Abstract
Since the last few decades, the promiscuous and uncontrolled use of plastics led to the accumulation of millions of tons of plastic waste in the terrestrial and marine environment. It elevated the risk of environmental pollution and climate change. The concern arises more due to the reckless and unscientific disposal of plastics containing high molecular weight polymers, viz., polystyrene, polyamide, polyvinylchloride, polypropylene, polyurethane, and polyethylene, etc. which are very difficult to degrade. Thus, the focus is now paid to search for efficient, eco-friendly, low-cost waste management technology. Of them, degradation of non-degradable synthetic polymer using diverse microbial agents, viz., bacteria, fungi, and other extremophiles become an emerging option. So far, very few microbial agents and their secreted enzymes have been identified and characterized for plastic degradation, but with low efficiency. It might be due to the predominance of uncultured microbial species, which consequently remain unexplored from the respective plastic degrading milieu. To overcome this problem, metagenomic analysis of microbial population engaged in the plastic biodegradation is advisable to decipher the microbial community structure and to predict their biodegradation potential in situ. Advancements in sequencing technologies and bioinformatics analysis allow the rapid metagenome screening that helps in the identification of total microbial community and also opens up the scope for mining genes or enzymes (hydrolases, laccase, etc.) engaged in polymer degradation. Further, the extraction of the core microbial population and their adaptation, fitness, and survivability can also be deciphered through comparative metagenomic study. It will help to engineer the microbial community and their metabolic activity to speed up the degradation process.
Keywords: Metagenomics, microbial community, plastic degrading microbes, microbiome engineering, prebiotics, probiotics, genetic engineering
1. INTRODUCTION
With the increase in the world’s population and changing lifestyles, the demand for easily plastic products continues to grow. Thus, whopping production of synthetic plastic covers the major share of the global industry. Now, it tends to generate about 350 to 400 million tones of plastic waste annually on a global scale [1] and expected to be tripled by 2050 [2]. Of them, around 40% of global plastic waste was processed and recycled, and the rest, 60% remains unprocessed [3]. The significant parts of the unprocessed plastic become naturally decomposed, but few fractions were left undecomposed for a longer period. Over the years, poor recycling and low reuses leave millions of tons of plastic waste to accumulate in the terrestrial and marine ecosystem. Thus, it leads to an elevated risk of environmental pollution and climate change.
Plastics are the synthetic or semi-synthetic polymeric compound composed of carbon, oxygen, hydrogen, nitrogen, silicon, chloride, etc. that can be used to design objects of different shapes. More than 80% of the annual plastic generation is shared by high molecular weight polymers such as polyamine (PA), polyethylene (PE), polyethylene terephthalate (PET), polypropylene (PP), polystyrene (PS), polyurethane (PU) and polyvinylchloride (PVC). It has become an omnipresent part of our environment and it seems that their biodegradation is extremely slow, often burns in the open air, which leads to the release of CO2, and poisonous chemicals as air pollutants. Further, leftover plastic allowed to deposit in the atmosphere, and fragmented into smaller particles, and finally reach into the aquatic environments, mainly ocean through multiple outlets like rivers. These contaminants are termed as the ‘microplastics’ [4]. Unfortunately, when macro and microplastics are mistakenly consumed by many birds, fish, seal, whale, along with various land animals, it causes blockage of the intestine, ultimately resulting in death due to starvation [5]. The macroplastic and microplastic pollution also causes ecological damage to aquatic milieu through the deliverance of toxic chemicals and gases, proliferation of pathogenic agents, lowering the oxygen levels, resulting in entrapment of marine species, including corals, fish, octopus, oysters, shrimp, etc. [6]. Thus, plastics are extremely hazardous, particularly to higher organisms.
The plastic items are highly resistant to biodegradation and persist in the environment for a long time. Currently, it is very difficult to make even a rough estimation of the time necessary for their biodegradation. They can take from 10 to 1,000 years to decompose in natural conditions [7]. Therefore, the adoption of suitable plastic waste management is prioritized in every country. Mainly four different approaches, viz., recycling, incineration, landfilling, and biodegradation, were adopted for plastic waste management. Among them, recycling and incineration are followed for economic value addition to plastic waste. But, only 9 percent of the plastic is estimated to be recycled worldwide and the rest 91% remain in the unrecycled form [8, 9]. Some parts (12%) can be reused through incineration, but always associated with environment and health-hazardous due to the release of toxic gases such as chlorofluorocarbon (CFC), vinyl monomers and dioxins [10, 11]. In some countries, landfilling of plastic waste (79% of total plastic) is practiced, but risks of exposure to the environment always exist if poorly executed. The concern arises more when the reckless and unscientific management of plastic waste is followed and disposed into rivers and ocean. For evidence, China, Indonesia, Philippines, and Vietnam dumped most of the plastic waste into the ocean [12, 13]. Thus, the focus is now shifted towards the technological innovation in plastic waste management that should be efficient, eco-friendly and cost-effective. In this direction, besides the production of biodegradable plastics, exploration and utilization of microbial resources for the biodegradation of non-degradable synthetic plastic wastes have gained momentum [14-16]. Thus, many scientists have already started to explore the microbial diversity worldwide, and are searching for the potential microbes that have spectacular ability to degrade polythene and synthetic polymers, and hunt the novel genes distributed within bacteria, fungi, and other extremophilic groups. So far, very few microbial agents have been identified and characterized for plastic degradation [17, 18]; this might be due to the difficulty in isolation and identification of major microbial species from the respective site. Further, it is also important to decipher the microbial community structure and its mechanism in the degradation of synthetic polymers. Unfortunately, very limited information is available in this aspect. Accordingly, this review focuses on the possible implication of metagenomics in the exploration of plastic degrading microbial communities and mining of novel enzymes from diverse milieu; so that they could be utilized for microbiome engineering to induce expeditious biodegradation of plastics. This information will offer promising insight for industrial applications to address the looming environmental threat posed by plastic waste.
2. PLASTIC DEGRADING MICROBES
Microbes are well popularized as the potential garbage cleaner. They have the tremendous ability to break down advanced polymers and complex molecules like chitin, lignin, pectin, keratin, and even polythene also. They can easily acclimatize to any environment on the planet and perform as the ‘natural ecosystem engineer’ for habitat restoration [19, 20]. They are highly skilled to degrade very complex carbon-based compounds into simpler forms with diverse catalytic weapons. Being the grand recycler, they may immaculate the environment through reclamation [21].
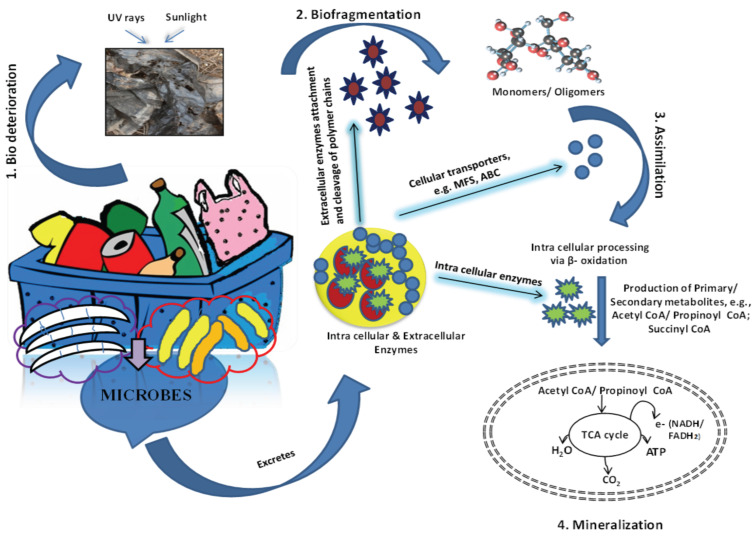
For the first time, microbial colonization on plastic was recorded in the marine ecosystem during the 1970s [22, 23]. But the systematic research on plastic biodegradation was started during the early 1980s. Thereafter, a large number of microbes spanning from prokaryotes to eukaryotes were identified and characterized. They are associated with the degradation of diverse plastics and polymers, including polyesters, nylon, polyethylene, etc. [24]. Around the plastic, a community of microbes grows as a thin layer of biofilm. The biofilm formation is the process of the congregation of surface-associated microbes on any surface and produces extracellular polymers that facilitate attachment and matrix formation, resulting in an alteration of physicochemical properties of the substrate. As a result, the life around the plastic surface is changed to form a unique biosphere, called ‘plastisphere’ [25]. The “Plastisphere” represents the whole life-sphere, including diverse metazoan and microbial communities that develop on the surface of any piece of plastic in aquatic or terrestrial environments. The microbial composition of the plastisphere considerably differs from the surrounding environmental microbial communities [26]. In spite of their taxonomic difference, they might be selected according to their metabolic functional redundancy (i.e., digestibility of plastic) [27]. The microbial community of a plastisphere is highly complex and is a congregation of the diverse multitude of microbes from autotrophs (e.g., cyanobacteria, algae) to heterotrophs (e.g., bacteria, fungi, protozoa) [26]. Therefore, the expedition of these plastic degrading microbes is very crucial to decipher their ecology, efficiency, and mechanism of plastic degradation as they involved in the different processes and metabolic pathways of plastic degradation (Fig. 1). After dumping off, the plastic materials undergo the initial deterioration due to some abiotic factors that destroy the polymeric structure and facilitate the microbial colonization and biofilm formation. The thickness and composition of the biofilm depend upon the chemical nature of the plastic and polymers [28]. Further, the environmental factors like moisture, pH also acts as the important determinant [29]. The hydrophobicity of plastic and polymer surface favours the growth of diverse microbial species to establish a stable biofilm. The microbial biofilm induces extensive physical and chemical degradation of plastics through the secretion of extracellular enzymes, polysaccharides, and other toxic acidic substances [30]. The physical deterioration and chemical degradation of plastic polymers lead to their fragmentation into simpler oligomeric and monomeric forms, viz., benzoic acid, benzyl alcohol, benzaldehyde, carboxylic acid, ethylene, ethylbenzene, propylene benzene, phenol, Poly-β-hydroxybutyrate, ketones, styrene, and vinyl chloride, etc. [31, 32]. The oligomers and monomers of plastic are assimilated by various microbes inside their cell and catabolized to produce the energy. The assimilation helps in the conversion of monomeric plastic into various secondary metabolites, which later on excreted into the environment [33]. These metabolites are either utilized by other microbes for further degradation or remain deposited in the environment
Fig. (1).
Mechanism of plastic biodegradation. The physical bio-deterioration by sunlight, UV-radiation predisposes plastic waste for microbial biodegradation that is carried out through enzymatic action. The conversion of complex polymer into monomers or oligomers facilitates their assimilation by cellular ATP binding cassette (ABC), Major facilitator superfamily (MFS) protein, and undergoes β-oxidation by intracellular enzymes to produce Acetyl CoA, Propinoyl CoA, or Succinyl CoA which are used as the substrate for TCA cycle to produce energy (ATP) in the form of Flavin adenine dinucleotide, (FADH2), Nicotinamide adenine dinucleotide (NADH). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
with other non-assimilative compounds. Through consecutive assimilation and degradation, these metabolites completely oxidized into minerals. For instance, the microbial degradation of styrene leads its bioconversion into degradative metabolites like Phenylacetyl-CoA, Pyruvate, Acetaldehyde 2-phenylethanol and 2-vinylmuconate. Of them, Phenylacetyl-CoA is utilized by P. putida via the tricarboxylic acid (TCA) cycle for energy production [34]. Similarly, biodegradation of PHB was initiated by various microbes viz., Ilyobacter delafeldii, Streptomyces ascomycinicus via., expression of extracellular PHB depolymerase, to convert into D-3-hydroxybutyrate monomer, which was further oxidized by 3-hydroxybutyrate dehydrogenase and the oxidized product was finally assimilated via TCA cycle [35].
It is now well established that microbes always function in consortia, especially in different phases of biofilm formation on the polymer surface. In this successive process, the heterogeneous microbial population was engaged. Of them, only a few microbial groups were identified and are mostly limited to culturable nature [36]. Whereas, the potentiality of huge numbers of unidentified and unculturable microbes remain underestimated [37]. To understand the significance of these unculturable microbial groups and their arsenals involved in the biodegradation process, and their interaction with well-characterized microbial groups, metagenomic analysis is advisable. The metagenomic analysis of microbial population engaged in the biodegradation of plastic will help to decipher the microbial community structure and biodegradation potentiality in situ. Moreover, the microbial mechanism that leads to the biochemical changes of complex plastic materials can also be predicted.
3. METAGENOMIC ANALYSIS OF PLASTIC DEGRADING MICROBES
So far, many plastic degrading microbial species have been identified from numerous dumping sites, and the enzymes produced by them were characterized for polymers degradation but with low efficiency and turn out. Environmental microbiologists estimate that only 2% of total microbial flora can be cultured in the laboratory leaving behind a huge proportion of uncultured fungi, bacteria, and other extremophiles as unexplored [38]. The recent advancements in next-generation sequencing technologies and bioinformatics tools allowed examining a huge amount of environmental samples through processing millions of DNA/RNA fragments and their successive analysis simultaneously [39, 40]. The metagenomic analysis of plastic degrading microbes is also feasible and it can proceed through deciphering the microbial community structure in ‘plastisphere’ and mining novel genes or enzymes responsible for degrading simple to complex polymer [25, 41]. In this way, the unexplored microbial gene pool can be revealed for biotechnological implication and further valorization.
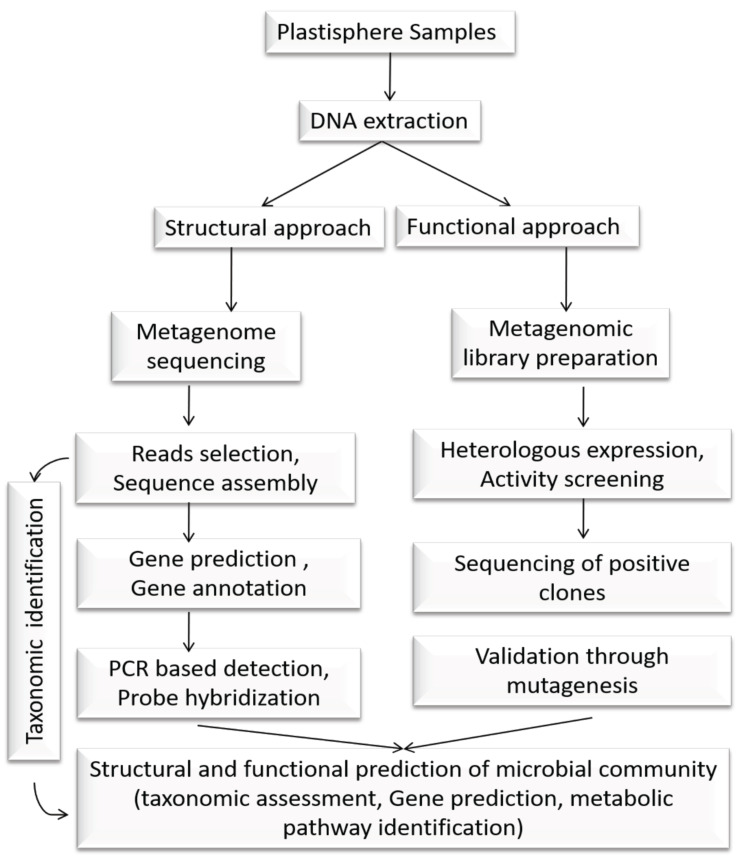
Nowadays, metagenomic analysis of any local microbial community is performed through two approaches: (1) structural approach, and (2) functional approach [42]. In a structural metagenomic approach, the main focus paid to unveil the microbial community structure of any defined ecology through the sequencing of environmental samples. Primarily, it will provide the taxonomic identity of the microbial population through a culture-independent manner (Table 1), and further can be utilized to explore other properties, such as the identification of novel genes, prediction of gene function with their possible engagement in the complex metabolic pathways (Fig. 2). This will also help to establish interaction between community members and their ecological preferences. It also shades lights on the microbial population dynamics of the specified ecology in different spatiotemporal scales and assigning minor or major geoecological roles of individual members in the community structure development. Differently, functional metagenomics help to hunt the gene function from its sequence or structural information. Starting with DNA extraction of environmental samples, it involves the prediction of the putative desirable genes from the metagenome library, followed by their heterologous expression for further activity-based screening and functional validation [43, 44]. Thus, the functional metagenomics approach is used as complementary to sequence-based structural metagenomics and helps in the annotation of genes from the huge metagenomic database [44, 45]. Furthermore, the comparative metagenomic study of different ‘plastisphere’ of diverse ecology will help to identify the ‘core’ microbial population, i.e., the certain microbial families that are common in ‘plastispheres’ across geographical locations [46-48] and consistently persist over a long period of time to perform a significant part of plastic degradation. In addition, their mechanism of adaptation, fitness, and survivability with plastic degrading potentiality in their respective ecological niche (marine to terrestrial) can also be explored. Therefore, engineering of the microbial community and their metabolic activity in the plastisphere will be feasible to speed up the degradation process.
Table 1.
Metagenomics approach adopted to decipher plastisphere microbiome.
| Platic Types |
Metagenomics
Approaches |
Abundant Microbes | Ecosystem (Location) | References |
|---|---|---|---|---|
| PE | Metataxononmics (V4-V6 16S rRNA sequencing) | Flavobacteriaceae (Flavobacterium), Rhodobacteraceae, Methylophilaceae (Methylotenera), Planctomycetaceae (Planctomyces,Pirellula), Hyphomonadaceae (Hyphomonas), Planctomycetaceae (Blastopirellula), Erythrobacteraceae (Erythrobacter), Sphingomonadaceae (Sphingopyxis) | North Atlantic and North Adriatic Sea (Sea floor) | [25, 120] |
| Metataxononmics (V4 18S rRNA sequencing) | Zalerion maritimum | Marine | [121] | |
| Metataxononmics (16S rRNA sequencing) | Cyanobacteria (Phormidium, Rivularia) | Sub-surface plastisphere | [26, 122] | |
| PS | Metataxononmics (V4-V6and V9 16S rRNA sequencing) | Flavobacteriaceae (Flavobacterium), Rhodobacteraceae, Methylophilaceae (Methylotenera), Planctomycetaceae (Planctomyces, Pirellula), Hyphomonadaceae (Hyphomonas), Erythrobacteraceae (Erythrobacter), Sphingopyxis (Sphingomonadacea), Verrucomicrobiaceae, Nocardiaceae (Rhodococcus), Pseudomonadaceae (Pseudomonas sp.) | North Atlantic and North Adriatic (Sea surface) | [25, 123] |
| Metataxononmics (V4 16S rRNA sequencing) | Flavobacteriaceae (Flavobacterium), Rhodobacteraceae, Methylophilaceae (Methylotenera), Planctomycetaceae (Planctomyces,), Hyphomonadaceae (Hyphomonas), Erythrobacteraceae (Erythrobacter), Sphingopyxis (Sphingomonadacea), Nocardiaceae (Rhodococcus), Pseudomonadaceae (Pseudomonas sp.) | Deep seawater of Tottori Prefecture | [120, 124] | |
| Metataxononmics (V4 16S rRNA sequencing) | Flavobacteriaceae (Flavobacterium), Rhodobacteraceae, Methylophilaceae (Methylotenera), Planctomycetaceae (Planctomyces, Pirellula), Hyphomonadaceae (Hyphomonas), Erythrobacteraceae (Erythrobacter),Sphingopyxis (Sphingomonadacea), Verrucomicrobiaceae, Nocardiaceae (Rhodococcus), Pseudomonadaceae (Pseudomonas sp.) | Off-shore in Toyama bay | [47, 80] | |
| Metataxononmics (V4-V6 16S rRNA sequencing) | Flavobacteriaceae (Flavobacterium), Rhodobacteraceae, Methylophilaceae (Methylotenera), Planctomycetaceae (Planctomyces,), Hyphomonadaceae (Hyphomonas), Erythrobacteraceae (Erythrobacter), Sphingopyxis (Sphingomonadacea), Nocardiaceae (Rhodococcus), Pseudomonadaceae (Pseudomonas sp.) | Deep-sea sediment (Kurile and Japan trenches) | [47, 80] | |
| HDPE | Metagenomics sequencing | Peregrinibacteria, Cellvibrionaceae, Flavobacteriaceae (Flavobacterium), Flammeovirgaceace, Hyphomonadaceae, Granulosicoccaceae, Nannocystaceae, Oceanospiralliceae, Parvularculaceae, Phycisphaeraceae, Phyllobacteriaceae, Rhodobacteraceae, Rhodospirillaceae (Thalassospira) | Plastic marine debris (North Pacific Subtropical Gyre) | [47, 64] |
| Metataxononmics (V3-V516S rRNA sequencing) | Peregrinibacteria, Cellvibrionaceae, Flavobacteriaceae (Flavobacterium), Flammeovirgaceace, Hyphomonadaceae, Granulosicoccaceae, Nannocystaceae, Oceanospiralliceae, Parvularculaceae, Phycisphaeraceae, Phyllobacteriaceae, Rhodobacteraceae, Rhodospirillaceae (Thalassospira) | Sea surface (Mediterranean Sea) | [47, 65] | |
| LDPE | Metataxononmics (V4 16S rRNA sequencing) | Peregrinibacteria, Cellvibrionaceae, Flavobacteriaceae (Flavobacterium), Vibrionaceae (Vibrio), Planctomycetes (Planctomyces, Pirellula), Alteromonadaceae (Marinobacter, Alteromonas), Rhodobacteraceae, Rhodospirillaceae (Thalassospira), Enterobacteriaceae (Rahnella aquatilis), Alcanivoracaceae (Alcanivorax borkumensis) | Marine (benthic zone) |
[77, 122, 124] |
| Platic Types |
Metagenomics Approaches |
Abundant Microbes | Ecosystem (Location) | References |
| PP | Metataxononmics (V4 16S rRNA sequencing) | Peregrinibacteria, Bacteroidetes (Chlorobia), Flavobacteriaceae (Flavobacterium), Idiomarinaceae, Flammeovirgaceace, Hyphomonadaceae, Granulosicoccaceae, Oceanospiralliceae, Cellvibrionaceae, Pseudoalteromonadaceae, Parvularculaceae, Phyllobacteriaceae | Coastal marine zone | [65, 119] |
| Metataxononmics (16S rRNA sequencing) | Bacteroidetes (Chlorobia), Flavobacteriaceae (Flavobacterium), Idiomarinaceae, Flammeovirgaceace, Hyphomonadaceae, Granulosicoccaceae, Oceanospiralliceae, Cellvibrionaceae, Pseudoalteromonadaceae, Parvularculaceae, Phyllobacteriaceae | Open ocean | [47, 64] | |
| PET | Metagenomics (shotgun sequencing) | Peregrinibacteria, Cellvibrionaceae, Alteromonadaceae (Marinobacter), Hyphomonadaceae, Granulosicoccaceae, Parvularculaceae, Phycisphaeraceae, Salinisphaeraceae, Sneathiellaceae (Sneathiella), Planctomycetes, Acidimicrobiales, Rhodobacteraceae, Rhodospirillaceae, Flavobacteriaceae (Flavobacterium), Commonadaceae (Ideonella sakaiensis), Nocardiosporaceae (Thermobifida fusca) | Coastal marine zone | [38, 47, 80] |
| PLA | Metataxononmics (V416S rRNA sequencing) | Peregrinibacteria, Caulobacteraceae, Alphaprotobacteria, Epsilonproteobacteria, Camphylobacteraceae, Cellvibrionaceae, Hyphomonadaceae, Flavobacteriaceae (Flavobacterium), Alteromonadaceae(Marinobacter), Flammeovirgaceace, Phycisphaeraceae, Planctomycetes, Pseudoalteromonadaceae, Rhizobiales, Spongiibacteraceae, Salinispharaceae | Ocean | [51, 122] |
| PVC | Metataxononmics (16S rRNA sequencing) | Peregrinibacteria, Bacteroidetes, Alphigiphilaceae, Flammeovirgaceace, Hyphomonadaceae,Caulobacteraceae, Sneathiellaceae, Oceanospiralliceae, Phycisphaeraceae, Phyllobacteriaceae, Alpigiphilaceae, Idiomarinaceae, Alcanivoraceae, Planctomycetes, Flavobacteriaceae (Flavobacterium), Idiomarinaceae | Cold marine habitat (sea surface) |
[47, 124] |
Flexithrix, Hirschia, Parvularcula, Phyllobactereacea, Roseovarius, Ulvibacter have some specific association the different types of plastics and play significant, but an undefined role in decomposition [50, 51, 54]. This specific association of microbiome with different plastic types is also reported by many other researchers. The members of the family Alcanivocareacea (Alcanivorax), Cryomophaceaea, Erythrobacter show higher abundance on the surface of Polyethylene (PE), Polyethylene terephthalate (PET) [50, 53]. Similarly, members of the family Oleiphilaceae (Oleiphilus) and Arenicellaceae are dominating on PE, and polypropylene (PP) [53, 59], whereas Bacteriodetes (Crocinitomix, Owenweeksia, Fluviicola, Tenacibaculum), Gammaproteobacteria (Acinetobacter), and Verrucomicrobia (Persicirhabdus) are directly associated with PET and PP degradation [50, 60], The members of Hyphomonadaceae and Erythrobacteraceae form biofilm on PE and Polystyrene (PS) surfaces [54], whereas, the members of Alteromonadaceae (Alteromonas), Cellvibrionaceae, Oceanospirillaceae are highly specific for PVC [53].
Fig. (2).
Metagenomics approach to decipher the structure and function of microbial community in plastisphere. The exploration of taxonomic identity of microbial population and the discovery of novel genes and their functional prediction in the complex metabolic pathways could be possible in different spatiotemporal scales.
3.1. Deciphering Microbial Community Structure Associated with Plastic Degradation
The microbial community structure of the plastisphere was depicted using a massive metagenome sequencing approach (Fig. 2). It is basically composed of Archaea, Bacteria, Fungi and other eukaryotic microbial species and significantly differs from the surrounding environmental microbial population [45, 49, 50]. Its composition and species richness are influenced by various spatiotemporal phenomenons like habitats/geographical location, ecosystem, (Table 2) and seasonal variation [51-53]. Further, the physiochemical nature of plastics like polyethylene, polypropylene, polystyrene, etc. also regulates a lot [43]. The microbial community composition associated with diverse plastics is significantly varying and it is also changing with the different phases of plastic degradation process [25]. Reportedly, the ‘specific’ assemblage of diverse organisms always remains constant with specific plastic types, and can also be distinguished from other communities. The composition and specificity of microbial assemblage associated with polyethylene (PE), and polystyrene (PS) in the marine aquatic ecosystem (coastal Baltic Sea) clearly indicates the abundance of Flavobacteriaceae (Flavobacterium), Rhodobacteraceae (Rhodobactor), Methylophilaceae (Methylotenera), Plactomycetaceae (Planctomyces, Pirellula), Hyphomonadaceae (Hyphomonas), Planctomycetaceae (Blastopirellula), Erythrobacteraceae (Erythrobacter), Sphingomonadaceae (Sphingopyxis), etc. as determined through 16S rRNAgene sequencing [54]. Within a defined community, there might have different microbial strata that are successively engaged in biofilm formation [53] and utilizes different byproducts as the substrate to channelize the total degradation of any specified plastic. But, very limited documents exist on the structural composition of the microbial biofilm and their successive development on the different plastic surface under the same environment. It was observed that the abundance of Roseobacter (class Alphaproteobacteria), and Alteromonas, Pseudoalteromonas, Vibrio, (class Gammaproteobacteria) is much more during the early stage of biofilm formation on polyurethane, and acrylic-based plastic surface [55, 56]; thus they are considered as the primary colonizers. For instance, the earliest (0∼9 h) colonization of γ- Proteobacteria (Pseudomonas, Acinetobacter, Alteromonas, and uncultured γ-Proteobac- teria), followed by the increasing abundance of α-Proteobacteria (Loktanella, Methylobacterium, Pelagibacter, and uncultured α-Proteobacteria) in the 24∼36 h indicates the successive development of microbial communities in a biofilm on three different solid surface in marine ecosystem (i.e., acryl, glass and steel), [56]. Accumulation of primary colonizers leads to the modification of the substratum, rendering it suitable for subsequent colonization by diverse secondary colonizers, viz., Acidobacteria, Actinobacteria (Acidimicrobium, Propionibacterium) Bacteroidetes (Polaribacter, Tenacibaculum), Betaproteobacteria (Comamonus), Cyanobacteria (Phormidium), Firmicutes (Streptococcus), Planctomycetes (Pirellula) Verrucomicrobia, etc. after 24∼36 h depending on the nature of the substrates and environments [57, 58]. These microbes appear during the later stage of biofilm formation and may perform separate functions that signify these bacterial classes as secondary colonizers. With the time duration, the microbial community composition changes, and the relative abundance of secondary colonizers like Bacteroidetes, Betaproteobacteria, etc. increases [58]. This dynamic shift in community structure from primary to secondary colonizers in different time frame depicts the progression of biofilm formation.
Table 2.
Diversity of microbes associated with plastic degradation in different ecological niche.
| Plastic Type | Enzyme Produced | Microbes in Different Ecology | References | |||||
|---|---|---|---|---|---|---|---|---|
| Terrestrial | Marine | - | ||||||
| Polyethylene (PE) | Unknown | Aspergillus niger | Aspergillus sp. | [124-127] | ||||
| Unknown | Bacillus cereus | - | [128] | |||||
| Unknown | Brevibacillus borstelensis | - | [85, 129] | |||||
| Lipase | Penicillium simplicissimum | - | [130] | |||||
|
Manganese peroxidase |
Phanerochaete chrysosporium |
- | [24, 131, 132] | |||||
| Laccase | - | Rhodococcus ruber | [133, 134] | |||||
| Unknown | - | Phormidium sp., Zalerion maritimum | [50, 121] | |||||
| Polyethylene terephthalate (PET) | Cutinase, Lipase |
Thermobifida fusca (Thermomonospora fusca) |
- | [135] | ||||
| MHETase, PETase | Ideonella sakaiensis | - | [38] | |||||
| Lipase | - | Pseudomonas sp. | [39, 47, 85] | |||||
| Unknown | - | Flavobacteriaceae, Cryomorphaceae, Saprospiraceae, Phormidium sp. | [47, 50] | |||||
| Cutinase | Fusarium sp., | - | [39, 136] | |||||
| Cutinase | Humicola sp. | - | [39, 137] | |||||
| Unknown | - | Diatoms (e.g. Coscinodiscophytina, Bacillariophytina). | [50] | |||||
| Polypropylene (PP) | - | - | Pseudophormidium sp. | [124] | ||||
| - | Alcaligenes, Pseudomonas, Vibrio | - | [138] | |||||
| - | Bacillus subtilis, B. flexus, Pseudomonas stutzeri | - | [139] | |||||
| Polystyrene (PS) | Alkane hydroxylase | - | Pseudomonas putida AJ, P. putida CA-3 | [140, 141] | ||||
| Styrene monooxygenase, Styrene oxide isomerase, Phenylacetaldehyde dehydrogenase | - | Mixed microbial communities (Bacillus, Micrococcus, Nocordia, Pseudomonas, Rhodococcus) |
[39, 47] | |||||
| Polyurethane/Polyester (PUR) | Serine hydrolase | Pestalotiopsis microspora | - | [142] | ||||
| Esterase | Pseudomonas aeruginosa | - | [143] | |||||
| Polyurethanase | Pseudomonas chlororaphis | - | [144] | |||||
| Protease | Pseudomonas fluorescen | - | [145] | |||||
| Lipase |
Pseudomonas chlororaphis,
P. protegens BC212 |
- | [39, 146] | |||||
| Aryl acylamidase | Rhodococcus equi | - | [147] | |||||
| Unknown | Actinetobacter gerneri P7 | - | [148] | |||||
| Unknown | Actinetobacter calcoaceticus | - | [149] | |||||
| Plastic Type | Enzyme Produced | Microbes in Different Ecology | References | |||||
| Terrestrial | Marine | |||||||
| PolyVinyl Chloride (PVC) | Unknown |
Poliporus versicolor, Pleurotus sajor caju, Thermomonospora fusca |
- | [147, 150] | ||||
| Unknown | - |
Alteromonadaceae(Alteromonas), Cellvibrionaceae Oceanospirillaceae Aestuariicela |
[39, 47] | |||||
| Others (Nylon,Polycaprolactone (PCL), Polyhudroxybutyrate/acetate (PHB/PHA)) |
Nylon hydrolase | Agromyces sp. | - | [151] | ||||
| Laccase | Tremetes versicolor | - | [152] | |||||
| Manganese peroxidase | White-rot fungus IZU-154, Amycolaptosis sp. | - | [153, 154] | |||||
| Polycaprolactone depolymerase | Alcaligenes faecalis | - | [47] | |||||
| Lipase | - |
Alcanivorax sp., Pseudomonas sp., Rhizopus delemar, R. arrhizus, Achromobactr sp., Candida cylindracea |
[155, 156] | |||||
| Unknown | - | Bacillus cereus, Bacillus sphaericus, Vibrio furnissii, Brevundimonas vesicularis | [157] | |||||
| Lipase | Rhodococcus arrizus | - | [47] | |||||
| Serine hydrolase | Acremonium sp., Cephalosporium sp., Pseudomonas stutzeri | - | [158-160] | |||||
for in situ microbiome engineering [87, 88]. But very little efforts have been paid for manipulating any ‘plastisphere’. Although a wealth of information is available on the microbial community composition, their useful and strategic engineering is still lagging behind. Further, the plenty of knowledge gained from the metagenomic analysis of the plastisphere also provides valuable information about enzymology and biosynthetic pathways; it opens up the scope of biocatalytic engineering for specific processes. Thus it is necessary to address the technological gaps that exist in these steps. The paramount importance is the complexity and spatiotemporal dynamics of the microbial community of the plastisphere in which individual groups have assigned separate functions [80]. They perform enzymatic biodegradation of plastics through an array of metabolic pathways [80, 81]. This biodegradative process is largely influenced by some biotic and abiotic factors. Both of these factors can be targeted to operate the microbiome composition and their function in a desirable manner. Therefore, manipulation of microbial community composition and engineering microbial genetic constitution are the two possible strategies that could be adopted for the microbiome engineering of ‘plastisphere’ (Table 3).
The identification and characterization of plastic ‘specific’ microbiome are relatively difficult. Their abundance relatively differs and may persist for short-term or long term scale; moreover, varying in different ecological contexts, i.e., from marine to terrestrial (Table 2). Therefore, time, location and depth of sampling are the crucial parameters to unveil the community structure of specific microbes associated with the degradation of diverse plastics. In a marine ecosystem, some bacterial genera, viz., Dokdonia, Erythrobacter,
Other than bacteria, many parasitic and saprophytic fungi, and autotrophic algae are also significant components of any biofilm. But their existence in the plastisphere and utmost importance on plastic degradation is overlooked [61]. Among the fungal assemblage, the members of Chytridiomycota, Cryptomycota and Ascomycota are dominating. Of them, Chytrids are most abundant on both PE and PS, especially in the aquatic ecosystem [50, 61]; however, in terrestrial ecosystem many Ascomycetes genera, viz., Aspergillus, Penicillium, Cladosporium, Fusarium, etc. efficient in degrading diverse types of plastics [62, 63]. Many photoautotrophic algal and cyanobacterial species can also digest plastics in the marine ecosystem. Of them, few cyanobacterial genera, viz., Phormidium and Rivularia are dominating in the marine ecosystem [26, 64, 65]; nevertheless, green microalga (Scenedesmus dimorphus), blue-green alga (Anabaena spiroides) and diatom (Navicula pupula) are common in moist terrestrial habitats [66]. Diatoms are also among the first colonizers of floating plastic in the sea surface [67]. These diatoms are also closely associated with bacterial communities in the marine plastisphere, and sometimes support their colonization, community development and biofilm formation of various plastic degrading bacterial genera, namely Alteromonas, Roseobacter, and Pseudoaltromonas Polaribacter, and Tenacibaculum [68-70]. Due to their versatile enzyme producing ability, both algae and fungi can break down recalcitrant plastic structures and speed up the process of micro-plastic digestion.
Whatever the polymer composition, in general, a core microbial population has a common existence in different plastisphere irrespective of plastic types, and their prevalence is much more than other specifically associative microbes. Different ‘plastisphere’ (varying with the type of the plastic polymers) shares a core microbiome that remains constant with the types of plastic polymers, and the ecosystem. As evidence, the core community of different plastic degrading microbes in the marine ecosystem is composed of Proteobacteria (Rhodobacter, Alcanivorax, Hyphomonas, Pseudoalteromonas), and Bacteroidetes (Flavobacterium) and cyanobacteria (Phormidium sp.) [25, 26, 58, 64, 65]; whereas Acinetobacter, Bacillus, Serratia form the core microbiome in the larval gut of plastic degrading insect Galleria mellonella [71, 72]. Although the contribution of the core microbial population is much higher to perform all vital ecological functioning [73] starting from substrate selection to biofilm formation [74], plastic degradation [75], and finally multi-nutrient cycling [76, 77], the importance of specific microbial species in plastic degradation cannot be overruled [25]. In a core population, the relative abundance of fungi and bacteria fluctuates significantly. Some reports hypothesized that certain fungi (Aspergillus sp.) are better degraders of low-density polyethylene type plastic than bacteria (Pseudomonas, Bacillus, Brevibacillus, Cellulosimicrobium, Lysinibacillus) in the terrestrial ecosystem, [78], whereas, others prioritized the role of bacteria, especially Alphaproteobacteria (Rhodobacter) Gammaproteobacteria (Alcanivorax, Marinobacter) and Bacteroidetes (Arenibacter, Tenacibaculum) in the degradation of low-density polyethylene type plastic in the marine ecosystem [25, 77]. Overall, the relative abundance of many species of genus Aspergillus and Penicillium among fungi and that of Bacillus and Pseudomonas among bacteria is always higher in a core population in terrestrial habitats, and the core microbiome of various marine ecosystem share some common taxa like Bacteroidetes, Firmibacteria, Proteobacteria along with many cyanobacteria, chytrids, and diatoms [26, 58, 64, 65]; will be a potential target for further biotechnological implication.
3.2. Mining Novel Genes or Enzymes in the Plastic Degradation Pathway
So far, the metagenomic exploration of the plastisphere remains restricted within the structural analysis of the microbial community. It helps to identify some new microbial species, quantify their richness in the local microbial niche, and make it easier to categorize them into ‘core’ to ‘specific’ and ‘rare’ species based on their abundance and specificity. But, their functional significance in plastic degradation is yet to define [25]. Apart from the core population, these ‘specific’ and ‘rare’ species could play a crucial role in biochemical interactions with various plastic types; many of them may harbor the novel genes or enzymes that can dissolve plastic polymers into minerals. One such gene is PETase genes, which encode hydrolases to degrade PET into oligomers and monomers. Various PET hydrolases, e.g., cutinases, lipases, possess a typical α/β hydrolase fold with serine, histidine, and aspartate catalytic triad [18, 24]. Some bacterial species such as Ideonella sakaiensis, Thermobifida fusca, etc. can produce PETase. However, none of them are adapted for the marine environment. Thus it will not be suitable to exploit these bacteria for the reclamation of marine plastic pollution. To resolve this issue, a functional metagenomic approach is followed, nowadays, to mine PET hydrolase homologs from various microbial resources [79, 80]. Based on the conserved amino acids, it was predicted that PET hydrolases were globally distributed in marine and terrestrial metagenomes, but interestingly Bacteroidetes and Actinobacteria are the main PET hydrolase producer in the marine and terrestrial ecosystem, respectively [80]. Similarly, identification of other novel enzymes, viz., alkane hydroxylase, carboxylase, esterases [81], lipase, tannase [82], etc. is also carried on from metagenomic libraries of diverse ecology like cold marine to hot spring, antarctic desert to contaminated terrestrial and oil spill, even from the gut microbes of invertebrates, etc [83-85]. Further, heterologous expression of such codon-optimized ‘synthetic’ genes enhances the prospects of new gene identification through functional metagenomics.
4. BIOTECHNOLOGICAL IMPLICATION OF METAGENOMIC INFORMATION
Metagenomics leads the way to mine the microbial treasure from any environmental samples. It can not only help to identify the microbial species and their community structure in a defined ecological niche but also enlighten about their functional significance. Further, the functional metagenomics approach speeds up the process of gene discovery and identification of diverse biocatalysts that have tremendous prospects in the field of agriculture, medicine, industry, and environment [42]. The biotechnological reconfiguration and refinement of these biocatalysts can also be possible to refabricate their function and optimization for a specific operation in the diverse metabolic pathways in the ecosystem [39]. Thus, various biotechnological approaches should be adopted to favour their biocatalytic transformation and subsequent practical implication.
So far, significant advances are made in the biotechnology application of metagenomic information for various microbial ecology, including human gut to plant rhizosphere [40, 86], and many microbes derived products are available
4.1. Strategy-1: Manipulating Microbial Community Composition
It is the most conventional and contemporary strategy and being well-practiced to manipulate the microbial community composition of different human, animal, plant and soil-based environments. It includes different chemical, cellular, and molecular methods for large scale manipulation with greater magnitude and specificity [88]. Similarly, for any plastisphere, prebiotic (chemical) and probiotic (cellular) approaches can suitably be applied. The prebiotic approach helps to modulate the environment for better acclimatization of microbes (Fig. 3). For that, different chemicals like oligosaccharides and polysaccharides can affect the microbiome composition and selectively favour the growth of plastic degraders. Chemical substrates like chitin, cellulose, starch, glycolipids, lipopeptides, etc. act as the biosurfactant on the plastic surface [89] and assist the biofilm formation [90]. But with the maturation of biofilm, the nutrient level gradually deprives, as the degradation of hydrocarbons increases the concentration of carbon over nitrogen. Thus external supplement of fresh nutrients like nitrogen and phosphorus induces the rate of microbial degradation [7]. Sometimes, certain chemical stimulants may be necessary to stimulate the
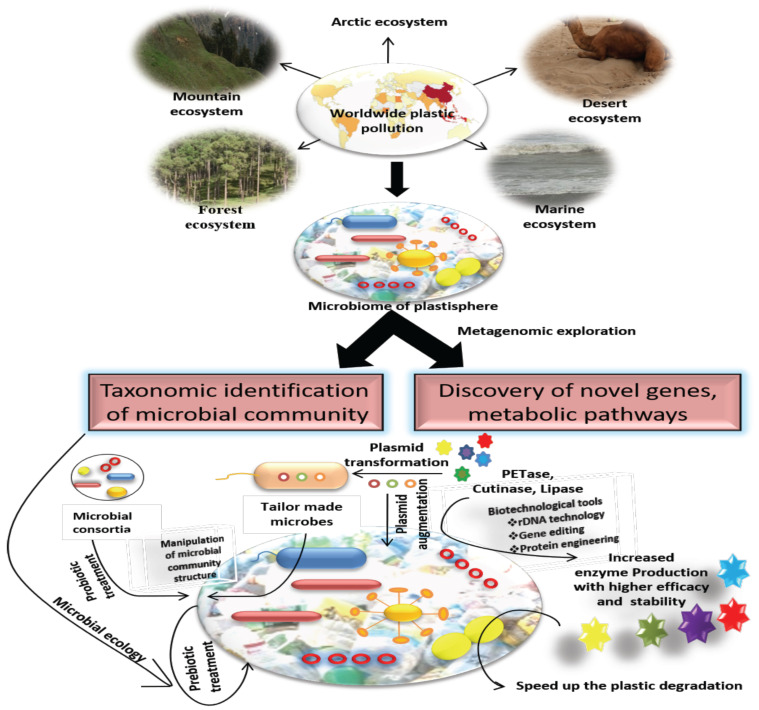
Fig. (3).
Approaches of microbiome engineering of plastisphere. The metagenomic study of different plastisphere from diverse ecosystem helps to identify the core and specific microbial population along with their ecological preferences. It opens up the scope of manipulating the microbial community composition via prebiotic and probiotic approaches. The discovery of novel genes, metabolic pathways leads the way for engineering microbial genetic constitution through plasmid augmentation, recombinant DNA technology, genome editing, protein engineering, etc. to enhance their metabolic activity and speed up the plastic degradation process. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
growth of microbes and to protect the microbial community from no harmful effects of toxic pollutants released from the degrading polymer surface [90, 91]. During the biodegradation process, the drastic change in pH value and abnormal high oxygen demand exerts a negative impact on the microbial activity [92, 93]. To avoid these environmental stresses, certain chemical stimulants with good buffering capacity can also be applied to regulate the pH, temperature and oxygen level in a suitable range for potential function of microbes.
Other than prebiotics, probiotics administration of microorganisms would be a plausible solution. The probiotics are living microbial cultures that are artificial and selectively grown in media and applied for better performance. The similar microbial probiotic culture of predominant microbial species associated with plastic degradation can also be prepared and would be useful for the bioaugmentation. Application of pre-grown microbial cultures enhances the beneficial microbial population in situ and accelerates the biological degradation or transformation of plastic polymers, thus preferred as one of the best methods for environmental cleanup in lesser time and cost. The success of bio-augmentation depends upon various biotic and abiotic factors, most importantly, the microbial strain [94] that should have extra-ordinary plastic biodegradation potential. Moreover, various edaphic factors like temperature, moisture, pH, osmotic pressure, nutrient availability in the environment, have a magnificent impact on the survival, fitness, and activity of microbes [21]. That’s way, microbial communities with wider environmental adaptability and higher affinity for diverse polymers and hydrocarbons are most commonly preferred [95]. For bioaugmentation, the core microbial population including Actinomycetes (Streptomyces, Thermoactinomyces), Bacteria (Psuedomonas, Streptococcus, Micrococcus, and Moraxella), and Fungi (Aspergillus, Penicillium) are favoured over the native and specific microbes due to the cultural dependence [32, 96]. But the significance of specific microbes in the complete degradation of specified plastic cannot be overlooked. For that, complete microbiome transplantation from one plastisphere to another having similar ecological preferences would be unique to eliminate the problem. The microbiota composed of the core as well as specific community represent the indigenous microbial population, thus easily be acclimatized to the new habitat. Distressfully, probiotic bioaugmentation and microbiome transplantation both generally remain unsuccessful initiative for mass application due to the slow growth, and limited distribution of microbial population along with low cellular viability and functionality. To overcome these issues, increasing the efficiency of the microbial population through genetic modification, and subsequent bioaugmentation is advisable.
4.2. Strategy-2: Engineering Microbial Genetic Constitution
With the advancement in microbial genetics and molecular biology, the development of genetically tailored microbes becomes practically feasible to degrade complex polymers in the environment. In the era of system biology, easy construction of biosynthetic units or genetic circuits makes it plausible to design programmable biological devices, i.e., synthetic cells or synthetic life with precise and novel functionalities [97]. These ‘synthetic’ microbial cells can be created through genetic engineering, protein/enzyme engineering, and genome editing tools, and further deployed for microbiome engineering in the plastisphere. The genetic manipulation of the microbiome can be achieved either through ex situ designing of tailored microbes or via in situ metagenome modification. The biodegradation of complex polymeric plastic waste proceeds through an array of oxidation steps to convert into monomers and finally enters into the TCA cycle via assimilation [47]. All these different stages of biodegradation could not be possible to carry out by any single species [98]. To resolve it, the genetic engineering tools can be implemented for the complementation of multiple genes of whole plastic metabolic pathways or complete gene circuit into different microbial species. The genetically engineered microbes could be designed for multiple enzyme production, regulating of quorum sensing mechanism for biofilm formation, etc. For evidence, genetic engineering makes it doable to express PETase, a key enzyme for PET degradation in several bacterial cell-system like Bacillus subtilis and Escherichia coli [99, 100], other than its natural host Ideonella sakaiensis [33]. The 3.8-fold expression of ‘active’ PETase enzyme was achieved in Bacillus subtilis via the Tat-independent secretory pathway using native signal peptide [99], where in Escherichia coli, PETase enzyme was expressed extracellularly via secretary (Sec) pathway-dependent manner [100]. The functionality of these heterogeneously expressed proteins was checked through PET film degradation assay [99, 100].
However, these bacterial systems are not able to function in the marine ecosystem, where the accumulation of plastic waste is more. Thus, efforts are made for the heterologous expression of PETase into photosynthetic microalga (Phaeodactylum tricornutum) for the salted marine environment [101]. But unfortunately, the majority of plastic degrading enzymes are highly temperature-sensitive and their functionality is environment-dependent (pH, pressure, oxygen, etc). For instance, some PET hydrolases, e.g., cutinases isolated from thermophilic microbes (like Thermobifida cellulosilytica, T. fusca, Saccharomonospora viridis) require higher operational temperature (~70-80ºC) [102], whereas, a novel PETase isolated from Ideonella sakaiensis are functional at ambient temperature (37ºC), but with less durability [103]. Thus, these candidate enzymes need to be further optimized according to their environmental preference, and their biocatalytic activities should be refabricated through protein engineering. The modification of enzymatic activity involves deciphering the crystal structure of protein followed by their rational designing through molecular docking and simulation dynamics for enhanced thermal stability and improved catalytic activity [104]. Previously, it was applied for improving the PET degrading activity of the cutinase enzyme from Thermobifida cellulosilytica [105] and T. fusca by mutagenesis [106, 107]. Now the target is shifted for rational designing of thermo-stable PETase from I. sakaiensis [103, 104, 108]. To resolve the inherent instability problem of the PETase from Ideonalla sakaiensis at ambient temperature, its computational redesigning, and subsequent protein engineering is done to improve the enzymatic/ catalytic activity (400 fold), and durability (10 days) with higher crystallinity at 40ºC [109]. In spite of much progress, some technical barriers still hinder their direct physical application on a wider scale. Primarily, the colonization of genetically modified microbial species or strain is always difficult in the ecologically competitive environment of any plastisphere, additionally functioning of individual microbial species or strain alone to degrade complex polymers is questionable. Thus, the practical implication may lead to some unwanted consequences.
In lieu of targeting individual species/strains, modifying the entire metagenome is approachable. Mostly the core microbial metagenome of any community is almost constant [42, 43], thus would be reasonable to manipulate rather than an individual one. Direct in situ metagenome modification of native microbial population is being possible through horizontal transfer of plasmid construct that is directly delivered via plasmid/genetic augmentation. Uniform distribution of plastic degrading genes (enzymes) among the indigenous microbial population would be accomplished easily via plasmid delivery. This can be exemplified as the bioaugmentation of conjugative plasmid pRO103 harboring the tfdA gene (encoding 2,4-dichlorophenoxyacetic acid/2-oxogluta- rate dioxygenase) through inoculation of E. coli HB101 (vector) leads to the horizontal transfer of into the indigenous microbial population in the soil for better phenol degradation [110]. Further, the receptor microbial cells harboring such genes could act as the donor cells for other microbes via horizontal gene transfer process [111]. Thus, bioaugmentation leads the increase in the numbers of indigenous PU-degrading fungal populations with the change in their microbial community composition of and enhanced biodegradation rate of polyurethane (PU) in soil [112]. In addition, virus-vector mediated gene delivery into the existing population is another option. The use of viruses or bacteriophages as the gene delivery vehicles is an emerging choice, has already been successfully applied in many clinical trials [113]. For direct gene or enzyme delivery, this can also be tested for plastic degradation. In these ways, the functional potential of the native microbial community can be engineered via direct delivery at the site.
conclusion, CHALLENGES AND FUTURE PROSPECTS
Nowadays, the genetic engineering of individual species or the entire metagenome based gene insertion, protein engineering, or plasmid/ virus augmentation remains restricted to laboratory scale due to their lack efficiency, and increasing environmental safety concern. The conventional genetic engineering tools were out spaced with recent advancements in genome editing and synthetic biology. Various genome editing tools like the CRISPR, TALEN, and ZFN enable the programmable modification of microbial genomes [114]. Other than deletion or gene insertion, these tools can be utilized for the overexpression or repression of certain genes [115]. The gene regulation by deactivated Cas (dCas) also improves environmental tolerance in the microbial species [116]. Genome editing also helps to customize microbes for the biosynthesis of novel enzymes and secondary metabolites [117]; thus, engineered microbes become the superior target as probiotics. The new approaches are heading for the biological designing or modeling of microbes in the whole genome-scale to obtain desirable phenotype. Accordingly, the introgression of synthetic regulatory gene circuits in living cells is required for broad-spectrum applications [118]. The combination of system biology and genome editing, along with sophisticated bioinformatics tools, may open up new avenues to solve the plastic problem.
Moreover, the application of cell-free methods for protein-expression is another convenient way to produce enzyme and their broad-spectrum application [119]. The expansion of metagenomic techniques not only helps to identify novel genes and their functional analysis through heterologous expression but also reach up to the development of proteins with improved characteristic with the aim of tailor-made solutions for specific problems. In these ways, metagenomics offers the extraction of more valuable information about the microbial world, and their biotechnological employment to resolve various social and environmental problems would be prioritized in the frontier of research.
Table 3.
Strategies commonly used for microbial degradation of plastic.
| Strategy | Approach | Components | Constitution | Mode of Action | References |
|---|---|---|---|---|---|
| I | Prebiotic | Prooxidant | Salt of iron, manganese, cobalt, titanium | Accelerate oxidation of polymer | [161-163] |
| Biosurfactants | Starch, cellulose, Glycoproteins, lipopeptides, and other polymeric biosurfactants, bacterial exopolysaccharide | Reduce surface tension of plastic substances and helps in the adhesion of microorganisms | [89, 164] | ||
| Stimulants | Organic compounds e.g. amino acids, cofactors, Citrate, succinate, etc. | Stimulate the growth of anaerobic and methanogenic bacteria | [146, 165] | ||
| Nutrients | Nitrogen, Potasium, Phosphorus, Sulphur | To avoid deficiency of certain essential elements, also acts as the biostimulent | [7, 166] | ||
| Protectants | KMnO4 | Reduces the toxicity of triclosan, leachates from degraded plastics on diatoms and other microbes | [91] | ||
| Probiotic | Bioauguments of microbial consortia | Actinobacteria, Bacteroidetes, Proteobacteria, Ascomycetous fungi | Helps in the colonization and complex polymer degradation | [167] | |
| Microbiota transplantation | Core and specific microbial population | Replacement of indigenous population, and establishment of new population | [168, 169] | ||
| II | Ex situ microbial genome engineering | Gene insertion | PETase | To genetically engineer microbes for complex polymer degradation | [101] |
| Gene/Protein engineering | PETase | To increase the bioefficacy and thermal stability | [103] | ||
| In situ metagenome engineering | Horizontal gene transfer | Plasmid, transposable element | To disseminate the genes (enzymes) among indigenous microbial population | [170] |
Acknowledgements
The authors acknowledge Dr. Mukesh Kumar, Dr. Sumit Raj for helpful discussions, and guidance during the preparation of this manuscript.
Consent for Publication
Not applicable.
Funding
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Ritchie H., Roser M. Plastic pollution. 2020 data.org/plastic-pollution
- 2.World Economic Forum Ellen MacArthur Foundation and McKinsey Company, The new plastics economy-rethinking the future of plastics; 2016. [Google Scholar]
- 3.Chalmers University of Technology . All plastic waste could become new, high-quality plastic through advanced steam cracking. ScienceDaily; 2019. [Google Scholar]
- 4.Kershaw P.J., Turra A., Galgani F. Guidelines or the monitoring and assessment of plastic litter and microplastics in the ocean (IMO/FAO/UNESCOIOC/UNIDO/WMO/IAEA/UN/UNEP/ UNDP/ISA-Joint group of experts on the scientific aspects of marine environmental protection). Rep. Stud. GESAMP. 2019;99:130. [Google Scholar]
- 5.Parker L. The world's plastic pollution crisis explained. 2019. geographic.com/ environment/habitats/plastic-pollution
- 6.Reddy S. Plastic pollution affects sea life throughout the ocean. 2018 https://www.pewtrusts.org/en/research-and-analysis/articles/2018/09/24/plastic-pollution-affects-sea-life-throughout-the-ocean
- 7.Webb H.K., Arnott J., Crawford R.J., Ivanova E.P. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers (Basel) 2013;5(1):1–18. doi: 10.3390/polym5010001. [DOI] [Google Scholar]
- 8.Hopewell J., Dvorak R., Kosior E. Plastics recycling: challenges and opportunities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364(1526):2115–2126. doi: 10.1098/rstb.2008.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017;3(7):e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayasekara R., Harding I., Bowater I., Lonergan G. Biodegradability of a selected range of polymers and polymer blends and standard methods for assessment of biodegradation. J. Polym. Environ. 2005;13:231–251. doi: 10.1007/s10924-005-4758-2. [DOI] [Google Scholar]
- 11.Crowley D., Staines A., Collins C., Bracken J., Bruen M., Fry J. Hrymak, Victor; Malone, D.; Magette, B.; Ryan, M.; Thunhurst, C. Health and environmental effects of landfilling and incineration of waste-a literature review. Reports; 2003. p. 3. [Google Scholar]
- 12.Jambeck J.R., Geyer R., Wilcox C., Siegler T.R., Perryman M., Andrady A., Narayan R., Law K.L. Marine pollution. Plastic waste inputs from land into the ocean. Science. 2015;347(6223):768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 13.Garcia de Oliveira B., Fang M.M., Lin J. Marine plastic pollution in Asia: all hands on deck! Chinese J. Env. Law. 2019;3(1):11–46. doi: 10.1163/24686042-12340034. [DOI] [Google Scholar]
- 14.Ghosh S.K., Pal S., Ray S. Study of microbes having potentiality for biodegradation of plastics. Environ. Sci. Pollut. Res. Int. 2013;20(7):4339–4355. doi: 10.1007/s11356-013-1706-x. [DOI] [PubMed] [Google Scholar]
- 15.El-Morsy E.M., Hassan H.M., Ahmed E. Biodegradative activities of fungal isolates from plastic contaminated soils. Mycosphere. 2017;8(8):1071–1087. doi: 10.5943/mycosphere/8/8/13. [DOI] [Google Scholar]
- 16.Alshehrei F. Biodegradation of synthetic and natural plastic by microorganisms. J. Appl. Envion. Microbiol. 2017;5(1):8–19. [Google Scholar]
- 17.Shah A.A., Hasan F., Hameed A., Ahmed S. Biological degradation of plastics: a comprehensive review. Biotechnol. Adv. 2008;26(3):246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Puglisi E., Romaniello F., Galletti S., Boccaleri E., Frache A., Cocconcelli P.S. Selective bacterial colonization processes on polyethylene waste samples in an abandoned landfill site. Sci. Rep. 2019;9(1):14138. doi: 10.1038/s41598-019-50740-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamar R.T., White R.B. Mycoremediation: commercial status and recent developments.; Proceedings sixth international symposium on in situ and on-site bioremediation; San Diego, USA. 2001. pp. 263–278. [Google Scholar]
- 20.Lawton J.H., Jones C.G. In: Linking species and ecosystems: organisms as ecosystem engineers. Linking species, ecosystems; Jones, C.G. Lawton J.H., editor. New York: Chapman & Hall; 1995. pp. 141–150. [Google Scholar]
- 21.Purohit J., Chattopadhyay A., Biswas M.K., Singh N.K. In: Mycoremediation of agricultural soil: bioprospection for sustainable development. Mycoremediation and environmental sustainability, Fungal Biology; Prasad, R. Nature S., editor. 2018. pp. 91–120. [Google Scholar]
- 22.Carpenter E.J., Smith K.L., Jr Plastics on the Sargasso sea surface. Science. 1972;175(4027):1240–1241. doi: 10.1126/science.175.4027.1240. [DOI] [PubMed] [Google Scholar]
- 23.Colton J.B., Jr, Burns B.R., Knapp F.D. Plastic particles in surface waters of the northwestern atlantic. Science. 1974;185(4150):491–497. doi: 10.1126/science.185.4150.491. [DOI] [PubMed] [Google Scholar]
- 24.Shimao M. Biodegradation of plastics. Curr. Opin. Biotechnol. 2001;12(3):242–247. doi: 10.1016/S0958-1669(00)00206-8. [DOI] [PubMed] [Google Scholar]
- 25.Kirstein I.V., Wichels A., Gullans E., Krohne G., Gerdts G. The Plastisphere - Uncovering tightly attached plastic “specific” microorganisms. PLoS One. 2019;14(4):e0215859. doi: 10.1371/journal.pone.0215859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 2013;47(13):7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 27.Louca S., Polz M.F., Mazel F., Albright M.B.N., Huber J.A., O’Connor M.I., Ackermann M., Hahn A.S., Srivastava D.S., Crowe S.A., Doebeli M., Parfrey L.W. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018;2(6):936–943. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 28.Glaser J.A. Biological degradation of polymers in the environment. In: Gomiero A., editor. Plastics in the environment. Intech open. 2019. pp. 1–22. [Google Scholar]
- 29.Lugauskas A., Levinskait L., Peciulyte D. Micromycetes as deterioration agents of polymeric materials. Int. Biodeter. Biodegr. 2003;52(4):233–242. doi: 10.1016/S0964-8305(03)00110-0. [DOI] [Google Scholar]
- 30.Dussud C., Ghiglione J.F. Bacterial degradation of synthetic plastics.; Proceedings of the CIESM workshop monograph, monaco city; 2014. pp. 43–48. [Google Scholar]
- 31.Gewert B., Plassmann M.M., MacLeod M. Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci. Process. Impacts. 2015;17(9):1513–1521. doi: 10.1039/C5EM00207A. [DOI] [PubMed] [Google Scholar]
- 32.Pathak V.M.N. Review on the current status of polymer degradation: a microbial approach. Bioresour. Bioprocess. 2017;4:15. doi: 10.1186/s40643-017-0145-9. [DOI] [Google Scholar]
- 33.Swapnil K.K., Deshmukh A.G., Dudhare M.S., Patil V.B. Microbial degradation of plastic: a review. J. Biochem. Technol. 2015;6(2):952–961. [Google Scholar]
- 34.Mooney A., Ward P.G., O’Connor K.E. Microbial degradation of styrene: biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl. Microbiol. Biotechnol. 2006;72(1):1–10. doi: 10.1007/s00253-006-0443-1. [DOI] [PubMed] [Google Scholar]
- 35.Sharma M., Dhingra H.K. Poly-β-hydroxybutyrate: a biodegradable polyester, biosynthesis and biodegradation. Br. Microbiol. Res. J. 2016;14(3):1–11. doi: 10.9734/BMRJ/2016/25430. [DOI] [Google Scholar]
- 36.Raziyafathima M., Praseetha P.K., Rimal Isaac R.S. Microbial degradation of plastic waste: a review. J. Pharm. Chem. Biol. Sci. 2016;4(2):231–242. [Google Scholar]
- 37.Hugenholtz P., Goebel B.M., Pace N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998;180(18):4765–4774. doi: 10.1128/JB.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science. 2016;351(6278):1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 39.Danso D., Chow J., Streit W.R. Plastics: environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019;85(19):e01095–e19. doi: 10.1128/AEM.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaiswal S., Sharma B., Shukla P. Integrated approaches in microbial degradation of plastics. Environ. Technol. Inno. 2020;17:100567. doi: 10.1016/j.eti.2019.100567. [DOI] [Google Scholar]
- 41.Jeffries T.C., Rayu S., Nielsen U.N., Lai K., Ijaz A., Nazaries L., Singh B.K. Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Front. Microbiol. 2018;9:147. doi: 10.3389/fmicb.2018.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alves L.F., Westmann C.A., Lovate G.L., de Siqueira G.M.V., Borelli T.C., Guazzaroni M.E. Metagenomic approaches for understanding new concepts in microbial science. Int. J. Genomics. 2018;2018:2312987. doi: 10.1155/2018/2312987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinnell L.J., Turner J.W. Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front. Microbiol. 2019;10:1252. doi: 10.3389/fmicb.2019.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam K.N., Cheng J., Engel K., Neufeld J.D., Charles T.C. Current and future resources for functional metagenomics. Front. Microbiol. 2015;6:1196. doi: 10.3389/fmicb.2015.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amaral-Zettler L.A., Zettler E.R., Slikas B., Boyd G.D., Melvin D.W., Morrall C.E., Proskurowski G., Mincer T.J. The biogeography of the Plastisphere: implications for policy. Front. Ecol. Environ. 2015;13(10):541–546. doi: 10.1890/150017. [DOI] [Google Scholar]
- 46.Quero G.M., Luna G.M. Surfing and dining on the “plastisphere”: Microbial life on plastic marine debris. Adv. Oceanol. Limnol. 2017;8(2):199–207. doi: 10.4081/aiol.2017.7211. [DOI] [Google Scholar]
- 47.Jacquin J., Cheng J., Odobel C., Pandin C., Conan P., Pujo-Pay M., Barbe V., Meistertzheim A.L., Ghiglione J.F. Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “plastisphere”. Front. Microbiol. 2019;10:865. doi: 10.3389/fmicb.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roager L., Sonnenschein E.C. Bacterial candidates for colonization and degradation of marine plastic debris. Environ. Sci. Technol. 2019;53(20):11636–11643. doi: 10.1021/acs.est.9b02212. [DOI] [PubMed] [Google Scholar]
- 49.Oberbeckmann S., Loeder M.G., Gerdts G., Osborn A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014;90(2):478–492. doi: 10.1111/1574-6941.12409. [DOI] [PubMed] [Google Scholar]
- 50.Oberbeckmann S., Osborn A.M., Duhaime M.B. Microbes on a Bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS One. 2016;11(8):e0159289. doi: 10.1371/journal.pone.0159289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirstein I.V., Wichels A., Krohne G., Gerdts G. Mature biofilm communities on synthetic polymers in seawater - Specific or general? Mar. Environ. Res. 2018;142:147–154. doi: 10.1016/j.marenvres.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 52.Martin O. Viktoria, Wenman.; Andreas, Barth.; Evelyne, H.B.; Sara D.; Elena, G. Microplastic intake, its biotic drivers, and hydrophobic organic contaminant levels in the baltic herring. Front. Environ. Sci. 2019;7:134. doi: 10.3389/fenvs.2019.00134. [DOI] [Google Scholar]
- 53.Pinto M., Langer T.M., Hüffer T., Hofmann T., Herndl G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One. 2019;14(6):e0217165. doi: 10.1371/journal.pone.0217165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oberbeckmann S., Kreikemeyer B., Labrenz M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018;8:2709. doi: 10.3389/fmicb.2017.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dang H., Lovell C.R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2000;66(2):467–475. doi: 10.1128/AEM.66.2.467-475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J.W., Nam J.H., Kim Y.H., Lee K.H., Lee D.H. Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 2008;46(2):174–182. doi: 10.1007/s12275-008-0032-3. [DOI] [PubMed] [Google Scholar]
- 57.Salta M., Wharton J.A., Blache Y., Stokes K.R., Briand J.F. Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 2013;15(11):2879–2893. doi: 10.1111/1462-2920.12186. [DOI] [PubMed] [Google Scholar]
- 58.De Tender C., Devriese L.I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., Dawyndt P., Ruttink T. Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ. Sci. Technol. 2017;51(13):7350–7360. doi: 10.1021/acs.est.7b00697. [DOI] [PubMed] [Google Scholar]
- 59.Golyshin P.N., Chernikova T.N., Abraham W.R., Lünsdorf H., Timmis K.N., Yakimov M.M. Oleiphilaceae fam. nov., to include Oleiphilus messinensis gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 2002;52(Pt 3):901–911. doi: 10.1099/00207713-52-3-901. [DOI] [PubMed] [Google Scholar]
- 60.Khanna N.D., Kaur I., Bhalla T.C., Gautam N. Effect of biodegradation on thermal and crystalline behavior of polypropylene-gelatin based copolymers. J. Appl. Polym. Sci. 2010;118(3):1476–1488. doi: 10.1002/app.32434. [DOI] [Google Scholar]
- 61.Kettner M.T., Rojas-Jimenez K., Oberbeckmann S., Labrenz M., Grossart H.P. Microplastics alter composition of fungal communities in aquatic ecosystems. Environ. Microbiol. 2017;19(11):4447–4459. doi: 10.1111/1462-2920.13891. [DOI] [PubMed] [Google Scholar]
- 62.Brunner I., Fischer M., Rüthi J., Stierli B., Frey B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS One. 2018;13(8):e0202047. doi: 10.1371/journal.pone.0202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sangale M.K., Shahnawaz M., Ade A.B. Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci. Rep. 2019;9(1):5390. doi: 10.1038/s41598-019-41448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryant J.A., Clemente T.M., Viviani D.A., Fong A.A., Thomas K.A., Kemp P., Karl D.M., White A.E., DeLong E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems. 2016;1(3):e00024–e16. doi: 10.1128/mSystems.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dussud C., Hudec C., George M., Fabre P., Higgs P., Bruzaud S., Delort A.M., Eyheraguibel B., Meistertzheim A.L., Jacquin J., Cheng J., Callac N., Odobel C., Rabouille S., Ghiglione J.F. Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front. Microbiol. 2018;9:1571. doi: 10.3389/fmicb.2018.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar R.V., Kanna G.R., Elumalai S. Biodegradation of polyethylene by green photosynthetic microalgae. J. Bioremediat. Biodegrad. 2017;8:381. [Google Scholar]
- 67.Cooksey K.E., Wigglesworth-Cooksey B. Adhesion of bacteria and diatoms to surfaces in the sea: a review. Aquat. Microb. Ecol. 1995;9:87–96. doi: 10.3354/ame009087. [DOI] [Google Scholar]
- 68.Abell G.C., Bowman J.P. Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on Southern Ocean seawater. FEMS Microbiol. Ecol. 2005;53(3):379–391. doi: 10.1016/j.femsec.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Amin S.A., Parker M.S., Armbrust E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012;76(3):667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eich A., Mildenberger T., Laforsch C., Weber M. Biofilm and Diatom succession on polyethylene (pe) and biodegradable plastic bags in two marine habitats: early signs of degradation in the pelagic and benthic zone? PLoS One. 2015;10(9):e0137201. doi: 10.1371/journal.pone.0137201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cassone B.J., Grove H.C., Elebute O., Villanueva S.M.P., LeMoine C.M.R. Role of the intestinal microbiome in low-density polyethylene degradation by caterpillar larvae of the greater wax moth, Galleria mellonella. Proc. Biol. Sci. 1922;2020(287):20200112. doi: 10.1098/rspb.2020.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lou Y., Ekaterina P., Yang S-S., Lu B., Liu B., Ren N., Corvini P.F-X., Xing D. Biodegradation of polyethylene and polystyrene by greater wax moth larvae (Galleria mellonella L.) and the effect of co-diet supplementation on the core gut microbiome. Environ. Sci. Technol. 2020;54(5):2821–2831. doi: 10.1021/acs.est.9b07044. [DOI] [PubMed] [Google Scholar]
- 73.Delgado-Baquerizo M., Oliverio A.M., Brewer T.E., Benavent-González A., Eldridge D.J., Bardgett R.D., Maestre F.T., Singh B.K., Fierer N. A global atlas of the dominant bacteria found in soil. Science. 2018;359(6373):320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 74.Syranidou E., Karkanorachaki K., Amorotti F., Franchini M., Repouskou E., Kaliva M., Vamvakaki M., Kolvenbach B., Fava F., Corvini P.F., Kalogerakis N. Biodegradation of weathered polystyrene films in seawater microcosms. Sci. Rep. 2017;7(1):17991. doi: 10.1038/s41598-017-18366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Tender C., Schlundt C., Devriese L.I., Mincer T.J., Zettler E.R., Amaral-Zettler L.A. A review of microscopy and comparative molecular-based methods to characterize “Plastisphere” communities. Anal. Methods. 2017;9:2132–2143. doi: 10.1039/C7AY00260B. [DOI] [Google Scholar]
- 76.Jiao S., Xu Y., Zhang J., Hao X., Lu Y. Core microbiota in agricultural soils and their potential associations with nutrient cycling. mSystems. 2019;4(2):e00313–e00318. doi: 10.1128/mSystems.00313-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delacuvellerie A., Cyriaque V., Gobert S., Benali S., Wattiez R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 2019;380:120899. doi: 10.1016/j.jhazmat.2019.120899. [DOI] [PubMed] [Google Scholar]
- 78.Muhonja C.N., Makonde H., Magoma G., Imbuga M. Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS One. 2018;13(7):e0198446. doi: 10.1371/journal.pone.0198446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sulaiman S., Yamato S., Kanaya E., Kim J.J., Koga Y., Takano K., Kanaya S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012;78(5):1556–1562. doi: 10.1128/AEM.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., Leggewie C., Li X., Hazen T., Streit W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)- degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018;84(8):e02773–e17. doi: 10.1128/AEM.02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hajighasemi M., Tchigvintsev A., Nocek B., Flick R., Popovic A., Hai T., Khusnutdinova A.N., Brown G., Xu X., Cui H., Anstett J., Chernikova T.N., Brüls T., Le Paslier D., Yakimov M.M., Joachimiak A., Golyshina O.V., Savchenko A., Golyshin P.N., Edwards E.A., Yakunin A.F. Screening and characterization of novel polyesterases from environmental metagenomes with high hydrolytic activity against synthetic polyesters. Environ. Sci. Technol. 2018;52(21):12388–12401. doi: 10.1021/acs.est.8b04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yao J., Fan X.J., Lu Y., Liu Y.H. Isolation and characterization of a novel tannase from a metagenomic library. J. Agric. Food Chem. 2011;59(8):3812–3818. doi: 10.1021/jf104394m. [DOI] [PubMed] [Google Scholar]
- 83.Tirawongsaroj P., Sriprang R., Harnpicharnchai P., Thongaram T., Champreda V., Tanapongpipat S., Pootanakit K., Eurwilaichitr L. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 2008;133(1):42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 84.Meier M.J., Paterson E.S., Lambert I.B. Use of substrate-induced gene expression in metagenomic analysis of an aromatic hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 2015;82(3):897–909. doi: 10.1128/AEM.03306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lewin A., Strand T., Haugen T., Klinkenberg G., Kotlar H., Valla S., Drablos F., Wentzel A. Discovery and characterization of a thermostable esterase from an oil reservoir metagenome. Adv. Enzyme Res. 2016;4(2):68–86. doi: 10.4236/aer.2016.42008. [DOI] [Google Scholar]
- 86.Ronda C., Chen S.P., Cabral V., Yaung S.J., Wang H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods. 2019;16(2):167–170. doi: 10.1038/s41592-018-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Foo J.L., Ling H., Lee Y.S., Chang M.W. Microbiome engineering: Current applications and its future. Biotechnol. J. 2017;12(3) doi: 10.1002/biot.201600099. [DOI] [PubMed] [Google Scholar]
- 88.Sheth R.U., Cabral V., Chen S.P., Wang H.H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016;32(4):189–200. doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaczorek E., Pacholak A., Zdarta A., Smułek W. The impact of biosurfactants on microbial cell properties leading to hydrocarbon bioavailability increase. Colloids Interfaces. 2018;2(3):35. doi: 10.3390/colloids2030035. [DOI] [Google Scholar]
- 90.Arkatkar A., Juwarkar A.A., Bhaduri S., Uppara P.V., Doble M. Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface. Int. Biodeter. Biodegr. 2016;4(6):530–536. doi: 10.1016/j.ibiod.2010.06.002. [DOI] [Google Scholar]
- 91.Ding T., Lin K., Yang M., Bao L., Li J., Yang B., Gan J. Biodegradation of triclosan in diatom Navicula sp.: Kinetics, transformation products, toxicity evaluation and the effects of pH and potassium permanganate. J. Hazard. Mater. 2018;344:200–209. doi: 10.1016/j.jhazmat.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 92.Fritz J., Sandhofer M., Stacher C., Braun R. Strategies for detecting ecotoxicological effects of biodegradable polymers in agricultural applications. Macromol. Symp. 2003;197:397–410. doi: 10.1002/masy.200350734. [DOI] [Google Scholar]
- 93.Haider T.P., Völker C., Kramm J., Landfester K., Wurm F.R. Plastics of the future? the impact of biodegradable polymers on the environment and on society. Angew. Chem. Int. Ed. Engl. 2019;58(1):50–62. doi: 10.1002/anie.201805766. [DOI] [PubMed] [Google Scholar]
- 94.Thompson I.P., van der Gast C.J., Ciric L., Singer A.C. Bioaugmentation for bioremediation: the challenge of strain selection. Environ. Microbiol. 2005;7(7):909–915. doi: 10.1111/j.1462-2920.2005.00804.x. [DOI] [PubMed] [Google Scholar]
- 95.Mrozik A., Piotrowska-Seget Z. Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol. Res. 2010;165(5):363–375. doi: 10.1016/j.micres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Bhatnagar S., Kumari R. Bioremediation: a sustainable tool for environmental management-a review. Ann. Rev. Res. Biol. 2013;3(4):974–993. [Google Scholar]
- 97.Trosset J.Y., Carbonell P. Synthetic biology for pharmaceutical drug discovery. Drug Des. Devel. Ther. 2015;9:6285–6302. doi: 10.2147/DDDT.S58049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghosh S., Qureshi A., Purohit H.J. In: Microbial degradation of plastics: Biofilms and degradation pathways. Contaminants in agriculture and environment: health risks and remediation; Kumar, V.; Kumar, R.; Singh, J.; Kumar, P. Media A.E., editor. Haridwar, India: 2019. pp. 184–199. [Google Scholar]
- 99.Huang X., Cao L., Qin Z., Li S., Kong W., Liu Y. Tat-independent secretion of polyethylene terephthalate hydrolase PETase in Bacillus subtilis 168 mediated by its native signal peptide. J. Agric. Food Chem. 2018;66(50):13217–13227. doi: 10.1021/acs.jafc.8b05038. [DOI] [PubMed] [Google Scholar]
- 100.Seo H., Kim S., Son H.F., Sagong H.Y., Joo S., Kim K.J. Production of extracellular PETase from Ideonella sakaiensis using sec-dependent signal peptides in E. coli. Biochem. Biophys. Res. Commun. 2019;508(1):250–255. doi: 10.1016/j.bbrc.2018.11.087. [DOI] [PubMed] [Google Scholar]
- 101.Moog D., Schmitt J., Senger J., Zarzycki J., Rexer K.H., Linne U., Erb T., Maier U.G. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb. Cell Fact. 2019;18(1):171. doi: 10.1186/s12934-019-1220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kawai F., Kawabata T., Oda M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019;103(11):4253–4268. doi: 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Son H.F., Cho I.J., Joo S., Seo H., Sagong H.Y., Choi S.Y., Lee S.Y., Kim K.J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient pet degradation. ACS Catal. 2019;9(4):3519–3526. doi: 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- 104.Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., Silveira R.L., Pollard B.C., Dominick G., Duman R., El Omari K., Mykhaylyk V., Wagner A., Michener W.E., Amore A., Skaf M.S., Crowley M.F., Thorne A.W., Johnson C.W., Woodcock H.L., McGeehan J.E., Beckham G.T. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA. 2018;115(19):E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herrero Acero E., Ribitsch D., Steinkellner G., Gruber K., Greimel K., Eiteljoerg I., Trotscha E., Wei R., Zimmermann W., Zinn M., Cavaco-Paulo A., Freddi G., Schwab H., Guebitz G. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules. 2011;44(12):4632–4640. doi: 10.1021/ma200949p. [DOI] [Google Scholar]
- 106.Wei R., Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb. Biotechnol. 2017;10(6):1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Furukawa M., Kawakami N., Tomizawa A., Miyamoto K. Efficient degradation of poly(ethylene terephthalate) with Thermobifida fusca cutinase exhibiting improved catalytic activity generated using mutagenesis and additive-based approaches. Sci. Rep. 2019;9(1):16038. doi: 10.1038/s41598-019-52379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma Y., Yao M., Li B., Ding M., He B., Chen S., Zhou X., Yuan Y. Enhanced poly (ethylene terephthalate) hydrolase activity by protein engineering. Engineering. 2018;4(6):888–893. doi: 10.1016/j.eng.2018.09.007. [DOI] [Google Scholar]
- 109.Cui Y., Chen Y., Liu X., Dong S., Tian Y., Qiao Y., Han J., Li C., Han X., Liu W., Chen Q., Du W., Tang S., Xiang H., Liu H. Computational redesign of PETase for plastic biodegradation by GRAPE strategy. bioRxiv. 2019;787069 doi: 10.1101/787069. [DOI] [Google Scholar]
- 110.de Lipthay J.R., Barkay T., Sørensen S.J. Enhanced degradation of phenoxyacetic acid in soil by horizontal transfer of the tfdA gene encoding a 2,4-dichlorophenoxyacetic acid dioxygenase. FEMS Microbiol. Ecol. 2001;35(1):75–84. doi: 10.1111/j.1574-6941.2001.tb00790.x. [DOI] [PubMed] [Google Scholar]
- 111.Ikuma K., Gunsch C.K. Genetic bioaugmentation as an effective method for in situ bioremediation: functionality of catabolic plasmids following conjugal transfers. Bioengineered. 2012;3(4):236–241. doi: 10.4161/bioe.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cosgrove L., McGeechan P.L., Handley P.S., Robson G.D. Effect of biostimulation and bioaugmentation on degradation of polyurethane buried in soil. Appl. Environ. Microbiol. 2010;76(3):810–819. doi: 10.1128/AEM.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seow Y., Wood M.J. Biological gene delivery vehicles: beyond viral vectors. Mol. Ther. 2009;17(5):767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chaudhary K., Chattopadhyay A., Pratap D. The evolution of CRISPR/Cas9 and their cousins: hope or hype? Biotechnol. Lett. 2018;40(3):465–477. doi: 10.1007/s10529-018-2506-7. [DOI] [PubMed] [Google Scholar]
- 115.Chattopadhyay A., Purohit J., Tiwari K.K., Deshmukh R. Targeting transcription factors for plant disease resistance: shifting paradigm. Curr. Sci. 2019;117(1):1598–1607. doi: 10.18520/cs/v117/i10/1598-1607. [DOI] [Google Scholar]
- 116.Otoupal P.B., Chatterjee A. CRISPR gene perturbations provide insights for improving bacterial biofuel tolerance. Front. Bioeng. Biotechnol. 2018;6:122. doi: 10.3389/fbioe.2018.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mohammad T., Hassan M.I. Modern approaches in synthetic biology: genome editing, quorum sensing, and microbiome engineering. In: Singh S., editor. Synthetic biology. Singapore: Springer; 2018. pp. 189–205. [Google Scholar]
- 118.Jusiak B., Cleto S., Perez-Piñera P., Lu T.K. Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 2016;34(7):535–547. doi: 10.1016/j.tibtech.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 119.Rolf J., Rosenthal K., Lütz S. Application of cell-free protein synthesis for faster biocatalyst development. Catalysts. 2019;9(2):190. doi: 10.3390/catal9020190. [DOI] [Google Scholar]
- 120.Oberbeckmann S., Labrenz M. Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Annu. Rev. Mar. Sci. 2020;12(12):209–232. doi: 10.1146/annurev-marine-010419-010633. [DOI] [PubMed] [Google Scholar]
- 121.Paço A., Duarte K., da Costa J.P., Santos P.S.M., Pereira R., Pereira M.E., Freitas A.C., Duarte A.C., Rocha-Santos T.A.P. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 2017;586:10–15. doi: 10.1016/j.scitotenv.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 122.Dussud C., Meistertzheim A.L., Conan P., Pujo-Pay M., George M., Fabre P., Coudane J., Higgs P., Elineau A., Pedrotti M.L., Gorsky G., Ghiglione J.F. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018;236:807–816. doi: 10.1016/j.envpol.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 123.Sekiguchi T., Ebisui A., Nomura K., Watanabe T., Enoki M., Kanehiro H. Biodegradation of several fibers submerged in deep sea waterand isolation of biodegradable plastic degrading bacteria from deep ocean water. Nippon Suisan Gakkaishi. 2009;75(6):1011–1018. doi: 10.2331/suisan.75.1011. [DOI] [Google Scholar]
- 124.Urbanek A.K., Rymowicz W., Mirończuk A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 2018;102(18):7669–7678. doi: 10.1007/s00253-018-9195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sekiguchi T., Saika A., Nomura K., Watanabe T., Watanabe T., Fujimoto Y. Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ɛ-caprolactone)-degrading bacteria. Polym. Degrad. Stabil. 2011;96(7):1397–1403. doi: 10.1016/j.polymdegradstab.2011.03.004. [DOI] [Google Scholar]
- 126.Kathiresan K. Polythene and plastic-degrading microbes in an Indian mangrove soil. Rev. Biol. Trop. 2003;51(3-4):629–633. [PubMed] [Google Scholar]
- 127.Sheik S., Chandrashekar K.R., Swaroop K., Somashekarappa H.M. Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int. Biodeter. Biodegr. 2015;105:21–29. doi: 10.1016/j.ibiod.2015.08.006. [DOI] [Google Scholar]
- 128.Suresh B., Maruthamuthu S., Palanisamy N., Ragunathan R., Pandiyaraj K.N., Muralidharan V.S. Investigation on biodegradability of polyethylene by Bacillus cereus strain Ma-Su isolated from compost soil. Int. Res. J. Microbiol. 2011;2(8):292–302. [Google Scholar]
- 129.Hadad D., Geresh S., Sivan A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 2005;98(5):1093–1100. doi: 10.1111/j.1365-2672.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 130.Yamada O.K., Mukumoto H., Katsuyaya Y., Saiganji A., Tani Y. Degradation of polyethylene by a fungus, Penicillium simplicissmum YK. Polym. Degrad. Stabil. 2001;72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- 131.Lee B., Pometto A.L., Fratzke A., Bailey T.B. Biodegradation of degradable plastic polyethylene by phanerochaete and streptomyces species. Appl. Environ. Microbiol. 1991;57(3):678–685. doi: 10.1128/AEM.57.3.678-685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liyoshi Y., Tsutsumi Y., Nishida T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 1998;44:222–229. doi: 10.1007/BF00521967. [DOI] [Google Scholar]
- 133.Gilan I., Sivan A. Effect of proteases on biofilm formation of the plastic-degrading actinomycete Rhodococcus ruber C208. FEMS Microbiol. Lett. 2013;342(1):18–23. doi: 10.1111/1574-6968.12114. [DOI] [PubMed] [Google Scholar]
- 134.Santo M., Weitsman R., Sivan A. The role of the copper-binding enzyme laccase in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeter. Biodegr. 2013;84:204–210. doi: 10.1016/j.ibiod.2012.03.001. [DOI] [Google Scholar]
- 135.Müller R-J., Schrader H., Profe J., Dresler K., Deckwer W-D. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolysis using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005;26:1400–1405. doi: 10.1002/marc.200500410. [DOI] [Google Scholar]
- 136.O’Neill A., Araújo R., Casal M., Guebitz G., Cavaco-Paulo A. Effect of the agitation on the adsorption and hydrolytic efficiency of cutinases on polyethylene terephthalate fibres. Enzyme Microb. Technol. 2007;40(7):1801–1805. doi: 10.1016/j.enzmictec.2007.02.012. [DOI] [Google Scholar]
- 137.Ronqvist Å.M., Xie W., Lu W., Gross R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules. 2009;42:5128–5138. doi: 10.1021/ma9005318. [DOI] [Google Scholar]
- 138.Cacciari I., Quatrini P., Zirletta G., Mincione E., Vinciguerra V., Lupattelli P., Giovannozzi Sermanni G. Isotactic polypropylene biodegradation by a microbial community: physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 1993;59(11):3695–3700. doi: 10.1128/AEM.59.11.3695-3700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arkatkar A., Arutchelvi J., Bhaduri S., Uppara P.V., Doble M. Degradation of unpretreated and thermally pretreated polypropylene by soil consortia. Int. Biodeter. Biodegr. 2009;63(1):106–111. doi: 10.1016/j.ibiod.2008.06.005. [DOI] [Google Scholar]
- 140.Danko A.S., Luo M., Bagwell C.E., Brigmon R.L., Freedman D.L. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl. Environ. Microbiol. 2004;70(10):6092–6097. doi: 10.1128/AEM.70.10.6092-6097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O’Leary N.D., O’Connor K.E., Ward P., Goff M., Dobson A.D. Genetic characterization of accumulation of polyhydroxyalkanoate from styrene in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 2005;71(8):4380–4387. doi: 10.1128/AEM.71.8.4380-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Russell J.R., Huang J., Anand P., Kucera K., Sandoval A.G., Dantzler K.W., Hickman D., Jee J., Kimovec F.M., Koppstein D., Marks D.H., Mittermiller P.A., Núñez S.J., Santiago M., Townes M.A., Vishnevetsky M., Williams N.E., Vargas M.P., Boulanger L.A., Bascom-Slack C., Strobel S.A. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011;77(17):6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peciulyte D. Microbial colonization and biodeterioration of plasticized polyvinyl chloride plastics. Ekologija (Liet. Moksl. Akad.) 2002;4:7–15. [Google Scholar]
- 144.Zheng Y., Yanful E.K., Bassi A.S. A review of plastic waste biodegradation. Crit. Rev. Biotechnol. 2005;25(4):243–250. doi: 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]
- 145.Howard G.T. Blake, R.C. Growth of Pseudomonas fluorescens on a polyester-polyurethane and the purification and characterization of a polyurethanase-protease enzyme. Int. Biodeter. Biodegr. 1998;42:213–220. doi: 10.1016/S0964-8305(98)00051-1. [DOI] [Google Scholar]
- 146.Hung C.S., Zingarelli S., Nadeau L.J., Biffinger J.C., Drake C.A., Crouch A.L. Carbon catabolite repression and impranil polyurethane degradation in Pseudomonas protegens strain Pf-5. Appl. Environ. Microbiol. 2016;82(20):6080–6090. doi: 10.1128/AEM.01448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kleeberg I., Hetz C., Kroppenstedt R.M., Müller R.J., Deckwer W.D. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates. Appl. Environ. Microbiol. 1998;64(5):1731–1735. doi: 10.1128/AEM.64.5.1731-1735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Howard G.T., Norton W.N., Burks T. Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme. Biodegradation. 2012;23(4):561–573. doi: 10.1007/s10532-011-9533-6. [DOI] [PubMed] [Google Scholar]
- 149.Devi R.S., Kannan V.R., Natarajan K., Nivas D., Kannan K., Chandru S., Anthony A.R. The role of microbes in plastic degradation. In: Chandra R., editor. Environmental waste management. Boca Raton: CRC Press; 2015. pp. 341–370. [Google Scholar]
- 150.Kirbaş Z., Keskin N., Güner A. Biodegradation of polyvinylchloride (PVC) by white rot fungi. Bull. Environ. Contam. Toxicol. 1999;63(3):335–342. doi: 10.1007/s001289900985. [DOI] [PubMed] [Google Scholar]
- 151.Negoro S., Shibata N., Tanaka Y., Yasuhira K., Shibata H., Hashimoto H., Lee Y.H., Oshima S., Santa R., Oshima S., Mochiji K., Goto Y., Ikegami T., Nagai K., Kato D., Takeo M., Higuchi Y. Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis. J. Biol. Chem. 2012;287(7):5079–5090. doi: 10.1074/jbc.M111.321992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fujisawa M., Hirai H., Nishida T. Degradation of polyethylene and nylon-66 by the Laccase-mediator system. J. Polym. Environ. 2001;9:103–108. doi: 10.1023/A:1020472426516. [DOI] [Google Scholar]
- 153.Deguchi T., Kitaoka Y., Kakezawa M., Nishida T. Purification and characterization of a nylon-degrading enzyme. Appl. Environ. Microbiol. 1998;64(4):1366–1371. doi: 10.1128/AEM.64.4.1366-1371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vijaya R., Reddy M. Impact of soil composting using municipal solid waste on biodegradation of plastics. Indian J. Biotechnol. 2008;7:235–239. [Google Scholar]
- 155.Zadjelovic V., Chhun A., Quareshy M., Silvano E., Hernandez-Fernaud J.R., Aguilo-Ferretjans M.M., Bosch R., Dorador C., Gibson M.I., Christie-Oleza J.A. Beyond oil degradation: enzymatic potential of Alcanivorax to degrade natural and synthetic polyesters. Environ. Microbiol. 2020;22(4):1356–1369. doi: 10.1111/1462-2920.14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tokiwa Y., Calabia B.P., Ugwu C.U., Aiba S. Biodegradability of plastics. Int. J. Mol. Sci. 2009;10(9):3722–3742. doi: 10.3390/ijms10093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Krueger M.C., Harms H., Schlosser D. Prospects for microbiological solutions to environmental pollution with plastics. Appl. Microbiol. Biotechnol. 2015;99(21):8857–8874. doi: 10.1007/s00253-015-6879-4. [DOI] [PubMed] [Google Scholar]
- 158.Mergaert J., Webb A., Anderson C., Wouters A., Swings J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl. Environ. Microbiol. 1993;59(10):3233–3238. doi: 10.1128/AEM.59.10.3233-3238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Matavulj M., Molitoris H.P. Fungal degradation of polyhydroxyalkanoates and a semiquantitative assay for screening their degradation by terrestrial fungi. FEMS Microbiol. Rev. 1992;9(2-4):323–331. doi: 10.1111/j.1574-6968.1992.tb05854.x. [DOI] [PubMed] [Google Scholar]
- 160.Muhamad W., Naimatul Asiah W., Othman R., Shaharuddin R.I. Microorganism as plastic biodegradation agent towards sustainable environment. Adv. Environ. Biol. 2015;9(13):8–13. [Google Scholar]
- 161.Koutny M., Lemaire J., Delort A.M. Biodegradation of polyethylene films with prooxidant additives. Chemosphere. 2006;64(8):1243–1252. doi: 10.1016/j.chemosphere.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 162.Konduri M.K.R., Koteswarareddy G., Rohini K.D.B., Venkata R.B., Lakshmi N.M. Effect of pro‐oxidants on biodegradation of polyethylene (LDPE) by indigenous fungal isolate, Aspergillus oryzae. J. Appl. Polym. Sci. 2011;120:3536–3545. doi: 10.1002/app.33517. [DOI] [Google Scholar]
- 163.Abrusci C., Pablos J., Corrales T., López-Marín J., Marín I., Catalina F. Biodegradation of photo-degraded mulching films based on polyethylenes and stearates of calcium and iron as pro-oxidant additives. Int. Biodeter. Biodegr. 2011;65(3):451–459. doi: 10.1016/j.ibiod.2010.10.012. [DOI] [Google Scholar]
- 164.Parthipan P., Preetham E., Machuca L.L., Rahman P.K.S.M., Murugan K., Rajasekar A. Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Front. Microbiol. 2017;8:193. doi: 10.3389/fmicb.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yadav G.D., Sontakke J.B. In: Methods for separation, recycling and reuse of biodegradation products. Biodegradation-engineering and technology; Chamy, R.; Rosenkranz, F. Open I., editor. 2013. pp. 277–311. [Google Scholar]
- 166.Rosa A.P., Triguis J.A. Bioremediation process on Brazil shoreline. Laboratory experiments. Environ. Sci. Pollut. Res. Int. 2007;14(7):470–476. doi: 10.1065/espr2007.02.377. [DOI] [PubMed] [Google Scholar]
- 167.Syranidou E., Karkanorachaki K., Amorotti F., Repouskou E., Kroll K., Kolvenbach B., Corvini P.F., Fava F., Kalogerakis N. Development of tailored indigenous marine consortia for the degradation of naturally weathered polyethylene films. PLoS One. 2017;12(8):e0183984. doi: 10.1371/journal.pone.0183984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McCormick A., Hoellein T.J., Mason S.A., Schluep J., Kelly J.J. Microplastic is an abundant and distinct microbial habitat in an urban river. Environ. Sci. Technol. 2014;48(20):11863–11871. doi: 10.1021/es503610r. [DOI] [PubMed] [Google Scholar]
- 169.Herlemann D.P., Labrenz M., Jürgens K., Bertilsson S., Waniek J.J., Andersson A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5(10):1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Garbisu C., Garaiyurrebaso O., Epelde L., Grohmann E., Alkorta I. Plasmid-mediated bioaugmentation for the bioremediation of contaminated soils. Front. Microbiol. 2017;8:1966. doi: 10.3389/fmicb.2017.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]