plants [35, 36]. Recent research showed that ‘omics’ is a promising tool for better understanding of plant-microbe interaction and also the discovery of new genes and proteins, which will be helpful in crop advancement for sustainable agriculture [37-39]. Whole-genome sequence analysis of Bacillus aryabhattai AB211 was done using an Illumina platform, HiSeq Illumina paired-end technology, with 151 bp of reads; and, the study performed focused mostly on the plant-microbe interaction elements. This study confirmed the presence of signature genes for plant growth promotion in AB211, such as, chemotaxis, siderophore production, phosphate solubilization, metal ion uptake, etc., which makes AB211 a potential candidate to be used as PGPR [40]. Similarly, other NGS projects have been undertaken to identify strains with potential PGPR properties, for example- S. marcescens UENF-22GI [41], Pantoeaagglomerans strain P5 [42], PGPR consortium of Bacillus cereus AR156, Bacillus subtilis SM21, and Serratia sp. XY21 for sweet pepper disease suppression [43].
3. Plant Defense Systems Against Pathogens
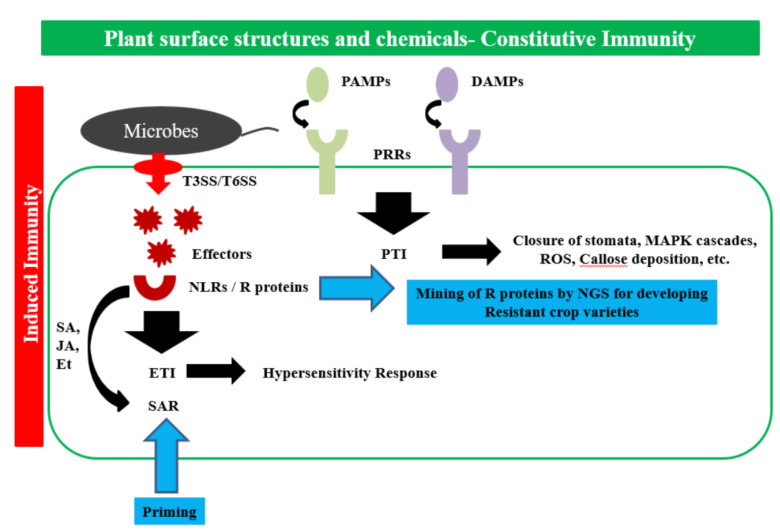
Plants are in constant contact with microorganisms that may be symbiotic or pathogenic. However, not all plants are infected or sick, because they have excellent defense mechanisms that effectively counteract pathogen attack. A pathogen can establish an infection in the plant only if the plant is susceptible and the environmental conditions favor the establishment of the disease [44]. Plant immunity is, however, innate in nature and does not possess mobile immune cells or antibodies [45]. The defense mechanism in plants can be broadly categorized as (i) constitutive- which includes the physical and chemical barriers that are inherently present in the plants, but none the less act as the first line of defense; and (ii) induced which is the second line of defense response and is the active response mounted upon the perception of pathogen attack by the plant [46]. The plant defense response has explained time and again with many models and each model accommodates the findings of that era in which the model or hypothesis was built. We are going to briefly give an overview of the different models and introduce the terms, concepts, and mechanisms of the plant defense response (Fig. 1). Previously the genetic basis of plant-microbe interaction was described in the gene for gene hypothesis [47]. It suggests that obligate parasites and hosts evolve parallelly, and there are specific resistance (R) genes in the host that match specific virulence factors (Avr) in the pathogen. The interaction of compatible R gene and Avr causes the plant to be resistant to the particular pathogen. On the other hand, the interaction of the incompatible R gene and Avr causes the establishment of infection by the pathogen. The zig-zag model was an extension of the gene for gene hypothesis and it proposed that plant immunity has two branches-(i) pathogen-associated molecular pattern triggered immunity (PTI), and (ii) effector-triggered immunity (ETI) [48]. Plant cell surfaces have receptors called pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs), also known as microbe-associated molecular patterns (MAMPs). PAMPs are specific pathogen signatures like the flg22 (a 22 amino acid peptide of the conserved flagellin domain) [49], chitin of the fungal cell wall [50], elongation factor EFTu [51], etc. that the plant cell perceives through PRRs as non- self [48]. Pathogens also modify certain plant molecules, e.g., oligogalactouronides [52] or cellobiose, upon infection or release plant molecules like ATP [53], NAPH, etc.; these molecules are perceived by the plant PRRs as damage-associated molecular patterns (DAMPs) as described elaborately in the danger model [54]. The immune response mounted in response to PAMPs and DAMPs is called the Pattern Triggered Immunity (PTI). The early responses mounted in PTI include- closure of stomata, activation of mitogen-activated protein kinase (MAPK) cascades, reactive oxygen species (ROS) generation, callose deposition and transcriptional activation of resistance-related genes [55]. PTI response is a moderate response and helps to prevent the spread of the pathogen from the site of infection. Phytopathogens that are capable of evading the PTI inject effectors through sophisticated machinery like the type III secretion system and the type VI secretion system into the plant cells to manipulate the plant defense response mechanism [56]. The plant cells then respond to the effectors by mounting the effector-triggered immunity (ETI), which is categorized as a more severe reaction and mostly culminates in hypersensitivity response (HR) or programmed cell death. Apart from PTI and ETI, pathogen perception by the host induces the production of a myriad of toxic substances, phenolic compounds, phytoalexins, proteins, and enzymes, which help to inactivate the pathogen enzymes and toxins. The induced plant response to MAMPs and damage-associated molecular patterns (DAMPs) perception by the receptors on the plant surface starts with the transphosphorylation of the cytoplasmic domains of the receptors, which finally leads to the cascade of chemical changes and changes in the transcriptional regulation in the host plant cell. The chemical response induced includes increasing alkalinity of the growth medium by the release of H+, Ca2+, K+, Cl- ions. Alleviated Ca2+ levels are important for plant immunity as it triggers salicylic acid (SA) and reactive oxygen species (ROS) production, and leads to stomatal closure. The chemicals produced during ETI are SA, jasmonic acid-JA, ethylene and various antimicrobial and cell wall strengthening compounds. The SA produced binds to the NPR3 protein, which is a resistance (R) protein and mediates the degradation of infected cells in a process called HR. HR is characterized by rapid cell death in and around the site of infection and stops the disease progression by the pathogen and is characterized by disease resistance in the plants [57]. Systemic acquired resistance (SAR) is similar to HR; however, it involves translocation of the resistance to distance parts of the plants and involves plant hormones like SA, JA, ethylene, etc. Furthermore, SAR results in prolonged resistance in plants to a broad range of pathogens [58]. The invasion model was then proposed to fill the lacunae of the zig-zag model; in this model, there is no dichotomy in the classification of PTI and ETI and the plant receptors and pathogen triggers. This model broadly categorizes the microbial immunogenic elicitors as invasion patterns (IPs), the receptors as invasion pattern receptors (IPRs) and the response as invasion pattern triggered response (IPTRs) [59]. The latest model that has been proposed is called the spatial immunity model and is an enhancement of the invasion model; in this, the immunogenic response has been categorized on the basis of the site as- extracellular triggered response (ExTR) and intracellular triggered response (InTR) [60]. CRISPR-Cas-9 (Clustered regularly interspaced short palindromic repeats- CRISPR-associated protein 9) technology has been gaining increasing popularity because of the ease, efficiency and reduced cost of the technique. Development of disease-resistant plant varieties is the new age eco-friendly response to the detrimental use of pesticides and chemicals against plant pathogens. Traditional breeding techniques used for constructing disease resistant crop varieties are cumbersome, time taking (sometimes extending for several years), and, have the drawback of undesirable traits being accumulated in the progeny [61]. The plant immune response and the interaction of the plant with pathogens are an extensive field of research; and with the new advances in omics, NGS and molecular biology techniques, new approaches for developing resistant crop varieties and manipulation of the pathogens can be made possible for overall crop improvement. Xanthomonasoryzae.pv.oryzae (Xoo) is the causative agent of bacterial blight (BB) and amounts to a 10-20% loss in rice yields [62]. Xoo injects TALEs (transcription activator-like effectors) through the type III secretion apparatus into the host cell, TALEs bind to specific targets regions in the promoters of the SWEET genes. SWEET genes are involved in the efflux of sugars out of the plant cells; and so, by upregulating the expression of these genes, the pathogen creates a favorable environment for itself for the establishment of the disease. Researchers used CRISPR–Cas9-mediated genome editing technique for creating mutations in the SWEET genes of different rice varieties and through field trials showed that these plants displayed robust, broad-spectrum resistance [63]. Apart from this, various studies have used CRISPR–Cas9 technology for building resistant varieties of different crops such as barley, wheat, tomato, banana, papaya, tomato, etc. [62, 64]. Another alternative of inducing the host plant immune response has been the modification of the pathogens to make it avirulent and use it as a biocontrol agent. There have been studies on mutants of different plant pathogenic fungi that have attenuated virulence; moreover, when applied to plants, they confer plants with heightened resistance to a broad spectrum of diseases. The CRISPR/Cas-9 induced mutations may be used for the development of attenuated fungal avirulent strains [65]. Identification of novel resistant plant genes, the knowledge of which can be used for the development of resistant crop varieties, has been recently studied through the proteomics approach. A comparative analysis of the proteome of the xylem sap of the resistant tomato plant (with R gene- I2); and, the susceptible tomato plant with the endophyte- Fusariumoxysporm (that confers endophyte mediated resistance) was done by nLC-MS/MS quantification of the proteome using MaxQuant software. This led to the enrichment of the protein PR5 in the plants, thus opening doors for the overexpression of these proteins for conferring resistance in plants that lack these resistance genes [66]. RNA-seq analysis of the resistant wild diploid banana variety- Musa acuminate Pahang; and, the triploid susceptible cultivar Brazillian variety; against the Fusarium wilt disease pathogen - Fusarium oxysporm; provided significant insights into the resistant candidate genes in the wild resistant variety that play a potential role in the resistance and can be used as potential candidates for building resistant banana cultivars [43].
Fig. (1).
Conceptual model, on plant immune response. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. Genome-wide Association Studies (GWAS) for Understanding Plant-Microbe Interactions
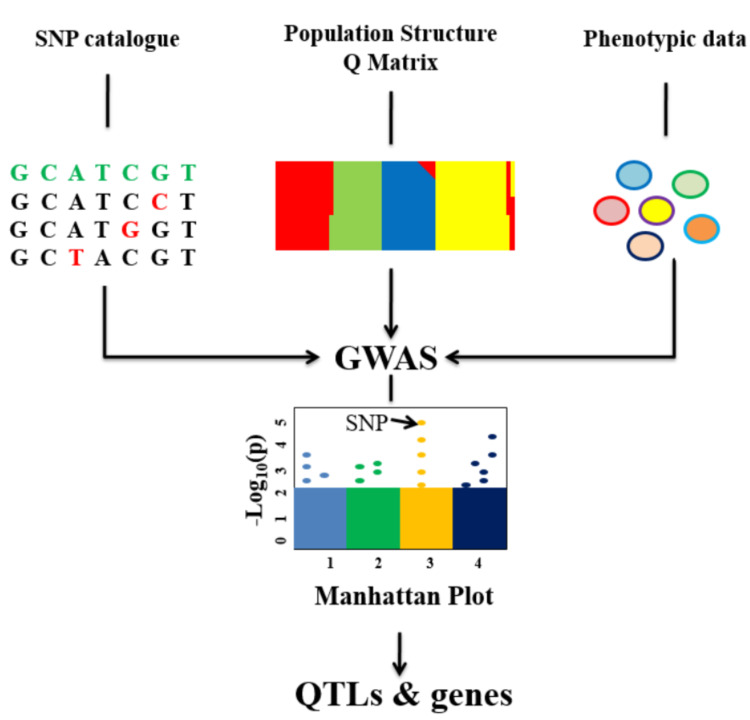
The quality of large-scale comparative genomics in the genomic era is now standard practice for detecting single nucleotide polymorphisms (SNPs) and evaluating their association with important phenotypes using GWAS. The GWAS approach enables the statistical determination of potential SNPs over a population of individuals with a shared evolutionary history [67]. GWAS is currently a powerful tool for detecting genomic regions associated with natural variations in disease resistance in both wild and cultivated plants (Fig. 2) [68]. GWAS allows the correlation of traits in a genetically diverse population by utilizing pre-existing cumulative recombination events in natural populations. However, GWAS dependency on a reference genome complicates the recognition of sequences such as resistance (R) genes, that have greatly diverged from the reference. This limitation was overcome by conducting trait-dependent subsequences (k-mers) based genetics in combination with R gene enrichment sequencing (AgRenSeq), which would enable the discovery and cloning of R genes from a panel of plant diversity. The ability to rapidly clone agronomically important R genes by AgRenSeq could be used for engineering resistance or as specific molecular markers for use in breeding programs [69].
Fig. (2).
Conceptual diagram on GWAS. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Many plant-microbe interaction studies involved GWAS that focused on how a plant genotype influences interactions with a single microbial taxon in pairs [70, 71]. An aggregate of 340 accessions of japonica and indica background was evaluated by GWAS, which recognized 16 loci related with blast resistance, out of which two loci in japonica and a single locus in indica were significantly associated with rice blast resistance [72]. A board of 162 rice cultivars from African nations was dissected with 44 000 SNP chip and 31 genomic regions were found related to rice blast resistance [73]. Plant GWAS has been shown to be effective in identifying genomic locations associated with disease resistance, while microbial GWAS reports genomic regions associated with pathogenicity, but these studies are still in the early stages. It is interesting to note that GWAS mapping was never carried out either independently or jointly on the two equivalents of the plant pathosystem. Characterizing the molecular environment of plant-pathogen interactions will significantly increase our knowledge of the co-evolutionary processes leading to the acceptance of adaptive dynamics of plant species in plant communities [74, 75], thereby strengthening our understanding and forecasts of Emerging Diseases (ED) [76].
On the other hand, plant pathogens GWASs are intended to establish the genes that are responsible for the different phenotypes, including those that the microbial community modulates. Current research on pathogen GWAS found intraspecific variability in characteristics such as aggressiveness in Fusarium graminearum (Fg), a widespread fungal pathogen of wheat, barley, and maize. They used Restriction site-associated DNA sequencing (RADseq) method to perform population genomics which analyzes 213 pathogen isolates from 13 German field populations of Fg and found that high gene flow between these field populations would allow this pathogen to adapt quickly to changes in its environment, including the deployment of resistant cultivars, applications of fungicides and a warming climate [77]. Dalman in 2013 used the GWA mapping method to classify the genetic components that underlie virulence in the Heterobasidion annosum (Ha), a fungal necrotrophic pathogen that is responsible for severe damages in forest conifers. Based on 23 haploid whole-genome sequenced Ha isolates collected in different geographic European countries, 33,018 non-singleton SNPs were used to carry out GWA mapping on virulence scored on both Scots pine and Norway spruce in controlled condition; in both host species, 12 SNPs are strongly associated with virulence [78]. In the study by Gao et al. (2016) on Phaeosphaeria nodorum (Pn), 191 isolates were phenotyped for virulence on two wheat lines and genotyped about 3,000 SNPs distributed throughout the genome, in addition to genetic markers for candidate genes, the discovery SNP of two previous cloned effector genes SnToxA and SnTox3 showed the ability of GWA mapping in Pn to map the fine virulence factors [79]. Recently, a hybrid approach of GWA mapping and comparative genomics has been used for 20 newly sequenced Puccinatriticina isolates from Australia, based on 306,474 SNPs, a polygenic architecture corresponding to 302 genes containing at least one SNP associated with leaf rust virulence on wheat was identified [80]. In the future, more analysis is required to illustrate the potential of GWAS to identify novel determinants of virulence. Through advancements in NGS technology, DNA sequencing has become an attractive alternative to genotyping SNP arrays, expanding GWAS beyond common variants and keeping the possibility of identifying rare alleles and structural variations. Genome sequencing-based strategy for GWAS has found wide-ranging uses in rice [81] including crops, sorghum [82], foxtail millet [83], soybean [84], maize [85, 86], etc. In addition, combining GWAS and gene-based association analysis accompanied by haplotype analysis is a successful means of identifying candidate genes for diverse traits [87]. Furthermore, GWAS studies and related experimental verification should be performed to explore the resistance mechanism and susceptibility mechanism for the plants, which will provide the new strategies for plant breeding against the disease.
5. Plant-microbe Interaction for Stress Management
Abiotic and biotic stress, which directly affects crop productivity, soil health, and fertility, continually affects the agricultural environment. Abiotic and biotic stress can be either natural or triggered by humans. Abiotic factors include drought, air pollution, low or high temperature, moisture and salinity, whereas biotic factors include fungi, bacteria, nematodes, and viruses. These stressors have a significant impact on plant physiological and metabolic changes and gene regulation [88]. Many plants have the ability to alter gene expression and cope with these stresses through acclimatization and adaptation, while others cannot. One of the positive alternatives is the deployment plant-associated microbial population such as mycorrhizal fungi and plant growth-promoting bacteria (PGPB), which help plants, grow and develop under various abiotic and biotic stresses [89]. PGPB has been considered a cost-effective and environmentally friendly means of disease control by activating the cellular component and accumulating secondary metabolites [90]. PGPR has been known to play an essential role in plant growth and metabolism in order to rescue plant growth in stress conditions. Most PGPRs contain the enzyme 1-aminocyclopropane -1- carboxylate (ACC) deaminase and facilitate plant growth by sequestering and cleaving ACC, the immediate precursor of plant hormone ethylene, thus reducing plant ethylene levels induced by stress (cold) [12]. PGPR, such as Pseudomonas reactans, Chryseobacterium humi, improve soil productivity, and plant growth. PGPR have a competitive advantage over fungi for iron uptake due to the production of siderophores. Such siderophores have a very high affinity for iron; iron–siderophore complex can be taken up bacteria. By using this method, PGPR retard pathogen growth by reducing iron availability and thus protecting the plant against diseases [91]. Recent developments in plant biotechnology, including structural and functional genomics, can provide important tools in developed and developing countries for agronomic improvement and also for stress management, for example, the use of molecular markers has enabled the identification, mapping and transfer of many disease resistance genes into tomatoes [92]. Moreover, the recent advancement of NGS identified PGPRs genes that can be attributed to their ability to improve nutrient availability, suppress pathogenic fungi, resist oxidative stress, quorum sensing and ability to break down aromatic and toxic compounds and other abiotic stress [93]. Likewise, gene-editing tools such as Transcription activator-like effector nucleases (TALENs) and CRISPER-Cas have been described to control the pathogen interactions with plants to obtain modified plants [94].
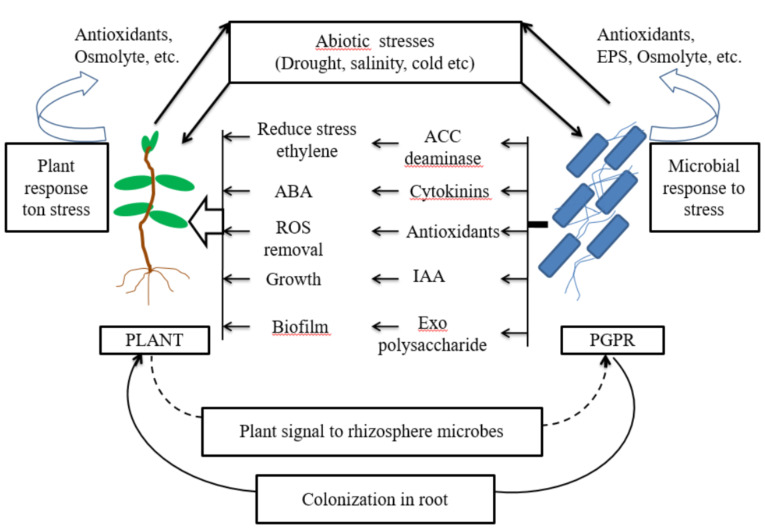
On the other hand, microbes such as Rhizobium, Bacillus, Pseudomonas, Methylobacterium, Variovorax, Enterobacter, etc. have been reported to provide tolerance to host plants under different abiotic stress such as drought, salinity, etc. [95]. The conditions associated with drought stress are limited water content, reduction in cell size, reduced membrane integrity, production of reactive oxygen species and increased leaf senescence, which lead to decreased crop productivity. Microbes evolved, adapt or develop a tolerance mechanism to survive under low water potential. They may form thick walls (Biofilm) or enter the dormant stage, can accumulate osmolytes, produce exopolysaccharides (EPS). Additionally, PGPR have the ability to synthesize plant hormones (IAA, cytokinins, etc), that stimulate plant growth and division under drought conditions [96]. Many PGPR strains produce antioxidants and cytokinin, which result in abscisic acid (ABA) accumulation and degradation of reactive oxygen species. For example, Azospirillum brasilense ameliorates the response of the plant to drought mainly via enhancement of ABA levels to tolerate drought stress [97]. Salinity causes low water potential in soil and it is difficult for the plant to uptake water and nutrients from the soil, which results in osmotic stress. PGPB ameliorate salt stress by potentially accumulating osmolytes in their cytoplasm, which counteract on osmotic stress and maintain cell turgor and plant growth. Microbial EPS induce resistance against salinity by binding with cations, thus making it unavailable to plants under stress conditions [98]. Thus the development of superior or novel PGPB strains by improving traits can be possible using genetic manipulations in the post-genomic era and can be exploited as a low-input, sustainable and environment-friendly technology for the management of plant stress (Fig. 3).
Fig. (3).
Conceptual diagram, on the plant-microbe interactions under stress. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
6. Applications of Beneficial Microbes for Crop Improvement
The green revolution that started in the 1970s greatly enhanced the agricultural yield and produce through the use of chemical fertilizers and pesticides. However, over the years, there have been reports about the potential hazards of the chemicals on soil, ecology and human health. Only 50% of the nitrogenous fertilizers are assimilated by the plant, the rest is lost through evaporation, drainage or leaching. This leads to a very high level of NO3- and NH4+ in the groundwaters that cause potential threats to human health [99]. This predicament has highlighted the potential use of microbes for crop improvement and this idea has been gaining momentum over many decades, for the sole reason that conventional organic farming alone will not suffice to produce crops with higher yield and greater resistance to disease. Effective microbes (EMs) or the plant growth-promoting microorganisms (PGPM) comprise PGPB, PGPR and vesicular-arbuscular mycorrhizae (AMF) fungi [100]. Studies have implicated various microorganisms with befitting mechanisms for crop improvement and we will be citing a few examples of the same. Biofertility inoculants are the inoculation of microorganisms applied to the soil that greatly enhance plant growth through nutrient acquisition and solubilization. Nitrogen and phosphorous are the sparsely available nutrients in the soil. Microorganisms with enhanced abilities to acquire these nutrients are being studied as potential candidates for nutrient acquisition [101]. PGPR Bacillus sp.1 (FOW1) and Lysinibacillus sp. (FOW7) have been reported to be tolerant to pesticides while still possessing bioremediation activity of enhancing crop yields through proper soil aeration, soil water holding property and boosting the plant growth [102]. Six strains of nitrogen-fixing- endophytic bacteria were tested for their ability to enhance the growth of Piceaglauca x trees, and it was proven that they were indeed able to cause a significant increase in plant biomass and seedling length and possessed enhanced ability to fix atmospheric nitrogen [103]. Fungi have been studied to have more phosphate solubilizing activity than bacteria. Many fungi like Penicillium bilaiae are being commercially marketed (brand name- Jumpstart); this organism solubilizes phosphate by using citric and oxalic acid as phosphate-solubilizing agents [104]. Biocontrol organisms, on the other hand, are antagonists to harmful pathogenic organisms and are being extensively studied and used for field applications [101]. Consortia of endophytic nodule forming bacteria- (Pseudomonas sp.), UFLA 02-286 (Bacillus sp.), and UFLA 04-227 (Burkholderiafungorum)
when applied along with Rhizobium tropici (CIAT 899) were able to enhance the growth of common bean and also control the pathogen Rhizoctonia solani [105]. Recent studies have also exploited the antagonistic activity of the endophytic bacterium Bacillus velezensis OEE1 against Verticillium dahlia, the causal agent of verticillium wilt of olive plants [106]. The metagenomic approach of studying entire genomes of all organisms (both culturable and con-culturable) present in different niches is spiking the interest of many researchers, as it taps on the humongous knowledge of all the beneficial microbes that can be used for plant growth promotion and as biocontrol agents [107]. Instead of concentrating solely on the rhizosphere microbial consortium, researchers have studied that the microbiomes of lichens, alpine mosses, and prime rose were able to promote growth and enhance stress tolerance in economically viable plants like maize and sugarbeet [108]. Functional analysis of the different rhizosphere, phyllosphere, and endosphere metagenomes has provided highly useful insights into species diversity of different microorganisms associated with different plants and habitats. Furthermore, functions like nutrient acquisition (nitrogen fixation, phosphorus utilization, and iron mobilization), as well as stress tolerance, have been associated with these microbiomes in these studies [109-111]. Metagenome analysis of the phenol adapted refinery wastewater yielded a novel genome of a novel member from Bradyrhizobiaceae family with unique properties like nitrogen fixation, nitrate uptake and conversion to nitrite, sulfate utilization, iron uptake and aromatic compound (phenol) utilization [112]. In the recent years, there have been tremendous advancements in non-culturing techniques like genomics, proteomics, metabolomics, and molecular biology; which include techniques like- DNA cloning, Sanger sequencing, denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (TRFLP), fluorescence in situ hybridization (FISH), stable isotope probe (SIP), the most recent next-generation sequencing (NGS), etc. These techniques provide impressive insights into non-culturable metagenomes and also help in the functional characterization of these microorganisms [113]. The use of these metagenomic consortia for field applications has now been gaining momentum, however, it has its own roadblocks like-pathogenesis caused by unknown microorganisms in the consortium, food contamination that could be hazardous to human health, and a more practicality based problem of the inability of the microorganisms to be cultured [113]. Strategies to culture non-culturable members of the microbiome are currently being studied and need new media and culture techniques to be developed. These strategies will help provide greater insights into the beneficial as well as the harmful microbes associated with the consortium and help in creating better formulations for use in agricultural fields [114].
Conclusion, Future Prospects and Challenges
Beneficial microbial-plant interaction can lead to promising solutions for environmentally sustainable farming. Moreover, plant-microbe interaction has played a vital role in developing the biofertilizer, biocontrol and bioremediation agents in sustainable agriculture. Although there is plenty of literature on plant-microbe interaction, the molecular mechanism underlying genes function and signal transduction during beneficial and pathogenic interaction are lacking. Therefore, understanding the genetic basis of plant-microbe relationship with the next-generation sequencing technology along with various ‘omics’ technologies will be the emerging tool to provide extensive and in-depth knowledge on the biological phenomenon to improve plant health, disease control, improve food quality and enhance plant stress (both biotic and abiotic) management [115, 116]. The near future faces many challenges in this area of research that need to be addressed for an integrated understanding of plant-pathogen interactions. These challenges are mainly, identification of key factors involved in such interaction during plant immune responses, detection and effective management of new emerging and re-emerging plant pathogens and development of pathogen-resistant crops. In the post-genomic era, understanding the mechanism of plant microbe-interaction could help mitigate these challenges, thus enhancing sustainable agriculture. Genomics tools mentioned in this review, such as GWAS, will continue to provide us novel disease resistance or defense-related genes that can be incorporated into crops through biotechnological approaches, which will become increasingly popular in the next few years and will further advance our understanding towards crop advancement.
Acknowledgements
Author RKA is thankful to MEXT: Monbukagakusho scholarship. Author PS is thankful to University Grants Commission, India, for Junior and Senior Research Fellowships.
Consent for Publication
Not applicable.
Funding
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Johansson J.F., Paul L.R., Finlay R.D. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 2004;48(1):1–13. doi: 10.1016/j.femsec.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Bennett R.A., Lynch J.M. Colonization potential of bacteria in therhizosphere. Curr. Microbiol. 1981;6:137–138. doi: 10.1007/BF01642386. [DOI] [Google Scholar]
- 3.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003;69(4):1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith K.P., Goodman R.M. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 1999;37:473–491. doi: 10.1146/annurev.phyto.37.1.473. [DOI] [PubMed] [Google Scholar]
- 5.Berg G., Krechel A., Ditz M., Sikora R.A., Ulrich A., Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 2005;51(2):215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Farrar K., Bryant D., Cope-Selby N. Understanding and engineering beneficial plant-microbe interactions: plant growth promotion in energy crops. Plant Biotechnol. J. 2014;12(9):1193–1206. doi: 10.1111/pbi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson L.M. Plant growth promoting rhizobacteria (PGPR): Prospects for new inoculants. Crop Manag. 2004;3(1) doi: 10.1094/CM-2004-0301-05-RV. [DOI] [Google Scholar]
- 8.Bhattacharyya P.N., Goswami M.P., Bhattacharyya L.H. Perspective of beneficial microbes in agriculture under changing climatic scenario: a review. J. Phytol. 2016;8:26–41. doi: 10.19071/jp.2016.v8.3022. [DOI] [Google Scholar]
- 9.Singh J.S., Pandey V.C., Singh D.P. Efficient soil microorganisms: a new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 2011;140:339–353. doi: 10.1016/j.agee.2011.01.017. [DOI] [Google Scholar]
- 10.Glick B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014;169(1):30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton C.E., Bever J.D., Labbe J., Yang X., Yin H. Mitigating climate change through managing constructed microbial communities in agriculture. Agric. Ecosyst. Environ. 2016;216:304–308. doi: 10.1016/j.agee.2015.10.006. [DOI] [Google Scholar]
- 12.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 13.Thynne E., McDonald M.C., Solomon P.S. Phytopathogen emergence in the genomics era. Trends Plant Sci. 2015;20(4):246–255. doi: 10.1016/j.tplants.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Withers S., Gongora-Castillo E., Gent D., Thomas A., Ojiambo P.S., Quesada-Ocampo L.M. Using next-generation sequencing to develop molecular diagnostics for Pseudoperonospora cubensis, the cucurbit downy mildew pathogen. Phytopathology. 2016;106(10):1105–1116. doi: 10.1094/PHYTO-10-15-0260-FI. [DOI] [PubMed] [Google Scholar]
- 15.Cai M., Qiu D., Yuan T., Ding X., Li H., Duan L., Xu C., Li X., Wang S. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2008;31(1):86–96. doi: 10.1111/j.1365-3040.2007.01739.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007;17(20):1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Olukolu B.A., Tracy W.F., Wisser R., De Vries B., Balint-Kurti P.J. A genome-wide association study for partial resistance to maize common rust. Phytopathology. 2016;106(7):745–751. doi: 10.1094/PHYTO-11-15-0305-R. [DOI] [PubMed] [Google Scholar]
- 18.Barea J.M. Future challenges and perspectives for applying microbial biotechnology in sustainable agriculture based on a better understanding of plant-microbiome interactions. J. Soil Sci. Plant Nutr. 2015;15:261–282. [Google Scholar]
- 19.Davison J. Plant beneficial bacteria. Nat. Biotechnol. 1988;6:282–286. doi: 10.1038/nbt0388-282. [DOI] [Google Scholar]
- 20.Whipps J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001;52(Spec Issue):487–511. doi: 10.1093/jxb/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 21.Veresoglou S.D., Rillig M.C. Suppression of fungal and nematode plant pathogens through arbuscular mycorrhizal fungi. Biol. Lett. 2012;8(2):214–217. doi: 10.1098/rsbl.2011.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith K.P., Handelsman J., Goodman R.M. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. USA. 1999;96(9):4786–4790. doi: 10.1073/pnas.96.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lugtenberg B.J., Chin-A-Woeng T.F., Bloemberg G.V. Microbe-plant interactions: principles and mechanisms. Antonie van Leeuwenhoek. 2002;81(1-4):373–383. doi: 10.1023/A:1020596903142. [DOI] [PubMed] [Google Scholar]
- 24.Akhond M.A.Y., Machray G.C. Biotech crops: technologies, achievements, and prospects. Euphytica. 2009;166(1):47–59. doi: 10.1007/s10681-008-9823-1. [DOI] [Google Scholar]
- 25.Gust A.A., Brunner F., Nürnberger T. Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 2010;21(2):204–210. doi: 10.1016/j.copbio.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Miller S.A., Beed F.D., Harmon C.L. Plant disease diagnostic capabilities and networks. Annu. Rev. Phytopathol. 2009;47:15–38. doi: 10.1146/annurev-phyto-080508-081743. [DOI] [PubMed] [Google Scholar]
- 27.Adesemoye A.O., Torbert H.A., Kloepper J.W. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb. Ecol. 2009;58(4):921–929. doi: 10.1007/s00248-009-9531-y. [DOI] [PubMed] [Google Scholar]
- 28.Adesemoye A.O., Kloepper J.W. Plant-microbes interactions in enhanced fertilizer-use efficiency. Appl. Microbiol. Biotechnol. 2009;85(1):1–12. doi: 10.1007/s00253-009-2196-0. [DOI] [PubMed] [Google Scholar]
- 29.Haggag W.M., Abouziena H.F., Abd-El-Kreem F., El Habbasha S. Agriculture biotechnology for management of multiple biotic and abiotic environmental stress in crops. J. Chem. Pharm. Res. 2015;7(10):882–889. [Google Scholar]
- 30.Allwood J.W., Clarke A., Goodacre R., Mur L.A.J. Dual metabolomics: a novel approach to understanding plant-pathogen interactions. Phytochemistry. 2010;71(5-6):590–597. doi: 10.1016/j.phytochem.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Abdin M.Z., Khan M.A., Ali A., Alam P., Ahmad A., Sarwat M. In: Signal transduction and regulatory networks in plant-pathogen interaction: a proteomics perspective. Stress Signaling in Plants: Genomics and Proteomics Perspective; Sarwat, M.; Ahmad, A. Abdin M., editor. Vol. 1. New York: Springer; 2013. pp. 69–90. [Google Scholar]
- 32.Seo E., Choi D. Choi. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genomics. 2015;14(4):260–267. doi: 10.1093/bfgp/elv011. [DOI] [PubMed] [Google Scholar]
- 33.Maciá-Vicente J.G., Jansson H.B., Talbot N.J., Lopez-Llorca L.V. Real-time PCR quantification and live-cell imaging of endophytic colonization of barley (Hordeum vulgare) roots by Fusarium equiseti and Pochonia chlamydosporia. New Phytol. 2009;182(1):213–228. doi: 10.1111/j.1469-8137.2008.02743.x. [DOI] [PubMed] [Google Scholar]
- 34.Tshikhudo P.P., Ntushelo K., Mudau F.N., Salehi B., Sharifi-Rad M., Martins N., Martorell M., Sharifi-Rad J. Understanding Camellia sinensis using omics technologies along with endophytic bacteria and environmental roles on metabolism. Appl. Sci. (Basel) 2019;9(2):281. doi: 10.3390/app9020281. [DOI] [Google Scholar]
- 35.Mark G.L., Dow J.M., Kiely P.D., Higgins H., Haynes J., Baysse C., Abbas A., Foley T., Franks A., Morrissey J., O’Gara F. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA. 2005;102(48):17454–17459. doi: 10.1073/pnas.0506407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bregar O., Mandelc S., Celar F., Javornik B. Proteome analysis of the plant pathogenic fungus Monilinialaxa showing host specificity. Food Technol. Biotechnol. 2012;50:326–333. [Google Scholar]
- 37.Swarupa V., Pavitra K., Shivashankara K.S., Ravishankar K.V. In: Omics-driven approaches in plant-microbe interaction. Microbial inoculants in sustainable agricultural productivity; Singh, D.P.; Singh, H.B.; Prabha, R. India S., editor. New Delhi: 2016. pp. 61–84. [Google Scholar]
- 38.Cox D.E., Dyer S., Weir R., Cheseto X., Sturrock M., Coyne D., Torto B., Maule A.G., Dalzell J.J. ABC transporter genes ABC-C6 and ABC-G33 alter plant-microbe-parasite interactions in the rhizosphere. Sci. Rep. 2019;9(1):19899. doi: 10.1038/s41598-019-56493-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandra A.K., Kumar A., Bharati A., Joshi R., Agrawal A., Kumar S. 2020.
- 40.Bhattacharyya C., Bakshi U., Mallick I., Mukherji S., Bera B., Ghosh A. Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB21. Front. Microbiol. 2017;8:411. doi: 10.3389/fmicb.2017.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matteoli F.P., Passarelli-Araujo H., Reis R.J.A., da Rocha L.O., de Souza E.M., Aravind L., Olivares F.L., Venancio T.M. Genome sequencing and assessment of plant growth-promoting properties of a Serratia marcescens strain isolated from vermi compost. BMC Genomics. 2018;19(1):750. doi: 10.1186/s12864-018-5130-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shariati J.V., Malboobi M.A., Tabrizi Z., Tavakol E., Owilia P., Safari M. Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 2017;7(1):15610. doi: 10.1038/s41598-017-15820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L.N., Wang D.C., Hu Q., Dai X.Q., Xie Y.S., Li Q., Liu H.M., Guo J.H. Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 2019;10:1668. doi: 10.3389/fmicb.2019.01668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surico G. The concepts of plant pathogenicity, virulence/avirulence and effector proteins by a teacher of plant pathology. Phytopathol. Mediterr. 2013;52(3):399–417. [Google Scholar]
- 45.Hammond-Kosack K.E., Jones J.D.G. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 46.Doughari J.H. An overview of plant immunity. Plant Pathol. Microbiol. 2015;6:322. [Google Scholar]
- 47.Flor H.H. Host-parasite interaction in flax rust-its genetics and other implications. Phytopathology. 1955;45:680–685. [Google Scholar]
- 48.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 49.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18(2):465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckardt N.A. Chitin signaling in plants: insights into the perception of fungal pathogens and rhizobacterial symbionts. Plant Cell. 2008;20(2):241–243. doi: 10.1105/tpc.108.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16(12):3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligo galacturonides. Proc. Natl. Acad. Sci. USA. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., Lee S.Y., Stacey G. Identification of a plant receptor for extracellular ATP. Science. 2014;343(6168):290–294. doi: 10.1126/science.343.6168.290. [DOI] [PubMed] [Google Scholar]
- 54.Boller T., Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 55.Hou S., Yang Y., Wu D., Zhang C. Plant immunity: evolutionary insights from PBS1, Pto, and RIN4. Plant Signal. Behav. 2011;6(6):794–799. doi: 10.4161/psb.6.6.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puhar A., Sansonetti P.J. Type III secretion system. Curr. Biol. 2014;24(17):R784–R791. doi: 10.1016/j.cub.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 2019;20(8):1163–1178. doi: 10.1111/mpp.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandadi K.K., Scholthof K.B. Plant immune responses against viruses: how does a virus cause disease? Plant Cell. 2013;25(5):1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook D.E., Mesarich C.H., Thomma B.P.H.J. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015;53(1):541–563. doi: 10.1146/annurev-phyto-080614-120114. [DOI] [PubMed] [Google Scholar]
- 60.van der Burgh A.M., Joosten M.H.A.J. Plant immunity: thinking outside and inside the box. Trends Plant Sci. 2019;24(7):587–601. doi: 10.1016/j.tplants.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Borrelli V.M.G., Brambilla V., Rogowsky P., Marocco A., Lanubile A.M.G., Brambilla V., Rogowsky P., Marocco A., Lanubile A. The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 2018;9:1245. doi: 10.3389/fpls.2018.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmad S., Wei X., Sheng Z., Hu P., Tang S. CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations and future prospects. Brief. Funct. Genomics. 2020;19(1):26–39. doi: 10.1093/bfgp/elz041. [DOI] [PubMed] [Google Scholar]
- 63.Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.S., Li C., Nguyen H., Liu B., Auguy F., Sciallano C., Luu V.T., Dossa G.S., Cunnac S., Schmidt S.M., Slamet-Loedin I.H., Vera Cruz C., Szurek B., Frommer W.B., White F.F., Yang B. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019;37(11):1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang T., Zhang H., Zhu H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019;6:77. doi: 10.1038/s41438-019-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muñoz I.V., Sarrocco S., Malfatti L., Baroncelli R., Vannacci G. CRISPR-Cas for fungal genome editing: A new tool for the management of plant diseases. Front. Plant Sci. 2019;10:135. doi: 10.3389/fpls.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Lamo F.J., Constantin M.E., Fresno D.H., Boeren S., Rep M., Takken F.L.W. Xylem Sap Proteomics reveals distinct differences between R gene and endophyte-mediated resistance against Fusarium wilt disease in tomato. Front. Microbiol. 2018;9:2977. doi: 10.3389/fmicb.2018.02977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broberg M., Dubey M., Sun M.H., Ihrmark K., Schroers H.J., Li S.D., Jensen D.F., Brandström D.M., Karlsson M. Out in the cold: identification of genomic regions associated with cold tolerance in the biocontrol fungus clonostachysrosea through genome-wide association mapping. Front. Microbiol. 2018;9:2844. doi: 10.3389/fmicb.2018.02844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartoli C., Roux F. Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front. Plant Sci. 2017;8:763. doi: 10.3389/fpls.2017.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arora S., Steuernagel B., Gaurav K., Chandramohan S., Long Y., Matny O., Johnson R., Enk J., Periyannan S., Singh N., Asyraf M.H.M., Athiyannan N., Cheema J., Yu G., Kangara N., Ghosh S., Szabo L.J., Poland J., Bariana H., Jones J.D.G., Bentley A.R., Ayliffe M., Olson E., Xu S.S., Steffenson B.J., Lagudah E., Wulff B.B.H. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019;37(2):139–143. doi: 10.1038/s41587-018-0007-9. [DOI] [PubMed] [Google Scholar]
- 70.Kim S.M., Reinke R.F. A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS One. 2019;14(2):e0211775. doi: 10.1371/journal.pone.0211775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao Y., Liu H., Wu L., Warburton M., Yan J. Genome-wide association studies in maize: praise and stargaze. Mol. Plant. 2017;10(3):359–374. doi: 10.1016/j.molp.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 72.Raboin L.M., Ballini E., Tharreau D., Ramanantsoanirina A., Frouin J., Courtois B., Ahmadi N. Association mapping of resistance to rice blast in upland field conditions. Rice (N. Y.) 2016;9(1):59. doi: 10.1186/s12284-016-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mgonja E.M., Balimponya E.G., Kang H., Bellizzi M., Park C.H., Li Y., Mabagala R., Sneller C., Correll J., Opiyo S., Talbot N.J., Mitchell T., Wang G.L. Genome-wide association mapping of rice resistance genes against Magnaporthe oryzae isolates from four African countries. Phytopathology. 2016;106(11):1359–1365. doi: 10.1094/PHYTO-01-16-0028-R. [DOI] [PubMed] [Google Scholar]
- 74.Karasov T.L., Kniskern J.M., Gao L., DeYoung B.J., Ding J., Dubiella U., Lastra R.O., Nallu S., Roux F., Innes R.W., Barrett L.G., Hudson R.R., Bergelson J. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature. 2014;512(7515):436–440. doi: 10.1038/nature13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roux F., Bergelson J. The genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: the challenges of linking evolutionary genetics and community ecology. Curr. Top. Dev. Biol. 2016;119:111–156. doi: 10.1016/bs.ctdb.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Lambrechts L. Dissecting the genetic architecture of host-pathogen specificity. PLoS Pathog. 2010;6(8):e1001019. doi: 10.1371/journal.ppat.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talas F., McDonald B.A. Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genomics. 2015;16:996. doi: 10.1186/s12864-015-2166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalman K., Himmelstrand K., Olson Å., Lind M., Brandström-Durling M., Stenlid J. A genome-wide association study identifies genomic regions for virulence in the non-model organism Heterobasidion annosum s.s. PLoS One. 2013;8(1):e53525. doi: 10.1371/journal.pone.0053525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Y., Liu Z., Faris J.D., Richards J., Brueggeman R.S., Li X., Oliver R.P., McDonald B.A., Friesen T.L. Validation of genome-wide association studies as a tool to identify virulence factors in Parastagonospora nodorum. Phytopathology. 2016;106(10):1177–1185. doi: 10.1094/PHYTO-02-16-0113-FI. [DOI] [PubMed] [Google Scholar]
- 80.Wu J.Q., Sakthikumar S., Dong C., Zhang P., Cuomo C.A., Park R.F. Comparative genomics integrated with association analysis identifies candidate effector genes corresponding to Lr20 in phenotype‐paired Puccinia triticina isolates from Australia. Front. Plant Sci. 2017;8:148. doi: 10.3389/fpls.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H., Xu X., Vieira F.G., Xiao Y., Li Z., Wang J., Nielsen R., Chu C. The power of inbreeding: NGS‐based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant. 2016;9(7):975–985. doi: 10.1016/j.molp.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 82.Morris G.P., Ramu P., Deshpande S.P., Hash C.T., Shah T., Upadhyaya H.D., Riera-Lizarazu O., Brown P.J., Acharya C.B., Mitchell S.E., Harriman J., Glaubitz J.C., Buckler E.S., Kresovich S. Population genomic and genome-wide association studies of agro climatic traits in sorghum. Proc. Natl. Acad. Sci. USA. 2013;110(2):453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jia G., Huang X., Zhi H., Zhao Y., Zhao Q., Li W., Chai Y., Yang L., Liu K., Lu H., Zhu C., Lu Y., Zhou C., Fan D., Weng Q., Guo Y., Huang T., Zhang L., Lu T., Feng Q., Hao H., Liu H., Lu P., Zhang N., Li Y., Guo E., Wang S., Wang S., Liu J., Zhang W., Chen G., Zhang B., Li W., Wang Y., Li H., Zhao B., Li J., Diao X., Han B. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013;45(8):957–961. doi: 10.1038/ng.2673. [DOI] [PubMed] [Google Scholar]
- 84.Zhou Z., Jiang Y., Wang Z., Gou Z., Lyu J., Li W., Yu Y., Shu L., Zhao Y., Ma Y., Fang C., Shen Y., Liu T., Li C., Li Q., Wu M., Wang M., Wu Y., Dong Y., Wan W., Wang X., Ding Z., Gao Y., Xiang H., Zhu B., Lee S.H., Wang W., Tian Z. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015;33(4):408–414. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]
- 85.Li H., Peng Z., Yang X., Wang W., Fu J., Wang J., Han Y., Chai Y., Guo T., Yang N., Liu J., Warburton M.L., Cheng Y., Hao X., Zhang P., Zhao J., Liu Y., Wang G., Li J., Yan J. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013;45(1):43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 86.Wen W., Li D., Li X., Gao Y., Li W., Li H., Liu J., Liu H., Chen W., Luo J., Yan J. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X., Pang Y., Zhang J., Wu Z., Chen K., Ali J., Ye G., Xu J., Li Z. Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci. Rep. 2017;7(1):17203. doi: 10.1038/s41598-017-17347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramegowda V., Senthil-Kumar M. The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015;176:47–54. doi: 10.1016/j.jplph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 89.Tank N., Saraf M. Salinity-resistant plant growth promoting rhizobacteria ameliorates sodium chloride stress on tomato plants. J. Plant Interact. 2010;5(1):51–58. doi: 10.1080/17429140903125848. [DOI] [Google Scholar]
- 90.Fahad S., Hussain S., Bano A., Saud S., Hassan S., Shan D., Khan F.A., Khan F., Chen Y., Wu C., Tabassum M.A., Chun M.X., Afzal M., Jan A., Jan M.T., Huang J. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: consequences for changing environment. Environ. Sci. Pollut. Res. Int. 2015;22(7):4907–4921. doi: 10.1007/s11356-014-3754-2. [DOI] [PubMed] [Google Scholar]
- 91.Penyalver R., Oger P., López M.M., Farrand S.K. Iron-binding compounds from Agrobacterium spp.: biological control strain Agrobacterium rhizogenes K84 produces a hydroxamate siderophore. Appl. Environ. Microbiol. 2001;67(2):654–664. doi: 10.1128/AEM.67.2.654-664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Foolad M.R. Genome mapping and molecular breeding of tomato. Int. J. Plant Genomics. 2007;2007:64358. doi: 10.1155/2007/64358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta A., Gopal M., Thomas G.V., Manikandan V., Gajewski J., Thomas G., Seshagiri S., Schuster S.C., Rajesh P., Gupta R. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One. 2014;9(8):e104259. doi: 10.1371/journal.pone.0104259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar V., Baweja M., Singh P.K., Shukla P. Recent developments in systems biology and metabolic engineering of plant microbe interactions. Front. Plant Sci. 2016;7:1421. doi: 10.3389/fpls.2016.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumar A., Verma J.P. Does plant-Microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 2018;207:41–52. doi: 10.1016/j.micres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 96.Grover M., Ali S.Z., Sandhya V., Rasul A., Venkateswarlu B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 2010;27(5):1231–1240. doi: 10.1007/s11274-010-0572-7. [DOI] [Google Scholar]
- 97.Cohen A.C., Bottini R., Pontin M., Berli F.J., Moreno D., Boccanlandro H., Travaglia C.N., Piccoli P.N. Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 2015;153(1):79–90. doi: 10.1111/ppl.12221. [DOI] [PubMed] [Google Scholar]
- 98.Choudhary D.K., Kasotia A., Jain S., Vaishnav A., Kumari S., Sharma K.P., Varma A. Bacterial-mediated tolerance and resistance to plants under abiotic and biotic stresses. J. Plant Growth Regul. 2015;35:276–300. doi: 10.1007/s00344-015-9521-x. [DOI] [Google Scholar]
- 99.Savci S. An agricultural pollutant: chemical fertiliser. Int. J. Environ. Sci. Technol. 2012;3:77–80. [Google Scholar]
- 100.Naik K., Mishra S., Srichandan H., Singh P.K., Sarangi P.K. Plant growth promoting microbes: Potential link to sustainable agriculture and environment. Biocatal. Agric. Biotechnol. 2019;21:101356 [Google Scholar]
- 101.Parnell J.J., Berka R., Young H.A., Sturino J.M., Kang Y., Barnhart D.M., DiLeo M.V. andDiLeo, M.V. From the lab to the farm: an industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 2016;7:1110. doi: 10.3389/fpls.2016.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nathiya S., Janani R., Kannan V.R. Potential of plant growth promoting rhizobacteria to overcome the exposure of pesticide in Trigonellafoenum–graecum (fenugreek leaves). Biocatal. Agric. Biotechnol. 2020;140:101493. doi: 10.1016/j.bcab.2020.101493. [DOI] [Google Scholar]
- 103.Puri A., Padda K.P., Chanway C.P. Can naturally-occurring endophytic nitrogen-fixing bacteria of hybrid white spruce sustain boreal forest tree growth on extremely nutrient-poor soils? Soil Biol. Biochem. 2020;140:107642. doi: 10.1016/j.soilbio.2019.107642. [DOI] [Google Scholar]
- 104.Cunningham J.E., Kuiack C. Production of citric and oxalic acids and solubilization of calcium phosphate by Penicillium bilaii. Appl. Environ. Microbiol. 1992;58(5):1451–1458. doi: 10.1128/AEM.58.5.1451-1458.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferreira L.D.V.S.M., Carvalho F.D., Andrade J.F.C., Oliveira D.P., Medeiros F.H.V.D., Moreira F.M.D.S. Co-inoculation of selected nodule endophytic rhizobacterial strains with Rhizobium tropici promotes plant growth and controls damping off in common bean. Pedosphere. 2020;30(1):98–108. doi: 10.1016/S1002-0160(19)60825-8. [DOI] [Google Scholar]
- 106.Azabou M.C., Gharbi Y., Medhioub I. Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M. A. The endophytic strain Bacillus velezensis OEE1: An efficient biocontrol agent against Verticillium wilt of olive and a potential plant growth promoting bacteria. Biol. Control. 2020;143:104168. doi: 10.1016/j.biocontrol.2019.104168. [DOI] [Google Scholar]
- 107.Müller C.A., Obermeier M.M., Berg G. Bioprospecting plant-associated microbiomes. J. Biotechnol. 2016;235:171–180. doi: 10.1016/j.jbiotec.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 108.Zachow C., Müller H., Tilcher R., Donat C., Berg G. Catch the best: novel screening strategy to select stress protecting agents for crop plants. Agronomy (Basel) 2013;3:794–815. doi: 10.3390/agronomy3040794. [DOI] [Google Scholar]
- 109.Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., von Mering C., Vorholt J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA. 2009;106(38):16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., von Mering C., Vorholt J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6(7):1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mendes L.W., Kuramae E.E., Navarrete A.A., van Veen J.A., Tsai S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014;8(8):1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tikariha H., Purohit H.J. Assembling a genome for novel nitrogen-fixing bacteria with capabilities for utilization of aromatic hydrocarbons. Genomics. 2019;111(6):1824–1830. doi: 10.1016/j.ygeno.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 113.Hao D.C., Xiao P. Rhizosphere microbiota and microbiome of medicinal plants: from molecular biology to omics approaches. Chin. Herb. Med. 2017;9(3):199–217. doi: 10.1016/S1674-6384(17)60097-2. [DOI] [Google Scholar]
- 114.Sarhan M.S., Hamza M.A., Youssef H.H., Patz S., Becker M., ElSawey H., Nemr R., Daanaa H.A., Mourad E.F., Morsi A.T., Abdelfadeel M.R., Abbas M.T., Fayez M., Ruppel S., Hegazi N.A. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media - a review. J. Adv. Res. 2019;19:15–27. doi: 10.1016/j.jare.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Knief C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014;5:216. doi: 10.3389/fpls.2014.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kankanala P., Nandety R.S., Mysore K.S. Genomics of plant disease resistance in legumes. Front. Plant Sci. 2019;10:1345. doi: 10.3389/fpls.2019.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]