Abstract
Gynostemma yixingense, an important medicinal member of the Cucurbitaceae family, is an endemic herbaceous species distributed in East China. It is morphologically similar to the plants in the same genus, which resulted in some confusion in identification and application. Meanwhile, there are still some controversies in taxonomy. Herein, the complete chloroplast genome sequence of G. yixingense was obtained by Illumina paired-end sequencing technology and compared to other chloroplast genome sequences of congeneric species. The complete chloroplast genome of G. yixingense is 157,910 bp in length with 36.94% GC content and contains a large single-copy (LSC) region of 86,791 bp, a small single-copy (SSC) region of 18,635 bp and a pair of inverted repeat (IR) regions of 26,242 bp. The whole genome contains 133 unique genes, including 87 protein-coding genes, 37 tRNA genes, eight rRNA genes and one pseudogene. In addition, 74 simple sequence repeats (SSRs) were identified, most of which were A/T rich. The phylogenetic analysis indicated that G. yixingense had the closest relationship to G. laxiflorum. The result of this study provided an important theoretical basis for chloroplast genome and phylogenetic analysis of G. yixingense.
Keywords: Chloroplast genome, Cucurbitaceae; Gynostemma; phylogeny; simple sequence repeats
Gynostemma yixingense, a member of the Gynostemma genus belonging to the family Cucurbitaceae, is an endemic species in East China. It occurs in forests or thickets with an altitude of below 100 m, mainly produced in Anhui, southern Jiangsu and Zhejiang Province, China (Chen et al., 2011). In 1990, G. yixingense var. trichocarpum was identified as a variant of G. yixingense (Ding, 1990). Chen (1995) classified the Gynostemma into two subgenera and two groups according to fruit type and style number. The berry type is of the Gynostemma subgenus, and the capsule type of the Trirostellum subgenus. The Trirostellum subgenus is further divided into two groups according to style number. The styles (4-) 5 is the Pentastylos group, and the styles 3 is the Trirostellum group. The original G. yixingense and G. yixingense var. trichocarpum were combined to G. yixingense in Flora of China (FOC). However, Jeffrey believed that G. laxiflorum and G. yixingense should be the same species in FOC. Today, the genus Gynostemma has 14 species (nine endemic) in China (Chen et al., 2011; Zhang et al., 2017).
For the existence of ginsenosides, G. pentaphyllum, the same genus of G. yixingense is known as ‘Panax ginseng of Southern China’ (Qin et al., 2015). It is an important medicinal plant with a variety of therapeutic effects that include enhancing immunity, lowering cholesterol, regulating blood pressure, anti-inflammatory and anticancer (Wang et al., 2017). G. yixingense contains similar chemical components to G. pentaphyllum, even containing higher ginsenosides (Zhang et al., 2015). In addition, G. yixingense has a sweeter taste that makes it appreciated by consumers (Xiang et al., 2010). G. yixingense is morphologically similar to G. pentaphyllum, which are often confused in identification and application.
Chloroplasts have relatively independent genetic material, the chloroplast genome. Compared with nuclear genes, the chloroplast genome is often more conserved, which is great significance in plant phylogeny and species identification (Zhou et al., 2017; Meng et al., 2018). Within Gynostemma, the complete chloroplast genomes of several plant species have been published (Zhang et al., 2017, 2018; Shi et al., 2019), nevertheless, no chloroplast genome of G. yixingense has been reported until now. Therefore, in order to provide a reference for systematic evolution and rational use of G. yixingense, the complete chloroplast genome of G. yixingense was sequenced and analyzed in this study. Phylogenetic analyses were performed with other nine plants in the same genus.
Fresh leaves of G. yixingense were collected from Tongwu village, Hangzhou City, Zhejiang Province, China, in bushes, 30°12’10’’ N, 120°03’04’’ E, elevation 237 m. Voucher specimens were deposited in the Center of Herbarium, China Pharmaceutical University, Nanjing, China, under accession number WL20191005. Whole genomic DNA was extracted from fresh leaves using Rapid Plant Genomic DNA Isolation Kit, Sangon Biotech (Shanghai). Then, the quality and integrity of DNA were checked using BioPhotometer Plus (Nucleic acid and protein detector, Eppendorf, Germany) and 1% agarose gels. High quality of DNA was used to construct the library. These raw reads were deposited in NCBI Sequence Read Archive (SRA) with the accession number PRJNA598798. Sequencing was performed on Illumina Xten platform (GENEWIZ Suzhou, China). Using the G. pentaphyllum chloroplast genome as reference (NCBI accession No. KX852298), clean reads of the matched reference genes were extracted. The obtained reads were assembled with NOVOPlasty v2.7.2 (Dierckxsen et al., 2016), and the assembled genome was annotated and analyzed using the GeSeq tool (Tillich et al., 2017). The chloroplast genome data was compared to the NCBI database by BLAST, searching for protein coding genes, rRNA genes and tRNA genes, while the tRNA genes were further confirmed using tRNAscan-SE v2.0 program (Lowe and Chan, 2016). The finally annotated chloroplast genome was deposited in GenBank with the accession number MT028489. The chloroplast genome map was drawn using OGDRAW v1.2 based on the annotated results (Lohse et al., 2007).
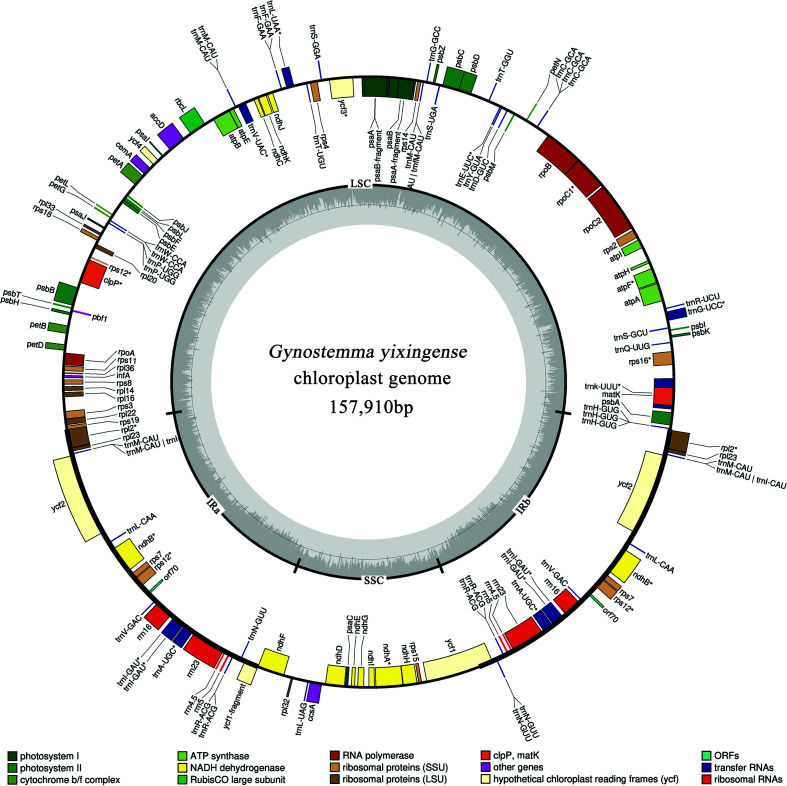
After quality filtering, a total number of 65,730,414 clean reads (>Q20) were obtained, and the whole chloroplast genome was assembled using these clean reads. The length of the chloroplast genome of G. yixingense was 157,910 bp, which had the cyclic tetrad structure of chloroplast genomes typical in angiosperms, including large and small single copy regions (LSC and SSC) of 86,791 bp and 18,635 bp, respectively, and a pair of inverted repeats (IRa and IRb) of 26,242 bp. The overall GC content was 36.94%, and the GC content of the LSC, SSC and IR regions were 34.76%, 30.61% and 42.79%, respectively. In addition, a total of 133 genes were annotated, including 87 protein-coding genes, eight ribosomal RNA genes, 37 tRNA genes and one pseudogene (infA). Among them, 19 genes were duplicated in the IR regions, which contain eight protein-coding genes, seven tRNA genes and four rRNA genes (Figure 1, Table 1). The rps12 gene had a trans-spliced structure. The 5’ and 3’ ends of rps12 were located in the LSC region and IR region, respectively, which were divided into two independent transcription units. Furthermore, 15 genes possessed introns. Thirteen genes contained one intron, and two genes (ycf3 and clpP) contained two introns. These introns ranged in length from 535 bp to 2,489 bp, of which TrnK-UUU gene had the longest intron, 2,489 bp (Table S1 (106.9KB, pdf) ).
Figure 1. Chloroplast genome map of Gynostemma yixingense. Genes drawn inside the circle are transcribed clockwise, genes outside are transcribed counterclockwise. Genes are color coded by their function in the legend. The area in darker gray and lighter gray in the inner circle indicates GC content and AT content, respectively.
Table 1. Gene content in the chloroplast genome of of Gynostemma yixingense.
| Category of genes | Group of genes | Name of genes |
|---|---|---|
| Photosynthesis | ATP synthase gene | atpA, atpB, atpE, atpF * , atpH, atpI |
| NADH dehydrogenase | ndhA * , ndhB * (x2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Cytochrome b/f complex | petA, petB, petD, petG, petN, petL | |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ ycf4, ycf3 * * | |
| Photosystem II | psbH, psbN, psbT, psbE, psbZ, psbK, psbC, psbA, psbJ, psbL, psbI, psbM, psbF, psbB, psbD | |
| Large chain of rubisco | rbcL | |
| Self-replication | Large subunit of ribosome | rpl2 * (x2), rpl14, rpl16, rpl20, rpl22, rpl23(x2) , rpl32, rpl33, rpl36 |
| RNA polymerase subunits | rpoA, rpoB, rpoC1 * , rpoC2 | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7(x2), rps8, rps11, rps12 * (x2) , rps14, rps15, rps16 * , rps18, rps19 | |
| rRNA genes | rrn23(x2), rrn4.5(x2), rrn5(x2), rrn16(x2) | |
| tRNA genes | trnR-ACG(x2), trnN-GUU(x2), trnV-GAC(x2), trnL-CAA(x2), trnE-UUC, trnY-GUA, trnD-GUC, trnR-UCU, trnI-CAU(x2), trnP-UGG, trnM-CAU, trnF-GAA, trnH-GUG, trnC-GCA, trnS-UGA, trnV-UAC * , trnT-GGU, trnQ-UUG, trnG-GCC, trnS-GGA, trnG-UCC * , trnI-GAU * (x2), trnA-UGC * (x2), trnk-UUU * , trnfM-CAU, trnS-GCU, trnT-UGU, trnL-UAA * , trnL-UAG, trnW-CCA | |
| Other genes | Acetyl-CoA carboxylase | accD |
| Cytochrome c biogenesis | ccsA | |
| Membrane protein | cemA | |
| ATP-dependent protease | clpP * * | |
| Maturase | matK | |
| Translational initiation factor | ψ infA | |
| Unknown function | Conserved open reading frames | ycf2(x2), ycf1(x2) |
| hypothetical chloroplast protein | orf70(x2) |
Indicates the genes containing one intron
Indicates the genes containing two introns, (x2) Indicates genes duplicated in the IR regions.
Indicates the pseudogene.
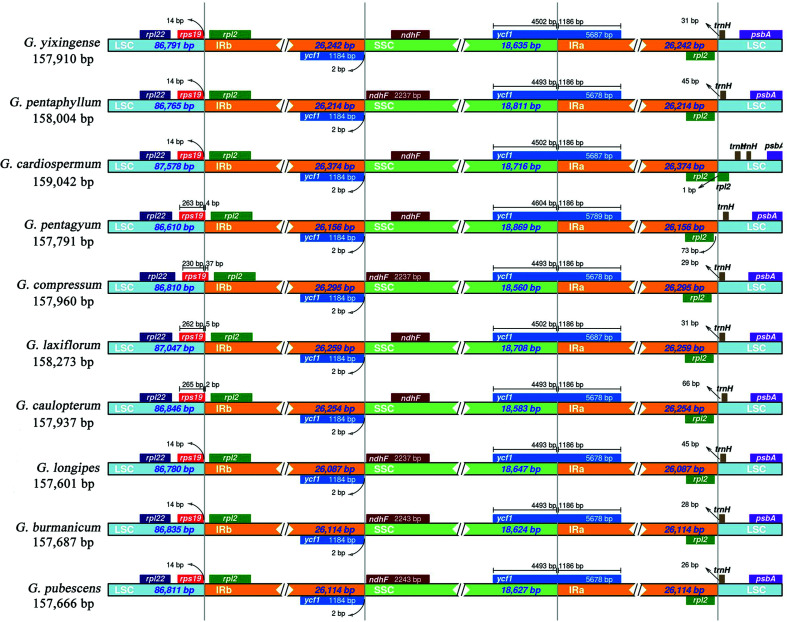
The length of angiosperm chloroplast genomes is variable primarily due to expansion and contraction of IR region (Zhang et al., 2014). Hence, the IR/SC boundary regions of the ten Gynostemma chloroplast genomes were compared in this study (Figure 2). The results showed, except some boundary differences, all the ten Gynostemma chloroplast genomes exhibited striking similarities on the IR borders. For example, rps19 across the IRb/LSC boundary in G. pentagyum, G. compressum, G. laxiflorum and G. caulopterum in IRb/LSC region, while the other six species were situated in the LSC region. The SSC/IRb boundary of all plants were located in the ycf1 gene, resulting in the production of the ycf1 pseudogene in the IRa region. In the IRa/SSC region, ycf1 gene across all the IRa/SSC boundaries, but the fragment length of ycf1 genes in SSC region were different.
Figure 2. Comparison of the LSC, IR and SSC junction positions among ten Gynostemma chloroplast genomes. Colored boxes for genes represent the gene position.
Simple sequence repeats (SSRs), also known as microsatellites, are short repeat sequences with length of 1-6 bp, which are widely used in phylogenetic analysis and population genetics (Cavalier-Smith, 2002). In this study, the microsatellite identification tool MISA (https://webblast.ipk-gatersleben.de/misa/) was used to detect SSRs (parameter setting: the minimum repeat number of 10, 5, 4, 3, 3 and 3 for mono-, di-, tri-, tetra-, penta- and hexanucleotide repeats, respectively). The maximum length of sequence between two SSRs to register as compound SSR was 100 bp. A total of 74 SSRs were identified in the chloroplast genome of G. yixingense, including 46 mononucleotide repeats (62.2%), 16 dinucleotide repeats (21.62%), three trinucleotide repeats (4.05%), and nine tetranucleotide repeats (12.16%). There were 67 SSRs made up of A or T (90.54%), which indicates that the composition of SSRs tends to use A/T (Table S2 (102.8KB, pdf) ).
To construct a phylogenetic tree, 13 chloroplast genomes from Cucurbitaceae were employed, including G. yixingense, nine other Gynostemma species and three outgroups (Table S3 (54.6KB, pdf) ). The criterion for selection of outgroups was that they should be medicinal plants in a different genus of Cucurbitaceae and plants with relatively similar morphology. The chloroplast genomes of the selected species were downloaded from NCBI. Phylogenetic inference was performed using 77 common protein-coding genes (Table S4 (136.3KB, pdf) ). MAFFT (Katoh and Standley, 2013) was employed to sequence alignment, and BioEdit v7.0.9.0 (Hall, 1999) was also used to examine and manually adjust the sequence alignment result. Phylogenetic trees were constructed using Maximum Parsimony (MP) and Bayesian Inference (BI) analysis. MP analysis was performed in PAUP* v4.0 beta 10 (Swofford, 2002), and BI was performed in MrBayes 3.2.6 (Ronquist et al., 2012). For the MP analysis, the bootstrap probability was determined with 1000 replicates. For BI analysis, the best-fit model (GTR+I+G) in the analysis was selected by Akaike information criterion (AIC) in MrModeltest v2.3 (Nylander, 2004). Four Markov Chains Monte Carlo (MCMC) samples were run for 1 × 106 generations. The convergence of MCMC runs was additionally confirmed by two independent runs, and trees were sampled every 100 generations. The burn-in was set to discard 25% of the trees to produce consensus tree of all remaining trees.
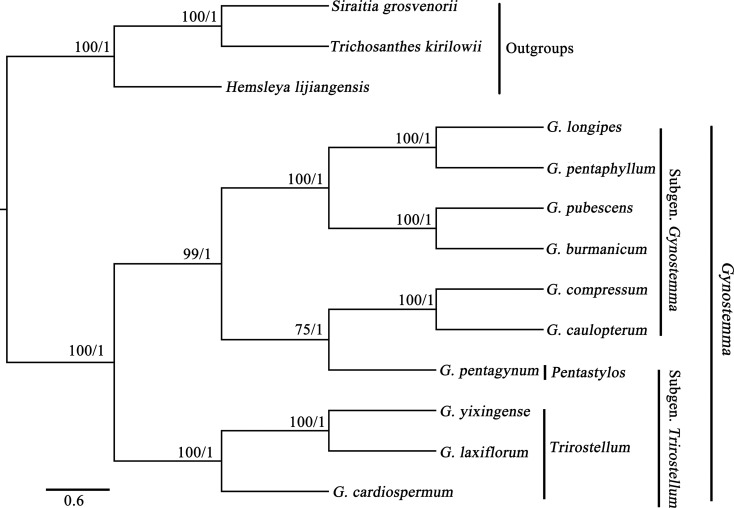
The results of molecular analysis based on MP and BI methods showed the same topology (Figure 3). All species of Gynostemma were clustered into one monophyletic clade with a high bootstrap support value, which were divided into two subclades. The first subclade consisted of the Subgen. Gynostemma except G. pentagynum, while the other subclade consisted of the Subgen. Trirostellum. It is basically consistent with the morphological classification by Chen (1995). G. pentagynum has styles (4-) 5, which differs from other species of Gynostemma. G. yixingense had the closest phylogenetic relationship to G. laxiflorum, which formed a clade and had a close phylogenetic relationship to the subclade of G. cardiospermum. We had already done a number of field resource surveys, and did not discover G. laxiflorum. G. laxiflorum and G. yixingense could be the same species in FOC. Combining the geographical distribution, morphological characteristics and molecular phylogeny, we consider that the taxonomy of G. laxiflorum and G. yixingense still needs further study.
Figure 3. The Bayesian Phylogenetic tree based on ten Gynostemma species. Numbers above the branches represent MP bootstrap (BS) and Bayesian posterior probabilities (PP) values.
Overall, the complete chloroplast genome sequence of G. yixingense was reported and analyzed. Comparing with chloroplast genomes of other Gynostemma, the chloroplast genome of G. yixingense was conserved and very similar to other Gynostemma species. Phylogenetic analysis indicated that G. yixingense has the closest phylogenetic relationship to G. laxiflorum. The repeat sequences could be usted for developing genetic markers. The data in this study provided a useful tool for molecular identification and evolutionary studies in Gynostemma.
Acknowledgments
This work was supported by the National Key Research and Development Program of China(2018YFC1707300), the National Natural Science Foundation of China (NSFC: 81973414), Natural Science Foundation of Jiangsu Province (Grant No. BK20191319), Fundamental Research Funds for the Central Universities(2632019ZD15), “Double First-Class” University project (CPU2018GY09; CPU2018GY11) and Natural Science Foundation of Hunan Province (2018JJ3008). We also thank Yucheng Zhao in China Pharmaceutical University for his help in data analysis of complete chloroplast genome in this study.
Supplementary Material
The following online material is available for this article:
Footnotes
Associate Editor: Rogerio Margis
Conflict of interest
The authors declare that they have no conflict of interest.
Author Contributions
XL and PL conceived, designed the study and reviewed draft of the manuscript, LW performed the experiments, analyzed the data and wrote the manuscript, GYL and LJH performed the experiments, HL and WMJ analyzed the data. All authors read and approved the final version.
References
- Chen SK, Lu AM, Jeffrey C. Flora of China. Vol. 19. Science press; Beijing: 2011. p. 12. Missouri Botanical Garden Press, St. Louis. [Google Scholar]
- Chen SK. A classificatory system and geographical distribution of the genus Gynostemma BL. (Cucurbitaceae) Acta Phytotaxonomica Sinica. 1995;33:403–410. [Google Scholar]
- Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- Ding JG. A new variety of Gynostemma yixingese . Bulletin of botanical research. 1990;10:71–72. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016;45:1–9. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Meng J, Li XP, Li HT, Yang JB, Wang H, He J. Comparative analysis of the complete chloroplast genomes of four aconitum medicinal species. Molecules. 2018;23:1015–1017. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MRMODELTEST version 2.1. Computer program distributed by the author. Uppsala University; Uppsala: 2004. [Google Scholar]
- Qin SS, Li HT, Wang ZY, Cui ZH, Yu LY. Analysis phylogenetic relationship of Gynostemma (Cucurbitaceae) China J Chin Mater Med. 2015;40:1681–1687. [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van DMP, Ayres DL, Darling A, Sebastian H, Larget B, Liu L, Suchard MA, Huelsrnbeck JP. Mrbayes3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YC, Zou R, Liu BB. Complete chloroplast genome sequence of Gynostemma pentaphyllum (Cucurbitaceae), a perennial medicinal herb. Mitochondrial DNA B. 2019;4:3967–3968. doi: 10.1080/23802359.2019.1688726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis using Parsimony (* and Other Methods) 2002.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang JL, Zhou PP, Meng XH, Shi YP. Further New Gypenosides from Jiaogulan (Gynostemma pentaphyllum) J Agric Food Chem. 2017;65:5926–5934. doi: 10.1021/acs.jafc.7b01477. [DOI] [PubMed] [Google Scholar]
- Xiang WJ, Guo CY, Ma L, Hu LH. Dammarane-type glycosides and long chain sesquiterpene glycosides from Gynostemma yixingense . Fitoterapia. 2010;81:248–252. doi: 10.1016/j.fitote.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Zhang T, Liu WP, Li H, Xiao YP. Ecological distribution and utilization of Gynostemma pentaphyllum germplasm resources in China. Shaanxi J Agric Sci. 2015;61:55–59. [Google Scholar]
- Zhang X, Li HM, Zhou T, Yang YC, Zhao GF. Characterization of the complete chloroplast genome sequence of Gynostemma compressum (Cucurbitaceae), an endemic plant in China. Conserv Genet Resour. 2018;10:141–144. [Google Scholar]
- Zhang X, Zhou T, Kanwal N, Zhao YM, Bai GQ, Zhao GF. Completion of Eight Gynostemma BL. (Cucurbitaceae) Chloroplast Genomes: Characterization, Comparative Analysis, and Phylogenetic Relationships. Front Plant Sci. 2017;8:1–13. doi: 10.3389/fpls.2017.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li L, Liang T, Liu Q. Complete chloroplast genome sequences of Praxelis (Eupatorium catarium Veldkamp), an important invasive species. Gene. 2014;549:58–69. doi: 10.1016/j.gene.2014.07.041. [DOI] [PubMed] [Google Scholar]
- Zhou JG, Chen XL, Cui YX, Sun W, Li YH, Wang Y, Song JY, Yao H. Molecular structure and phylogenetic analyses of genomes of two aristolochia medicinal species. Int J Mol Sci. 2017;18:1839. doi: 10.3390/ijms18091839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.